Abstract
A rapid method, using PCR-restriction fragment length and single-strand conformation polymorphism (SSCP), was applied to screen for mutations of the fluoroquinolone resistance determinants in Streptococcus pneumoniae. One hundred nonduplicate Streptococcus pneumoniae isolates with ciprofloxacin MICs of ≥4.0 μg/ml from the Prince of Wales Hospital, Hong Kong, years 2000 to 2003, were examined. For each isolate, PCR amplicons of quinolone resistance-determining regions (QRDRs) of gyrA, gyrB, parC, and parE genes were digested with AluI, HinfI, Sau3AI, and MspI, respectively, and analyzed by SSCP. Each SSCP pattern was given a number, and each isolate obtained a four-digit code, e.g., 1111, that represented the SSCP profile. The SSCP patterns were correlated to mutations characterized from sequence analyses of PCR amplicons. The most common SSCP profile obtained was no. 5232 (40%), which included strains with two amino acid substitutions in the ParC (Lys-137-Asn) and ParE (Ile-460-Val) genes, followed by the SSCP profile 5223 (17%), which included strains with amino acid substitutions in the ParE (Ile-460-Val) gene only. Ten isolates (10%) with amino acid substitutions at GyrA and ParE (±ParC) genes were resistant to levofloxacin with a MIC of ≥16 μg/ml. Other SSCP profiles were unique in distinguishing the common amino acid substitutions in GyrA (Ser-81-Phe) and ParC (Lys-137-Asn, Ser-79-Phe plus Lys-137-Asn, Asp-83-Asn plus Lys-137-Asn, Ser-79-Phe, and Glu-96-Asp). SSCP analysis of restricted fragments generated patterns that were highly discriminative for mutations present in the QRDRs of gyrA, gyrB, parC, and parE. This method provides a database of high resolution profiles on these mutations and allows rapid screening for new mutations of the fluoroquinolone resistance genes.
Antibiotic-resistant Streptococcus pneumoniae has evolved to be a worldwide problem in the last decade. Fluoroquinolones are an important class of antibiotics, and agents such as levofloxacin or moxifloxacin are incorporated in guidelines for the empirical treatment of community-acquired pneumonia (17, 18). The prevalence of levofloxacin nonsusceptibility (MIC of ≥4.0 μg/ml) varies by region and has remained low, ranging from 0% in some European countries (24) to 1.3% in the United States (25). Higher rates have been documented in some countries, e.g., South Korea (2.9%) (4), and in Hong Kong, a rate of 13% has been reported in 2001 (9) and the spread hypothesized to be a result of clonal dissemination of the Spanish 23F strain of S. pneumoniae (10). However, there is increasing evidence that interspecies transfer of the parE-parC gene region arising from other viridans group streptococci occurs (7, 28). It is thus important to identify and monitor the spread of fluoroquinolone resistance determinants among S. pneumoniae strains.
Fluoroquinolone resistance in S. pneumoniae is primarily due to mutations in the quinolone resistance-determining regions (QRDRs) of the genes encoding the A and B subunits of DNA gyrase and topoisomerase IV, in particular, the parC and gyrA genes (3, 6, 14). Often, the level of resistance increases as more amino acids are substituted for by additional mutations. Mutations in parE and gyrB have also been reported, but to a lesser extent (2, 23, 30). Reduced susceptibility to fluoroquinolone may also be due to altered accumulation of the drug or efflux, but this plays a less significant role (31).
Screening for mutations in the QRDRs is performed by sequencing of the gyrA, gyrB, parC, and parE genes of the fluoroquinolone-resistant strains. Methods for detection of known mutations in the QRDRs have included PCR-restriction fragment length polymorphism (PCR-RF) (1), oligonucleotide probe assay (6), TaqMan assay (8), and single-strand conformational polymorphism (SSCP) (27). Often, these methods are not designed to have, or do not have, a sufficiently high sensitivity or resolution to identify new mutations. Tawata et al. (29) introduced a modified method of using a combination of PCR-restriction fragment length polymorphism and single-strand conformational polymorphism (PCR-RF-SSCP) as a tool for mass screening of the genome. We thus sought to identify and monitor the spread of fluoroquinolone resistance determinants in S. pneumoniae using a modified method of SSCP to examine and screen for new mutations at the QRDRs of the respective fluoroquinolone resistance genes in S. pneumoniae.
MATERIALS AND METHODS
Bacterial isolates and susceptibility tests.
One thousand eighty-one nonduplicate S. pneumoniae isolates from patients admitted to the Prince of Wales Hospital, a 1,350-bed teaching and tertiary hospital in Hong Kong, from 2000 to 2003 were screened for fluoroquinolone nonsusceptibility. The MICs of ciprofloxacin, levofloxacin, moxifloxacin, and gatifloxacin were determined by the microdilution broth method as described by NCCLS (21). One hundred isolates that were nonsusceptible to ciprofloxacin (CIP) with MICs of ≥4 μg/ml were further examined on the mutations and amino acid substitutions at the QRDRs of the respective fluoroquinolone resistance genes using PCR-RF-SSCP.
Analysis by PCR-RF-SSCP.
Chromosomal DNA from the isolates was obtained by melting a small piece of DNA plug in 150 μl double-distilled H2O at 65°C. DNA plug was prepared for pulsed-field gel electrophoresis according to the method previously described (11, 12). For each isolate, the QRDRs of gyrA, gyrB, parC, and parE genes were amplified by PCR using the primers listed in Table 1 and conditions as previously described (19, 20, 22). For the restriction digestion, the PCR amplicons of gyrA, gyrB, parC, and parE genes were digested with AluI, HinfI, Sau3AI, and MspI enzymes (Amersham Biosciences), respectively, each in a 10-μl reaction mixture containing reaction buffer and incubated at 37°C overnight as recommended by the manufacturer. For example, AluI was the restriction enzyme used for the digestion of the PCR amplicons of the gyrA gene, HinfI for gyrB, and so forth. A quantity of 3.5 μl (approximately 100 to 150 ng) of the digested product was mixed with an equal volume of denaturing solution (94% formamide, 0.05% xylene cyanol solution, 0.4 mg/ml bromophenol blue) according to the manufacturer's instructions (ExcelGel DNA Analysis Kit; Amersham Biosciences), the mixture was denatured at 95°C for 8 min in a thermocycler, and the mixture was placed on ice immediately. The denatured mixture was electrophoresed on precast gels for SSCP (ExcelGel DNA Analysis Kit;, Amersham Biosciences) at 600 V, with a current of 50 mA and 30 W power for 90 min at 4°C using the Multiphor II electrophoresis unit (Amersham Biosciences). The precast gels were 12.5% acrylamide gels and contained 48 wells for sample loading. The DNA gel was stained by silver stain according to the manufacturer's instructions (DNA silver staining kit; Amersham Biosciences) to visualize and permanently stain the discrete DNA bands.
TABLE 1.
PCR primers for QRDRs of the fluoroquinolone resistance genes
| Gene | Primer (5′ to 3′)a | Amplicon size (bp) | Reference |
|---|---|---|---|
| gyrA | CCG TCG CAT TCT TTA CG | 382 | 22 |
| AGT TGC TCC ATT AAC CA | |||
| gyrB | TTC TCC GAT TTC CTC ATG | 457 | 19 |
| AGA AGG GTA CGA ATG TGG | |||
| parC | TGG GTT GAA GCC GGT TCA | 366 | 20 |
| TGC TGG CAA GAC CGT TGG | |||
| parE | AAG GCG CGT GAT GAG AGC | 289 | 22 |
| TCT GCT CCA ACA CCC GCA |
For each gene, the first primer listed is forward, and the second is reverse.
Analysis of SSCP patterns.
Each SSCP pattern was given a number and each isolate obtained a four-digit code that represented the SSCP profile. For example, 1234 represented the SSCP profile of pattern 1 for gyrA, pattern 2 for gyrB, pattern 3 for parC, and pattern 4 for the parE gene. The SSCP patterns were correlated to the mutations characterized from the sequence analyses of the respective PCR amplicons, and any amino acid substitutions were noted. At least three pairs of forward and reverse sequences of the PCR amplicons, if available, for each corresponding SSCP pattern were sequenced for confirmation. Sequencing was performed with an ABI 310 sequencer and an ABI Prism dRhodamine terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.). The sequences were aligned and compared with the corresponding QRDR regions of the four fluoroquinolone resistance genes of the S. pneumoniae R6 strain from the GenBank.
RESULTS
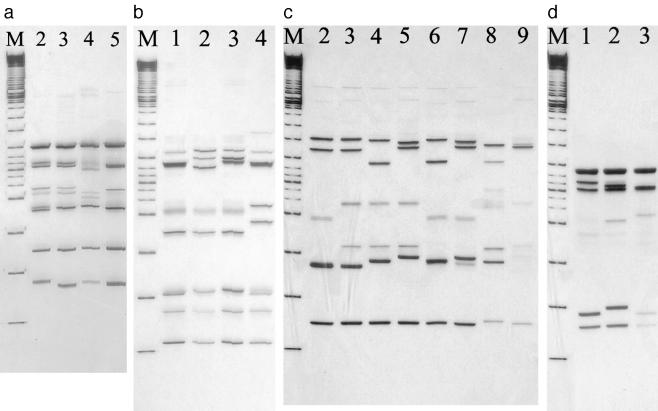
The different SSCP patterns obtained from PCR-RF-SSCP of the QRDRs of the gyrA, gyrB, parC, and parE genes are shown in Fig. 1a to d, respectively. Four SSCP patterns were observed for gyrA, four patterns for gyrB, eight for parC, and three for parE. The corresponding nucleotide change and amino acid substitution for each of the SSCP patterns are shown in Tables 2 to 5. The sequencing results confirmed that the same SSCP pattern represented the same sequence of the QRDR of the PCR amplicons of the specific fluoroquinolone-resistant genes.
FIG. 1.
SSCP patterns of DNA amplicons of gyrA (a), gyrB (b), parC (c), and parE (d) after restriction digestion with AluI, HinfI, Sau3AI, and MspI, respectively. (a) SSCP patterns 2 to 5 of the gyrA gene after digestion with AluI restriction enzyme. Lane M represents a 100-bp DNA ladder. (b) SSCP patterns 1 to 4 of the gyrB gene after restriction digestion using HinfI. Lane M represents a 100-bp DNA ladder. (c) SSCP patterns 2 to 9 of the parC gene after restriction digestion using Sau3AI. Lane M represents a 100-bp DNA ladder. (d) SSCP patterns 1 to 3 of the parE gene after restriction digestion using MspI. Lane M represents a 100-bp DNA ladder.
TABLE 2.
Correlation of SSCP patterns with nucleotide sequences and amino acid substitutions in gyrA gene, compared to that of S. pneumoniae R6
| SSCP pattern | Description of patternb | No. isolates (%) | MIC range (μg/ml)a
|
75d
|
81d
|
83d
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LEV | nte | aaf | nt | aa | nt | aa | |||
| 1c | R6 | 0 | TAC | Tyr | TCC | Ser | ATT | Ile | ||
| 2 | 1 sub + 1 var | 9 (9) | 16-64 | >16 | --T | -T- | Phe | |||
| 3 | 1 sub | 1 (1) | 8 | 16 | -T- | Phe | ||||
| 4 | 1 var | 1 (1) | 4 | 2 | --C | |||||
| 5 | 1 var | 89 (89) | 4->16 | 0.5-16 | --T | |||||
CIP, ciprofloxacin; LEV, levofloxacin.
sub, no. of amino acid substitution; var, no. of base pair variation compared to R6.
S. pneumoniae R6 strain.
Numeral indicates amino acid position. nt, nucleotide; aa, amino acid.
Nucleotide sequence changes relative to S. pneumoniae R6.
Amino acid substitution corresponding to nt change.
TABLE 5.
Correlation of SSCP patterns with nucleotide sequences and amino acid substitutions in parE compared to that of S. pneumoniae R6
| SSPC pattern | Description of patternb | No. isolates (%) | MIC range (μg/ml)a
|
460d
|
476d
|
|||
|---|---|---|---|---|---|---|---|---|
| CIP | LEV | nte | aaf | nt | aa | |||
| 1c | R6 | 4 (4) | 4-16 | 1-2 | ATC | Ile | ATC | Ile |
| 2 | 1 sub + 1 var | 54 (54) | 4-32 | 0.5->16 | G-- | Val | --T | |
| 3 | 1 sub | 42 (42) | 4-64 | 1-16 | G-- | Val | ||
CIP, ciprofloxacin; LEV, levofloxacin.
sub, no. of amino acid substitution; var, no. of base pair variation compared to R6.
S. pneumoniae R6 strain.
Numeral indicates amino acid position. nt, nucleotide; aa, amino acid.
Nucleotide sequence change relative to S. pneumoniae R6.
Amino acid substitution corresponding to nt change.
Four SSCP patterns were obtained for gyrA (Fig. 1a). The majority of isolates (90%) belonged to pattern 5 and were found to have one base pair difference from S. pneumoniae R6 (Table 2). Only one amino acid substitution (Ser-81-Phe) was found, represented in both SSCP patterns 2 and 3, and was present in 10 (10%) of the isolates. The CIP and levofloxacin (LEV) MICs of these isolates were ≥8 μg/ml and ≥16 μg/ml, respectively. Four SSCP patterns were obtained for gyrB (Fig. 1b), and these included a number of silent mutations, but there was no amino acid substitution. Fourteen (14%) of the sequences were identical to that of the wild-type strain, R6, while 86% had two or three nucleotide changes (patterns 2 to 4) (Table 3). Nine SSCP patterns were obtained for parC (Fig. 1c). SSCP pattern 2 included 40 (40%) isolates that differed from the R6 strain by a base pair, while the other 60 (60%) isolates had one, two, or three amino acid substitutions, as shown in SSCP patterns 3 to 9. These amino acid substitutions included Ser-79-Phe, Asp-83-Asn/Tyr, Glu-96-Asp, and Lys-137-Asn. The most common substitution is represented by SSCP pattern 3, with 49 (49%) of the isolates with an amino acid substitution of Lys-137-Asn (Table 4). Ninety-six (96%) isolates had an amino acid substitution of Ile-460-Val in parE, as represented by SSCP patterns 3 and 4 (Table 5). These isolates have various MICs ranging 4 to 64 μg/ml and 0.5 to ≥16 μg/ml for CIP and LEV, respectively.
TABLE 3.
Correlation of SSCP patterns with nucleotide sequences and amino acid substitutions in gyrB compared to that of S. pneumoniae R6
| SSCP pattern | Description of patternb | No. isolates (%) | MIC range (μg/ml)a
|
381d
|
384d
|
386d
|
461d
|
472d
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LEV | nte | aaf | nt | aa | nt | aa | nt | aa | nt | aa | |||
| 1c | R6 | 14 (14) | 4 | 1-2 | GTA | Vla | GGA | Gly | TTG | Leu | AAC | Asn | GCT | Ala |
| 2 | 3 var | 80 (80) | 4-64 | 0.5->16 | --G | --G | --A | |||||||
| 3 | 2 var | 5 (5) | 4-16 | 1-4 | --G | --A | ||||||||
| 4 | 2 var | 1 (1) | 4 | 2 | --T | --C | ||||||||
CIP, ciprofloxacin; LEV, levofloxacin.
sub, no. of amino acid substitution; var, no. of base pair variation compared to R6.
S. pneumoniae R6 strain.
Numeral indicates amino acid position. nt, nucleotide; aa, amino acid.
Nucleotide sequence changes relative to S. pneumoniae R6.
Amino acid substitution corresponding to nt change.
TABLE 4.
Correlation of SSCP patterns with nucleotide sequences and amino acid substitutions in parC compared to that of S. pneumoniae R6
| SSCP pattern | Description of patternb | No. isolates (%) | MIC range (μg/ml)a
|
41d
|
79d
|
83d
|
96d
|
128d
|
137d
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LEV | nte | aaf | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |||
| 1c | R6 | 0 | CAA | Gln | TCT | Ser | GAT | Asp | GAG | Glu | GGC | Gly | AAG | Lys | ||
| 2 | 1 var | 40 (40) | 4-16 | 1-16 | --G | |||||||||||
| 3 | 1 sub + 1 var | 49 (49) | 4->16 | 0.5->16 | --G | --T | --T | Asn | ||||||||
| 4 | 2 sub + 2 var | 2 (2) | 8->16 | 2-16 | --G | A-- | Asn | --T | --T | Asn | ||||||
| 5 | 2 sub + 2 var | 2 (2) | >16 | >16 | --G | -T- | Phe | --T | --T | Asn | ||||||
| 6 | 1 sub + 1 var | 1 (1) | 4 | 2 | --G | T-- | Tyr | |||||||||
| 7 | 1 sub + 1 var | 4 (4) | 8-64 | 2-16 | --G | -T- | Phe | |||||||||
| 8 | 1 sub + 2 var | 1 (1) | 4 | 1 | --G | --T | --T | Asn | ||||||||
| 9 | 3 sub + 2 var | 1 (1) | 32 | 16 | --G | -T- | Phe | --T | Asp | --T | --T | Asn | ||||
CIP, ciprofloxacin; LEV, levofloxacin.
sub, no. of amino acid substitution; var, no. of base pair variation compared to R6.
S. pneumoniae R6 strain.
Numeral indicates amino acid position. nt, nucleotide; aa, amino acid.
Nucleotide sequence change relative to S. pneumoniae R6.
Amino acid substitution corresponding to nt change.
The correlation of the different SSCP profiles with the amino acid substitutions at GyrA, ParC, and ParE and the fluoroquinolone MICs are summarized in Table 6. The most common SSCP profile was 5232 (40%), which included strains with two amino acid substitutions in the ParC (Lys-137-Asn) and ParE (Ile-460-Val) genes, followed by the SSCP profile of 5223 (17%), which included strains with amino acid substitutions in the ParE (Ile-460-Val) gene only. Ten isolates (10%) with amino acid substitutions at the GyrA and ParE (±ParC) genes are clearly resistant to levofloxacin with MICs of ≥16 μg/ml.
TABLE 6.
Correlation of different SSCP profiles with amino acid substitutions of GyrA, ParC, and ParE and fluoroquinolone MICs
| Gene(s) with amino acid substitution | SSCP profile | Position of amino acid substitution
|
No. of isolates (n = 100) | Fluoroquinolone MIC (μg/ml)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GyrA | ParC
|
ParE
|
CIP
|
LEV
|
||||||||||||||
| Ser-81- | Ser-79- | Asp-83- | Glu-96- | Lys-137- | Ile-460- | 4 | 8 | 16 | >16 | 0.5 to 1 | 2 | 4 | 8 | 16 | >16 | |||
| No substitution | 5121 | 1 | 1 | 1 | ||||||||||||||
| 5221 | 2 | 2 | 1 | 1 | ||||||||||||||
| parE | 4223 | Val | 1 | 1 | 1 | |||||||||||||
| 5123 | 11 | 11 | 4 | 7 | ||||||||||||||
| 5222 | 3 | 3 | 1 | 2 | ||||||||||||||
| 5223 | 17 | 16 | 1 | 8 | 8 | 1 | ||||||||||||
| 5323 | 4 | 3 | 1 | 1 | 2 | 1 | ||||||||||||
| parC, parE | 5372 | Phe | Val | 1 | 1 | 1 | ||||||||||||
| 5273 | 1 | 1 | 1 | |||||||||||||||
| 5263 | Tyr | Val | 1 | 1 | 1 | |||||||||||||
| 5232 | Asn | Val | 40 | 35 | 4 | 1 | 20 | 18 | 2 | |||||||||
| 5233 | 2 | 2 | 2 | |||||||||||||||
| 5132 | 2 | 2 | 2 | |||||||||||||||
| 5433 | 1 | 1 | 1 | |||||||||||||||
| 5283 | 1 | 1 | 1 | |||||||||||||||
| 5242 | Asn | Asn | Val | 1 | 1 | 1 | ||||||||||||
| parC | 5231 | Asn | 1 | 1 | 1 | |||||||||||||
| gyrA, parE | 2223 | Phe | Val | 1 | 1 | 1 | ||||||||||||
| gyrA, parC, parE | 2273 | Phe | Phe | Val | 2 | 2 | 2 | |||||||||||
| 2232 | Phe | Asn | Val | 2 | 2 | 2 | ||||||||||||
| 3232 | 1 | 1 | 1 | |||||||||||||||
| 2252 | Phe | Phe | Asn | Val | 2 | 2 | 1 | 1 | ||||||||||
| 2242 | Phe | Asn | Asn | Val | 1 | 1 | 1 | |||||||||||
| 2292 | Phe | Asp | Asn | Val | 1 | 1 | 1 | |||||||||||
DISCUSSION
PCR-RF-SSCP has been described as a useful tool for mass screening for DNA polymorphism analysis on other genes (15, 26) and is able to detect a nucleotide substitution, deletion, or insertion in up to a 22,000-base-pair amplicon (29). We applied this method for the analysis of the QRDRs of the gyrA, gyrB, parC, and parE genes of S. pneumoniae. The restriction digestion of the PCR amplicons generates a higher degree of polymorphism that could be readily detected by SSCP. Each restriction enzyme was chosen to cut the respective PCR amplicons in regions to produce fragments in the range of 100 to 200 bp. Up to nine bands in the SSCP pattern were observed by using this method. However, using PCR-SSCP alone, only two to three bands were produced, and the method was unable to discriminate single nucleotide changes (results not shown). The SSCP patterns were able to detect a single base difference (for example, patterns 3, 4, and 5 in gyrA); therefore, the patterns produced from PCR-RF-SSCP were highly discriminative for mutations present in gyrA, gyrB, parC, and parE.
Among the hundred isolates screened, the SSCP profiles distinguished the common amino acid substitutions in GyrA (Ser-81-Phe), ParC (Ser-79-Phe, Asp-83-Asn/Tyr, and Lys-137-Asn) and ParE (Ile-460-Val) (5, 6, 14). Of the 36 isolates that possessed an amino acid substitution (Ile-460-Val) at ParE alone, 34 (94%) strains had a CIP MIC of 4 μg/ml and a LEV MIC of 0.5 to 2 μg/ml, similar to isolates that had no amino acid substitution detected, supporting that the ParE substitution did not contribute to significant fluoroquinolone resistance (14). However, two isolates had higher fluoroquinolone MICs (LEV MIC of 4 or 8 μg/ml) that may be attributed to other mutations in regions that were not studied here or some other mechanism, such as those that alter the drug permeation.
ParC substitution has been reported as contributing to low-level fluoroquinolone resistance (13, 16, 20). Although CIP MICs may be increased, the LEV MIC generally remained low, in the range of 0.5 to 2 μg/ml. Many reports showed that ParC and GyrA substitutions are responsible for high-level fluoroquinolone resistance and ParC substitution was a first-step prerequisite in the development of high-level resistance (13, 16, 20). Our findings supported that all nine of our isolates with ParC and GyrA substitutions had LEV MICs of ≥16 μg/ml. These amino acid substitutions have previously been described to be responsible for high levels of resistance (13, 16, 20). An exception, Glu-96-Asp, is an unusual substitution found in ParC, and its importance remains to be determined.
Our data are similar to the previous findings from Hong Kong in which Ser-81-Phe (9) was the substitution found in GyrA that contributed to high-level resistance to fluoroquinolones. However, an additional number of new substitutions in ParC (Asp-83-Tyr/Asn and Glu-96-Asp) were identified in the present study.
PCR-RF-SSCP provides a database of high-resolution profiles on these mutations and allows rapid screening for any new mutation. The analysis and comparison of the pattern with the known sequences provide an alternative method for detecting mutation in the gene studied other than direct nucleotide sequencing. The four-digit code that represents the SSCP profile for an isolate also provides an objective way of recognizing the QRDR mutations associated with that strain. If the SSCP patterns for a fluoroquinolone-resistant gene, e.g., gyrA, exceed 10, then the single-digit number could be replaced by a single alphabet letter, e.g., A to indicate the eleventh pattern, B for the twelfth pattern, etc. The method is sensitive and cheaper and easier to perform compared to direct nucleotide sequencing of all sequences. In addition, this method is rapid, and up to 48 samples could be run per gel. The method could readily be adaptable to a clinical laboratory if the laboratory has a PCR facility. The only additional requirement is an electrophoresis system for SSCP and the gels and reagents are readily available commercially. It is discriminatory and could readily be applied as a screening method to examine for known and new mutations of the fluoroquinolone resistance genes.
Acknowledgments
The work described in this paper was supported by an earmarked grant from the Research Grants Council of the Hong Kong Special Administrative Region (Project No. CUHK 4432/03 M).
REFERENCES
- 1.Alonso, R., M. Galimand, and P. Courvalin. 2004. An extended PCR-RFLP assay for detection of parC, parE and gyrA mutations in fluoroquinolone-resistant Streptococcus pneumoniae. J. Antimicrob. Chemother. 53:682-683. [DOI] [PubMed] [Google Scholar]
- 2.Bast, D. J., D. E. Low, C. L. Duncan, L. Kilburn, L. A. Mandell, R. J. Davidson, J. C. de Azavedo, et al. 2000. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob. Agents Chemother. 44:3049-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brueggemann, A. B., S. L. Coffman, P. Rhomberg, H. Huynh, L. Almer, A. Nilius, R. Flamm, and G. V. Doern. 2002. Fluoroquinolone resistance in Streptococcus pneumoniae in United States since 1994-1995. Antimicrob. Agents Chemother. 46:680-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 5.Davies, T. A., A. Evangelista, S. Pfleger, K. Bush, D. F. Sahm, and R. Goldschmidt. 2002. Prevalence of single mutations in topoisomerase type II genes among levofloxacin-susceptible clinical strains of Streptococcus pneumoniae isolated in the United States in 1992 to 1996 and 1999 to 2000. Antimicrob. Agents Chemother. 46:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, T. A., and R. Goldschmidt. 2002. Screening of large numbers of Streptococcus pneuomoniae isolates for mutations associated with fluoroquinolone resistance using an oligonucleotide probe assay. FEMS Microbiol. Lett. 17:219-224. [DOI] [PubMed] [Google Scholar]
- 7.de la Campa, A. G., L. Balsalobre, C. Ardanuy, A. Fenoll, E. Perez-Trallero, J. Linares, et al. 2004. Fluoroquinolone resistance in penicillin-resistant Streptococcus pneumoniae clones, Spain. Emerg. Infect. Dis. 10:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giles, J., J. Hardick, J. Yuenger, M. Dan, K. Reich, and J. Zenilman. 2004. Use of Applied Biosystems 7900HT sequence detection system and Taqman assay for detection of quinolone-resistant Neisseria gonorrhoae. J. Clin. Microbiol. 42:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, P. L., R. W. Yung, D. N. Tsang, T. L. Que, M. Ho, W. H. Seto, T. K. Ng, W. C. Yam, and W. W. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of a Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 10.Ho, P. L., T. L. Que, S. S. Chiu, R. W. H. Yung, T. K. Ng, D. N. C. Tsang, W. H. Seto, and Y. L. Lau. 2004. Fluoroquinolone and other antimicrobial resistance in invasive pneumococci, Hong Kong, 1995-2001. Emerg. Infect. Dis. 10:1250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ip, M., D. J. Lyon, R. W. H. Yung, C. Chan, and A. F. Cheng. 1999. Evidence of clonal dissemination of multidrug-resistant Streptococcus pneumoniae in Hong Kong. J. Clin. Microbiol. 37:2834-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ip, M., D. J. Lyon, R. W. H. Yung, L. Tsang, and A. F. Cheng. 2002. Introduction of new clones of penicillin-nonsusceptible Streptococcus pneumoniae in Hong Kong. J. Clin. Microbiol. 40:1522-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janoir, C., V. Zeller, M.-D. Kitzis, N. J. Moreau, and L. Gutmann. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, M. E., D. F. Sahm, N. Martin, S. Scheuring, P. Heisig, C. Thornsberry, K. Köhrer, and F. J. Schmitz. 2000. Prevalence of gyrA, gyrB, parC, and parE mutations in clinical isolates of Streptococcus pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from worldwide surveillance studies during the 1997-1998 respiratory season. Antimicrob. Agents Chemother. 44:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurihara, A., M. Tawata, Y. Ikegishi, K. Aida, and T. Onaya. 1999. The procedure of polymerase chain reaction-restriction fragment-single strand conformation polymorphism analysis by Hha I/Hinc II to detect mitochondrial DNA mutations. Life Sci. 64:1223-1230. [DOI] [PubMed] [Google Scholar]
- 16.Lim, S., D. Bast, A. McGeer, J. de Azavedo, and D. E. Low. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg. Infect. Dis. 9:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandell, L. A., T. J. Marrie, R. F. Grossman, A. W. Chow, R. H. Hyland, Canadian Infectious Disease Society, and Canadian Thoracic Society. 2000. Summary of Canadian guidelines for the initial management of community-acquired pneumonia: an evidence-based update by the Canadian Infectious Disease Society and the Canadian Thoracic Society. Can. Respir. J. 7:371-382. [DOI] [PubMed] [Google Scholar]
- 19.Muñoz, R., M. Bustamante, and A. G. de la Campa. 1995. Ser-127-to-Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J. Bacteriol. 177:4166-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz, R., and A. G. de la Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing. M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Pan, X. S., and L. M. Fisher. 1999. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolone. Antimicrob. Agents Chemother. 43:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perichon, B., J. Tankovic, and P. Courvalin. 1997. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1166-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinert, R. R., S. Reinert, M. van der Linden, M. Y. Cil, A. Al-Lahham, and P. Appelbaum. 2005. Antimicrobial susceptibility of Streptococcus pneumoniae in eight European countries from 2001 to 2003. Antimicrob. Agents Chemother. 49:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter, S. S., K. P. Heilmann, S. E. Beekmann, N. J. Miller, C. L. Rice, and G. V. Doern. 2005. The molecular epidemiology of Streptococcus pneumoniae with quinolone resistance mutations. Clin. Infect. Dis. 15:225-235. [DOI] [PubMed] [Google Scholar]
- 26.Sato, Y., and T. Nishio. 2003. Mutation detection in rice waxy mutants by PCR-RF-SSCP. Theor. Appl. Genet. 107:560-567. [DOI] [PubMed] [Google Scholar]
- 27.Sougakoff, W., N. Lemaitre, E. Cambau, M. Szpytma, V. Revel, and V. Jarlier. 1997. Nonradioactive single-strand conformation polymorphism analysis for detection of fluoroquinolone resistance in mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 16:395-398. [DOI] [PubMed] [Google Scholar]
- 28.Stanhope, M. J., S. L. Walsh, J. A. Becker, M. J. Italia, K. A. Ingraham, M. N. Gwynn, T. Mathie, J. A. Poupard, L. A. Miller, J. R. Brown, and H. Amrine-Madsen. 2005. Molecular evolution perspectives on intraspecific lateral DNA transfer of topoisomerase and gyrase loci in Streptococcus pneumoniae, with implications for fluoroquinolone resistance development and spread. Antimicrob. Agents Chemother. 49:4315-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tawata, M., E. Iwase, K. Aida, and T. Onaya. 1996. A mass screening device of genome by polymerase chain reaction-restriction fragment-single strand conformation polymorphism analysis. Genet. Anal. 12:125-127. [DOI] [PubMed] [Google Scholar]
- 30.Weigel, L. M., G. J. Anderson, R. R. Facklam, and F. C. Tenover. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:3517-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeller, V., C. Janoir, M. D. Kitzis, L. Gutmann, and N. J. Moreau. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1973-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]