Abstract
During the present study three type 1 poliovirus strains isolated in Greece during the 1996 poliomyelitis outbreak in Albania were retrospectively investigated and determination of their relationship with other epidemic strains isolated in Albania or elsewhere during previous epidemics was attempted. SimPlot analysis revealed that the three Greek strains are the result of a recombination event in the VP2 coding region.
Three non-Sabin-like poliovirus type 1 (PV1) isolates similar to the serotypes that circulated in the epidemic in Albania in 1996 were isolated from members of a Gypsy community in Greece between the end of June 1996 and September 1996 (15). The outbreak in Albania was caused by a wild-type 1 poliovirus and lasted from May till November 1996 (6, 14). The three non-Sabin-like type 1 poliovirus strains were isolated in and recorded by the National Enterovirus Reference Laboratory for Greece at the Hellenic Pasteur Institute in Athens (15). The purpose of the present study was to further characterize in retrospect the specific poliovirus isolates throughout 5′ untranslated region (UTR) and coding regions VP4, VP2, VP3, VP1, and 2A, in an attempt to investigate their relationship with strains isolated in Albania at the same period or elsewhere in the world during previous or more recent epidemics. Since the outbreak in Albania started soon after the national immunization days in Albania (April to May 1996) there was also an attempt to define whether these isolates had a vaccine origin and, if they did have, to elucidate the mutation and recombination events that may have contributed to the reversion of the attenuated phenotype to a neurovirulent one.
The three non-Sabin-like type 1 poliovirus strains were isolated from stool samples at the Enterovirus Reference Laboratory for Greece from cases of acute flaccid paralysis diagnosed in nonvaccinated Gypsy children, aged 7 months old, 9 [1/2] months old, and 2 [1/2] years old, in 1996. Initial isolation and identification of the strains by cell culture and seroneutralization with mixed, equine antiserum pools (supplied by the National Institute for Public Health and the Environment, Bilthoven, The Netherlands) took place in summer 1996, simultaneously with a serious poliomyelitis outbreak in Albania (15).
Enterovirus RNA was extracted from 200 μl of the inoculated Rd cell cultures (4). Six different primer sets (72437 and 216616 [2, 12]; EUG3a, EUG3b, EUG3c, and EUC2 [3]; and Z752 and Z1461, Z1196 and Z1941, Z1814 and Z2478, and Z2378 and Z3021 [present study]) were used in reverse transcription-PCR, and sequence information from a part of the 5′ UTR and VP4 capsid region and complete VP2, VP3, VP1, and 2A coding regions was obtained. Table 1 shows details of the primers designed in this study. The isolated RNA was reversed transcribed with all primer pairs (7). PCR was performed with primers EUC2, EUG3a, EUG3b, and EUG3c, while reverse transcription was performed with primers EUC2a and EUC2b, as previously described (3). The produced cDNA was amplified by PCR. Thirty-five cycles of denaturation (95°C, 20 s), annealing (57°C, 20 s, for primer pairs Z752- Z1461 and Z1814-Z2478 and 60°C, 10 s, for primer pairs Z1196-Z1941 and Z2378-Z3021), and extension (74°C, 20 s), followed by incubation for 15 min at 78°C were performed in an Eppendorf master cycler.
TABLE 1.
Designed primers
| Primer | Position | Polarity | Sequence (5′-3′) | Gene |
|---|---|---|---|---|
| Z752 | 752-774 | Sense | CAGGTCTCATCCCAGAAAGTTG | VP4 |
| Z1461 | 1461-1483 | Antisense | GGGTGTCGTTTGGTTATTGTCT | VP2 |
| Z1196 | 1196-1216 | Sense | ACCGAACACACTAAGGGACA | VP2 |
| Z1941 | 1941-1963 | Antisense | GGTAGCACTCAAATCAAAAGGA | VP3 |
| Z1814 | 1814-1836 | Sense | GACAACTTTCAGTCTCCGTGTG | VP3 |
| Z2478 | 2478-2500 | Antisense | GCCTTGTGCTATTGCTTTTTGT | VP3 |
| Z2378 | 2378-2400 | Sense | GAAATGGATATTCTCGGGTTTG | VP3 |
| Z3021 | 3021-3043 | Antisense | TTGATGGGTTTGATGAAGTCTG | VP1 |
Initial identification of the obtained genomic sequences was carried out by comparing them with all available sequences in the database using BLAST software. Pairwise comparison of the sequences of the three PV1 strains (accession numbers AY956405, AY956408, AY960848, AY956406, AY956409, AY960849, AY956407, AY956410, and AY960850) and the sequences of the respective genomic regions of reference and wild-type poliovirus strains, reference human enterovirus C strains (HEV-Cs), and other HEV representative species (CBV5, CAV9, CAV2, CAV12, and EV70) for which such data are available in GenBank was made with the aid of ClustalW software. Finally, plots of nucleotide similarity between poliovirus strains were created with the aid of SimPlot software (9), in an attempt to define genomic regions that display significant percentages of nucleotide sequence identity among the PV strains, providing indications of possible recombination events throughout the genome.
Sequencing alignment of the three PV1 strains referred to as KatGRE1996, HalGRE1996, and KarGRE1996 with ClustalW revealed the great degree of relatedness of these strains, with sequence identity of 99% in all sequenced genomic regions: nucleotides (nt) 43 to 565 of the 5′ UTR; the 3′ end of VP4; and complete VP2, VP3, VP1, and 2A. Sequencing of the VP1 coding region verified the serotype of the three poliovirus type 1 strains (13). The nucleotide comparison of VP1 regions of the isolates with the respective regions of the poliovirus Sabin 1 strain and reference strain Mahoney showed the same homology (81%).
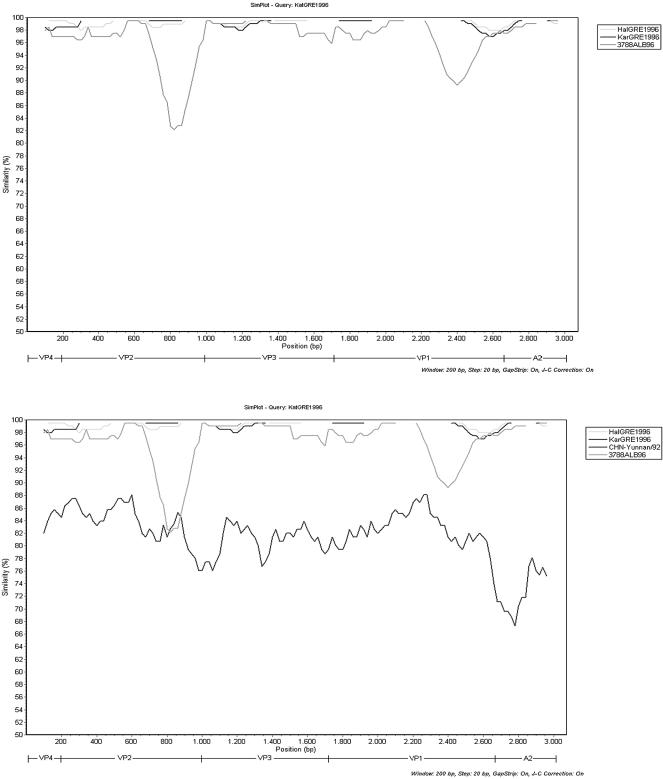
The nucleotide sequence comparison of the three PV1 isolates with other poliovirus strains using the BLAST software showed that the closest strains were those isolated from the epidemic in Albania in 1996, in all sequenced regions. Figure 1, top, shows the SimPlot comparison of the 3′ ends of the VP4, VP2, VP3, VP1, and 2A coding regions of the three PV1 Greek isolates and the Albanian strain 3788ALB96, whose almost-complete sequence is available in GenBank (11). The similarity between the three PV1 strains and the Albanian strains ranges between 95% and 100% in different genomic regions. Obviously, the epidemic PV1 strain that circulated in Albania in 1996 was transferred in Greece, as has also been suggested by Fiore et al. (6). Nevertheless, as represented in Fig. 1, top, there is a region containing part of the almost 200 nt in the VP2 coding region that differs a lot from the respective region of the Albanian strain. Sequence analysis of this part of VP2 with BLAST software indicated a high degree of homology with PV1 strains isolated in China in 1991 (CHN-Yunnan/92). Multiple alignment of nt 550 to 700 of VP2 of the Greek isolates with the respective sequences of Chinese and Albanian strains revealed 89% and 78% homology, respectively. There is also another region of approximately 240 nt in the VP1 coding region (nt 620 to 860 of VP1) where the similarity to the Albanian strains falls to 92%.
FIG. 1.
SimPlot analysis of the 3′ ends of VP4, VP2, VP3, VP1, and 2A genomic regions. KatGRE1996 is compared against (top) HalGRE1996, KarGRE1996, and the Albanian strain 3788ALB96 and (bottom) HalGRE1996, KarGRE1996, the Albanian strain 3788ALB96, and the Chinese isolate CHN-Yunnan/92.
The above result leads to the assumption that this small part of VP2 is the product of a recombination event. Natural genetic exchange within the capsid region appears to be rare (1, 10), but a recombination event in the VP2 gene was never mentioned before. Sequence analysis of this part of the genome with BLAST software indicated a high degree of homology (81%) with PV1 strain isolated in China in 1991 (CHN-Yunnan/92) (5). As shown in Fig. 1, bottom, the homology of the Greek and the Chinese strains ranges between 67% and 88% in different genomic regions and the only part of the genome where this homology exceeds the homology with the Albanian strains is the region of the 200 nt of VP2. According to these results we can assume that there was a recombination event in the VP2 gene, but the 81% similarity is not high enough to prove that the Chinese strains are the donors of the inserted sequence. Potentially, the donor is a descendant of the Chinese strains.
As shown by the Simplot analysis there is also another genomic region in the VP1 gene (nt 620 to 860 of VP1) where the similarity of the Greek strains with the Albanian strains falls to 92%. Nucleotide substitutions accumulate at a rate of approximately 1 to 2% per year during an epidemic (8). For this reason, this 8% nucleotide difference couldn't be attributed to the accumulation of nucleotide substitutions, as the Greek and the Albanian isolates circulated at the same period.
In conclusion the nucleotide sequence analysis revealed that there has been a recombination event in the VP2 coding region. The Greek isolates seem to be the product of recombination of the Albanian strains with an unknown virus. The similarity of the inserted sequence to the respective sequence of the type 1 polioviruses isolated in China in 1991 to 1993 can lead to the hypothesis that an offspring of the Chinese isolates is the donor of the unknown sequence.
Acknowledgments
The present work was cofunded by the European Union (75%) and the Greek Ministry of Education (25%) under the framework of the Education and Initial Vocational Training Program “Pythagoras II.”
This is research project “Molecular Detection of Enteroviruses in Clinical Samples and the Environment: Implications for the Public Health,” code 52213.11.
REFERENCES
- 1.Blomqvist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 2.Blomqvist, S., A. Skyttä, M. Roivainen, and T. Hovi. 1999. Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. J. Clin. Microbiol. 37:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for “serotyping” of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 4.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2005. Progress toward interruption of wild poliovirus transmission—worldwide, January 2004-March 2005. Morb. Mortal. Wkly. Rep. 54:408-412. [PubMed] [Google Scholar]
- 6.Fiore, L., D. Genovese, E. Diamanti, S. Catone, B. Ridolfi, B. Ibrahimi, R. Konomi, H. G. van der Avoort, T. Hovi, R. Crainic, P. Simeoni, and C. Amato. 1998. Antigenic and molecular characterization of wild type 1 poliovirus causing outbreaks of poliomyelitis in Albania and neighboring countries in 1996. J. Clin. Microbiol. 36:1912-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakasiliotis, I., P. Markoulatos, and T. Katsorchis. 2004. Site analysis of recombinant and mutant poliovirus isolates of Sabin origin from patients and from vaccinees. Mol. Cell. Probes. 18:103-109. [DOI] [PubMed] [Google Scholar]
- 8.Kinnunen, L., A. Huovilainen, T. Poyry, and T. Hovi. 1990. Rapid molecular evolution of wild type 3 poliovirus during infection in individual hosts. J. Gen. Virol. 71:317-324. [DOI] [PubMed] [Google Scholar]
- 9.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin, J., E. Samoilovich, G. Dunn, A. Lackenby, E. Feldman, A. Heath, E. Svirchevskaya, G. Cooper, M. Yermalovich, and P. D. Minor. 2002. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J. Virol. 76:10921-10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marturano, J., and L. Fiore. 2002. Investigation of the presence of recombinant polioviruses in the hit population in Albania during the 1996 outbreak. J. Clin. Microbiol. 40:316-317. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulders, M. N., J. H. Reimerink, M. Stenvik, I. Alaeddinoglu, H. G. van der Avoort, T. Hovi, and M. P. Koopmans. 1999. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. J. Gen. Virol. 80:907-916. [DOI] [PubMed] [Google Scholar]
- 13.Oberste, M. S., W. A. Nix, K. Maher, and M. A. Pallansch. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375-377. [DOI] [PubMed] [Google Scholar]
- 14.Prevots, D. R., M. L. Ciofi degli Atti, A. Sallabanda, E. Diamante, R. B. Aylward, E. Kakariqqi, L. Fiore, A. Ylli, H. van der Avoort, R. W. Sutter, A. E. Tozzi, P. Panei, N. Schinaia, D. Genovese, G. Oblapenko, D. Greco, and S. G. Wassilak. 1998. Outbreak of paralytic poliomyelitis in Albania, 1996: high attack rate among adults and apparent interruption of transmission following nationwide mass vaccination. Clin. Infect. Dis. 26:419-425. [DOI] [PubMed] [Google Scholar]
- 15.Siafakas, N., A. Georgopoulou, P. Markoulatos, and N. Spyrou. 2000. Isolation of polioviruses and other enteroviruses in south Greece between 1994 and 1998. J. Clin. Lab. Anal. 14:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]