Abstract
Formalin-fixed lung or trachea tissue specimens from four infants and one adolescent who died of respiratory illness were tested for Bordetella pertussis by conventional and real-time PCR assays. B. pertussis was confirmed in all cases. PCR can be an invaluable retrospective diagnostic tool for evaluating archival tissues from patients with suspected fatal pertussis.
Bordetella pertussis, a gram-negative coccobacillus, is the primary etiologic agent of pertussis (whooping cough). Pertussis is an important cause of morbidity among children of <4 months of age (10), and some studies of sudden infant death syndrome (SIDS) have shown an epidemiologic association with B. pertussis cases (1-4). Laboratory diagnosis of Bordetella sp. infections has traditionally been determined by culture of the agent; however, because of the enhanced sensitivity of PCR, this assay is increasingly being used for the diagnosis of B. pertussis infection.
Minimal DNA can be recovered from formalin-fixed tissues; thus, the use of a multicopy target for PCR detection, such as IS481, is advantageous. This study demonstrates that PCR assays targeting IS481 and the pertussis toxin gene can be used to identify B. pertussis in formalin-fixed, paraffin-embedded autopsy tissues from patients with unexplained deaths and in fixed tissues from suspected pertussis patients for whom fresh samples are not available.
(Part of this work was presented at the 105th meeting of the American Society for Microbiology, Atlanta, Ga., 6 June 2005.)
Study samples consisted of formalin-fixed autopsy tissues from three patients suspected of having Bordetella infection, one suspected of having influenza, and one whose death was attributed to SIDS. Four patients ranged in age from 35 days to 3 months, and one patient was 13 years old. The clinical histories of the five patients varied, but all had a cough and other respiratory symptoms; one patient was immunosuppressed. Four patients had an epidemiologic link with an ill family member. Except for the SIDS patient (patient 4), all patients were on antibiotics and had laboratory evidence of pertussis by culture or PCR.
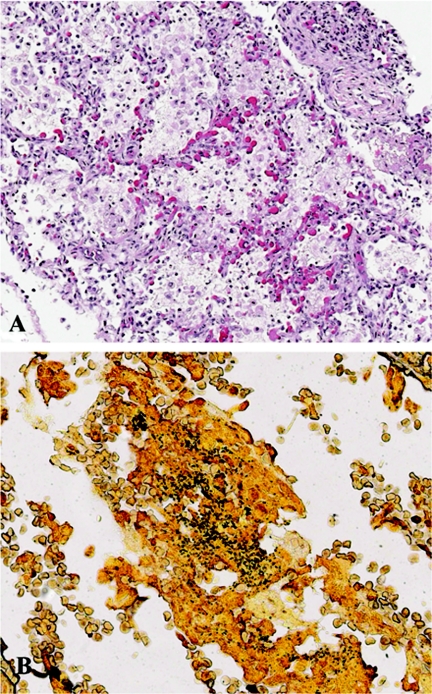
Hematoxylin and eosin staining of lung tissues from the patients showed bronchopneumonia comprised of intra-alveolar infiltrates of neutrophils and macrophages (Fig. 1A), with foci of necrosis and intra-alveolar hemorrhage as the predominant pulmonary pathologies. The trachea of patient 4 exhibited focal areas of denuded epithelium accompanied by mild to moderate submucosal mononuclear cell infiltrates. Silver staining (Steiner's method) demonstrated coccobacilli in all patients (Fig. 1B), while Gram's staining (Brown and Hopps method) showed gram-negative bacilli in only patients 2 and 4.
FIG. 1.
Hematoxylin and eosin (A) and special (B) staining of lung tissues from selected patients with B. pertussis infection. (A) Lung from patient 2 showing intra-alveolar infiltrates comprised predominantly of macrophages and neutrophils, accompanied by fibrin and necrotic debris. Magnification, ×100. (B) Steiner silver staining of lung from patient 3 showing abundant coccobacilli in a bronchiole. Magnification, ×158.
Clinical isolates of Bordetella spp. (Table 1) were fixed in formalin, minced with uninfected tissues, and embedded in paraffin. DNA was extracted from one 10-μm section of each formalin-fixed tissue by use of a QIAamp DNA Mini kit (QIAGEN, Valencia, CA) and was eluted in 100 μl of 10 mM Tris buffer (pH 7.5). To determine whether the extracted DNAs could be amplified by PCR, a 167-bp region of the β-globin gene was amplified from the tissue specimens (6). The PCR amplification assay for B. pertussis was performed using primers IS481F (5′-GATTCAATAGGTTGTATGCATGGTT) and IS481R (5′-TTCAGGCACACAAACTTGATGGGCG), which were designed by Templeton et al. (8). Each PCR mixture consisted of a 300 nM concentration of each primer, 10 μl of DNA extract, high-fidelity PCR master mix (containing 1.5 mM MgCl2 and a 0.2 mM concentration of each deoxynucleoside triphosphate), and an enzyme mixture of Taq and Tgo DNA polymerases in a 50-μl volume. The DNA was denatured for 3 min at 94°C and then subjected to 40 cycles of amplification (94°C for 15 s, 60°C for 20 s, and 72°C for 30 s) followed by a final extension of 10 min at 72°C. The PCR results for the assay of IS481 from different clinical isolates of Bordetella spp. demonstrated that this assay detects both B. pertussis and B. holmesii but not B. parapertussis or B. bronchiseptica (Table 1). This result correlates with previous studies that showed that B. holmesii contains the insertion sequence IS481 at 8 to 10 copies per cell (5). Since IS481 is present at one copy only in some B. bronchiseptica strains (S. Gladbach, S. Hanauer, and U. Reischl, Abstr. 7th Int. Symp. Pertussis, abstr. 34, 2002), the IS481 sequence may not have been present in these particular test strains or the conventional assay may not have detected the sequence because the total DNA concentration was nominal. When we used DNAs extracted from clinical samples of lung tissue infected with Bacillus anthracis, group A Streptococcus, group B Streptococcus, Haemophilus influenzae, Legionella pneumophila, Staphylococcus aureus, Streptococcus pneumoniae, or Yersinia pestis and from liver tissue infected with spotted-fever-group Rickettsia, the IS481 PCR assay did not generate an amplicon. The IS481 PCR assay amplified a 181-bp fragment from lung or trachea specimens from the five patients, suggesting that either B. pertussis or B. holmesii was present.
TABLE 1.
PCR results for clinical isolates of Bordetella spp. used in this study
We next sequenced the 181-bp amplicons of the IS481 gene from the clinical isolates and the five patients. Comparison of the DNA sequences of the 181-bp amplicons of B. pertussis and B. holmesii illustrated one consistent difference between the two species: at nucleotide 100 in all five clinical isolates of B. holmesii, a mixture of nucleotide bases C and A occurred, with C slightly more predominant than A (data not shown). This mixture of C and A has been documented previously (5) and is indicated as “m” at nucleotide 133 in the B. holmesii IS481 insertion sequence (accession number AF349431). In contrast, an A was always present at the same nucleotide position in all five patient and clinical isolates of B. pertussis. An analysis of the sequence from the reverse strand confirmed that a mixture of nucleotide bases G and T occurred at the homologous position in all five B. holmesii isolates, while only a T occurred in the B. pertussis isolates and the five patient isolates. Although the mixture of nucleotides at this position in IS481 appears to be indicative of B. holmesii isolates, an amplicon with an A at this nucleotide position can only be used as suggestive information that a patient might be infected with B. pertussis. The patient history and clinical information are beneficial in differentiating between B. holmesii and B. pertussis. Although B. holmesii is known to cause septicemia (11) and, in some instances, respiratory illnesses (7) in adolescents and adults, the respiratory illness caused by B. holmesii is mild compared with that caused by B. pertussis, and no deaths have been attributed to B. holmesii.
All real-time PCR assays were performed using a LightCycler instrument (Roche Applied Science, Indianapolis, IN). To quantify the amounts of DNA extracted from human tissues, a real-time assay to detect an 80-bp region of the human RNase P gene was performed (Table 2). The real-time IS481 assay targets a region downstream from the inverted repeat that generates a 66-bp amplicon (K.-H. Wu and G. N. Sanden, unpublished data). The region targeted by the real-time IS481 PCR assay is different from that targeted by the conventional IS481 PCR assay. A real-time assay specific for a 55-bp region of the pertussis toxin gene was developed (Wu and Sanden, unpublished data). The weakest PCR signal for the pertussis toxin gene was obtained from patient 5, the same patient with the negative Gram's and Steiner stains (Table 2). The results from real-time IS481 PCRs substantiated the results of the conventional IS481 assay, verifying that all five patients were infected with either B. pertussis or B. holmesii, while the specific real-time pertussis toxin assay, which targets a single-copy gene, demonstrated that all specimens from the five patients were positive for B. pertussis (Table 2). An analysis of the real-time data illustrated that cycle threshold (CT) values for the real-time IS481, pertussis toxin, and RNase P assays are concordant. The IS481 assay generates relatively higher CT values because the insertion element is present in multiple copies, and assay results for specimens with low DNA concentrations suggested that the IS481 assay may be more sensitive than single-copy targets at detecting B. pertussis (Table 2).
TABLE 2.
Analysis of real-time data for the five patients analyzed in this study
| Case no., tissue | RNase P CTa | IS481 CT | IS481 CT/RNase P CT | Pertussis toxin CT | Pertussis toxin CT/RNase P CT |
|---|---|---|---|---|---|
| 1, lung | 27 | 22.2 | 0.82 | 28.4 | 1.05 |
| 2, lungb | 26.8, 30.8 | 19.57, 22.88 | 0.73, 0.74 | 26.43, 29.86 | 0.99, 0.97 |
| 3, lung | 31 | 25.62 | 0.83 | 32.33 | 1.04 |
| 4, tracheac | 30.1, 31.15 | 28.52, 28.89 | 0.95, 0.93 | 34.53, 34.79 | 1.15, 1.12 |
| 5, lungb | 34.2, 33 | 32.51, 29.26 | 0.95, 0.89 | 37.03, 35.11 | 1.08, 1.06 |
The CT value is the cycle threshold, the time at which the fluorescence intensity is greater than the background fluorescence.
Two different blocks of lung tissues (indicated by the two CT values) were analyzed for cases 2 and 5.
Two independent assays (indicated by the two CT values) were performed with the same block of tracheal tissue.
This study demonstrates the utility of various PCR assays for the detection of B. pertussis DNA in formalin-fixed tissue specimens. For further analysis, lung tissue samples were obtained from an additional 10 infants whose deaths were suspected to be due to pertussis. PCR analysis utilizing the assays described in this study demonstrated that the infants were infected with B. pertussis (data not shown). Of the five patients diagnosed with B. pertussis infection in this study, the epidemiologic data support the PCR results for four who were in contact with ill family members. Patient 2 presented as a laboratory-confirmed influenza case without an epidemiologic link but was determined to be infected with B. pertussis by hospital PCR tests and the PCR tests in this study. Patient 4 presented as a presumed SIDS-related fatality; however, conventional and real-time PCR tests, an immunohistochemical assay (C. D. Paddock, unpublished data), and an epidemiologic link to a known B. pertussis case established that the infant was infected with B. pertussis. Studies by several groups using PCR analysis (1, 2) or surveys of statistical records and epidemiologic studies (3, 4) have suggested that between 5% and 18% of SIDS cases may be attributable to B. pertussis. Future studies using molecular and immunohistochemical assays are planned to explore the role of B. pertussis in SIDS deaths.
Acknowledgments
We thank Laurie Mueller for her assistance with graphics, Jeanine Bartlett for her special stains, Pamela Cassiday for growing strains of the Bordetella spp., and Claudia Chesley for her editorial expertise. In addition, we thank the following people for submitting tissue specimens for analysis and consultation: B. Fulton, Pathology and Laboratory Medicine Spectrum Health, Grand Rapids, MI; J. Wyatt-Ashmead, The University of Mississippi Medical Center; S. Ladd-Wilson, Oregon Health Division/EIP Department of Human Services; D. Drehner, Children's Hospital and Clinic, Minneapolis, MN; and K. Lemon, Children's Hospital Medical Center, Boston, MA.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
REFERENCES
- 1.Heininger, U., K. Stehr, G. Schmidt-Schlapfer, R. Penning, R. Vock, W. Kleemann, and J. D. Cherry. 1996. Bordetella pertussis infections and sudden unexpected deaths in children. Eur. J. Pediatr. 155:551-553. [DOI] [PubMed] [Google Scholar]
- 2.Heininger, U., W. J. Kleemann, J. D. Cherry, and Sudden Infant Death Syndrome Study Group. 2004. A controlled study of the relationship between Bordetella pertussis infections and sudden unexpected deaths among German infants. Pediatrics 114:9-15. [DOI] [PubMed] [Google Scholar]
- 3.Lindgren, C., J. Milerad, and H. Lagercrantz. 1997. Sudden infant death and prevalence of whooping cough in the Swedish and Norwegian communities. Eur. J. Pediatr. 156:405-409. [DOI] [PubMed] [Google Scholar]
- 4.Nicoll, A., and A. Gradner. 1988. Whooping cough and unrecognized postperinatal mortality. Arch. Dis. Child. 63:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reischl, U., N. Lehn, G. N. Sanden, and M. J. Loeffelholz. 2001. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J. Clin. Microbiol. 39:1963-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saiki, R. K., D. Gelfand, S. Stoffel, S. Scharf, R. Higuchi, G. Horn, K. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 7.Tang, Y.-W., M. K. Hopkins, C. P. Kolbert, P. A. Hartley, P. J. Severance, and D. H. Persing. 1998. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin. Infect. Dis. 26:389-392. [DOI] [PubMed] [Google Scholar]
- 8.Templeton, K. E., S. A. Scheltinga, A. van der Zee, B. M. W. Diederen, A. M. Kruijssen, H. Goossens, E. Kuijper, and E. C. J. Claas. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J. Clin. Microbiol. 41:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Zee, A., C. Agterberg, M. Peeters, J. Schellekens, and F. R. Mooi. 1993. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J. Clin. Microbiol. 31:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitek, C. R., F. B. Pascual, A. L. Baughman, and T. V. Murphy. 2003. Increase in deaths from pertussis among young infants in the United States in the 1990s. Pediatr. Infect. Dis. J. 22:628-634. [DOI] [PubMed] [Google Scholar]
- 11.Weyant, R. S., D. G. Hollis, R. E. Weaver, M. F. Amin, A. G. Steigerwalt, S. P. O'Connor, A. M. Whitney, M. I. Daneshvar, C. W. Moss, and D. J. Brenner. 1995. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]