Abstract
Variations in time and space of a clonal group of Escherichia coli O165:H25 on a cattle farm were monitored. The virulence marker pattern (stx genes, eae gene, hlyEHEC gene, katP gene, espP gene, efa gene) suggests that E. coli O165:H25 of bovine origin may represent a risk for human infection.
Shiga toxin-producing Escherichia coli (STEC) is a group of zoonotic enteric pathogens (29). Human infections with some STEC serotypes, also designated enterohemorrhagic Escherichia coli (EHEC), result in hemorrhagic or nonhemorrhagic diarrhea, which may be complicated by hemorrhagic colitis (HC) and several renal sequelae, including the hemolytic-uremic syndrome (HUS) (20, 34, 41, 52). The mechanisms by which EHEC strains cause disease are not completely understood. The virulence factors include production of two major phage-encoded toxins, Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2), which can be produced alone or in combination. Stx1 and Stx2 are thought to cause the vascular endothelial damage observed in patients with HC and HUS (53). In addition to Stx, EHEC strains possess other virulence factors, such as the ability to cause attaching and effacing (eae) lesions in the large intestine (26). They often contain a plasmid that carries other potential virulence genes, such as an enterohemolysin gene (hlyEHEC), a catalase peroxidase gene (katP), and an extracellular serine protease gene (espP) (9, 10, 11, 45). The efa1 (E. coli factor for adherence) gene represents another intestinal colonization factor in an EHEC O111:H- strain (30). The efa1 gene of O111:H- is 99.9% homologous to the lifA gene in enteropathogenic E. coli. The efa1 locus is not physically linked to the locus for enterocyte effacement pathogenicity island (51).
Ruminants, especially cattle, are considered the primary reservoir for human EHEC infections (23). In this study, 38 bovine E. coli O165:H25 isolates were characterized to assess their potential to cause EHEC disease in humans. These isolates were detected over a 4-month period in eight different animals (Table 1) from a single group of beef cattle during a long-term study (19). Sporadic cases of human infections with O165:H25 and O165:H- in Europe and Canada have been described previously (6, 15, 17, 24). The four German O165:H25 and O165:H- strains of human origin (kindly supplied by H. Tschäpe, Robert Koch-Institute, Wernigerode, Germany) used in our study for comparison were associated with diarrhea in patients (54). Human HUS cases caused by O165 isolates have been reported from Denmark (15) and Germany (17).
TABLE 1.
Strains examined
| Sampling day or yeara | Source | Strain | Serotype | Virulence profile
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1 gene | stx2 gene | Stx1 (toxin) | Stx2 (toxin) | stx2- Subtype | eae Subtype | efa1/lifA gene | hlyEHEC gene | katP gene | espP gene | Plasmid(s) (bp) | Genetic subcluster | ||||
| Day 1 | Cattle 17 | 02/17/009-9 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 7 |
| Cattle 24 | 02/24/007-1 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | |
| 02/24/007-2 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/24/007-3 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/24/007-4 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/24/007-5 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/24/007-6 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 3 | ||
| 02/24/007-7 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/24/007-9 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/24/007-10 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| Cattle 26 | 02/26/006-1 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | |
| 02/26/006-2 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/26/006-3 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/26/006-4 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 9 | ||
| 02/26/006-5 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 9 | ||
| 02/26/006-6 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | − | + | 67, 55 | 9 | ||
| 02/26/006-7 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | − | − | − | 55 | 4 | ||
| 02/26/006-8 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 5 | ||
| 02/26/006-9 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | − | − | − | 55 | 6 | ||
| 02/26/006-10 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| Day 28 | Cattle 25 | 02/25/007-1 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 |
| 02/25/007-4 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/25/007-5 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | − | − | − | 55 | 1 | ||
| 02/25/007-6 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/25/007-7 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/25/007-8 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/25/007-9 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| 02/25/007-10 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | ||
| Day 56 | Cattle 9 | 02/09/010-1 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 8 |
| 02/09/010-2 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 8 | ||
| Cattle 18 | 02/18/011-1 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70 | 10 | |
| 02/18/011-5 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 10 | ||
| 02/18/011-6 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 10 | ||
| Cattle 25 | 02/25/008-1 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 1 | |
| 02/25/007-3 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 2 | ||
| Day 119 | Cattle 19 | 02/19/013-7 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | − | + | + | + | 70, 55 | 11 |
| 02/19/013-8 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 8 | ||
| 02/19/013-10 | O165:H25 | − | + | − | + | stx2-EDL933 | ɛ | + | + | + | + | 70, 55 | 8 | ||
| 1998 | Human, Germany, diarrhea | 98-4982b | O165:H- | − | + | − | + | stx2/2c | ɛ | + | + | + | + | 75 | |
| 1998 | Human, Germany, diarrhea | 98-8419-1b | O165:H- | − | + | − | + | stx2/2c | ɛ | + | + | + | + | 95, 75 | |
| 1999 | Human, Germany, diarrhea | 99-2258b | O165:H25 | − | + | − | + | stx2/2c | ɛ | + | + | + | + | 95, 75 | |
| 2002 | Human, Germany, diarrhea | 02-11228b | O165:H25 | − | + | − | + | stx2/2c | ɛ | + | + | + | + | 95, 75 | |
On days −120 to 0 O165:H25 was not detected on eight occassions, on day 91 O165:H25 was not detected, and on days 147 to 511 O165:H25 was not detected by 17 investigations.
Kindly provided by H. Tschäpe.
Typing and subtyping of genes (stx1 and/or stx2, eae, hlyEHEC, katP, and espP) associated with STEC were performed by LightCycler fluorescence PCR (40) and different Block cycler PCRs (Tables 2 and 3). A complete pattern of virulence markers was detected in most bovine isolates examined. An stx2 gene, but not an stx1 gene, was present in all O165 strains (Table 1). EHEC strains with stx2 genes are significantly more frequently associated with HUS and other severe diseases than isolates with an stx1 gene, which are more often associated with uncomplicated diarrhea or healthy individuals (6, 36). Stx2 is closely related to a family of Stx2 variants or alleles, which includes Stx2c (48), Stx2d (36), Stx2e (56), and Stx2f (47), although Stx2c and Stx2d are produced by STEC strains isolated from humans (36, 38, 43, 44, 48, 50). Additional genetic variants of the stx2 gene have been described (5, 14, 28, 55). In contrast to STEC strains harboring stx2 gene variants, however, STEC strains with the stx2 genotype were significantly associated with HUS (17). An stx2 gene with the stx2-EDL933 genotype was found in all O165 isolates tested (Table 1). The nucleotide sequences of the A and B subunits of the stx2 gene of the bovine O165:25 strain 02/09/010-1 (GenBank accession number AY652745) were identical to the sequences of the stx2 gene of EHEC type strain EDL933 (35), a typical O157:H7 strain isolated from a HUS patient, with the sequences of the gene of bacteriophage 933W (37), and with the sequences of stx2 genes of other E. coli O157:H7 strains of human origin isolated from EHEC outbreaks (25, 27). All bovine O165:H25 strains produced an Stx2 with high cytotoxicity for Vero cells as determined by an Stx enzyme-linked immunosorbent assay and by a Vero cell neutralization assay (49).
TABLE 2.
Oligonucleotide primers and LightCycler hybridization probes used for LightCycler PCR
| Target(s) | Primer | Sequence (5′-3′)a | Reference |
|---|---|---|---|
| stx1 and stx2 | STEC-1 | GA(AG) C(AG)A AAT AAT TTA TAT GTG | 40 |
| STEC-2 | TGA TGA TG(AG) CAA TTC AGT AT | 33 | |
| stx1 | STEC-I HP-1 | TTT ACG TTT TCG GCA AAT ACA GAG GGG AT-(FL) | 40 |
| STEC-I HP-2 | (Red 640)-TCG TAC AAC ACT GGA TGA TCT CAG TGG G-Ph | ||
| stx2 | STEC-II HP-1 | TCA GGC ACT GTC TGA AAC TGC TCC TGT GTA-(FL) | 40 |
| STEC-II HP-2 | (Red 705)-ACC ATG ACG CCG GGA GAC GTG GAC CT-Ph | ||
| eae | eaeAF | GAC CCG GCA CAA GCA TAA GC | 32 |
| eaeAR | CC ACCT GCA GCA ACA AGA GG | ||
| eae | eae HP1 | ACA GTT CTG AAA GCG AAA TGA TGA AGG c-(FL) | 40 |
| eae HP2 | (Red 640)-CCT GGT CAG CAG ATC ATT TTG CCA CT-Ph | ||
| hlyEHEC | hlyAF | GCA TCA TCA AGC GTA CGT TCC | 32 |
| hlyAR | AAT GAG CCA AGC TGG TTA AGC T | ||
| hlyEHEC | hlyA HP1 | GCA TGG CTC TTG ATG AAT TGC T-(FL) | 40 |
| hlyA HP2 | (Red 705)-CAA CGG GAA GGA GAG GAT ATA AGT CAG-Ph |
FL, fluorescein; Red 640, LC Red 640-N-hydroxy-succinimide ester; Red 705, LC Red 705-phosphoramidite; Ph, 3-phosphate.
TABLE 3.
PCR primers used for detection and characterization of STEC by conventional PCR
| Target | Primer | Sequence (5′-3′) | PCR conditions
|
Product length (bp) | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Denaturation
|
Annealing
|
Extension
|
No. of cycles | ||||||||
| Temp (°C) | Time (s) | Temp (°C) | Time (s) | Temp (°C) | Time (s) | ||||||
| stxB2/2c | GK3 | ATG AAG AAG ATG TTT ATG | 94 | 30 | 52 | 60 | 72 | 40 | 35 | 260 | 22 |
| GK4 | TCA GTC ATT ATT AAA CTG | ||||||||||
| stxB2d | VT2-cm | AAG AAG ATA TTT GTA GCG G | 94 | 30 | 52 | 60 | 72 | 60 | 35 | 256 | 36 |
| VT2-f | TAA ACT GCA CTT CAG CAA AT | ||||||||||
| stxB2e | FK1 | CCG GAT CCA AGA AGA TGT TTA TAG | 94 | 30 | 55 | 60 | 72 | 40 | 35 | 280 | 16 |
| FK2 | CCC GAA TTC TCA GTT AAA CTT CAC C | ||||||||||
| stxB2f | 128-1 | AGA TTG GGC GTC ATT CAC TGG TTG | 94 | 30 | 57 | 60 | 72 | 60 | 35 | 428 | 47 |
| 128-2 | TAC TTT AAT GGC CGC CCT GTC TCC | ||||||||||
| α-eae | SK1 | CCC GAA TTC GGC ACA AGC ATA AGC | 94 | 30 | 55 | 60 | 72 | 120 | 30 | 2,807 | 31 |
| LP2 | CCC GAA TTC GGC ACA AGC ATA AGC | ||||||||||
| β-eae | SK1 | CCC GAA TTC GGC ACA AGC ATA AGC | 94 | 30 | 55 | 60 | 72 | 120 | 30 | 2,287 | 31 |
| LP4 | CCC GTG AT ACCA GTA CCA ATT ACG GTC | ||||||||||
| γ-eae | SK1 | CCC GAA TTC GGC ACA AGC ATA AGC | 94 | 30 | 45 | 60 | 72 | 120 | 3 | 2,792 | 31 |
| LP3 | CCC GAA TTC TTA TTC TAC ACA AAC CGC | 94 | 30 | 52 | 60 | 72 | 120 | 28 | |||
| δ-eae | Int-d | TAC GGA TTT TGG GGC AT | 95 | 20 | 45 | 60 | 72 | 60 | 30 | 544 | 1 |
| Int-Ru | TTT ATT TGC AGC CCC CCA T | ||||||||||
| ɛ-eae | SK1 | CCC GAA TTC GGC ACA AGC ATA AGC | 94 | 30 | 55 | 60 | 72 | 120 | 30 | 2,608 | 31 |
| LP5 | AGC TCA CTC GTA GAT GAC GGC AAG CG | ||||||||||
| katP | wkat-B | CTT CCT GTT CTG ATT CTT CTG G | 94 | 30 | 56 | 60 | 72 | 150 | 30 | 2,125 | 10 |
| wkat-F | AAC TTA TTT CTC GCA TCA TCC | ||||||||||
| espP | EspA | AAA CAG CAG GCA CTT GAA CG | 94 | 30 | 56 | 60 | 72 | 150 | 30 | 1,830 | 11 |
| EspB | GGA GTC GTC AGT CAG TAG AT | ||||||||||
| lifA/efa1 | lifA1 | AGC CAT TCC ATC AAT CCG AT | 95 | 30 | 50 | 60 | 72 | 60 | 30 | 532 | 7 |
| lifA2 | TCC CTG CCA AAC TAC CGA CAC | ||||||||||
| lifA/efa1 | lifA3 | CAG CTA CAG GAG ACC GTT TTT | 95 | 30 | 50 | 60 | 72 | 60 | 30 | 560 | 7 |
| lifA4 | CAA TAT CAG GCC AAT CAA | ||||||||||
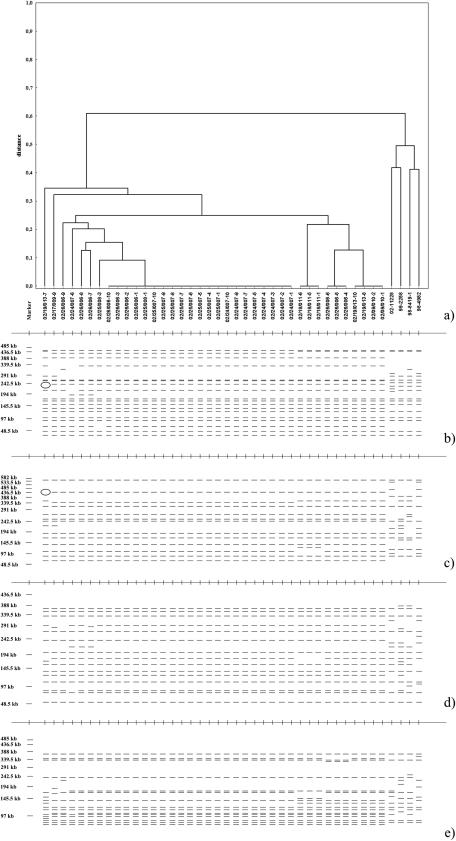
Not only factors that influence basal and inducible Stx production are important in STEC pathogenesis. In previous studies, it has been suggested that the eae and hlyEHEC genes likely contribute to STEC pathogenicity (3, 6, 42). Ritchie et al. (42) found both of these genes in all HUS-associated STEC isolates that they analyzed. The stx2 genes were present in combination with eae genes in all O165 isolates that we obtained (Table 1). To date, 10 distinct variants of eae have been described (13, 21, 31, 39). Some serotypes were closely associated with a particular intimin variant (4, 12, 13, 55). Our study confirmed these associations. Like the O103 isolates, all bovine and human E. coli O165 strains were grouped into the ɛ-eae subgroup. Also, nucleotide sequencing of the bovine O165:H25 strain 02/09/010-1 (GenBank accession number AF479581) revealed a high level of sequence homology (99.7%) to the eae gene of an O103:H2 strain (31). E. coli O103:H2 strains have frequently been associated with human HUS cases in Europe. Like the eae gene, the hlyEHEC gene was found in association with severe disease in humans (45, 46). In our study, the hlyEHEC gene was detected in the O165 strains in which a 70-kb plasmid was also found (Table 1). The presence of a 70-kb plasmid was associated with the occurrence of additional virulence markers, such as the espP gene, and all but one isolate contained the katP gene (10, 11). The reason for the slightly smaller size (67 kb) of the plasmid in this isolate may be linked to fact that the katP gene was not present in this isolate (Table 1). The efa1 genes were detected in 37 of 38 bovine O165:H25 isolates with two DNA probes by colony hybridization, and the results were confirmed by Southern hybridization after pulsed-field gel electrophoresis (PFGE); in this analysis the DNA probes were labeled with digoxigenin (DIG), primers lifA1 and lifA2 and primers lifA3 and lifA4 (Table 3) were used with a PCR DIG probe synthesis kit (Roche Diagnostics, Mannheim, Germany), and DIG Easy Hyb solution (Roche) was used for prehybridization and hybridization. The efa-1 gene was located on an approximately 240-kb XbaI fragment or on an approximately 440-kb NotI fragment. These fragments were missing in the isolate that was negative as determined by colony hybridization (Fig. 1). To determine this, slices of the plugs were digested for 4 h with XbaI, NotI (New England Biolabs GmbH, Frankfurt am Main, Germany), BlnI (AvrII), or SpeI (Amersham Biosciences Inc., Buckinghamshire, United Kingdom). The resulting fragments were separated in a 1.0% agarose gel (SeaKem Gold agarose; Cambrex) in 0.5× Tris-borate-EDTA at 10°C with a CHEF Mapper XA system. The pulse times for XbaI and NotI digests were increased from 5 to 50 s (gradient, 6 V/cm) during 25 h at a constant angel of 120°. The switch time values for BlnI and SpeI were set using the Auto Algorithm function of the CHEF Mapper XA to separate fragments in the range from 50 to 450 kb (BlnI) or from 30 to 350 kb (SpeI). All fragments larger than 45 kb (up to 27 fragments with XbaI, up to 24 fragments with NotI, up to 25 fragments with BlnI, and up to 29 fragments with SpeI) were included in the clonal analysis of the isolates.
FIG. 1.
(a) Dendrogram of DNA divergence (distance) generated by using the Dice coefficient for pairwise comparisons of banding patterns for XbaI, NotI, BlnI, and SpeI restriction of potential EHEC O165:H25 and O165:H- isolates. (b to e) Schematic representations of restriction patterns of bovine and human E. coli O165:H25 and O165:H- strains after digestion with XbaI (b), NotI (c), BlnI (d), and SpeI (e). Ellipses indicate the missing XbaI and NotI fragments in strain 02/19/013-7. The efa1 gene is located at this position in the other bovine strains.
We also analyzed the spatial and temporal behavior of the clonal group of O165:H25 strains in the herd by genomic typing with PFGE (Fig. 1). During a 3-year monitoring program on four cattle farms (19), the O165:H25 clone was detected on only one farm for 4 months. This serotype was not detected before or after this period, although many other potential EHEC strains belonging to other serotypes were found in this herd (19). Twenty O165:H25 isolates were found in four different cattle on the first date of detection. Different subclusters were already present and were even isolated from a single animal (Table 1). These results suggest that the O165 clone either had been introduced into the farm shortly before the first detection or was the result of recombination due to horizontal gene transfer (8, 18). The occurrence of different subclusters at the same time could have been the result of interactions between different bacteria or between bacteria and the host in the bovine intestine. At later sampling dates the number of O165:H25 isolates was reduced. The dominant subcluster, subcluster 1, could still be found, but many other subclusters were detected at the same time. The genetic distance between the O165:H25 isolates increased, and in some isolates plasmid-encoded virulence markers or complete virulence plasmids were missing. Variations in the O165:H25 clonal group might have been caused by increasing competition between the bacterial populations of various subtypes in the bovine intestine or by potential interactions between the O165:H25 EHEC and the host. The O165:H25 clonal group finally disappeared from the herd after it had persisted for 4 months. Perhaps the loss of the efa-1 gene in one isolate obtained on the last date when O165:H25 was isolated from the herd can help us understand why the clone disappeared. Efa1 is considered an E. coli factor for colonization of the bovine intestine by non-O157 STEC (51). Also, this O165:H25 strain exhibited the greatest genetic distance compared to the remaining strains in the clonal group (Fig. 1). The distances were calculated from the fragmentation patterns produced by each of the four PFGE enzymes by using the RAPDistance program, version 1.04 (2). The Euclidean distance in three dimensions was not calculated because all O165:H25 strains were isolated from the same farm (i.e., the geographic distance was zero for all isolates). A cluster analysis, using the unweighted pair group method with arithmetic averages, was performed for PFGE by using Statistica 6.1 for Windows (StatSoft Inc., Tulsa, OK). In any case, our results suggest that the persistence of a distinct clone of EHEC may be limited in time and space (i.e., in a cattle herd). This is apparently not a unique property of O165:H25, since we obtained similar results for clonal groups of other EHEC serotypes (O26:H11 and O157:H7) in cattle. The four human O165H25/H- isolates belonged to different other clonal groups (Fig. 1).
In conclusion, bovine O165:H25 isolates can carry virulence factors of EHEC that are associated with EHEC-related disease in humans, particularly HC and HUS. Therefore, strains of bovine origin may represent a considerable potential risk for human infection.
Acknowledgments
We thank J. Bockemühl, H. Tschäpe, T. Kuzius, and A. Fruth for serotyping the E. coli strains and H. Tschäpe for providing E. coli O165:H25 and O165:H- strains of human origin.
This study was funded by the German Federal Ministry of Consumer Protection, Food and Agriculture.
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. S., Gibbs, A. J., Peakall, R., and Weiller, G. 1994. The RAPDistance package, version 1.04. [Online.] http://life.anu.edu.au/molecular/software/rapd.html.
- 3.Barrett, T. J., J. B. Kaper, A. E. Jerse, and I. K. Wachsmuth. 1992. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J. Infect. Dis. 165:979-980. [DOI] [PubMed] [Google Scholar]
- 4.Bertin, Y., K. Boukhors, V. Livrelli, and C. Martin. 2004. Localization of the insertion site and pathotype determination of the locus of enterocyte effacement of Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 70:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertin, Y., K. Boukhors, N. Pradel, V. Livrelli, and C. Martin. 2001. Stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böhnke, U. 2002. Untersuchung zur Lokalisation und Regulation des als “lymphocyte inhibitory factor” (lifA) oder “EHEC factor for adherence” (efa1) bezeichneten Gens des bovinen enterohämorrhagischen E. coli-Stammes RW1374 (O103:H2). Diplomarbeit. Freie Universität Berlin, Berlin, Germany.
- 8.Brunder, W., and H. Karch. 2000. Genome plasticity in Enterobacteriaceae. Int. J. Med. Microbiol. 290:153-165. [DOI] [PubMed] [Google Scholar]
- 9.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 10.Brunder, W., H. Schmidt, and H. Karch. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305-3315. [DOI] [PubMed] [Google Scholar]
- 11.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7, cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 12.China, B., F. Goffaux, V. Pirson, and J. Mainil. 1999. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 178:177-182. [DOI] [PubMed] [Google Scholar]
- 13.China, B., E. Jacquemin, A. C. Devrin, V. Pirson, and J. Mainil. 1999. Heterogeneity of the eae genes in attaching/effacing Escherichia coli from cattle: comparison with human strains. Res. Microbiol. 150:323-332. [DOI] [PubMed] [Google Scholar]
- 14.De Baets, L., D. T. Van, I., M. De Filette, D. Pierard, L. Allison, H. De Greve, J. P. Hernalsteens, and H. Imberechts. 2004. Genetic typing of Shiga toxin 2 variants of Escherichia coli by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70:6309-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ethelberg, S., K. E. Olsen, F. Scheutz, C. Jensen, P. Schiellerup, J. Enberg, A. M. Petersen, B. Olesen, P. Gerner-Smidt, and K. Molbak. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke, S., D. Harmsen, A. Caprioli, D. Pierard, L. H. Wieler, and H. Karch. 1995. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J. Clin. Microbiol. 33:3174-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 18.Fruth, A., R. Prager, A. Friedrich, T. Kuczius, P. Roggentin, H. Karch, A. Ammon, J. Bockemühl, and H. Tschäpe. 2002. Infektionen des Menschen durch enterohämorrhagische Escherichia coli (EHEC) in der Bundesrepublik Deutschland von 1998 bis 2001—Prävalenz und Typenspektrum. Bundesgesundheitsblatt Gesundheitsforsch. Gesundheitsschutz 45:715-721. [DOI] [PubMed] [Google Scholar]
- 19.Geue, L., M. Segura-Alvarez, F. J. Conraths, T. Kuczius, J. Bockemuhl, H. Karch, and P. Gallien. 2002. A long-term study on the prevalence of shiga toxin-producing Escherichia coli (STEC) on four German cattle farms. Epidemiol. Infect. 129:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 21.Jores, J., K. Zehmke, J. Eichberg, L. Rumer, and L. H. Wieler. 2003. Description of a novel intimin variant (type zeta) in the bovine O84:NM verotoxin-producing Escherichia coli strain 537/89 and the diagnostic value of intimin typing. Exp. Biol. Med. 228:370-376. [DOI] [PubMed] [Google Scholar]
- 22.Karch, H., H. I. Huppertz, J. Bockemühl, H. Schmidt, A. Schwarzkopf, and R. Lissner. 1997. Shiga toxin-producing Escherichia coli infections in Germany. J. Food Prot. 11:1454-1457. [DOI] [PubMed] [Google Scholar]
- 23.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keskimaki, M., M. Saari, T. Heiskanen, and A. Siitonen. 1998. Shiga toxin-producing Escherichia coli in Finland from 1990 through 1997: prevalence and characteristics of isolates. J. Clin. Microbiol. 36:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, K., K. Yokoyama, Y. Kubota, C. H. Yutsudo, S. Kimura, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, T. Iida, K. Yamamoto, M. Onishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 26.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muniesa, M., M. de Simon, G. Prats, D. Ferrer, H. Panella, and J. Jofre. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakao, H., K. Kimura, H. Murakami, T. Maruyama, and T. Takeda. 2002. Subtyping of Shiga toxin 2 variants in human-derived Shiga toxin-producing Escherichia coli strains isolated in Japan. FEMS Immunol. Med. Microbiol. 34:289-297. [DOI] [PubMed] [Google Scholar]
- 29.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 31.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paton, A. W., J. C. Paton, P. N. Goldwater, and P. A. Manning. 1993. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J. Clin. Microbiol. 31:3063-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 36.Pierard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandran, V., M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2001. The common ovine Shiga toxin 2-containing Escherichia coli serotypes and human isolates of the same serotypes possess a Stx2d toxin type. J. Clin. Microbiol. 39:1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 40.Reischl, U., M. T. Youssef, J. Kilwinski, N. Lehn, W. L. Zhang, H. Karch, and N. A. Strockbine. 2002. Real-time fluorescence PCR assays for detection and characterization of Shiga toxin, intimin, and enterohemolysin genes from Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 40:2555-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remuzzi, G., and P. Ruggenenti. 1998. The hemolytic uremic syndrome. Kidney Int. Suppl. 66:S54-S57. [PubMed] [Google Scholar]
- 42.Ritchie, J. M., P. L. Wagner, D. W. Acheson, and M. K. Waldor. 2003. Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 69:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russmann, H., E. Kothe, H. Schmidt, S. Franke, D. Harmsen, A. Caprioli, and H. Karch. 1995. Genotyping of Shiga-like toxin genes in non-O157 Escherichia coli strains associated with haemolytic uraemic syndrome. J. Med. Microbiol. 42:404-410. [DOI] [PubMed] [Google Scholar]
- 44.Russmann, H., H. Schmidt, A. Caprioli, and H. Karch. 1994. Highly conserved B-subunit genes of Shiga-like toxin II variants found in Escherichia coli O157 strains. FEMS Microbiol. Lett. 118:335-340. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, H., C. Kernbach, and H. Karch. 1996. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:907-914. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segura-Alvarez, M., H. Richter, F. J. Conraths, and L. Geue. 2003. Evaluation of enzyme-linked immunosorbent assays and a PCR test for detection of Shiga toxins for Shiga toxin-producing Escherichia coli in cattle herds. J. Clin. Microbiol. 41:5760-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephan, R., and L. E. Hoelzle. 2000. Characterization of Shiga toxin type 2 variant B-subunit in Escherichia coli strains from asymptomatic human carriers by PCR-RFLP. Lett. Appl. Microbiol. 31:139-142. [DOI] [PubMed] [Google Scholar]
- 51.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su, C., and L. J. Brandt. 1995. Escherichia coli O157:H7 infection in humans. Ann. Intern. Med. 123:698-714. [DOI] [PubMed] [Google Scholar]
- 53.Tesh, V. L., and A. D. O'Brien. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 5:1817-1822. [DOI] [PubMed] [Google Scholar]
- 54.Tschäpe, H. 2003. Personal communication.
- 55.Vernozy-Rozand, C., M. P. Montet, Y. Bertin, F. Trably, J. P. Girardeau, C. Martin, V. Livrelli, and L. Beutin. 2004. Serotyping, stx2 subtyping, and characterization of the locus of enterocyte effacement island of Shiga toxin-producing Escherichia coli and E. coli O157:H7 strains isolated from the environment in France. Appl. Environ. Microbiol. 70:2556-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]