Abstract
We developed single-point genome signature tags (SP-GSTs), a generally applicable, high-throughput sequencing-based method that targets specific genes to generate identifier tags from well-defined points in a genome. The technique yields identifier tags that can distinguish between closely related bacterial strains and allow for the identification of microbial community members. SP-GSTs are determined by three parameters: (i) the primer designed to recognize a conserved gene sequence, (ii) the anchoring enzyme recognition sequence, and (iii) the type IIS restriction enzyme which defines the tag length. We evaluated the SP-GST method in silico for bacterial identification using the genes rpoC, uvrB, and recA and the 16S rRNA gene. The best distinguishing tags were obtained with the restriction enzyme Csp6I upstream of the 16S rRNA gene, which discriminated all organisms in our data set to at least the genus level and most organisms to the species level. The method was successfully used to generate Csp6I-based tags upstream of the 16S rRNA gene and allowed us to discriminate between closely related strains of Bacillus cereus and Bacillus anthracis. This concept was further used successfully to identify the individual members of a defined microbial community.
A variety of comprehensive DNA-based fingerprinting techniques have been developed to characterize and compare whole genomes of organisms, either independently or as members of communities. These techniques include amplified fragment length polymorphism (31), terminal restriction fragment length polymorphism (17), denaturing gradient gel electrophoresis (19), amplified rRNA gene restriction analysis (27), restriction landmark genome scanning (12), and automated ribosomal intergenic spacer analysis (11). The disadvantages of these techniques are that they perform poorly when comparing data from different experiments and when identifying novel organisms.
An emerging alternative approach to studying microbial communities is the use of microarrays designed to detect specific sequences from important lineages of microorganisms known or suspected to be present in a particular population (16, 21, 22). While this approach can provide a comprehensive quantitative survey for the presence or absence of a particular sequence, the technique has a closed architecture; i.e., it cannot identify novel sequences, nor can it easily distinguish between two or more closely related sequences in mixed populations. For microbial community analysis to be meaningful, the ability to identify previously uncharacterized members and to discriminate between closely related organisms in a population is essential.
The improvement of sequencing technologies has made metagenome shotgun sequencing of an environmental sample feasible; however, most environmental communities are far too complex to be fully sequenced in this manner. Reconstruction of community metagenomes was initially attempted for viral communities in the ocean and in human feces (2-4) and has since been applied on samples from the Sargasso Sea (29) and an acid mine drainage biofilm (25). Most marine communities, however, are far richer in species diversity, on the order of 100 to 200 species per ml of water (8, 9), further complicating sequencing and assembly efforts. Soil communities are even more complex, with an estimated species richness on the order of 4,000 species per gram of soil (8, 9, 24). Sequencing a soil community's metagenome will require technological developments aimed at increasing sequencing capacity and data processing, along with more cost-effective sequencing chemistries.
Recently, serial analysis of ribosomal sequence tags (SARST) was developed as a novel technique for characterizing microbial community composition. The SARST method captures sequence information from concatenates of short PCR amplicons (tags) derived from either the V1 (20) or V6 hypervariable regions (15) of 16S rRNA genes from complex bacterial populations. The major advantage of the SARST method is the high-throughput generation of sequence data that can be directly used for species identification and comparisons between different experiments.
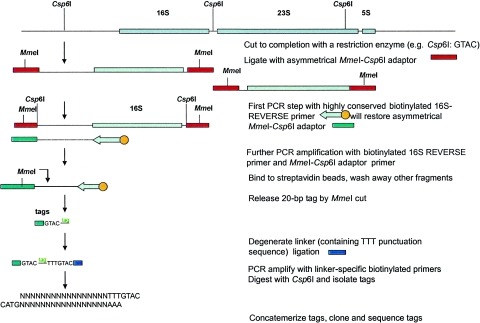
Genome signature tags (GSTs) were developed for use in a cost-effective sequencing-based method to identify and quantitatively analyze genomic or mixtures of genomic DNA (10). In silico analysis of the 168 entries in the current NCBI database of completely sequenced genomes indicates that in many cases the individual GST sequences provided sufficient specificity for species identification. This result prompted us to look for fragmenting enzymes that would generate only one or a few informative tags per organism, which in turn would reduce the complexity of the tag libraries and decrease the amount of sequencing required to characterize complex microbial communities. Since we were unable to identify a universal fragmenting enzyme that would generate a limited number of tags from all the listed genomes, we decided to devise a modified approach that uses conserved gene sequences in place of the requirement for a fragmenting enzyme. Based on the position of the conserved region and the orientation of the primer, single-point GSTs (SP-GSTs) can be generated internally or externally for any gene of interest, such as the 16S rRNA, rpoC, recA, and uvrB genes. This new approach is schematically outlined for the 16S rRNA gene in Fig. 1. In this paper we describe the application of this method to discriminate between closely related strains of Bacillus cereus and Bacillus anthracis and to identify the individual members of a defined microbial community.
FIG. 1.
Schematic representation of the SP-GST approach on the 16S rRNA gene. Tags are generated upstream of a conserved domain (e.g., position 8 to 27 in the 16S rRNA gene). DNA is first cleaved to completion with Csp6I, the anchoring enzyme. The free cohesive ends are ligated with an asymmetrical oligonucleotide cassette that restores the recognition sequence for the anchoring enzyme and places an MmeI recognition sequence immediately adjacent to the restored sequence. A biotinylated primer specific for the region of position 8 to 27 in the 16S rRNA gene and pointing outward of this gene is used in a first PCR cycle to linearly amplify the region between this specific domain and the most proximal site for the anchoring enzyme. This will result in the synthesis of the complementary strand of the linker fragment. The resulting single-stranded fragment is then exponentially amplified using a primer unique to the restored sequence of the MmeI cassette and the domain-specific primer. The biotinylated products are bound to streptavidin-coated magnetic beads and then digested with MmeI to release the tags, which are further treated as described in our original GST protocol (10).
MATERIALS AND METHODS
In silico SP-GST surveys on conserved genes.
SP-GSTs for any organism are determined by three parameters: (i) the primer designed to recognize a conserved gene sequence, (ii) the anchoring enzyme recognition sequence, and (iii) the type IIS restriction enzyme which defines the tag length.
For the selection of anchoring enzymes, we surveyed the restriction enzyme database REBASE (http://rebase.neb.com) for enzymes that met the following criteria: are commercially available, recognize a palindromic sequence, create cohesive overhangs, are insensitive to inhibition by DNA methylation, and contain no ambiguity codes. Of the 3,816 enzymes in REBASE, 479 met these criteria and recognized a total of 59 unique sequences as their restriction sites, which we considered as candidates in our in silico survey.
The type IIS restriction enzymes MmeI and EcoP15I were considered for tag generation, yielding tags of 21 bp and 27 bp, respectively. The number of possible sequences for each tag is represented by the expression 4(m−n+o), where m is the overhang length of the type IIS restriction enzyme, n is the length of the anchoring enzyme's recognition site, and o is the overlap in nucleotide sequence between recognition sites of the type IIS restriction site and the recognition site of the fragmenting enzyme. To design the best SP-GST protocol, 168 unique prokaryotic genomes were surveyed from the NCBI database (ftp://ftp.ncbi.nih.gov/genomes/bacteria) for the in silico generation of SP-GSTs from conserved domains present in the 16S rRNA, rpoC, recA, and uvrB genes. In cases where the sequences of several strains of the same species were available, we selected the strain with the larger genome.
DNA isolation, DNA fragmentation, and linker ligation.
Genomic DNA was isolated from all bacterial strains as described in Bron et al. (5). Before a DNA sample was used for the SP-GST protocol, its quality was checked via PCR using the 16S rRNA gene-specific primers 8F and 1392R (1) (Table 1) as previously described (6), while DNAs from clinical B. cereus isolates were also compared using BOX-PCR (18, 26, 30).
TABLE 1.
Table of primersa
| Primer name | Base positionb | Sequence 5′→3′ |
|---|---|---|
| 8F | 8-27 | AGAGTTTGATCCTGGCTCAG |
| 8F-Bio | 8-27 | Bio-AGAGTTTGATCCTGGCTCAG |
| 27R | 27-8 | CTGAGCCAGGATCAAACTCT |
| 27R-Bio | 27-8 | Bio-CTGAGCCAGGATCAAACTCT |
| 1392R | 1392-1372 | ACGGGCGGTGTGTRC |
| Csp6l cas1 | NA | TTTGGATTTGCTGGTCGAATTCAACTA GGCTTAATCCGACG |
| Csp6l cas2 | NA | TACGTCGGATTAAGCCTAGTTGAATT |
| Deg cas1 | NA | Pho-TTTGTACGGCGGAGACGTCCGCCA CTAGTGTCGCAACTGACTA-AmMC7 |
| Deg cas2 | NA | TAGTCAGTTGCGACACTAGTGGCGGAC GTCTCCGCCGTACAAANN |
| GST1 | NA | GGATTTGCTGGTCGAATTCAAC |
| GST2 | NA | TAGTCAGTTGCGACACTAGTGGC |
Abbreviations: Pho, 5′ phosphate; AmMC7, 3′ amino modification; Bio, 5′ biotin; NA, not applicable.
E. coli numbering.
Based on the outcome of the in silico analysis, Csp6I was chosen as the anchoring enzyme, and 1 μg of each genomic DNA was digested in 100 μl of Fermentas 1× B+ buffer (10 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 0.1 mg/ml BSA) with 10 U of Csp6I (Fermentas Life Sciences, Hanover, MD) for 5 h at 37°C. Csp6I was subsequently heat inactivated by incubation of the digestion mixture for 20 min at 65°C, and the product was checked on a 0.8% agarose gel. For tags generated from the defined consortium, equal DNA quantities (0.5 μg of DNA [each] of Arthrobacter globiformis DSM 20124, Bacillus licheniformis B-6-4J, Deinococcus radiodurans R1, and Pseudomonas stutzeri strains Stanier 221 and BRW1) were mixed. The consortium DNA was then purified with phenol-chloroform (equal mixture, vol/vol), ethanol precipitated overnight at −20°C, and resuspended in 34 μl of sterile distilled H2O.
A nonphosphorylated Csp6I-compatible, asymmetric oligonucleotide cassette was created by mixing 3,600 pmol of Csp6I Cas1 (sense strand) and Csp6I Cas2 (antisense strand) (Table 1) with 10 μl of OFA buffer (10 mM Tris-acetate, pH 7.5, 10 mM Mg acetate, 50 mM K acetate; Amersham Biosciences, Piscataway, NJ) and 18 μl of TESL buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA-Na3). The mixture was heated at 95°C for 2 min and then for 10 min at 65°C, 10 min at 37°C, and finally for 20 min at room temperature, and it was then placed on ice. Subsequently, ∼600 pmol was ligated to the fragmented DNA in a total volume of 50 μl of 1× ligase buffer containing 3 Weiss units of T4 DNA ligase (Takara, Pittsburgh, PA). The reaction mixture was incubated overnight at 16°C, purified by using a GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ) per the manufacturer's instructions, and eluted in 50 μl of double-distilled water (ddH2O).
Amplification of DNA/adapter product: extended tags.
PCR was performed on the ligation product using a 0.4 μM final concentration of both the 27R-Bio and GST1 primers (Table 1), in 1× Promega buffer (catalog no. M190G; Madison, WI) containing 2 mM Mg sulfate, a 0.3 mM concentration of each deoxynucleoside triphosphate, 5 μl of ligation product, and 1 unit of high fidelity platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) in a total volume of 50 μl. Only fragments that have the bound asymmetric linker cassette and that contain the annealing site for the 27R-Bio primer will be amplified during this PCR; these fragments are referred to as extended tags. The reaction was carried out with an initial denaturing step for 2 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 52°C, and 3 min at 72°C, with a final extension step for 8 min at 72°C.
Binding biotinylated fragments to streptavidin beads and MmeI digestion.
A total of 100 μl of thoroughly suspended streptavidin MagneSphere paramagnetic particles (Promega, Madison, WI) was transferred to a 1.5-ml Eppendorf tube and bound to a magnetic stand. The storage buffer was removed; the beads were washed three times with 400 μl of 1× B&W buffer (10 mM Tris-HCl, pH 8.0, 2 M NaCl, 1 mM EDTA) and resuspended in 100 μl of 1× B&W buffer. A total of 50 μl of 2× B&W buffer was added to 50 μl of the PCR mixture, which was then added to the beads. The PCR tube was washed with 200 μl of 1× B&W buffer and pooled to the beads. The sample was mixed gently and incubated at room temperature for 1 h with occasional mixing. Unbound DNA fragments were removed by washing the beads once with 400 μl of 1× B&W buffer, twice with TE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA-Na3), and once with 100 μl of MmeI digestion buffer (100 mM HEPES, pH 8.0, 25 mM K acetate, pH 8.0, 50 mM Mg acetate, pH 8.0, 20 mM dithiothreitol, 4 mM S-adenosylomethionine-HCl). The beads were finally resuspended in 100 μl of 1× MmeI digestion buffer containing 8 U of MmeI (New England Biolabs, Beverly, MA) and incubated for 3 h at 37°C. The beads were collected, and the supernatant containing the released tags was removed to a clean 1.5-ml Eppendorf tube. The beads were washed with 100 μl of TESL buffer, which was combined with the first MmeI supernatant. The pooled MmeI digest was extracted with phenol-chloroform (equal mixture, vol/vol) and precipitated overnight at −20°C with 1 ml of ethanol after the addition of 133 μl of 7.5 M ammonium acetate and 2 μl of GlycoBlue (Ambion, Austin, TX). The resulting pellet was washed with cold 75% ethanol, dried, and resuspended in 29.5 μl of TESL buffer plus 4 μl of 10× T4 DNA ligase buffer (Takara, Pittsburgh, PA).
Degenerate linker ligation and GST amplification.
A degenerated linker containing a Csp6I site preceded by a TTT triplet (serving as punctuation mark to orient the GST toward the 16S rRNA gene) was prepared by annealing Deg.cas1 (sense strand) and Deg.cas2 (antisense strand) (Table 1) as described above. A total of 35 pmol of the degenerate linker (in 3.5 μl) was added to 29.5 μl of suspended tag solution, along with 3 μl of DNA ligase (8 Weiss units; Takara, Pittsburgh, PA), after which the reaction mixture was incubated overnight at 16°C. The ligation product was then subjected to PCR amplification, and the cycling programs and reaction mixture composition (50 μl) were as previously described (10) with the primers used being GST1 and GST2 (Table 1).
Linear amplifications to reduce heteroduplexes.
The homology of the adapter sequences results in the formation of heteroduplexes. These were resolved, the unincorporated primers were digested, and the final sample was purified using previously described methods (10) with the same primer modification mentioned above. The only exception is that the 500 μl of amplified product was purified using the GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions, and eluted in 240 μl of ddH2O.
Csp6I digestion, concatenation, cloning, and sequencing.
A total of 240 μl of the product of linear amplification to reduce heteroduplexes was digested at 37°C for 3 h with 20 units of Csp6I in a final volume of 400 μl. The digest was purified via phenol-chloroform extraction (equal mixture, vol/vol), ethanol precipitated in the presence of Na acetate and GlycoBlue (Ambion, Austin TX) carrier, and resuspended in 20 μl of TESL buffer. The sample was then run on a 12% polyacrylamide gel with a 20-bp DNA ladder (Sigma, St. Louis, MO) and the 25-bp band corresponding to the tags was cut out. SP-GSTs were eluted from the pulverized gel by adding 250 μl of TESL buffer and 50 μl of 7.5 M ammonium acetate and by incubating the sample at 37°C for 6 h. The tags were purified using a GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ) column without the chaotrophic agent, thus trapping the polyacrylamide on the column and permitting the small tags to pass through. The tags were then precipitated by adding 2.5 volumes of ethanol and 2.5 μl of GlycoBlue (Ambion, Austin, TX); they were washed twice with ice-cold 80% ethanol, resuspended in 12.5 μl of TESL buffer, and concatenated as previously described (10). The concatenated tags were then purified using a GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ), and the sample was eluted in 20 μl of ddH2O. Five microliters of this product was cloned into NdeI-digested pGEM5 vector (Promega, Madison, WI). Recombinant clones, obtained after electroporation of competent Escherichia coli TOP10 cells (Invitrogen, Carlsbad, CA), were selected on LB plates containing 100 μg/ml ampicillin supplemented with 0.4 mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Plasmid preps, DNA sequencing, and data analysis were carried out as previously described (10). The SP-GST analysis software we developed is now publicly available at (http://genome.bio.bnl.gov:16080/16S_defined_GSTs/).
Real-time PCR.
After sequencing the extended tags of each isolate, primer pairs were designed (see supplemental material) to determine the number of 16S rRNA genes linked to each tag. This was carried out via quantitative real-time PCR (qRT-PCR) using an iCycler and iQ SYBR Green Supermix kit (Bio-Rad, Hercules, CA) chemistry according to the manufacturer's instructions. The qRT-PCR consisted of an initial hot-start activation step at 80°C for 30 s, followed by a denaturation step at 95°C for 30 s, followed by 35 cycles at 95°C for 15 s, 55°C for 30 s, and 72°C for 1.5 min; the final extension was for 4 min at 72°C. It should be noted that for all Pseudomonas samples, qRT-PCR results obtained with 27R were normalized relative to sequence length to obtain true quantification values.
Software programs to extend the SP-GST concept to other functions.
Restriction enzyme candidate sequences were obtained via SQL queries on a PostgreSQL database containing relevant information downloaded from REBASE. A program written in C of our own making was used to produce tables of tag sequences and their respective distances from adjacent restriction enzyme sites for each bacterial genome and candidate enzyme. Primer sequences and positions were identified in each genome using a different C program which finds patterns and allows for substitution mismatches. To simulate the various protocols described in this work, we wrote a series of PERL scripts to collate the tag and primer site files and then summarize uniqueness and degeneracies across genomes. Phylogenetic assignments (based on Bergey's taxonomy) were made for each bacterial genome by automatically querying the Ribosomal Database Project website (http://rdp.cme.msu.edu/index.jsp) with 1,500-bp sequences extracted downstream of the 8F (Table 1) priming sites in each genome sequence.
RESULTS
In silico SP-GST surveys on conserved genes.
Primers were selected by back-translating conserved domains of the RpoC (13), UvrB (23), and RecA (7) proteins into their corresponding nucleotide sequences using standard prokaryotic codon usage, including appropriate codon degeneracy when needed (Table 2). Resulting primer sequences were subsequently analyzed for their copy number within the selected microbial genomes (Table 2). Tag sequences, generated in silico upstream or downstream of the primer's annealing position, were examined for their discriminating power using the NCBI genome data set. Tags located more than 3 kb from the primer's annealing position were excluded in order to reflect potential PCR biases when tags were generated from large fragments. Selected examples for MmeI in combination with anchoring enzyme HpyCH4IV, Csp6I, Sau3AI, or BamHI are presented in Table 3 (the complete data set of this in silico analysis is available in the supplemental online materials available at http://genome.bnl.gov/SP-GSTs/).
TABLE 2.
Overview of primer sequences designed for the in silico generation of unique identifier tagsa
| Protein | Conserved amino acid sequence | Primer sequence | No. of genomes for which the primer sequence has a copy number of:
|
||
|---|---|---|---|---|---|
| 0 | 1 | ≥2 | |||
| RpoC | FDGDQMA | TTYGAYGGNGAYCARATGGC | 22 | 146 | 0 |
| UvrB | DYYQPE | GAYTAYTAYCARCCNGAR | 32 | 134 | 2 |
| RecA | EG(E/D)(I/M)GD | GARGGNGANATNGGNGA | 55 | 86 | 27 |
Primers were designed by reversed translation of highly conserved regions of the RpoC, UvrB, and RecA proteins located at positions 461, 94, and 157, respectively, in their E. coli homologues. The numbers of primer sequence occurrences in the 168 NCBI microbial genomes are based on a 100% match between the primer and the microbial genome sequence.
TABLE 3.
Numbers of rpoC-, uvrB-, and recA-derived tags and the phylogenetic level at which they are able to discriminate the 168 sequenced microbial genomes
| Gene and tag sequence location | Enzyme | Recognition sequence | No. of tags at the level ofa:
|
No. of nonidentified organisms | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Domain | Phylum | Class | Order | Family | Genus | Species | ||||
| rpoC, upstream | HpyCH4IV | ACGT | 0 | 1 (4) | 0 | 1 (2) | 1 (3) | 7 (16) | 119 (119) | 2 |
| Sau3AI | GATC | 2 (8) | 4 (16) | 1 (2) | 1 (3) | 0 | 8 (18) | 99 (99) | 0 | |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 0 | 0 | 21 (21) | 125 | |
| Csp6I | GTAC | 4 (15) | 0 | 2 (5) | 1 (3) | 1 (4) | 6 (13) | 105 (105) | 1 | |
| rpoC, downstream | HpyCH4IV | ACGT | 8 (36) | 1 (4) | 3 (6) | 0 | 0 | 5 (11) | 86 (86) | 1 |
| Sau3AI | GATC | 0 | 3 (7) | 0 | 0 | 0 | 10 (24) | 115 (115) | 0 | |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 1 (3) | 7 (15) | 61 (61) | 67 | |
| Csp6I | GTAC | 6 (18) | 0 | 1 (3) | 1 (2) | 0 | 7 (17) | 104 (104) | 2 | |
| uvrB, upstream | HpyCH4IV | ACGT | 0 | 0 | 0 | 0 | 0 | 8 (17) | 113 (111) | 8 |
| Sau3AI | GATC | 0 | 1 (2) | 0 | 0 | 0 | 9 (20) | 114 (112) | 0 | |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 0 | 2 (4) | 31 (31) | 101 | |
| Csp6Ib | GTAC | 1 (9) | 1 (2) | 1 (4) | 0 | 0 | 4 (8) | 109 (107) | 4 | |
| uvrB, downstream | HpyCH4IVb | ACGT | 0 | 2 (7) | 1 (4) | 1 (2) | 0 | 5 (10) | 109 (107) | 3 |
| Sau3AI | GATC | 2 (7) | 1 (4) | 1 (2) | 1 (2) | 0 | 9 (19) | 104 (102) | 0 | |
| BamHI | GGATCC | 0 | 0 | 1 (2) | 0 | 0 | 2 (4) | 36 (36) | 94 | |
| Csp6I | GTAC | 0 | 1 (4) | 0 | 1 (3) | 0 | 5 (10) | 115 (113) | 6 | |
| recA, upstream | HpyCH4IV | ACGT | 1 (NE)c | 0 | 1 (2) | 0 | 0 | 10 (16) | 122 (93) | 2 |
| Sau3AI | GATC | 3 (7) | 0 | 0 | 2 (5) | 0 | 7 (11) | 117 (89) | 1 | |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 0 | 5 (10) | 59 (51) | 52 | |
| Csp6I | GTAC | 0 | 0 | 0 | 0 | 0 | 10 (17) | 122 (91) | 5 | |
| recA, downstream | HpyCH4IV | ACGT | 2 (4) | 0 | 0 | 0 | 0 | 7 (12) | 123 (94) | 3 |
| Sau3AI | GATC | 1 (4) | 0 | 1 (NE) | 1 (NE) | 0 | 9 (13) | 123 (96) | 0 | |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 0 | 1 (2) | 45 (36) | 75 | |
| Csp6I | GTAC | 0 | 0 | 0 | 0 | 1 (2) | 10 (17) | 123 (92) | 2 | |
Tags were generated in silico with an MmeI-containing linker cassette from the first position of the anchoring enzyme located upstream or downstream of the gene-specific primer annealing site. Data are presented for HpyCH4IV, Sau3AI, BamHI, and Csp6I as anchoring enzymes. Numbers in parentheses indicate the number of species that can be identified by SP-GSTs at a given phylogenetic level. In the case of multiple gene copies per species, as is the case for uvrB and recA with 138 and 151 occurrences in 136 and 113 strains, respectively, tag numbers at a phylogenetic level can be higher than the number of species distinguished at that level. Tags located at more than 3,000 nucleotides from the primer annealing sites were discarded, as a result of which some organisms were not identified.
For this specific restriction enzyme, tags were generated that did not distinguish between two or more organisms at the domain level (between Archaea and Bacteria).
NE, no effect. At this level the tag had no effect on the final identification of the species, as additional tags were generated from the same species that allowed for identification at a lower phylogenetic level.
The discriminating power depends strongly on the choice of target gene, the anchoring enzyme, and the orientation of the primer. Of the three conserved genes and related primers, the best results were obtained with tags upstream of uvrB in conjunction with HpyCH4IV and Sau3AI as the anchoring enzymes. These tags offered maximal discrimination of species and missed a minimum number of organisms due to the 3-kb cutoff for PCR length. For tags that failed to distinguish between organisms, we determined their phylogenetic level of discrimination based on Bergey's taxonomy (Ribosomal Database Project, http://rdp.cme.msu.edu/index.jsp).
HpyCH4IV yields a nondiscriminating tag downstream of the uvrB primer, which was present in Streptomyces coelicolor, Thermus thermophilus, and the archaeon Haloarcula marismortui (results not shown in Table 3). Csp6I also yields one upstream tag unable to distinguish the phylogenetic domain of two organisms: H. marismortui, an archaeon, and Nocardia farcinica, a bacterium. In all these cases the tags were located immediately adjacent (20 nucleotides) to the conserved priming sites.
For rpoC, tags generated upstream with HpyCH4IV and Sau3AI gave the best results (Table 3). The worst case for HpyCH4IV was a single upstream tag unable to discriminate at the phylum level between three Bordetella species, Bordetella bronchiseptica, Bordetella parapertussis, and Bordetella pertussis, and Caulobacter crescentus. However, in the complete data set (see supplemental material) tags generated with TasI (/AATT) as the anchoring enzyme were able to discriminate to at least the family level.
Many of the genomes examined contained more than one copy of the recA priming site, in some cases yielding multiple tags; however, tags generated with Csp6I discriminated all organisms to at least the genus level and most to the species level. More than one different tag per genome can be helpful for phylogenetic identification: HpyCH4IV sites upstream and Sau3AI sites downstream of the primer annealing position yielded some tags shared across phylogenetic domains, classes, and orders, but these organisms had additional recA-linked tags that permitted their identification at a lower phylogenetic level.
From this survey we can conclude that anchoring enzymes that yield excellent discrimination can be chosen for each conserved primer. However, there is not one choice that is optimal for all primers. Interestingly, we found that EcoP15I-generated tags (27 bp) in general did not provide much more information than the MmeI-generated tags (21 bp) in this data set.
SP-GSTs on the 16S rRNA gene: in silico analysis.
Although rpoC, uvrB, and recA can function as phylogenetic identifiers, their number of entries in current sequence databases is marginal. Given this limitation, the 16S rRNA gene is an ideal alternative. Though typically present in multiple copies, it is found in all prokaryotes and has several highly conserved regions. An in silico survey was performed on this gene, as previously described on the NCBI genomes, to examine how unique and informative 21-bp MmeI-generated tags would be for species identification. All 59 anchoring enzyme candidates were examined; only the exemplars HpyCH4IV, Csp6I, Sau3AI, and BamHI are presented in Table 4. The conserved sequence from position 8 to 27 was chosen as the optimal primer annealing site. Tags generated downstream of the priming site were largely located within the rRNA operon, and their uniqueness was compared to those generated from the V1 hypervariable region by SARST (20). Using SARST, several organisms were not discriminated below the family level and many downstream 16S-derived SP-GSTs yielded even less information. The best results using the 16S rRNA gene were obtained with Csp6I upstream-derived tags, which discriminated all organisms to at least the genus level and most organisms to the species level.
TABLE 4.
Numbers of 16S rRNA gene-derived tags and the phylogenetic level at which they are able to discriminate the 140 sequenced bacterial genomes
| Identifier and tag location | Enzyme | Recognition sequence | No. of tags at the level ofa:
|
No. of nonidentified organisms | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Domain | Phylum | Class | Order | Family | Genus | Species | ||||
| 16S rRNA gene, | HpyCH4 IV | ACGT | 0 | 1 (NE)c | 0 | 0 | 2 (6) | 19 (10) | 328 (120) | 4 |
| upstream | Sau3AI | GATC | 2 (8) | 3 (12) | 5 (8) | 1 (NE) | 1 (NE) | 17 (8) | 233 (104) | 0 |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 1 (2) | 5 (5) | 101 (62) | 71 | |
| Csp6I | GTAC | 0 | 0 | 0 | 0 | 2 (NE) | 33 (9) | 374 (129) | 2 | |
| 16S rRNA gene, | HpyCH4 IV | ACGT | 4 (45) | 0 | 1 (2) | 0 | 2 (5) | 10 (23) | 73 (65) | 0 |
| downstream | Sau3AI | GATC | 5 (7) | 6 (7) | 3 (3) | 0 | 3 (6) | 14 (24) | 126 (93) | 0 |
| BamHI | GGATCC | 0 | 0 | 0 | 0 | 0 | 6 (14) | 26 (22) | 104 | |
| Csp6I | GTAC | 1 (2) | 0 | 3 (10) | 5 (19) | 3 (6) | 13 (25) | 83 (78) | 0 | |
| SARST, internalb | V1 region | 0 | 0 | 2 (3) | 1 (4) | 1 (2) | 9 (16) | 162 (124) | 0 | |
Tags were generated in silico with an MmeI-containing linker cassette from the first position of the anchoring enzyme located upstream or downstream of the 27R primer annealing site. Data are presented for HpyCH4IV, Sau3AI, BamHI, and Csp6I as anchoring enzymes. Numbers in parentheses indicate the number of species that can be identified by SP-GSTs at a given phylogenetic level. Since the 16S rRNA gene often has multiple copies per species, tag numbers at a phylogenetic level can be higher than the number of species distinguished at that level. Tags located at more than 3,000 nucleotides from the primer annealing sites were discarded, as a result of which some organisms were not identified.
SARST data for the V1 hypervariable region, which was also present in nine Archaea, are presented as comparison.
NE, no effect. At this level the tag had no effect on the final identification of the species, as additional tags were generated from the same species that allowed for identification at a lower phylogenetic level.
Comparison between closely related B. cereus and B. anthracis strains.
To investigate the application of this technology, we determined whether the 16S SP-GSTs generated would allow us to discriminate between closely related strains of B. cereus and B. anthracis. The rRNA operons of B. anthracis strains Ames, Ames 0581, and Sterne are virtually identical; therefore, none of the 59 chosen anchoring enzymes yielded, in silico, internal or upstream SP-GSTs from 16S rRNA genes able to distinguish between them. Internal 16S SP-GSTs and SARST (20) also failed to discriminate between B. cereus and B. anthracis on the species level. However, Csp6I-based identifier tags generated upstream of the 16S rRNA gene clearly distinguished between B. cereus and B. anthracis species, as well as between different B. cereus strains (Table 5). This was confirmed on a set of five closely related, clinically isolated B. cereus strains. Initial profiling of B. cereus strains H27141, H52652, F65185, F69977, and SB460 with BOX-PCR was unsuccessful at discriminating between all strains, indicating that they are very closely related (results not shown). As an alternative to using BOX-PCR, we also analyzed the banding profiles of the extended tags on a gel. Although all five strains showed common bands, each strain also possessed a number of unique fragments (results not shown). Analysis of the tags generated upstream of the 16S rRNA gene showed that each strain provided a number of both unique tags and tags in common with other B. cereus and B. anthracis strains (Table 5). The B. cereus clinical isolates did not generate any tags that were previously identified as unique for B. anthracis, indicating that tags generated upstream of the 16S rRNA gene can be successfully used to distinguish between B. cereus and B. anthracis. Based on tag profiles, our data suggest that the five clinical B. cereus isolates are closely related and that they share the largest numbers of tags with the genomes from the sequenced strains B. cereus ZK (also referred to as B. cereus E33L) and B. cereus ATCC 10987.
TABLE 5.
Comparison of the Csp6I-generated SP-GSTs located upstream of the 16S rRNA gene for B. cereus and B. anthracis speciesa
| GST | Presence (+) of the tag in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| B. anthracis strains | B. cereus ZK | B. cereus ATCC 10987 | B. cereus ATCC 14579 | B. cereus H27141 | B. cereus H52652 | B. cereus F65185 | B. cereus F69977 | B. cereus SB460 | |
| TTGCATTTGAAAATGTA | + | + | + | + | + | + | + | + | |
| TGCATGATATATTAATA | + | + | + | + | + | + | |||
| AACAACAATCCAATATG | + | + | |||||||
| AACAACCCTCTAATTAT | + | ||||||||
| AACAATAAAACAAATTA | + | ||||||||
| AGGTCATTCATAAGGAG | + | ||||||||
| TACATATGGCGATGGTA | + | ||||||||
| TCCGATTGATGAATATC | + | ||||||||
| TGATATACAATTTAAAT | + | ||||||||
| TAGCAGGAACACGAATA | + | + | |||||||
| CTTCAAAAGAACAATAG | + | + | + | + | + | ||||
| AACAACCCCCTAATTAT | + | ||||||||
| AGGTCATTCATAGGGAG | + | ||||||||
| AACAAGTTTGACTACGA | + | ||||||||
| CGCAGGCAGAAGAGCAT | + | ||||||||
| TATGATATATTATAAAA | + | ||||||||
| TGGTATACAATTTAAAT | + | ||||||||
| TTATAATTTCTAGAGAG | + | ||||||||
| TTGTATTGGAAATAAGT | + | ||||||||
| AACCACTTTTTTGGCTC | + | + | + | + | |||||
| TATTATTCCCTGCTATG | + | + | + | + | + | + | |||
| AACAAGTTTTACTGCGA | + | ||||||||
| AGGAGTGTAATATAGAA | + | ||||||||
| CGCAAGCAGAAGAGCAT | + | ||||||||
| CGTCTACAAAGCCGTGG | + | ||||||||
| GTCTTTTCTTACTATAT | + | ||||||||
| TGCAACAATCACAAGTT | + | ||||||||
| TTTAGAGGTGTAATATA | + | ||||||||
| TTGTGTTGGAAATAAGT | + | + | |||||||
| ACCGATTGATGAATACC | + | + | + | + | |||||
| AACAAGTTTCACAGCGA | + | ||||||||
| AGCAGCAATAACGAGTT | + | ||||||||
| AGCCGCTTTTTTGACTC | + | ||||||||
| CAGTTGTTCTGCCAAGG | + | ||||||||
| CCCATACTACCGATTTC | + | ||||||||
| CTTGTGGAATCAATGAC | + | ||||||||
| GATTTCTTTTTCAATTT | + | ||||||||
| GGGTCACCACTTCGGAG | + | ||||||||
| GGTATGCCTCCTACGGG | + | ||||||||
| TAAAAGAAAAAATACTA | + | ||||||||
| TGATGGAAGTTGTTCGG | + | ||||||||
| CGCAAGCGGAAGAGCAT | + | + | |||||||
| TCCAGTTGAAGAATAT | + | ||||||||
| TTACGTATCAAGTGGC | + | ||||||||
| TTGTTATTTCGAAATC | + | ||||||||
| AGCGACAGTAACAAGT | + | ||||||||
| AGAAGTGTAATATAGA | + | + | |||||||
| TATTATTACCCTGCTA | + | ||||||||
| TTTGTTCTTTGAAAAT | + | ||||||||
| TGAATAGAGGGGGCAGG | + | ||||||||
| TTACGTATCGAGCGG | + | ||||||||
| CCCATAGATAGTTCTG | + | ||||||||
| ACACTTGCGGATGGTA | + | ||||||||
| GCCAATTGATGAATAC | + | ||||||||
| TTGGCATTTGAAAATG | + | ||||||||
| AACAACTCTCTAATTA | + | ||||||||
| AGCGGCAATAACGAGT | + | ||||||||
| TCCAGTTGAAGAGTAT | + | ||||||||
Tags were obtained using the 27R primer against position 8-27 (Fig. 1) of the 16S rRNA gene (1) and Csp6I (GTAC) as anchoring enzyme. All three B. anthracis strains share identical internal and upstream GSTs, making it impossible to distinguish between individual strains. Furthermore, it was impossible to distinguish between B. anthracis and B. cereus strains using internal tags (results not shown). The Csp6I sequence (GTAC) at the 5′ end of the tags was omitted, resulting in 16- or 17-bp sequences.
Deconvoluting microbial community composition.
As the in silico analysis showed that tags generated from the variable region upstream of the 16S rRNA gene have a better discriminating power for species comparison than sequence tags obtained from internal regions, we tested this approach to identify the individual members of a defined microbial community. The members of this community were D. radiodurans R1, whose genome has been sequenced (32), B. licheniformis B-6-4J, whose close relative ATCC 14580 (also referred to as B. licheniformis DSM 13) was sequenced (28), P. stutzeri strains Stanier 221 and BRW1, and A. globiformis DSM 20124.
Using Csp6I, sequence analysis of the resulting library of concatenated tags demonstrated that we were successful in obtaining 16S-linked tags from all species (Table 6). We accurately found the two tags adjacent to the Csp6I sites upstream of three 16S rRNA genes of D. radiodurans: GST-DR1, which is present in both sections 8 and 213 of the complete chromosome 1 sequence, and GST-DR2 from section 198 of the chromosome 1 sequence. These two D. radiodurans tags were present in a ratio of approximately 2:1, demonstrating that tag frequency can provide quantitative information concerning the relative abundance of the target sequence from which they were derived. We also obtained an unexpected tag, GST-MP1, with the sequence GTACAGCGAGGAATGGCTCA from the D. radiodurans R1 177-kb megaplasmid. PCR amplification with the GST-MP1 and 27R primers and sequence analysis of the obtained amplicon showed that the 27R primer annealed to a region of the megaplasmid, which resulted in the generation of the GST-MP1 tag.
TABLE 6.
16S SP-GST identifier tags obtained from a microbial consortium comprised of D. radiodurans R1, B. licheniformis B-6-4J, A. globiformis DSM 20124, and the P. stutzeri strains Stanier 221 and BRW1a
| Sequence 5′→3′ | Species | No. of occurrences | Tag no. | Upstream distance (bp) | Copy no. |
|---|---|---|---|---|---|
| GTACTATTTCTGAGCCTCGA | D. radiodurans | 53 | GST-DR1 | 238 | 2 |
| GTACAGCGAGGAATGGCTCA | D. radiodurans | 29 | GST-MP1 | 26 | 1c |
| GTACGGCGCGGACGCTCTGC | D. radiodurans | 26 | GST-DR2 | 379 | 1 |
| GTACATGCAAGTGTGCGTAG | B. licheniformis | 46 | GST-BL1 | 79 | 2 |
| GTACATGCGAATGTGCGTAG | B. licheniformis | 40 | GST-BL2 | 79 | 2 |
| GTACCTGTTAATTCATTTTT | B. licheniformis | 28 | GST-BL3 | 107 | 1 |
| GTACCTGTTAATTCATTATA | B. licheniformis | 28 | GST-BL4 | 104 | 1 |
| GTACCTGTTAATTCATTAAA | B. licheniformis | 24 | GST-BL5 | 44 | 1 |
| GTACCGGCGCGGTGATAGAG | P. stutzeri | 19 | GST-PS1 | 450 | 2 |
| GTACGGCGCAGGAGCGCGAT | P. stutzeri | 11 | GST-PS2 | 750 | 1 |
| GTACGCGAAAGAACAAAGTT | P. stutzeri | 7 | GST-PS3 | 600 | 1 |
| GTACGGCCAGCCTTCCCAGT | P. stutzeri | 7 | GST-PS4 | 1,200 | 1 |
| GTACAAGTCCACGCCGGCAC | A. globiformisb | 16 | GST-AG1 | 930 | 8 |
| GTACGTGTCGACGACCGGGG | A. globiformis | 2 | GST-AG2 | 1,236 | 4 |
| GTACTGCACCCGGGAGGGTG | A. globiformis | NDd | GST-AG3 | 1,105 | 2 |
| GTACTGCCGCCGAGCGGGGT | A. globiformis | ND | GST-AG4 | 1,236 | 1 |
Tags were obtained using the 27R primer against position 8-27 (Fig. 1) of the 16S rRNA gene (1) and Csp6I (GTAC) as anchoring enzyme. For each tag, its occurrence in the sequenced tag library, its distance to the 16S rRNA gene from which it was derived, and copy number in its host strain are indicated. The tag copy numbers were confirmed by qRT-PCR. For both P. stutzeri strains, Stanier 221 and BRW1, identical qRT-PCR results were obtained.
For A. globiformis DSM 20124, GST-AG3 and GST-AG4 were only found after conducting SP-GST analysis specifically on purified A. globiformis DNA, after which qRT-PCR was used to determine their respective copy numbers.
On plasmid.
ND, not detected.
As SP-GSTs can be converted into PCR primers (10), we ordered oligonucleotides corresponding to the tags that were not derived from D. radiodurans R1 and then used them in combination with the conserved 1392R reverse primer on the 16S rRNA gene to amplify and clone their corresponding 16S rRNA gene. Sequence analysis allowed us to link each SP-GST to its 16S rRNA gene and thus to identify the species from which it was derived. In this way, all species present in the consortium were identified. Quantitative PCR (QPCR) was used to determine the copy numbers of the 16S rRNA gene to which the individual GSTs were linked (Table 6).
As was the case for the D. radiodurans R1 tags, tag frequencies for B. licheniformis B-6-4J reflected the relative abundances of the target sequences from which they were derived. QPCR showed that GST-BL3, GST-BL4, and GST-BL5 were present once in the B. licheniformis B-6-4J genome, while GST-BL1 and GST-BL2 were observed twice as frequently. This suggests that the B. licheniformis B-6-4J genome contains, like strain ATCC 14580, seven copies of its 16S rRNA gene. These tag frequencies were compared to that of the fully sequenced genome of B. licheniformis ATCC 14580 (GenBank accession no. AE017333) and proved that these two species had four tags in common although their frequencies differed between strains. Three copies of GST-BL2, two copies of GST-BL3, and one copy of both GST-BL4 and GST-BL5 were identified in B. licheniformis ATCC 14580, while GST-BL1 turned out to be a tag unique to B. licheniformis B-6-4J.
Tag frequencies for P. stutzeri also reflected the relative abundances of the target sequences from which they were derived. QPCR showed that GST-PS2, GST-PS3, and GST-PS4 were present once in the P. stutzeri genome, while GST-PS1 was observed twice as frequently. These data were consistent for both P. stutzeri Stanier 221 and BRW1 strains and indicate that both P. stutzeri strains contain five copies of the 16S rRNA genes, one more than previously found for this species (http://rrndb.cme.msu.edu/rrndb/servlet/controller).
SP-GST distributions in A. globiformis suggested that this species has three copies of a 16S rRNA gene with two copies of GST-AG1 and a single copy of GST-AG2 (Table 6). Tags for A. globiformis DSM 20124 may possibly have been harder to obtain due to the high genomic GC content of this species. Due to the small number of tags recovered from this species, tagging using SP-GSTs was specifically carried out on A. globiformis DNA to determine if these results were accurate. Two additional tags were discovered belonging to this species which were linked to two additional copies of the 16S rRNA gene: GST-AG3, GTACTAGAGGGGCCCAAGAT, and GST-AG4, GTACTGCACCCGGGAGGGTG. QPCR on A. globiformis DSM 20124 confirmed that GST-AG1 was present twice as frequently on the genome as GST-AG2. QPCR further suggested that A. globiformis DSM 20124 has a total of 15 copies of its 16S rRNA gene, 8 of which were linked to GST-AG1, 4 to GST-AG2, 2 to GST-AG3, and 1 to GST-AG4.
DISCUSSION
The tagging method using SP-GSTs, which we developed to analyze closely related species and to study changes in microbial community composition, provides a generally applicable sequencing-based method that addresses specific genes of interest to generate identifier tags from well-defined loci within a genome(s). The major advantage of SP-GSTs over other whole-genome fingerprinting techniques, such as amplified fragment length polymorphism (31), terminal restriction fragment length polymorphism (17), denaturing gradient gel electrophoresis (19), amplified rRNA gene restriction analysis (27), restriction landmark genome scanning (12), and automated ribosomal intergenic spacer analysis (11), is that a “digital” image of the strain or community is obtained in the form of tag sequences. This provides a very straightforward way to compare data from individual experiments, something which is very difficult for methods where gel electrophoresis is used to determine fragment sizes. In addition, tag sequences can be used for species identification, either via sequence comparison or via an additional PCR step.
Due to differences in codon usage, especially among unrelated species, it is not always easy (or reliable) to translate conserved protein domains into their corresponding DNA sequences. The use of SP-GSTs has the advantage over other PCR based methods in that only one conserved DNA domain, rather than two, is required for primer annealing. In addition to taxonomic identification, this method promises to be very useful for examining the distribution of specific functional genes that share only one conserved domain, which are inaccessible to SARST (15, 20) or other related techniques. Other advantages of the SP-GST method are as follows: (i) the number of tags, defined by the copy number of the target gene, is small and minimizes the amount of required sequencing; (ii) the output is actual DNA sequence data, making it easy to make comparisons between experiments; and (iii) different anchoring enzymes can be used to tailor the sampling depth to the community in question. This also avoids complications that would arise where a recognition site for an anchoring enzyme is present in a specific target domain, as was the case, for instance, with Sau3AI tags generated from the 16S rRNA gene.
The large number of 16S rRNA gene entries in databases has reinforced their extensive use for the culture-independent identification of prokaryotes by PCR and cloning. 16S rRNA gene-based tags thus have the advantage that they can be easily used to identify more organisms from which they were derived, making them preferable to those generated by other conserved genes. SP-GSTs located within the 16S rRNA gene have the advantage that the sequence is already tied to phylogenetic identification for many thousands of species. Since many tags (between 10 and 20, depending on the efficiency of the concatenation) are sequenced concomitantly, the SP-GSTs provide a major reduction in sequencing effort compared to 16S rRNA gene libraries for community analysis. However, their discriminatory power is reduced, given that they can also be located in regions conserved across species. Identifier tags upstream of the 16S rRNA gene are typically located in more variable regions and have a better discriminating power for species identification. A disadvantage of the upstream 16S SP-GST approach is that the identifier tags are not yet directly tied to species identification unless they are derived from species with sequenced genomes; this is also the case for tags derived from rpoC, uvrB, and recA. It is possible, however, to use the tag sequence as a primer in combination with a primer against a conserved domain in the 16S rRNA gene, such as the 1392R reverse primer, to amplify and subsequently identify by sequencing the 16S rRNA gene and, thus, the organism from which the tag was derived. Using this approach, databases of SP-GSTs can be established. This approach also helps to exclude false tags: as expected, the GST-MP1 tag derived from the D. radiodurans R1 177-kb megaplasmid in combination with the 1392R primer failed to provide a PCR amplicon (results not shown).
The best results using the 16S rRNA gene were obtained with Csp6I upstream-derived tags, which discriminated all organisms to at least the genus level and most organisms to the species level. Csp6I has the following additional characteristics that make this restriction enzyme a suitable choice: the enzyme frequently cuts all known microbial genomes (theoretically, once per 256 nucleotides); it is insensitive to Dam methylation; the in silico analysis showed that the average position of its first recognition site is approximately 400 to 600 nucleotides upstream of the 16S priming site, which is well within the range of a PCR; the enzyme generates a 2-nucleotide 5′ cohesive end; and, unlike the case for Sau3AI, e.g., none of the highly conserved domains of the 16S rRNA gene contains a Csp6I site.
The discriminating power of identifier tags generated from the variable regions upstream of the 16S rRNA gene was further demonstrated in comparisons of Csp6I-based tags generated from closely related B. cereus and B. anthracis species. Although none of the generated tags could distinguish between the closely related B. anthracis strains, Csp6I-based tags upstream of the 16S rRNA gene were often found to be specific for the different B. cereus strains. From the three B. cereus strains whose genomes have been sequenced to completion, strain ZK was the most closely related to B. anthracis. This strain shared the highest number of tags with B. anthracis, including a unique internally generated identifier tag from one of its 16S rRNA genes (Table 4). The second closest strain is B. cereus ATCC 10987, and strain ATCC 14579 shares the lowest number of tags and is phylogenetically the most distant from B. anthracis. This was confirmed by determining the percentage of exactly shared sequences between the genomes of the individual species using MUMmer version 3.0 (14). Compared to the B. anthracis Ames reference strain, these percentages were 79.7%, 59.1%, and 44.4% for B. cereus ZK, B. cereus ATCC 10987, and B. cereus ATCC 14579, respectively. We conclude that tags upstream of the 16S rRNA gene can be used to rapidly provide information on the phylogenetic relationship between closely related Bacillus strains and species without the need of whole-genome sequencing. A prerequisite is that a sufficiently large number of unique identifier tags can be generated. This was also experimentally observed when we obtained tags from other clinical B. cereus isolates and compared them with tags found in the sequenced B. cereus and B. anthracis strains. Based on the tag profiles, our data suggest that these clinical isolates are more closely related to each other than to the fully sequenced strains. The fact that the majority of them share the largest numbers of tags with the genomes from B. cereus ZK and B. cereus ATCC 10987 would suggest that they are evolutionarily closer to these two strains than to B. cereus ATCC 14579 and the B. anthracis strains.
The SP-GST method successfully produced tags from all member species of a defined microbial consortium. Within a species, tag frequencies reflected the relative abundances of the target sequences from which they were derived and allowed for the determination of 16S rRNA gene copy numbers within a species. As has been documented for other PCR-based methods, amplification biases lead to a misrepresentation of the overall community composition. It was concluded that the great strength in this technology lies in its discriminatory power. Given its open architecture, diverse application, and the facility with which we can link tags to any gene of interest, the use of SP-GSTs has great potential and application for identifying and analyzing closely related species or strains and simple microbial communities.
Supplementary Material
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Science, project number DE-AC02-98CH10886, entitled “Composition of Microbial Communities Used for In Situ Radionuclide Immobilization Projects.” Portions of this work were supported by NIH grant U01 AI056480-01 to J.D. D.V.D.L., C.L., and S.T. are presently being supported by Laboratory Directed Research and Development funds at the Brookhaven National Laboratory under contract with the U.S. Department of Energy.
We specially thank Diane Heiser, who received a Student Undergraduate Laboratory Internship from the Department of Energy's Office of Science, for her role in primer design. We also thank George T. Tortora for providing us with the clinical B. cereus isolates. Judi Romeo and Mike Blewitt are acknowledged for sequencing the SP-GSTs.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. Lond. B 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, S., and G. Venema. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat. Res. 15:1-10. [DOI] [PubMed] [Google Scholar]
- 6.Chandler, D. P., F. J. Brockman, T. J. Bailey, and J. K. Fredrickson. 1998. Phylogenetic diversity of Archaea and Bacteria in a deep subsurface paleosol. Microb. Ecol. 36:37-50. [DOI] [PubMed] [Google Scholar]
- 7.Cox, M. M. 2003. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 57:551-577. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, J. J., S. R. McCorkle, L. A. Praissman, G. Hind, D. Van Der Lelie, W. F. Bahou, D. V. Gnatenko, and M. K. Krause. 2002. Genomic signature tags (GSTs): a system for profiling genomic DNA. Genome Res. 12:1756-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirotsune, S., I. Hatada, H. Komatsubara, H. Nagai, K. Kuma, K. Kobayakawa, T. Kawara, A. Nakagawara, K. Fujii, T. Mukai, et al. 1992. New approach for detection of amplification in cancer DNA using restriction landmark genomic scanning. Cancer Res. 52:3642-3647. [PubMed] [Google Scholar]
- 13.Ishihama, A., and R. Fukuda. 1980. Autogenous and post-transcriptional regulation of RNA polymerase synthesis. Mol. Cell. Biochem. 31:177-196. [DOI] [PubMed] [Google Scholar]
- 14.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kysela, D. T., C. Palacios, and M. L. Sogin. 2005. Serial analysis of V6 ribosomal sequence tags (SARST-V6): a method for efficient, high-throughput analysis of microbial community composition. Environ. Microbiol. 7:356-364. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 17.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 18.Martin, B., O. Humbert, M. Camara, E. Guenzi, J. Walker, T. Mitchell, P. Andrew, M. Prudhomme, G. Alloing, R. Hakenbeck, et al. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neufeld, J. D., Z. Yu, W. Lam, and W. W. Mohn. 2004. Serial analysis of ribosomal sequence tags (SARST): a high-throughput method for profiling complex microbial communities. Environ. Microbiol. 6:131-144. [DOI] [PubMed] [Google Scholar]
- 21.Schena, M., R. A. Heller, T. P. Theriault, K. Konrad, E. Lachenmeier, and R. W. Davis. 1998. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 16:301-306. [DOI] [PubMed] [Google Scholar]
- 22.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 23.Theis, K., M. Skorvaga, M. Machius, N. Nakagawa, B. Van Houten, and C. Kisker. 2000. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat. Res. 460:277-300. [DOI] [PubMed] [Google Scholar]
- 24.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 26.van Belkum, A., M. Sluijuter, R. de Groot, H. Verbrugh, and P. W. Hermans. 1996. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 34:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaneechoutte, M., R. Rossau, P. De Vos, M. Gillis, D. Janssens, N. Paepe, A. De Rouck, T. Fiers, G. Claeys, and K. Kersters. 1992. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA). FEMS Microbiol. Lett. 72:227-233. [DOI] [PubMed] [Google Scholar]
- 28.Veith, B., C. Herzberg, S. Steckel, J. Feesche, K. H. Maurer, P. Ehrenreich, S. Baumer, A. Henne, H. Liesegang, R. Merkl, A. Ehrenreich, and G. Gottschalk. 2004. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 7:204-211. [DOI] [PubMed] [Google Scholar]
- 29.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 30.Vinuesa, P., J. L. Rademaker, F. J. de Bruijn, and D. Werner. 1998. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl. Environ. Microbiol. 64:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.