Abstract
The secreted proteolytic activity of Aspergillus fumigatus is of potential importance as a virulence factor and in the industrial hydrolysis of protein sources. The A. fumigatus genome contains sequences that could encode a five-member gene family that produces proteases in the sedolisin family (MEROPS S53). Four putative secreted sedolisins with a predicted 17- to 20-amino-acid signal sequence were identified and termed SedA to SedD. SedA produced heterologously in Pichia pastoris was an acidic endoprotease. Heterologously produced SedB, SedC, and SedD were tripeptidyl-peptidases (TPP) with a common specificity for tripeptide-p-nitroanilide substrates at acidic pHs. Purified SedB hydrolyzed the peptide Ala-Pro-Gly-Asp-Arg-Ile-Tyr-Val-His-Pro-Phe to Arg-Pro-Gly, Asp-Arg-Ile, and Tyr-Val-His-Pro-Phe, thereby confirming TPP activity of the enzyme. SedB, SedC, and SedD were detected by Western blotting in culture supernatants of A. fumigatus grown in a medium containing hemoglobin as the sole nitrogen source. A degradation product of SedA also was observed. A search for genes encoding sedolisin homologues in other fungal genomes indicates that sedolisin gene families are widespread among filamentous ascomycetes.
Aspergillus fumigatus, the main causative agent of invasive aspergillosis in neutropenic patients, is a saprophyte that grows and sporulates in a humid environment on decaying organic materials. Secreted proteolytic activity could be an important contributor to this virulence capability. Like many other ascomycetous fungi, A. fumigatus grows well in a medium containing protein as the sole nitrogen and carbon source and secretes endoproteases and exopeptidases (for a review of Aspergillus endo- and exoproteases, see reference 20). According to the MEROPS list (http://merops.sanger.ac.uk) (1, 31a), A. fumigatus secreted endopeptidases may be classified into aspartic proteases of the pepsin family (MEROPS A1) (17, 34, 35), serine proteases of the subtilisin subfamily (MEROPS S8A) (14, 24, 32), and metalloproteases of two different families (MEROPS M35 and M36) (13, 18, 23, 30, 39). Aspergillus secreted exoproteases include aminopeptidases (MEROPS M28), carboxypeptidases (MEROPS S10), and dipeptidyl peptidases (MEROPS S9B and S9C).
Recently, a new endoprotease of the sedolisin family (MEROPS S53) was isolated from a commercial protease extract of Aspergillus oryzae and termed aorsin (16). This enzyme was homologous to a tripeptidyl-peptidase (TPP) secreted by the same fungal species (GenBank accession no. AB089196) and another TPP from Aspergillus niger (12). The term sedolisin was recently introduced (46) for a group of similar acidic serine proteases of various origins. In particular, this family contains the human lysosomal TPP involved in hydrolysis of hydrophobic proteins. A hereditary deficiency of human lysosomal TPP results in infantile neuronal ceroid lipofuscinosis (31, 40). A BLAST analysis based on the A. fumigatus genome (www.tigr.org/tdb/e2k1/afu1) revealed the presence of an apparently five member gene family encoding sedolisins. Four of these enzymes have a predicted 17- to 20-amino-acid signal sequence for putative secretion and were named SedA, SedB, SedC, and SedD.
The objective of this study was to provide a detailed description of these four putative sedolisin proteases secreted by A. fumigatus and to validate predictions from the A. fumigatus genome. This is the first time outside the patent literature (12) that TPPs secreted by fungi have been identified and characterized. SedA, SedB, SedC, and SedD, together with the aspartic protease Pep1 (35), constitute a set of proteases that could be used by A. fumigatus to degrade proteins at acidic pHs.
MATERIALS AND METHODS
Strains and plasmids.
Aspergillus fumigatus NCPF 7367 from the genome-sequencing project was used in this study. All plasmid subcloning experiments were performed in Escherichia coli XL1-Blue using plasmids pMTL21 (7), pUC18, and pET11-a. The Pichia pastoris strains used were GS115 and KM71 (Invitrogen, Carlsbad, CA). The expression vectors pKJ111 (21), pKJ113 (5), and pPICZαA (Invitrogen) were used to express heterologous peptidases.
Aspergillus fumigatus growth media.
Aspergillus fumigatus was grown on Sabouraud agar or, to promote production of proteolytic activity, in a liquid medium containing 1.2% yeast carbon base (Difco, Detroit, MI), 10 g/liter glucose, and 5 g/liter hemoglobin as the sole nitrogen source. The pH of this solution was adjusted to between 4 and 5. One-liter flasks containing 200 ml of medium were inoculated with approximately 109 spores and incubated for 48 h at 37°C on an orbital shaker at 200 rpm.
Genomic and cDNA libraries.
An A. fumigatus λgt11 cDNA library was previously constructed using mRNA from the CHUV192-88 strain grown at 30°C for 40 h in a liquid medium containing collagen as the sole nitrogen and carbon source (24). Total RNA was extracted as described previously (19) and the mRNA purified by using oligo(dT) cellulose (Sigma, Taufkirchen, Germany) according to standard protocols (37). The cDNA library was prepared from this mRNA using phage λgt11 (Promega, Madison, WI) and protocols supplied by the manufacturer. Alternatively, another λ-phage cDNA library made from an A. fumigatus D141 culture growing in the logarithmic phase (minimal medium, 37°C, 16 h) was used (33).
DNA fragments for production of heterologous proteins.
The cDNA coding for putative SedB and SedD was amplified by PCR. Target DNA was prepared from 106 clones of the cDNA libraries. PCR was performed with homologous primers derived from genomic DNA sequences (P17 to P24 [Tables 1 and 2]). Two hundred nanograms of target DNA, 10 μl each of the sense and antisense oligonucleotides at a concentration of 42 mM, and 8 μl of deoxynucleotide mix (containing 10 mM of each deoxynucleoside triphosphate) were dissolved in 100 μl PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, and 1.5 mM MgCl2). To each reaction mixture, 2.5 U of AmpliTAQ DNA polymerase (Perkin Elmer, Boston, MA) was added. The reaction mixtures were incubated for 5 min at 94°C, then subjected to 25 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C, and finally incubated for 10 min at 72°C.
TABLE 1.
Primers used in this studya
| Primer | Oligonucleotide sequenceb | Location | PCR product size (bp) (cloning sites) |
|---|---|---|---|
| P1 | 5′-GTTGAATTCAGTCCCACCCCGAACGACTATG-3′ | sedA | 179 (EcoRI-BamHI) |
| P2 | 5′-CTTGGATCCGATATCTCCATCAACAAGTCATG-3′ | Complement of sedA | |
| P3 | 5′-CTTGGATCCGCGCTCGTCACGCTATGGAC-3′ | sedA | 262 (BamHI-SphI) |
| P4 | 5′-TCGGCATGCAATGTGTGACTTTCCTGAACC-3′ | Complement of sedA | |
| P5 | 5′-ATTGCATGCCGAGAATACCATGTCCCCCACTCA-3′ | sedA | 244 (SphI-SspI) |
| P6 | 5′-GTTTGTACAGAGCTGATATACACAGTGGTGT-3′ | Complement of sedA | |
| P7 | 5′-CTCTGTACAACATTACTCGCGGCTCAA-3′ | sedA | 143 (SspI-Asp718) |
| P8 | 5′-GTTGGTACCCTGGGGAATTTGCTGTGCAAATGTTGAAAAGAA CAGG-3′ | Complement of sedA | |
| P9 | 5′-GTTGAATTCGTAACTCGAGCTAGTCCCACCCCGAACGACTATG-3′ | pSedA bp47-826 | 835 (EcoRI-SacI) |
| P10 | 5′-CTTGAGCTCCGTCGACGGCCTTCAGGATGGGATGGGTACCCT GGGGAATTTGC-3′ | pSedA bp47-826 | |
| P11 | 5′-GACGGAGCTCAAGCCCCAACCAGCGTGACC-3′ | sedA | 1,038 (SacI-HindIII) |
| P12 | 5′-GTTAAGCTTGCGGCCGCCTACGGCAGGCTCATGAACAAGTC-3′ | Complement of sedA | |
| P13 | 5′-GTTAAGCTTAGGCCTGCTGCAGAGCCCAGCTACGCGG-3′ | sedC | 1,449 (HindIII-Asp718) |
| P14 | 5′-AGACCTGGTACCGCCTACCCTCAACG-3′ | Complement of sedC | |
| P15 | 5′-GTTGGTACCAGTGCATCTGCGCCTGTATTT-3′ | sedC | 321 (Asp718-BglII) |
| P16 | 5′-CTTGAGATCTAGGCCTCATGGGGTACTTGAAACACTCG-3′ | Complement of sedC | |
| P17 | 5′-GTTCTCGAGCTAGTCCCACCCCGAACGACT-3′ | sedA | 2,002 (XhoI-NotI) |
| P18 | 5′-CTTGCGGCCGCCTACGGCAGGCTCATGAACAAGTC-3′ | Complement of sedA | |
| P19 | 5′-GTTCTCGAGCTGCCGAGGTTTTTGAGAAG-3′ | sedB | 1,760 (XhoI-BamHI) |
| P20 | 5′-CTTGGATCCCTATCTCCGCATAGCAAGAGAC-3′ | Complement of sedB | |
| P21 | 5′-GTTAGATCTGCTGCAGAGCCCAGCTACGCG-3′ | sedC | 1,743 (BglII-XbaI) |
| P22 | 5′-CTGTCTAGATCATGGGGTACTTGAAACACTCGT-3′ | Complement of sedC | |
| P23 | 5′-GTTGTCGACTTGCCCCAGCCCTTTGTGTGGTGC-3′ | sedD | 1,684 (SalI-NotI) |
| P24 | 5′-CTTGCGGCCGCTTACAACGACAGAACCAACTC-3′ | Complement of sedD | |
| P25 | 5′-GTTCCATGGGTCCTACTGCAGCGAGATC-3′ | sedA | 845 (NcoI-BamHI) |
| P26 | 5′-GTTGGATCCCTATCCCGTCGCAGCGGAGAACCC-3′ | Complement of sedA | |
| P27 | 5′-GGTCCATGGGCTGAACGACCAGAACTCCGCC-3′ | sedB | 1,002 (NcoI-BamHI) |
| P28 | 5′-AACAGGATCCCTATCTCCGCATAGCAAGAGAC-3′ | Complement of sedB | |
| P29 | 5′-GTTCCATGGGAGAACGCTTGCGTATTCCGTTCC-3′ | sedC | 1,048 (NcoI-BamHI) |
| P30 | 5′-GTTGGATCCCTATGTGCCGCCTACCCTCAA-3′ | Complement of sedC | |
| P31 | 5′-GGTCCATGGGAAGCTTCCAGCTCTTCCTAGG-3′ | sedD | 1,121 (NcoI-BamHI) |
| P32 | 5′-AACAGGATCCCTAGGCAGTAGCGTTCCAGTGAGCG-3′ | Complement of sedD | |
| s1-1 | 5′-TATCTAGAAGTCCCACCCCGAACGACTATG-3′ | sedA | 1,027 (XbaI-HindIII) |
| s1-2 | 5′-ATAAGCTTGGCCCTGCATTGGTCACGC-3′ | Complement of sedA | |
| s2-1 | 5′-TCTAGATGAGAAGCTGTCCGCGGTGC-3′ | sedB | 829 (XbaI-HindIII) |
| s2-2 | 5′-AAGCTTCCGCTGTCACTGGCGGAGTTC-3′ | Complement of sedB | |
| s3-1 | 5′-TCTAGACGGTCGTTGAGCAGCTCAG-3′ | sedC | 767 (XbaI-HindIII) |
| s3-2 | 5′-AAGCTTGGTACTGCTCAGGGACGAGTCCTG-3′ | Complement of sedC | |
| s4-1 | 5′-GCCGGCGTGGTGCAGGAGAAGCTCTCAGC-3′ | sedD | 753 (NgoMIV-XbaI) |
| s4-2 | 5′-TCTAGAGCCGAGGGAAGCAGTGGCC-3′ | Complement of sedD | |
| s1-A | 5′-GTACACACAAGCTATTCTTGCG-3′ | Upstream sequence in sedA | 1,183 |
| s1-B | 5′-AAGGCGATTAAGTTGGGTAACG-3′ | pAN 7.1 sequence | |
| s1-C | 5′-CGTTTACCCAGAATGCACAGG-3′ | pAN 7.1 sequence | 1,143 |
| s1-D | 5′-AGAGAATGGAGTTCTGCGGCC-3′ | Downstream sequence in sedA | |
| s2-A | 5′-TCTCTTGTCCTCTTCCGTTGC-3′ | Upstream sequence in sedB | 896 |
| s2-B | 5′-ACGACGTTGTAAAACGACGGC-3′ | pAN 7.1 sequence | |
| s2-C | 5′-GTCGTTTACCCAGAATGCACAG-3′ | pAN 7.1 sequence | 906 |
| s2-D | 5′-TGCAGGTCGAGATTGGCCTC-3′ | Downstream sequence in sedB | |
| s3-A | 5′-ATCGTGTAGCTCACATGGCTCC-3′ | Upstream sequence in sedC | 932 |
| s3-B | 5′-TTAAGTTGGGTAACGCCAGGG-3′ | pAN 7.1 sequence | |
| s3-C | 5′-CTCTCCTATGAGTCGTTTACCC-3′ | pAN 7.1 sequence | 881 |
| s3-D | 5′-GCTTTATACGCCAAAGAATAGG-3′ | Downstream sequence in sedC | |
| s4-A | 5′-TTCGCAGTCCGTCCCGTAATGC-3′ | Upstream sequence in sedD | 865 |
| s4-B | 5′-GTGGGCGCTTACACAGTACACG-3′ | pAN 7.1 sequence | |
| s4-C | 5′-TGCATGGTTGCCTAGTGAATGC-3′ | pAN 7.1 sequence | 824 |
| s4-D | 5′-ATCAACTCCACGTCCAGATTGG-3′ | Downstream sequence in sedD |
Primers P1 to P16 were used to construct DNA segments encoding SedB and SedC. Primers P17 to P24 were used for sed gene expression in P. pastoris. Primers P25 to P32 were used for antigen production in E. coli. Primers s1-1 to s4-2 were used for construction of sed gene disruption vectors. Primers s1-A to s4-D were used for screening of sed-negative mutants. For the locations of those primers, e.g., in the sedA-negative mutant, compare Fig. 1.
Italicized and boldfaced nucleotides represent cloning sites.
TABLE 2.
Materials used for the expression of SedA to SedD in P. pastoris and yields of heterologous proteins
| Gene | Oligonucleotide primera | Encoded amino acid sequenceb | Targeted DNA | PCR product (nucleotide positions on cDNAc); cloning site | Vector (cloning site) | Yield of heterologous protein (μg ml−1) |
|---|---|---|---|---|---|---|
| sedA | P17 | (R)ASPTPND | pSedAd | sedA (52-1935); XhoI-NotI | pKJ113 (XhoI-NotI) | 30 |
| P18 | DLFMSLPstop | |||||
| sedB | P19 | (R)(A)AEVFEK | DNA prepared from 106 clones of the cDNA library | sedB (70-1809); XhoI-BamHI | pKJ113 (XhoI-BamHI) | 30 |
| P20 | SLAMRRstop | |||||
| sedC | P21 | (R)(S)AAEPSYA | pSedCd | sedC (67-1791); BglII-XbaI | pAC3e (BglII-XbaI) | 10 |
| P22 | TSVSSTPstop | |||||
| sedD | P23 | (R)(L)APALCVV | DNA prepared from 106 clones of the cDNA library | sedD (46-1704); Sa1-NotI | pKJ113 (XhoI-NotI) | 30 |
| P24 | ELVLSLstop |
See Table 1.
Amino acids in parentheses are encoded by the restriction site sequences and added to the N-terminal ends of heterologously expressed enzymes.
From the ATG start codon.
This work; see Materials and Methods.
Plasmid pAC3 is a pPICZαA derivative (M. Monod et al., unpublished results).
Amplified DNA segments encoding putative SedA and SedC proteins were constructed and cloned into pUC18 to generate plasmids pSedA and pSedC. For pSedA cloning, the following pairs of sense and antisense primers were used to amplify four fragments of A. fumigatus genomic DNA: P1-P2, P3-P4, P5-P6, and P7-P8 (Table 1). The PCR products were digested with EcoRI/BamHI, BamHI/SphI, SphI/SspI, and SspI/Asp718, respectively, and were inserted end-to-end into pUC18 digested with EcoRI/Asp718 to generate plasmid pSedAbp47-826. In a second step, the following pairs of sense and antisense primers were used to amplify pSedA bp47-826 and A. fumigatus genomic DNA, respectively: P9-P10 and P11-P12 (Table 1). The PCR products were digested with EcoRI/SacI and SacI/HindIII and were inserted end-to-end in pUC18 digested with EcoRI/HindIII to generate plasmid pSedA.
For pSedC construction, the following pairs of sense and antisense primers were used to amplify two contiguous fragments of A. fumigatus genomic DNA: P13-P14 and P15-P16 (Table 1). Subsequently, the two PCR products were digested with HindIII/Asp718 and Asp718/BglII and were inserted end-to-end into pUC18 digested with HindIII/BglII.
Production and analysis of heterologously expressed proteins.
Expression plasmids were constructed by cloning PCR products into Pichia pastoris expression vectors (Table 2). The PCR products were purified with a PCR purification kit (Roche Diagnostics, Mannheim, Germany) and then digested with restriction enzymes for which a site was previously designed at the 5′ end of the primers. Pichia pastoris transformation and selection of transformants were performed as previously described (4, 5). For enzyme production, transformants were grown to near-saturation (optical density at 600 nm, 10) at 30°C in 10 ml (or 11 ml for large preparations of SedB) of a glycerol-based yeast medium (0.1 M potassium phosphate buffer at pH 6.0, containing 10 g/liter yeast extract, 20 g/liter peptone, 13 g/liter yeast nitrogen base without amino acids [Becton Dickinson, Sparks, MD], 10 ml/liter glycerol, and 40 mg/liter biotin). Cells were harvested and resuspended in 2 ml (200 ml) of the same medium with 5 ml/liter methanol instead of glycerol and were incubated for 2 days. Then the culture supernatant was harvested after centrifugation (3,000 × g, 4°C, 5 min).
Salts and low-molecular-weight solutes were removed from 2.5 ml of P. pastoris culture supernatants by being passed through a PD10 column (Amersham Pharmacia, Dübendorf, Switzerland) with 20 mM citrate buffer (pH 6.0) before testing for proteolytic activity. Supernatants of P. pastoris GS115 and KM71 grown under the same conditions were used as negative controls for comparison.
Heterologously expressed proteins were identified and confirmed by a de novo sequencing analysis approach. The supernatant was treated with N-glycosidase F as described previously (8) and then subjected to regular sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% polyacrylamide). Protein bands stained with Coomassie brilliant blue R-250 (Bio-Rad, Munich, Germany) were excised from the gel, reduced, and alkylated, and then in-gel trypsin digestion was performed (38). The resulting peptides were extracted from the gel, dried, and dissolved in 0.1% formic acid. Peptide sequencing was performed on a Q-TOF Ultima Global mass spectrometer (Micromass, Manchester, United Kingdom) equipped with a nanoflow electrospray ionization (ESI) Z-spray source in positive-ion ESI-tandem mass spectrometry (MS/MS) mode. Samples were introduced with a CapLC autosampler (Waters) onto a C18 pepMap μ-Precolumn cartridge (300 μm by 5 mm; particle size, 5 μm) and then further separated through a C18 pepMap100 nano Series analytical column (75 μm by 15 cm; particle size, 3 μm; LC Packings-Dionex, Amsterdam, The Netherlands). The precursor ions were selected with a quadrupole analyzer, and fragmentation took place in a hexapole collision gas cell, with argon as the collision gas. Acquisition was performed with MassLynx (v. 4.0) and Windows NT on a personal computer, while data were further processed on a Protein Lynx Global Server (v, 2.1; Micromass, Manchester, United Kingdom). The resulting MS/MS ion data were searched against a mass spectrometry protein sequence database (MSDB) database through the MASCOT search engine (www.matrixsciences.com) for identification.
N-terminal Edman sequencing data for proteins were obtained with a Procise sequencer from Applied Biosystems (Foster City, CA) according to the protocols of the manufacturer. Sequencing was conducted by SEQLAB (Sequence Laboratories, Göttingen, Germany).
Purification of heterologously expressed SedB.
The secreted proteins from 250 ml of P. pastoris culture supernatants were concentrated by ultrafiltration to 6 ml by using an Amicon cell and an Ultracel Amicon YM30 membrane (30-kDa cutoff) (Millipore, Volketswil, Switzerland). The concentrate was washed with 10 mM sodium acetate, pH 5.0, and applied at 4°C to a Mono Q-Sepharose column (Amersham Pharmacia, Dübendorf, Switzerland) equilibrated with the same buffer at a flow rate of 0.5 ml/min. After the column was washed with 10 mM sodium acetate, pH 5.0, elution was performed with a linear gradient of 0 to 1.0 M NaCl over 96 min at a flow rate of 0.5 ml/min. The different fractions eluted from the Mono Q-Sepharose column were screened for enzymatic activity with Ala-Ala-Phe-p-nitroanilide (Ala-Ala-Phe-pNA) as a substrate, and TPP-containing fractions were pooled. After concentration in an Amicon ultrafiltration cell with an Ultracel Amicon YM30 membrane and washing with 10 mM sodium acetate, pH 5.0, the TPP extract was loaded onto a size exclusion Superose 6 fast protein liquid chromatography column (Amersham Pharmacia), and elution was performed at a flow rate of 0.2 ml/min using 10 mM sodium acetate, pH 5.0, as the eluant. The eluted active fractions were pooled. The TPP enzyme was concentrated (1.0 mg/ml) to a final volume of 0.4 to 1.0 ml in a Centricon concentrator with a 30-kDa cutoff (Millipore) and stored at −20°C prior to further functional characterization.
Proteolytic activities.
Endoproteolytic activity was measured with 50 μl of a P. pastoris culture supernatant and 50 μl of 0.2% resorufin-labeled casein at different pHs in sodium citrate buffer (50 mM final concentration; pH 2.0 to 7.0) in a total volume of 0.5 ml. After incubation at 37°C, the undigested substrate of the enzyme-substrate mixture was precipitated by trichloroacetic acid (4% final concentration) and separated from the supernatant by centrifugation. Five hundred microliters of Tris-HCl buffer (500 mM; pH 9.4) was added to the collected supernatant (neutralization step), and the A574 of the mixture (1 ml) was measured. A blank was performed with 50 μl of a P. pastoris GS115 culture supernatant. For practical purposes, one milliunit of activity was defined as producing an increase in absorbance of 0.001 per min in a proteolytic assay (1 ml) at the optimal pH for activity. The assays were performed in triplicate.
Exoproteolytic activity was tested with synthetic substrates supplied by Bachem (Bubendorf, Switzerland). Stock solutions were prepared at 100 mM concentration and stored at −20°C. Ala-pNA, Gly-Pro-pNA, Ala-Ala-pNA, Phe-Pro-Ala-pNA, Ala-Ala-Pro-pNA, and Ala-Ala-Pro-Leu-pNA were dissolved in ethanol. Ala-Ala-Phe-pNA was dissolved in dimethylformamide. The reaction mixture contained a concentration of 5 mM substrate and the enzyme preparation (between 0.1 to 1.0 μg per assay) in 100 μl of 50 mM citrate buffer at different pH values for each Sed (between pH 2.0 and pH 7.0). After incubation at 37°C for 10 min, the reaction was terminated by addition of 5 μl of glacial acetic acid, followed by 0.9 ml of water, to the mixture. The pNA released was measured by spectrometry as a change in A405. A control with a blank substrate and blank culture broth was carried out in parallel. The Sed TPP activities were expressed in milliunits (nanomoles of pNA released per minute) using Phe-Pro-Ala-pNA as the substrate.
For TPP kinetic analysis, purified heterologously produced SedB (see above) was tested with Phe-Pro-Ala-pNA as a substrate at various concentrations between 10−3 and 10−7 M at 20°C in 0.1 M sodium citrate buffer, pH 6.0. The final concentration of SedB in the reaction mixture was 2.5 × 10−9 M. The absorption of liberated p-nitroaniline was monitored photometrically at 405 nm (Ultrospec 1000 photometer; Amersham Pharmacia). The Michaelis constant (Km) and the turnover number (kcat) were calculated on the basis of a standard Lineweaver-Burk plot.
Peptide digestion by SedB was investigated with the synthetic peptide Ala-Pro-Gly-Asp-Arg-Ile-Tyr-Val-His-Pro-Phe (Sigma catalog no. A-0289). The peptide was dissolved in 50 mM sodium citrate buffer, pH 6.0, at a final concentration of 0.5 mM, and purified heterologously produced SedB was added to a final concentration of 5 μg/ml. A control without SedB was carried out in parallel. The reaction was stopped after a 2-h incubation at 37°C by addition of 1/20 volume of 100% formic acid, and samples were then analyzed with a Q-TOF Ultima Global mass spectrometer (Micromass) in MS scan mode. For this purpose, samples were diluted to 1 pmol/μl of the peptide starting material with acetonitrile-H2O (50:50) and subsequently injected into the mass spectrometer using a HAMMEL syringe at an approximate flow rate of 300 nl/min. Acquisition of data was started after a constant spray was obtained and was carried out for at least 10 min for each digested and undigested sample. Data were interpreted with MassLynx 4.0 SP4 software (Micromass).
Antigen preparation for immunization of rabbits.
Large peptides (200 to 300 aa) corresponding to sequences from SedA, SedB, SedC, and SedD were produced using the pET expression system from Novagen (Darmstadt, Germany). Plasmid pET-11a was changed to incorporate the sequence 5′-CATGCACCA TATGCACCATATGCACCATGGTAAGGATC-3′, encoding a Met-His6 amino acid sequence between the unique NheI and BamHI cloning sites. The sixth His residue in the newly generated plasmid pET-11aH6 was encoded by a unique NcoI cloning site. The following pairs of sense and antisense primers were used to amplify DNA from plasmids encoding heterologous SedA, SedB, SedC, and SedD, respectively: P25-P26, P27-P28, P29-P30, and P31-P32 (Table 1). The PCR products were digested with NcoI and BamHI and cloned into the NcoI and BamHI sites of pET-11aH6. The resulting plasmids were termed pAgSedA, pAgSedB, pAgSedC, and pAgSedD.
Heterologous His6-tagged peptides were produced in E. coli BL21 transformed with pAgSedA, pAgSedB, pAgSedC, or pAgSedD. Cells were grown at 37°C to an optical density at 600 nm of 0.6, and His6-tagged peptide expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. Incubation was continued for an additional 4 h at 37°C. Cells were collected by centrifugation (4,500 × g, 4°C, 15 min), and His6-tagged peptides were extracted with guanidine hydrochloride buffer and nickel-nitriloacetic acid resin (QIAGEN, Hilden, Germany) columns according to the manufacturer's instructions. Rabbit antisera were made by Eurogentec (Liège, Belgium) by using the purified SedA, SedB, SedC, or SedD polypeptide chains as antigens.
Extraction and Western blot analysis of A. fumigatus native proteins.
A. fumigatus was grown in hemoglobin medium (see above) for 2 days at 37°C under aerobic conditions (rotary culture, 200 rpm). After filtration, the fungal culture supernatant was precipitated at 80% ammonium sulfate saturation and centrifuged (15,000 × g, 20 min, 8°C). The precipitate was dissolved in 20 mM sodium phosphate buffer, pH 7.0, and dialyzed against the same buffer using Vivaspin concentrator systems (10,000-molecular-weight cutoff, polyethersulfone membranes) according to the manufacturer's protocols (Vivascience, Hanover, Germany). Protein concentrations were measured by the method of Bradford with a commercial reagent (Bio-Rad, München, Germany). SDS-PAGE (12.5% polyacrylamide) gels with samples with and without N-glycosidase F digestion (8) were stained with Coomassie brilliant blue R-250 (Bio-Rad).
Alternatively, proteins were blotted onto a nitrocellulose membrane. The membrane was blocked with phosphate-buffered saline (pH 7.4) plus 0.1% (vol/vol) Tween 20 plus 10% nonfat dry milk (wt/vol) for 2 h at room temperature (20 to 22°C) and then incubated for another 2 h in the blocking buffer with a 1:1,000 dilution of rabbit antisera raised against SedA, SedB, SedC, or SedD. After thorough washing of the membrane with phosphate-buffered saline (pH 7.4) plus 0.1% Tween 20, another incubation for 2 h with the blocking buffer containing a 1:10,000 dilution of peroxidase-conjugated mouse anti-rabbit monoclonal immunoglobulins (Sigma A2074) followed. This conjugate was detected by chemiluminescence with the ECL system (Amersham Pharmacia) as recommended by the manufacturer.
Gene disruption.
Gene disruption vectors were constructed using pAN7.1 (29) and 0.7- to 1.1-kb internal fragments of the respective sed gene. In detail, sed fragments were obtained by PCR using appropriate primers (Table 1, primers s1-1 to s4-2) and genomic A. fumigatus DNA as the template. The PCR products were first cloned into pCR-Script Amp SK(+) (Stratagene, La Jolla, CA). In a second step, the sed fragments were excised from the plasmid constructs with XbaI and HindIII or XbaI and NgoMIV, for which a site was introduced into the primers, and were ligated to the larger fragment of pAN7.1 digested with the same restriction enzymes. The plasmids generated were termed pΔsedA to pΔsedD. Undigested plasmids were used for subsequent gene-targeted disruption experiments. For generation of a sedB disruption mutant only, the pΔsedB plasmid was digested with BsaAI prior to a second transformation experiment. A single BsaAI site was present in pΔsedB, approximately in the middle of the sedB gene fragment.
A. fumigatus NCPF 7367 was transformed according to a protocol that has been used for Aspergillus nidulans and A. fumigatus (27, 44). Transformation of 107 protoplasts either with 5 μg of undigested pΔsedA, pΔsedB, pΔsedC, or pΔsedD or with BsaAI-digested pΔsedB plasmid DNA typically yielded 100 to 200 hygromycin-resistant colonies. After overnight expression of the hygromycin B phosphotransferase gene (HPH), the transformants were incubated on agar based on GYE medium (1% glucose, 0.5% yeast extract) containing 200 μg/ml hygromycin (Sigma) and were selected after 5 days of incubation at 20°C followed by overnight incubation at 42°C. Transformants initially identified as hygromycin resistant were picked and subcultured again on agar containing hygromycin.
The sed disruptants were identified by PCR of genomic DNA from various numbers of hygromycin-resistant colonies as a template and two pairs of specific primers (Table 1, primers s1-A to s4-D). Each primer pair yields a product of the predicted size when the respective plasmid is integrated at a homologous site. In each primer pair, one primer hybridized with the transformation plasmid and the other primer hybridized with genomic DNA near the desired homologous integration locus, as shown for sedA disruption (Fig. 1). At least one disruption mutant for each sed gene was identified.
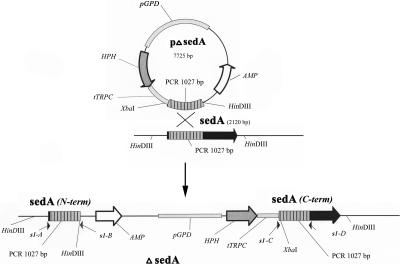
FIG. 1.
Plasmid construct and predicted outcome of pΔsedA integration event. A 1,027-bp internal PCR fragment of sedA was cloned into the pAN7.1 plasmid (29). pAN7.1 carries the hygromycin resistance gene (HPH) from Escherichia coli as a dominant selectable marker under the control of the GPD promoter (pGPD) and the TRPC terminator (tTRPC) from Aspergillus nidulans. Homologous recombination of the plasmid construct with sedA by a single-crossover event results in the generation of two incomplete copies of the A. fumigatus gene [labeled sedA (N-term) and sedA (C-term)] separated by the linearized sequence of pAN7.1. The primers used to screen for the incomplete sedA genes are s1-A and s1-B (3′ fragment) and s1-C and s1-D (5′ fragment). For further information on primers, see Table 1.
Phylogenetic analyses and genomic data for Aspergillus.
Amino acid sequences of Aspergillus fumigatus sedolisins (SedA to SedE) were first analyzed using the BLAST algorithm (blastp) (http://www.ncbi.nlm.nih.gov/BLAST/) with a BLOSUM62 substitution matrix to determine the existence of homologous proteins in other fungal species. All fungal protein sequences displaying a BLAST score higher than 150 were included in further phylogenetic analyses. The sequences were then aligned using Clustal W (43) as implemented in BioEdit Sequence Alignment Editor software (10).
Phylogenetic analyses of A. fumigatus sedolisins and homologous sequences from other fungi were performed in PAUP* v4.0b10 (42). Amino acid sequences were analyzed using maximum-parsimony (MP) and neighbor-joining (NJ) phylogenetic methods. The Dayhoff PAM model of protein evolution was used to compute the distances between the amino acid sequences. Analyses were performed using a heuristic search with the TBR branch swapping algorithm. The reliability of internal branches was assessed using the bootstrap method (9), with 1,000 replicates. Phylogenetic trees were edited using TreeView (26).
In general, genomic data for A. fumigatus were provided by The Institute for Genomic Research (www.tigr.org/tdb/e2k1/afu1) and The Wellcome Trust Sanger Institute (www.sanger.ac.uk/Projects/A_fumigatus), while genomic data for A. nidulans, Chaetomium globosum, and Stagonospora nodorum were provided by The Broad Institute (http://www.broad.mit.edu/annotation/fgi/). Coordination of the analyses of these data was enabled by an international collaboration involving more than 50 institutions from 10 countries and coordinated from Manchester, United Kingdom (www.cadre.man.ac.uk; www.aspergillus.man.ac.uk).
RESULTS
Production and structure of A. fumigatus sedolisins.
The nature of sedolisins secreted by A. fumigatus was investigated by a reverse genetics approach using enzymes produced heterologously in Pichia pastoris. The cDNAs encoding putative SedB and SedD could be specifically amplified using 5′ sense and 3′ antisense primers (Tables 1 and 2) and DNA extracted from a pool of 106 clones of the A. fumigatus cDNA libraries as the template. The intron-exon structures of the sedB and sedD genes were determined by comparing the cDNA sequences with the A. fumigatus genome sequence.
No sedA or sedC cDNA was obtained by PCR. However, the possible intron-exon structures of the sedA and sedC genes (Table 3) could be deduced following alignment with sedB and sedD cDNA sequences. DNA coding for putative SedA and SedC proteins was synthesized by adding end-to-end PCR-generated DNA fragments for the deduced exons and then cloning into pUC18. The resulting plasmids were termed pSedA and pSedC. The A. fumigatus cDNAs obtained by PCR and the sedA and sedC DNAs were cloned into P. pastoris expression vectors (Table 2) and expressed in P. pastoris grown in a methanol-inducing medium. Thirty micrograms of heterologously produced protein per milliliter was obtained for SedA, SedB, and SedD, while the yield for SedC was 10 μg ml−1 of culture supernatant. The identities of the heterologously produced proteins were confirmed as described in Materials and Methods by ESI-liquid chromatography-MS/MS de novo sequencing of trypsin-digested bands of SedA to SedD. When sequences were subjected to a database search, the relevant heterologous Sed protein was identified as the only candidate in each inquiry. The amino acid sequence coverage confirmed by de novo sequencing for mature SedA, SedB, and SedD was 44%, 40%, and 19%, respectively. The N terminus of SedC was not sequenced (see below), so no coverage value for this enzyme was determined.
TABLE 3.
A. fumigatus genes encoding sedolisins, and main characteristics of deduced translation products
| Parameter | sedA | sedB | sedC | sedD |
|---|---|---|---|---|
| Gene length (nt) with stop codon | 2,120 | 1,858 | 1,845 | 1,971 |
| No. of introns | 4 | 1 | 1 | 4 |
| Preproprotein (aa) | 644 | 602 | 596 | 568 |
| Signal peptide (aa)a | 18 | 24 | 24 | 20 |
| Mature domain of the protein (aa) | 448 | 397 | 381 | |
| Calculated mol mass of the polypeptide chain of the preproprotein (kDa) | 70.0 | 65.8 | 65.3 | 60.1 |
| Calculated mol mass of the polypeptide chain of the mature protein (kDa)b | 47.9 | 43.0 | 40.6 | |
| Mol mass of the native heterologous enzyme (kDa) (SDS-PAGE) | ≈55 | ≈60-90 | ≈65 | ≈70-100 |
| Mol mass of heterologous enzyme after deglycosylation (kDa) (SDS-PAGE) | ≈46 | ≈46 | ≈48 | ≈48 |
| No. of putative N-glycosylation sitesc (mature domain) | 4 | 4 | 8 | 4 |
| Calculated pI (mature domain)b | 4.24 | 5.17 | 5.12 | |
| GenBank accession no. | AJ585109 | AJ578478 | AJ581835 | AJ578479 |
Based on the method of von Heijne (45).
Calculated using VectorNTI suite 8 (InforMax, Inc.).
The putative glycosylation sites correspond to the NXT/S pattern (X stands for any amino acid except for P).
Heterologously expressed SedA, SedB, SedC, and SedD polypeptides were glycoproteins, as determined from reductions in their molecular weights following treatment with N-glycosidase F (Fig. 2, lanes 3 and 4). The apparent molecular mass of each deglycosylated heterologously expressed protein was lower than the calculated molecular mass of the complete polypeptide chain deduced from the nucleotide sequence of the encoding gene (Table 3). Based on the N-terminal amino acid sequence obtained by Edman degradation, the mature SedA, SedB, and SedD proteins start at Arg-Ser-Pro-Leu-Pro, Thr-Ser-Thr-Cys-Asp, and Ala-Ala-Thr-Asn-Ser, respectively, and thus prove the existence of a prosequence for each of these enzymes. After the signal and propeptide portions of the polypeptide chain were subtracted, the calculated molecular masses for the mature SedA, SedB, and SedD proteins were consistent with the molecular masses of deglycosylated proteins estimated by SDS-PAGE. The N terminus of SedC could not be determined by Edman degradation of the protein.
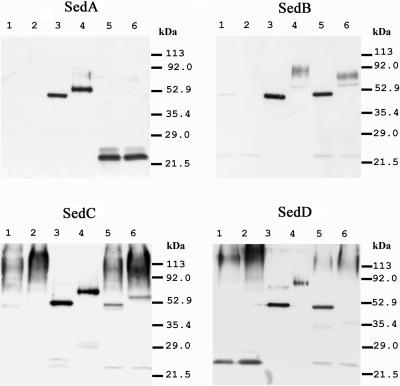
FIG. 2.
Western blot of A. fumigatus culture supernatant and heterologously expressed enzymes. A. fumigatus wild-type and mutated strains were grown in liquid hemoglobin medium at 37°C for 2 days on a rotatory shaker at 200 rpm. The proteins of Aspergillus culture supernatants (lanes 1 and 2, mutant strain; lanes 5 and 6, wild-type strain) were concentrated 150-fold, and 30 μl of this solution was loaded onto SDS-PAGE (12.5% polyacrylamide) gels. As a control, 1.0 μg of matching heterologous enzymes from P. pastoris culture supernatants was loaded in parallel (lanes 3 and 4). In all blots, supernatant preparations were used with (lanes 1, 3, and 5) and without (lanes 2, 4, and 6) prior treatment with N-glucosidase F. Western blots were revealed using antisera raised against parts of the respective heterologously expressed sedolisin (see Materials and Methods).
Enzymatic activities of heterologously produced A. fumigatus sedolisins.
Heterologously produced proteins were tested for endo- and exoproteolytic activity. Only SedA had endoproteolytic activity as described in Materials and Methods. This enzyme was active between pH 3.0 and 6.5, with optimal activity at pH 5.5. At this pH, SedA activity was 15 mU ml−1 of P. pastoris culture supernatant. SedA had no exoprotease activity, as evidenced by the fact that it did not release pNA from any of the mono-, di-, tri-, or tetrapeptide substrates tested in this study (see Materials and Methods).
The heterologously expressed SedB, SedC, and SedD proteins had no endoproteolytic activity, but they released pNA very efficiently when Phe-Pro-Ala-pNA or Ala-Ala-Phe-pNA was used as the substrate. Supernatants of P. pastoris GS115 and KM71 had no activity on these tripeptide pNA substrates. SedB and SedC were active between pH 3.0 and 7.0, with an optimum at pH 6.0, while SedD was active between pH 1.5 and 6.0, with an optimum peak at pH 5.0. At the optimal pH, SedB, SedC, and SedD activities were 1,560, 400, and 480 mU ml of P. pastoris culture supernatant−1, respectively. None of these three enzymes had activity toward either Ala-Ala-Pro-pNA or mono-, di-, or tetrapeptide pNA substrates. Since SedB, SedC, and SedD share a common specificity for tripeptide-pNA substrates, they were considered TPPs.
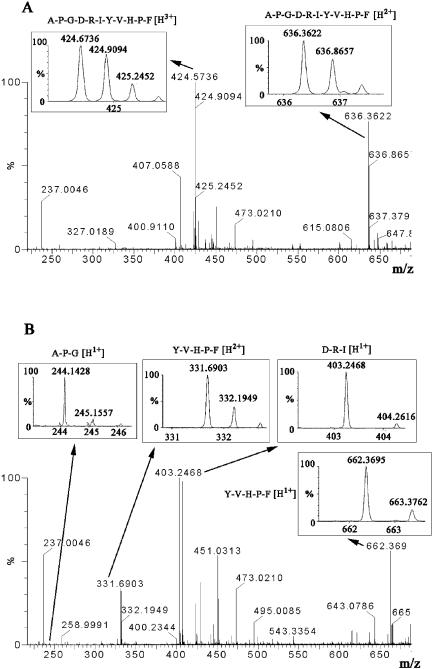
SedB was purified as described in Materials and Methods by ion-exchange chromatography and gel filtration for further characterization. At room temperature (20°C), with Phe-Pro-Ala-pNA as a substrate at the optimum pH, SedB had a kcat of 35 s−1 and a Km of 6.25 × 10−5 M, leading to a kcat/Km value of 5.6 × 105 M−1 s−1. The specific activity of SedB was 52 mU μg of protein−1. When purified SedB was incubated with the peptide Ala-Pro-Gly-Asp-Arg-Ile-Tyr-Val-His-Pro-Phe, mass peaks in accordance with the degradation products Ala-Pro-Gly, Asp-Arg-Ile, and Tyr-Val-His-Pro-Phe were detected by MS, thus confirming a TPP activity of the enzyme (Fig. 3).
FIG. 3.
MS scan analysis of protease activity of SedB. (A) MS spectrum representing the undigested 11-amino-acid peptide A-P-G-D-R-I-Y-V-H-P-F in the absence of SedB. The sample (1 pmol/μl of peptide in acetonitrile-water [50:50]) was analyzed at a flow rate of 0.3 μl/min. The chromatogram (not shown) and spectrum were recorded for 10 min and combined. Peaks at m/z 636.3 and m/z 424.6 represent double- and triple-charged whole peptides (insets). No digested di-, tri-, or pentapeptide fragments were observed. (B) MS spectrum of the SedB-digested 11-amino-acid peptide. Two peaks at m/z 244.1 and 403.2 were detected in accordance with the single-charged tripeptides A-P-G and D-R-I, respectively. Two other peaks at m/z 662.3 and 331.6 represent the single- and double-charged pentapeptide Y-V-H-P-F.
Detection of A. fumigatus sedolisins in the culture supernatant.
sed-negative mutants were constructed as described in Materials and Methods. The TPP activity of the A. fumigatus wild-type strain in comparison to that of sed-negative mutants was measured in hemoglobin liquid medium culture supernatants with Phe-Pro-Ala-pNA as a substrate at pH 5.0. At this pH, secreted A. fumigatus leucine aminopeptidase (Lap) and X-prolyl-peptidase (DppIV) (22) activities are not detectable, and thus interference by these enzymes was excluded. sedB-, sedC-, and sedD-negative mutants had 20, 90, and 80% of the TPP activity of the A. fumigatus wild-type strain NCPF 7367, respectively. Therefore, TPP activity in the A. fumigatus culture supernatant was due mainly to SedB. None of the sed-negative mutants had a visible phenotype on Sabouraud agar or in hemoglobin liquid medium.
Immunoblot analyses using polyclonal antibodies were performed to detect Seds in A. fumigatus culture supernatants. Antisera against SedA, SedB, and SedC were highly specific, while the anti-SedD antiserum cross-reacted slightly with SedA (data not shown). Western blot analysis unambiguously detected the presence of SedB, SedC, and SedD in A. fumigatus culture supernatants. Deglycosylated native enzymes and heterologously expressed enzymes from P. pastoris had the same electrophoretic mobility (Fig. 2). With the exception of glycosylated native SedD, which did not appear as a distinct band, the corresponding glycosylated heterologous and native Seds had approximately the same electrophoretic mobility. In contrast, only a 25-kDa protein was identified by anti-SedA or anti-SedD antibodies. This protein was absent in the sedA-negative mutant culture supernatant, leading to the conclusion that it is a degradation product of SedA that cross-reacts with the anti-SedD antibodies (Fig. 2).
Phylogenetic analysis of A. fumigatus sedolisins.
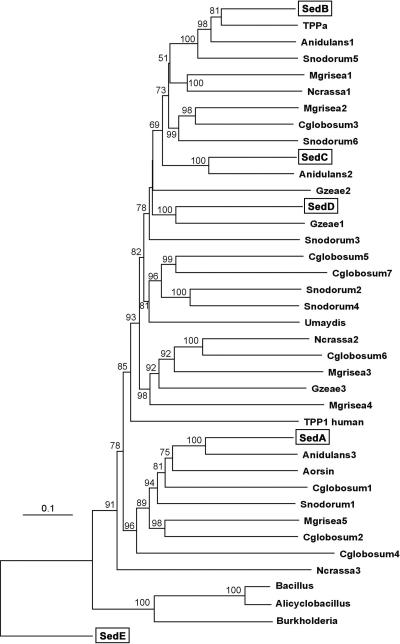
BLAST searches performed with SedA to SedD displayed hypothetical proteins from complete genome sequences of various fungi. Matching sequences were obtained from the genomes of the ascomycetes Magnaporthe grisea (five sequences), Gibberella zeae (three sequences), Neurospora crassa (three sequences), Aspergillus nidulans (three sequences), Chaetomium globosum (seven sequences), and Stagonospora nodosum (six sequences) and from the genome of the basidiomycete Ustilago maydis (one sequence) (Fig. 4). Additionally, A. oryzae TPPa and aorsin sequences are displayed. The best BLAST scores for nonfungal sequences were those for TPPs from vertebrates. The SedE sequence appeared to be highly divergent from SedA to SedD (18 to 21% sequence identity), and BLAST searches showed homology with the kumamolisin (sedolisin) precursors from a Bacillus sp., Alicyclobacillus sendaiensis, and Burkholderia pseudomallei. The sequences closest to SedA to SedD were Anidulans3 (55% identity), A. oryzae TPPa (69% identity), Anidulans2 (63% identity), and Gzeae1 (47% identity), respectively (Fig. 4).
FIG. 4.
NJ phylogenetic tree inferred from sedolisin amino acid sequences. Aspergillus fumigatus sedolisins are boxed. Bootstrap values (1,000 replicates) higher than 50% are given at nodes. Bar, number of substitutions per site. The aligned data set of SedA to SedE and putative orthologues from fungi and bacteria cited comprised 912 sites, among which 975 were variable (74%). Phylogenetic analyses performed using MP and NJ methods produced very similar tree topologies. The MP analysis produced a single most-parsimonious tree, which differed from the NJ tree mainly by the basal position of the human TPP1 sequence. The trees were rooted with the most divergent Sed sequence (SedE). Amino acid sequence accession numbers at NCBI are as follows: SedA, CAE51075; SedB, CAE17674; SedC, CAE46473; SedD, CAE17675; SedE, EAL86850; A. oryzae TPPa, BAC56232; A. oryzae aorsin, BAB97387; Anidulans1, XP411296; Anidulans2, XP407157; Anidulans3, XP411338; Mgrisea1, EAA53127; Mgrisea2, EAA49282; Mgrisea3, EAA51305; Mgrisea4, EAA53282; Mgrisea5, EAA47813; Ncrassa1, XP329464; Ncrassa2, XP324260; Ncrassa3, XP329780; Gzeae1, XP383248; Gzeae2, XP390519; Gzeae3, XP382642; Umaydis, XP403733; TPP1 human, O14773; Bacillus, BAB85637; Alicyclobacillus, BAC41257; Burkholderia, YP_111568. Amino acid sequence accession numbers at Broad Institute are as follows: Cglobosum1, CHG02181; Cglobosum2, CHG05416; Cglobosum3, CHG07661; Cglobosum4, CHG08553; Cglobosum5, CHG09918; Cglobosum6, CHG05162; Cglobosum7, CHG09915; Snodorum1, SNU10163; Snodorum2, SNU01084; Snodorum3, SNU12941; Snodorum4, SNU12886; Snodorum5, SNU09464; Snodorum6, SNU14758.
DISCUSSION
We identified and characterized four novel proteases secreted by A. fumigatus that belong to the sedolisin family (MEROPS S53). The catalytic triad of sedolisins (Glu, Asp, Ser) was conserved in the four enzymes described here (see Fig. S1 in the supplemental material). SedA is an endoprotease similar to the previously described aorsin from A. oryzae (16) (MEROPS S53.007). No 46-kDa polypeptide chain comparable to that of the heterologously expressed enzyme was detected in A. fumigatus culture supernatants by Western blot analyses. However, comparison of results obtained using a wild-type strain and sed mutants suggests that the 25-kDa protein identified by Western blotting was a SedA product following degradation of the mature enzyme by other endo- and exoproteases secreted by A. fumigatus into media that contained proteins as their sole nitrogen source (32). The other three A. fumigatus secreted sedolisins, SedB, SedC, and SedD, are TPPs like the human lysosomal enzyme of the sedolisin family. SedD activity clearly was more acidic than those of SedB and SedC.
The N-terminal amino acid sequences of heterologously expressed SedA, SedB, SedD (this work), and aorsin (16) indicate that the mature enzymes are made as preproproteins. N-terminal Edman degradation of SedC did not yield usable results. However, the presence of a prosequence was strongly supported by the molecular mass of deglycosylated SedC, which was comparable to those of the other Seds. Consistently, no peptide before alignment position 200 was detected by mass spectrometric de novo sequence analysis of any of the four secreted Seds (see Fig. S1 in the supplemental material).
Single TPPs from A. oryzae and A. niger were previously described in a patent (12). However, this is the first report of a fungal species secreting several TPPs of the sedolisin family. The finding of putative homologous proteins in all of the fungal genomes available for the ascomycete subphylum Pezizomycotina (A. nidulans, G. zeae, M. grisea, N. crassa, C. globosum, and S. nodorum) and in one basidiomycete genome (U. maydis) indicates that proteases of the S53 family are widespread among fungi. The sedolisin phylogenetic tree has a complex branching pattern of orthologues (versions of the same gene in different genomes that have been created by the splitting of taxonomic lineages) and paralogues (genes in the same genome that have been created by gene duplication events). The SedA sequence belongs to a basal, well-supported group that includes aorsin. This group is clearly distinguished from other groups, including all of the other sequences we analyzed, which emphasizes the unusual endoprotease activity of SedA and aorsin. Most of the ascomycete sedolisin paralogues are distributed in distinct clades, suggesting a common origin from ancient gene duplications. In contrast, only one putative sedolisin gene was found in the genome of the basidiomycete Ustilago maydis, and no putative sedolisin genes were detected in the yeast genomes (ascomycete subphylum Saccharomycotina) screened. This would suggest that the duplications of sedolisins occurred after the ascomycete-basidiomycete split, followed by loss of all genes in the Saccharomycotina. The current disparity in protease gene numbers among filamentous ascomycetes (subphylum Pezizomycotina) has been observed previously for other fungal proteases (15) and likely results from further duplication and/or loss of genes under selective pressure. The interaction of secreted proteases with the environment is believed to confer considerable selective functions and possibly to preadapt fungi for decomposition of organic matter in saprophytism or in pathogenesis.
Digestion of protein into amino acids and short peptides by Aspergillus spp. has been investigated extensively by the food fermentation industry. These fungi secrete various endo- and exoproteases that cooperate very efficiently. In Aspergillus, endoprotease cleavage of proteins at neutral pHs with the subtilisin Alp (24, 32) and metalloproteases (23, 30), Laps (22), and DppIV (2) synergistically digests large peptides into amino acids and X-Pro dipeptides (6, 25). Laps can degrade peptides from their N termini; however, X-Pro acts as a stop sequence. In a complementary manner, these X-Pro sequences can be removed by DppIV, thus allowing Laps access to the next residues. Synergistic action of A. oryzae Lap and DppIV at pH 7.5 leads to complete digestion of the Ala-Pro-Gly-Asp-Arg-Ile-Tyr-Val-His-Pro-Phe peptide into amino acids and X-Pro dipeptides (6). Using the same peptide, we have shown that SedB can bypass Pro residues by degrading large peptides from their N termini when these Pro residues are in the second position. Although the substrate specificity of A. fumigatus TPPs needs further investigation, these enzymes appeared to be active when the amino acid in the P1 or P′1 position (amino acids in positions 3 and 4 from the N terminus of the substrate peptide) is not a proline. Indeed, the remaining pentapeptide Tyr-Val-His-Pro-Phe was not digested by SedB, just as the synthetic substrate Ala-Ala-Pro-pNA was not digested by any of the TPPs tested.
Filamentous fungi, like bacteria, yeasts, and specialized cells of plants and animals, express membrane proteins for uptake of amino acids, dipeptides, and tripeptides (3, 11, 28, 36, 41). Large peptides cannot be used as nutrients. Therefore, SedA and the aspartic protease Pep1 (35) as endoproteases, together with SedB, SedC, and SedD as exoproteases, constitute a set of proteases that can be used by A. fumigatus to degrade proteins at acidic pHs and to generate assimilable nitrogen sources in decomposing organic matter and composts. In addition, as A. fumigatus regularly acidifies its culture supernatant in vitro (32), it is likely to acidify its microenvironment in the living host. Thus, the Seds may also play a role in proper nutrition of the fungus during infection. Growth studies on protein medium and in the living host comparing the wild type and a triple-knockout sedB sedC sedD mutant should therefore follow our investigations as an intriguing future project.
Supplementary Material
Acknowledgments
We thank Birgit Riemenschneider for technical assistance at various stages of this investigation and Victor W. Armstrong for helpful suggestions and correction of the manuscript.
This project was supported in part by Deutsche Forschungsgemeinschaft grant RE 953/3 and by the Swiss National Foundation for Scientific Research, grants 3100-043193 and 3100-105313/1.
Footnotes
Supplementary material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barrett, A. J., N. D. Rawlings, and J. F. Woessner. 2003. Handbook of proteolytic enzymes, 2nd ed. Elsevier Academic Press, San Diego, Calif.
- 2.Beauvais, A., M. Monod, J. Wyniger, J. P. Debeaupuis, E. Grouzmann, N. Brakch, J. Svab, A. G. Hovanessian, and J. P. Latge. 1997. Dipeptidyl-peptidase IV secreted by Aspergillus fumigatus, a fungus pathogenic to humans. Infect. Immun. 65:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, J. M., and F. Naider. 1995. Fungal peptide transport as a drug delivery system, p. 369-384. In M. Taylor and G. Amidon (ed.), Peptide based drug design: controlling transport and metabolism. American Chemical Society, Washington, D.C.
- 4.Beggah, S., B. Lechenne, U. Reichard, S. Foundling, and M. Monod. 2000. Intra- and intermolecular events direct the propeptide-mediated maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology 146:2765-2773. [DOI] [PubMed] [Google Scholar]
- 5.Borg-von Zepelin, M., S. Beggah, K. Boggian, D. Sanglard, and M. Monod. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28:543-554. [DOI] [PubMed] [Google Scholar]
- 6.Byun, T., L. Kofod, and A. Blinkovsky. 2001. Synergistic action of an X-prolyl dipeptidyl aminopeptidase and a non-specific aminopeptidase in protein hydrolysis. J. Agric. Food Chem. 49:2061-2063. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 8.Doumas, A., P. van den Broek, M. Affolter, and M. Monod. 1998. Characterization of the prolyl dipeptidyl peptidase gene (dppIV) from the koji mold Aspergillus oryzae. Appl. Environ. Microbiol. 64:4809-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 10.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 11.Hauser, M., V. Narita, A. M. Donhardt, F. Naider, and J. M. Becker. 2001. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 18:105-112. [PubMed] [Google Scholar]
- 12.Holm, K. A., G. Rasmussen, T. Halkier, and J. Lehmbeck. October 1998. Tripeptidyl aminopeptidase. U.S. patent 5,821,104.
- 13.Jaton-Ogay, K., S. Paris, M. Huerre, M. Quadroni, R. Falchetto, G. Togni, J. P. Latgé, and M. Monod. 1994. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol. Microbiol. 14:917-928. [DOI] [PubMed] [Google Scholar]
- 14.Jaton-Ogay, K., M. Suter, R. Crameri, R. Falchetto, A. Fatih, and M. Monod. 1992. Nucleotide sequence of a genomic and a cDNA clone encoding an extracellular alkaline protease of Aspergillus fumigatus. FEMS Microbiol. Lett. 71:163-168. [DOI] [PubMed] [Google Scholar]
- 15.Jousson, O., B. Lechenne, O. Bontems, B. Mignon, U. Reichard, J. Barblan, M. Quadroni, and M. Monod. 2004. Secreted subtilisin gene family in Trichophyton rubrum. Gene 339:79-88. [DOI] [PubMed] [Google Scholar]
- 16.Lee, B. R., M. Furukawa, K. Yamashita, Y. Kanasugi, C. Kawabata, K. Hirano, K. Ando, and E. Ichishima. 2003. Aorsin, a novel serine proteinase with trypsin-like specificity at acidic pH. Biochem. J. 371:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J. D., and P. E. Kolattukudy. 1995. Molecular cloning of the cDNA and gene for an elastinolytic aspartic proteinase from Aspergillus fumigatus and evidence of its secretion by the fungus during invasion of the host lung. Infect. Immun. 63:3796-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markaryan, A., I. Morozova, H. Yu, and P. E. Kolattukudy. 1994. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 62:2149-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monod, M., and L. A. Applegate. 1994. RNA extractions and Northern hybridization of Aspergillus fumigatus for the detection of alkaline and metalloprotease gene expression, p. 19-27. In B. Maresca and G. S. Kobayashi (ed.), Molecular biology of pathogenic fungi. A laboratory manual. Telos Press, New York, N.Y.
- 20.Monod, M., S. Capoccia, B. Lechenne, C. Zaugg, M. Holdom, and O. Jousson. 2002. Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 292:405-419. [DOI] [PubMed] [Google Scholar]
- 21.Monod, M., K. Jaton-Ogay, and U. Reichard. 1999. Aspergillus fumigatus secreted proteases as antigenic molecules and virulence factors, p. 182-192. In A. Brakhage, B. Jahn, and A. Schmidt (ed.), Aspergillus fumigatus. Biology, clinical aspects and molecular approaches to pathogenicity, vol. 2. Karger AG, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 22.Monod, M., B. Lechenne, O. Jousson, D. Grand, C. Zaugg, R. Stocklin, and E. Grouzmann. 2005. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology 151:145-155. [DOI] [PubMed] [Google Scholar]
- 23.Monod, M., S. Paris, D. Sanglard, K. Jaton-Ogay, J. Bille, and J. P. Latgé. 1993. Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect. Immun. 61:4099-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monod, M., G. Togni, L. Rahalison, and E. Frenk. 1991. Isolation and characterisation of an extracellular alkaline protease of Aspergillus fumigatus. J. Med. Microbiol. 35:23-28. [DOI] [PubMed] [Google Scholar]
- 25.O'Cuinn, G., R. FitzGerald, P. Bouchier, and M. McDonnell. 1999. Generation of non-bitter casein hydrolysates by using combinations of a proteinase and aminopeptidases. Biochem. Soc. Trans. 27:730-734. [DOI] [PubMed] [Google Scholar]
- 26.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 27.Paris, S. 1994. Isolation of protease negative mutants of Aspergillus fumigatus by insertion of a disrupted gene, p. 49-55. In B. Maresca and G. S. Kobayashi (ed.), Molecular biology of pathogenic fungi. A laboratory manual. Telos Press, New York, N.Y.
- 28.Payne, J. W., and M. W. Smith. 1994. Peptide transport by micro-organisms. Adv. Microb. Physiol. 36:1-80. [DOI] [PubMed] [Google Scholar]
- 29.Punt, S. J., P. O. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1987. Transformation of Aspergillus based on hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 30.Ramesh, M. V., T. D. Sirakova, and P. E. Kolattukudy. 1995. Cloning and characterization of the cDNAs and genes (mep20) encoding homologous metalloproteinases from Aspergillus flavus and A. fumigatus. Gene 165:121-125. [DOI] [PubMed] [Google Scholar]
- 31.Rawlings, N. D., and A. J. Barrett. 1999. Tripeptidyl-peptidase I is apparently the CLN2 protein absent in classical late-infantile neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta 1429:496-500. [DOI] [PubMed] [Google Scholar]
- 31a.Rawlings, N. D., F. R. Morton, and A. J. Barrett. 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34:D270-D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reichard, U., S. Büttner, H. Eiffert, F. Staib, and R. Rüchel. 1990. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. Med. Microbiol. 33:243-251. [DOI] [PubMed] [Google Scholar]
- 33.Reichard, U., G. T. Cole, R. Rüchel, and M. Monod. 2000. Molecular cloning and targeted deletion of PEP2 which encodes a novel aspartic proteinase from Aspergillus fumigatus. Int. J. Med. Microbiol. 290:85-96. [DOI] [PubMed] [Google Scholar]
- 34.Reichard, U., H. Eiffert, and R. Rüchel. 1994. Purification and characterization of an extracellular aspartic proteinase from Aspergillus fumigatus. J. Med. Vet. Mycol. 32:427-436. [DOI] [PubMed] [Google Scholar]
- 35.Reichard, U., M. Monod, and R. Rüchel. 1995. Molecular cloning and sequencing of the gene encoding an extracellular aspartic proteinase from Aspergillus fumigatus. FEMS Microbiol. Lett. 130:69-74. [DOI] [PubMed] [Google Scholar]
- 36.Rubio-Aliaga, I., and H. Daniel. 2002. Mammalian peptide transporters as targets for drug delivery. Trends Pharmacol. Sci. 23:434-440. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shipkova, M., H. Beck, A. Voland, V. W. Armstrong, H. J. Grone, M. Oellerich, and E. Wieland. 2004. Identification of protein targets for mycophenolic acid acyl glucuronide in rat liver and colon tissue. Proteomics 4:2728-2738. [DOI] [PubMed] [Google Scholar]
- 39.Sirakova, T. D., A. Markaryan, and P. E. Kolattukudy. 1994. Molecular cloning and sequencing of the cDNA and gene for a novel elastinolytic metalloproteinase from Aspergillus fumigatus and its expression in Escherichia coli. Infect. Immun. 62:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohar, I., D. E. Sleat, M. Jadot, and P. Lobel. 1999. Biochemical characterization of a lysosomal protease deficient in classical late infantile neuronal ceroid lipofuscinosis (LINCL) and development of an enzyme-based assay for diagnosis and exclusion of LINCL in human specimens and animal models. J. Neurochem. 73:700-711. [DOI] [PubMed] [Google Scholar]
- 41.Stacey, G., S. Koh, C. Granger, and J. M. Becker. 2002. Peptide transport in plants. Trends Plant Sci. 7:257-263. [DOI] [PubMed] [Google Scholar]
- 42.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, Mass.
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilburn, J., C. Scazzocchio, G. G. Taylor, J. H. Zabicky-Zissman, R. A. Lockington, and R. W. Davies. 1983. Transformation by integration in Aspergillus nidulans. Gene 26:205-221. [DOI] [PubMed] [Google Scholar]
- 45.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wlodawer, A., M. Li, A. Gustchina, H. Oyama, B. M. Dunn, and K. Oda. 2003. Structural and enzymatic properties of the sedolisin family of serine-carboxyl peptidases. Acta Biochim. Pol. 50:81-102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.