Abstract
Oxalic acid is found in dietary sources (such as coffee, tea, and chocolate) or is produced by the intestinal microflora from metabolic precursors, like ascorbic acid. In the human intestine, oxalate may combine with calcium, sodium, magnesium, or potassium to form less soluble salts, which can cause pathological disorders such as hyperoxaluria, urolithiasis, and renal failure in humans. In this study, an operon containing genes homologous to a formyl coenzyme A transferase gene (frc) and an oxalyl coenzyme A decarboxylase gene (oxc) was identified in the genome of the probiotic bacterium Lactobacillus acidophilus. Physiological analysis of a mutant harboring a deleted version of the frc gene confirmed that frc expression specifically improves survival in the presence of oxalic acid at pH 3.5 compared with the survival of the wild-type strain. Moreover, the frc mutant was unable to degrade oxalate. These genes, which have not previously been described in lactobacilli, appear to be responsible for oxalate degradation in this organism. Transcriptional analysis using cDNA microarrays and reverse transcription-quantitative PCR revealed that mildly acidic conditions were a prerequisite for frc and oxc transcription. As a consequence, oxalate-dependent induction of these genes occurred only in cells first adapted to subinhibitory concentrations of oxalate and then exposed to pH 5.5. Where genome information was available, other lactic acid bacteria were screened for frc and oxc genes. With the exception of Lactobacillus gasseri and Bifidobacterium lactis, none of the other strains harbored genes for oxalate utilization.
Oxalic acid is a strong dicarboxylic acid (pKa1 = 1.23; pKa2 = 3.83) and a toxic compound that irritates tissues. This effect was recognized in the 18th century, when oxalic acid was used for cleaning and bleaching. Oxalate at extremely high concentrations can cause death in humans and pathological disorders, including hyperoxaluria (an oxalate level exceeding the normal range), pyridoxine deficiency, urolithiasis (formation of calculi or uroliths), renal failure, and other disorders (16). The toxicity of oxalate has been related to its ability to generate reactive oxygen species (through the Fenton reaction) as hydroxyl or carbonate radicals during its interaction with hydrogen peroxide (30, 40). Oxalate occurs widely in nature and in many foods, such as boiled carrots (1.88 mg/g), tomatoes (0.04 mg/g), celery (0.17 mg/g), potatoes (0.02 mg/g), and corn (0.03 mg/g), as well as in other dietary sources, such as tea (0.11 mg/ml), coffee (0.05 mg/ml), and chocolate (1.17 mg/g) (15). Oxalic acid can also be produced by nonenzymatic degradation or from metabolic precursors (like ascorbic acid) by the intestinal microflora (28). In the intestine, oxalate may combine with calcium, sodium, magnesium, potassium, or iron to form nonsoluble salts. It has been proposed that bacteria that specifically degrade oxalate regulate the oxalate homeostasis of the host by catabolizing free oxalate, reducing its concentration in plasma and urine, and thereby preventing adsorption. A recent clinical study demonstrated that there was a correlation between low rates of intestine colonization by oxalate-degrading bacteria, specifically Oxalobacter formigenes, and an increased risk of hyperoxaluria (39). This organism is a natural inhabitant of the gastrointestinal tract (GIT) of vertebrates, including humans, and it is the best-characterized microorganism of the intestinal microbiota with an oxalate-degrading mechanism (11), which decarboxylates oxalate, yielding formic acid and CO2. This reaction generates a proton gradient that contributes to the generation of one ATP molecule when it is coupled with oxalate/formate transport. Two enzymes involved in the catabolism of oxalate have been identified in O. formigenes. The first enzyme is a formyl coenzyme A (CoA) transferase (EC 2.8.3.16), encoded by frc, which activates the oxalate molecule by cycling a CoA moiety from formyl-CoA (35). The second enzyme is an oxalyl coenzyme A decarboxylase (EC 4.1.1.8), encoded by oxc, which decarboxylates the activated oxalate molecule (27).
Lactobacillus acidophilus is a member of the lactic acid bacteria (LAB) that are used in the manufacture of fermented milk products. LAB, especially bifidobacteria and lactobacilli, constitute an important part of the human intestinal microbiota. The potential probiotic roles of these organisms have been reviewed extensively (13, 29), and their beneficial effects include reinforcement of natural defense mechanisms and protection against gastrointestinal disorders. Probiotics have been successfully used to manage infant diarrhea, food allergies, and inflammatory bowel disease (7). A recent study showed that feeding a mixture of freeze-dried LAB led to a significant reduction in urinary excretion in patients with idiopathic calcium-oxalate urolithiasis and mild hyperoxaluria (10). The presence of an oxalyl-CoA decarboxylase gene in Bifidobacterium lactis has recently been documented (12).
L. acidophilus NCFM has been widely used as a probiotic organism for over 30 years in fluid milk, yogurt, infant formulas, and dried dietary supplements (34). In the present study, genes potentially encoding a formyl-CoA transferase and an oxalyl-CoA decarboxylase were identified in the L. acidophilus NCFM genome (2). Predicted frc and oxc genes were transcriptionally and functionally analyzed to reveal a pathway for oxalate catabolism in L. acidophilus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was propagated at 37°C in Luria-Bertani (Difco Laboratories Inc., Detroit, MI) broth with shaking. When appropriate, E. coli cultures were plated onto brain heart infusion agar (Difco) supplemented with 150 μg/ml erythromycin. Lactobacilli were propagated statically at 37°C in MRS broth (Difco) or on MRS broth supplemented with 1.5% agar. Erythromycin (5 μg/ml) and/or chloramphenicol (5 μg/ml) was added to MRS broth or agar when it was appropriate. The semidefined medium (BM) contained 0.5% tryptone, 0.5% yeast extract, 0.5% meat extract, 0.25% sodium chloride, 0.1% Tween 80, 0.02% MgSO4, 0.005% MnSO4, 0.004% FeSO4, 0.2% ammonium citrate, 0.001% thiamine, 0.2% K2PO4, 0.01% CaCO3, ammonium oxalate, and 0.1% glucose. For determination of the maximum specific growth rates of L. acidophilus strains, standardized inocula were added to obtain an initial absorbance at 600 nm (A600) of approximately 0.1 (total volume, 200 μl BM per well). Plates were incubated at 37°C, and growth was automatically monitored by determining the changes in A600 as a function of time using a FLUOStar OPTIMA microtiter plate reader (BMG Labtech GmbH, Offenburg, Germany). The maximum specific growth rate was calculated from the slope of a linear regression line during exponential growth with a correlation coefficient (r2) of 0.99. Each point represented the mean of three independent cultures.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Characteristics | Reference |
|---|---|---|
| Strains | ||
| Escherichia coli EC1000 | RepA+ MC1000, Kmr; host for pORI28-based plasmids | 26 |
| Lactobacillus acidophilus NCFM | Human intestinal isolate | 6 |
| Lactobacillus acidophilus NCK1392 | NCFM containing pTRK669 | 32 |
| Lactobacillus acidophilus NCK1728 | NCFM containing deleted version of ORF LBA0395 (frc) | This study |
| Plasmids | ||
| pFrc | 1.42 kb containing ORF_LBA395 cloned into the BglII/XbaI sites of pORI28 | This study |
| pTRK837 | pFrc containing a 72-bp deletion introduced by inverted PCR and self-ligation | This study |
| Primers for gene replacementa | ||
| LFoX | GATCTCTAGA-ATGCGTTATAATTGG | This study |
| RFoB | GATCAGATCT-ATCGGCAACTTAAA | This study |
| LFoE | GATCGAATTC-TTGCCGTATTAAGT | This study |
| RFoE | GATCGAATTC-CACGTTGCATTAAA | This study |
| Primers for RT-QPCR | ||
| RTLact16SF | GTAGGGAATCTTCCACAATG | 8 |
| RTLact16R | TAGTTAGCCGTGACTTTCTG | 8 |
| Q-394-F | ATGACGGTTGCCGATACGAT | This study |
| Q-394-R | GCGCAATTATGACCGCCTTA | This study |
| Q-395-F | AAGGTCTGGAGCACGCTTAT | This study |
| Q-395-R | CCAGTTGGTCCTGTCCTTGA | This study |
| Q-396-F | TGATGCAGCAGATGCTGGAG | This study |
| Q-396-R | CAACCGTGCCGTGGAATTAT | This study |
| Q-397-F | TTGAGCCGCTCTTAGGCTGCAA | This study |
| Q-397-R | TAGCGAACGTCCTTCAGGGAAA | This study |
| Q-BSH-F | GTGAAGAGAGGAGGCTTGCATTT | This study |
| Q-BSH-R | AGGTATGGCCGGACTCAACTATC | This study |
| Q-BSH2-F | ACGCCGACGTTACTCCACATA | This study |
| Q-BSH2-R | GTGGAGTGTGTGCCAAGACAA | This study |
Dashes indicate introduction of restriction enzyme sites.
Standard DNA techniques.
E. coli plasmid preparation was done by using a QIAprep Spin Plasmid Minipreps kit (QIAGEN Inc., Valencia, CA). Chromosomal DNA from L. acidophilus was extracted by the method of Walker and Klaenhammer (41). Restriction enzymes and T4 DNA ligase were obtained from Roche Molecular Biochemicals (Indianapolis, IN) and New England Biolabs (Beverly, MA), respectively, and were used according to the suppliers' recommendations. Standard protocols were used for ligation, restriction endonuclease digestion, DNA modification, and transformation as described by Sambrook et al. (33). Electrotransformation of L. acidophilus was carried out as described previously (42). PCR was performed by using standard protocols.
Phylogenetic analysis and conserved domains.
Protein sequences obtained from the Entrez Protein Database at NCBI (http://www.ncbi.nlm.nih.gov/) were aligned and utilized to generate an unrooted phylogram tree using the neighbor-joining method (ClustalX software) (38).
Conserved domains in potential proteins encoded by the open reading frames (ORFs) of interest were inferred from the amino acid sequences by using the Protein Families Database of Alignments and HMMs (http://www.sanger.ac.uk/Software/Pfam/) as well as Clusters of Orthologous Groups of Proteins (http://www.ncbi.nlm.nih.gov/COG/).
RNA isolation, cDNA probe preparation, and microarray hybridization.
RNA isolation was carried out as described previously (5). Briefly, 10-ml aliquots of L. acidophilus cultures were centrifuged at 3,148 × g, and the cell pellets were immediately frozen in a dry ice-ethanol bath. Cell pellets were thawed and homogenized in 1 ml Trizol (Technologies, Rockville, MD) with a Mini-Beadbeater-8 cell disruptor (Biospec Products, Bartlesville, OK). The phases were separated by centrifugation (14,000 rpm, 15 min, 4°C). The aqueous phase was removed and placed in a fresh tube, and 0.4 ml of Trizol and 0.2 ml of chloroform were added. The mixture was vortexed for 15 s and centrifuged to separate the phases. RNA was precipitated by adding 1 volume of isopropanol. Identical amounts (25 μg) of total RNA were aminoallyl labeled by reverse transcription with random hexamers in the presence of aminoallyl dUTP (Sigma Chemical Co.), using Superscript II reverse transcriptase (Life Technologies) at 42°C overnight, followed by fluorescence labeling of aminoallylated cDNA with N-hydroxysuccinimide-activated Cy3 or Cy5 esters (Amersham Pharmacia Biotech). Labeled cDNA probes were purified using a PCR purification kit (QIAGEN). Coupling of the Cy3 and Cy5 dyes to the aminoallyl dUTP-labeled cDNA and hybridization of samples to microarrays were performed as described previously (5).
Data normalization and gene expression analysis.
Fluorescence intensities were acquired using a General Scanning ScanArray 4000 microarray scanner (Packard Biochip BioScience, Biochip Technologies LLC, Massachusetts) and were processed as TIFF images. Signal intensities were quantified using the QuantArray 3.0 software package (Packard BioScience). Two independent arrays (biological replicates) on slides containing each gene spotted in triplicate (technical replicates) were hybridized reciprocally to Cy3- and Cy5-labeled probes in each experiment (dye swap) as described previously (5). Spots were analyzed by adaptive quantitation. The local background was subsequently subtracted from the recorded spot intensities. Data were median normalized. The median of the six ratios for each gene was recorded. The ratio of the average absolute pixel value for the replicated spots of each gene with treatment to the average absolute pixel value for the replicated spots of each gene without treatment represented the fold change in gene expression. Confidence intervals and P values for the fold changes were also calculated by using a two-sample t test as described by Knudsen (25). P values of 0.05 or less were considered significant. The microarray platform and data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GPL1401 (platform), GSE2782 (series), and GSM60519 and GSM60522 (samples).
Construction of L. acidophilus frc mutant.
A 1.42-kb fragment containing frc was amplified using L. acidophilus NCFM chromosomal DNA as the template and primers LFoX and RFoB (Table 1). The fragment was cloned in the integrative vector pORI28 (26), generating pFrc. Subsequently, a 72-bp fragment of the cloned gene was removed by inverse PCR amplification of pFrc (using primers LfoE and RFoE) and posterior self-ligation of the created EcoRI site. The resulting 3-kb plasmid, pTRK837, was then introduced by electroporation into L. acidophilus NCFM harboring pTRK669 (32). Subsequent steps to facilitate the integration event and gene replacement were carried out by using the protocols described previously (9, 32). The suspected integrants were confirmed by PCR and Southern hybridization analysis, using standard procedures.
Survival of logarithmic-phase cells after acid challenge.
To determine the acid sensitivity of log-phase cells, cultures were grown to an A600 of 0.25 to 0.3 (pH >5.8) from a 2% inoculum (initial A600, ∼0.05) in MRS broth. The cultures were centrifuged at room temperature for 10 min at 3,148 × g, and the cells were resuspended in the same volume of MRS broth adjusted to pH 3.0, 3.5, or 4.0 with HCl, lactic acid, or 5% oxalic acid. After incubation for 2 h at 37°C, the number of CFU was determined by serial dilution in 10% MRS broth and enumeration on MRS agar using a Whitley automatic spiral plater (Don Whitley Scientific Limited, West Yorkshire, England).
RT-QPCR.
L. acidophilus was transferred three times in MRS broth or MRS broth containing 0.05% ammonium oxalate (pH 6.7) and then transferred to fresh media having the same composition. Cells were then grown to an A600 of 0.3 and transferred to (i) fresh MRS broth, (ii) MRS broth containing 0.5% oxgall (pH 6.5), (iii) MRS broth containing 0.5% ammonium oxalate (pH 6.8), or (iv) MRS broth (pH 5.5; acidified with lactic acid) containing 0.5% ammonium oxalate. Following incubation at 37°C, samples were taken at zero time and 1, 2, 4, and 6 h, and RNA was isolated, treated with DNase, quantified, and diluted to a concentration of 50 ng/μl. Primers meeting the standard criteria for reverse transcription-quantitative PCR (RT-QPCR) for the following genes were designed using CloneManager 7, version 7.10 and Primer Designer 5, version 5.10 (Scientific & Educational Software, Cary, NC): LBA0394, LBA0395 (frc), LBA0396 (oxc), LBA0397, LBA0892 (bsh1), and LBA1078 (bsh2) (Table 1).
RT and PCR were carried out with an iCycler iQ (Bio-Rad Laboratories Ltd.). The reaction mixtures (final volume, 20 μl) contained 2× QuantiTect SYBR Green (10 μl), each primer at a final concentration of 0.1 μM, a Quanti Tect RT mixture (0.2 μl), RNase-free H2O (1.8 μl), and 4 μl of template. The conditions for the RT and amplification reactions were one cycle at 50°C for 30 min and one cycle at 95°C for 15 min, followed by 40 cycles of 15 s at 94°C, 30 s at 49°C, and 30 s at 72°C for data acquisition. A melting curve analysis was conducted at 65°C, with increments set at 1°C for 10 s (31 cycles). Serial dilutions (from 102 to 1010 molecules) of a known PCR product (using the 16S primers [Table 1]) were included in each run to establish a standard curve. Each sample was included in triplicate in each run. Data were analyzed using the iCycler iQ software (version 3.0; Bio-Rad Laboratories Ltd.). The user-defined “PCR base line subtracted” and “threshold cycle calculation” options were used to obtain the number of threshold cycles per well. The linear equation for the standard curve (i.e., for preparations containing known quantities of DNA) was then used to interpolate the numbers of copies present in the unknown samples. The correlation coefficients for the standards were 0.99.
A reliable quantitative RT-PCR method requires correction for experimental variations in individual reverse transcription and PCR steps, since differences in the efficiency of each can result in a concentration of cDNA that does not correspond to the starting amount of RNA (14). For this study, the 16S rRNA gene was used for normalization.
Oxalate degradation activity.
Lactobacillus strains were transferred three times in BM broth without citrate (BMcit) containing 1% glucose plus 3.5 mM ammonium oxalate. After this, 100 μl of cells was inoculated into the same medium, grown to an A600 of 0.6, centrifuged, and resuspended in BMcit containing 0.1% glucose plus 35 mM ammonium oxalate (32 mM oxalate). The initial pH of BMcit was 6.5, and the pH was allowed to naturally fall during the 90-h incubation. Samples were taken over time, centrifuged, neutralized to obtain pH values between 5 and 7 (according to the manufacturer's instructions) with 1 N sodium hydroxide, and stored at −20°C. The oxalate concentrations in the supernatants were measured in triplicate using a diagnostic oxalate kit (Trinity Biotech, County Wicklow, Ireland). In this assay, oxalate is oxidized to carbon dioxide and hydrogen peroxide by oxalate oxidase. Hydrogen peroxide, 3-methyl-2-benzothiozolinone hydrazone, and 3-(dimethylamino)benzoic acid, in the presence of peroxidase, yield an indamine dye which has a maximum absorbance at 590 nm.
RESULTS
Analysis of the chromosomal region containing frc and oxc.
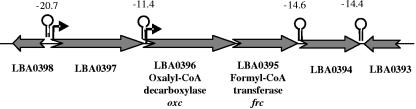
The genome sequence of L. acidophilus NCFM (2) revealed the presence of an operon putatively involved in oxalate catabolism (Fig. 1). The predicted operon consisted of two genes: the formyl-CoA transferase gene (LBA0395, frc) and the oxalyl-CoA decarboxylase gene (LBA0396, oxc). High-energy rho-independent terminators were predicted to be downstream of LBA0397 (ΔG, −11.4 kcal/mol) and frc (ΔG, −14.6 kcal/mol). Additionally, a typical ribosome binding sequence (AGAAGG; 7 nucleotides from the start codon) and a putative promoter were located upstream of oxc (data not shown).
FIG. 1.
Formyl-CoA transferase and oxalyl-CoA decarboxylase genes in L. acidophilus NCFM. Putative rho-independent terminators (lollipop symbols) and their corresponding free energies (in kcal/mol) are indicated. Potential promoter regions for ORFs LBA0396 and LBA0397 are indicated by bent arrows.
frc encoded a 445-amino-acid (aa) protein that was very similar to a predicted acyl-CoA transferase/carnitin dehydratase from Lactobacillus gasseri NCK334 (accession number ZP_00046082) and a putative formyl-CoA transferase from E. coli K-12 (accession number NP_416872). A conserved domain (pfam02515) belonging to a new family of CoA transferases is present in this protein. Most CoA transferases belong to two well-known enzyme families, but recently a third family of CoA transferases was described (17). The members of this enzyme family include oxalyl-CoA transferase, succinyl-CoA:(R)-benzylsuccinate CoA transferase, (E)-cinnamoyl-CoA:(R)-phenyllactate CoA transferase, and butyrobetainyl-CoA:(R)-carnitine CoA transferase. Additionally, the NCFM frc product exhibited 44% identity (61% similarity) with the protein encoded by O. formigenes frc. The gene encoding formyl-CoA transferase (35) was the first member of family III of CoA transferases to be characterized.
oxc encoded a 569-aa protein similar to the oxalyl-CoA decarboxylases (EC 4.1.1.8) from O. formigenes (53% identity and 71% similarity) (27) and B. lactis (46% identity and 63% similarity) (12). The protein encoded by oxc has a conserved domain that is present in thiamine pyrophosphate (TPP)-requiring enzymes (COG0028). This domain is also present in several other enzymes, including acetolactate synthase, pyruvate dehydrogenase (cytochrome), glyoxylate carboligase, and phosphonopyruvate decarboxylase. In the oxc product, an N-terminal TPP-binding domain (pfam02776) starts at residue 20 and spans 171 aa, and the central TPP domain (pfam00205) starts at residue 210 and spans 154 aa.
The potential product of the gene downstream of frc, LBA0394, was a 395-aa protein which was virtually identical (90% identity; E value, 0.0) to the predicted acyl-CoA transferase from L. gasseri and exhibited 44% identity to the formyl-CoA transferase from E. coli K-12 and 44% identity to putative protein F (accession number BAA16242) encoded by a bile acid-inducible operon in E. coli. Interestingly, the frc product exhibited 30% identity (48% similarity) with the putative product of LBA0394. The latter, however, did not exhibit significant similarity to the formyl-CoA transferase from O. formigenes, indicating that although LBA0394 might encode a CoA transferase, the enzyme is not necessarily a formyl-CoA transferase.
LBA0397, upstream of oxc, encodes a 639-aa protein having the conserved COG0488 domain Uup, which corresponds to ATPase components of ABC transporters with duplicated ATPase domains (21). High levels of identity (more than 75%) were observed with nearly equivalent proteins in L. gasseri and Lactobacillus johnsonii.
Other members of the lactic acid bacteria were screened in silico for frc- and oxc-related genes. L. gasseri NCK334 (accession number ZP_00046991) and B. lactis DSM 10140 (formerly Bifidobacterium animalis) (12) harbored genes for oxalate utilization, whereas Lactobacillus plantarum WCFS1 (24) and L. johnsonii NCC553 (31) did not. Figure 2 shows the phylogenetic relationships of several putative oxalyl-CoA decarboxylases from organisms whose protein sequences were available. As expected, the decarboxylases from L. gasseri and L. acidophilus clustered together and, interestingly, clustered closer to the enzyme from B. lactis.
FIG. 2.

Unrooted phylogram tree of oxalyl-CoA decarboxylase sequences from diverse organisms. Proteins were aligned by CLUSTALX. Alignments were used for tree reconstruction. The organisms used were L. acidophilus NCFM, L. gasseri ATCC 3323 (GenBank accession number ZP_00046991), B. lactis (BAD11779), Bradyrhizobium japonicum USDA110 (BAC48422.1), E. coli CFT073 (NP_754791.1), Mycobacterium tuberculosis CDC1551 (NP_334536.1), O. formigenes (P40149), Mycobacterium bovis (NP_853789), Oryza sativa (BAB33274.1), Schizosaccharomyces pombe (CAA22176), Mycobacterium leprae (CAA15478), Saccharomyces cerevisiae (AAB64497), and Arabidopsis thaliana (CAC19854).
Transcriptional analysis of the oxc operon using microarrays.
Antiport of oxalate/formate in O. formigenes is coupled to oxalate decarboxylation and generates a proton motive gradient (1). We were not able to identify a putative oxalate permease/antiporter by in silico analysis of the L. acidophilus genome. Therefore, microarray experiments were conducted in an attempt to identify a candidate that might be responsible for the specific transport of oxalate into the cell.
During growth of L. acidophilus in MRS medium, the pH of a culture starting at pH 6.5 typically decreases to less than 4.0 due to fermentation and lactic acid production. In a previous study (Gene Expression Omnibus accession numbers GPL1401 [platform] and GSE1976 [series]) (5), a whole-genome array containing 97.4% of the NCFM annotated genes was used to identify genes that were differentially expressed when log-phase cells were exposed to MRS medium at pH 5.5 and pH 4.5 acidified with lactic acid. After exposure to pH 5.5 (adjusted with lactic acid) for 30 min, we observed induction of frc (3.2-fold) and oxc (4.5-fold) encoding the putative formyl-CoA transferase and oxalyl-CoA decarboxylase, respectively. No statistically significant differences in the levels of expression of frc or oxc were observed between the control (pH 6.8) and the samples exposed to pH 4.5.
In the present study, we studied gene expression at pH 6.8 in an attempt to separate the specific effect of the oxalate salt from the effect of the pH. The L. acidophilus whole-genome array was used to analyze the global gene expression after cells were exposed to 1% (70 mM) ammonium oxalate for 30 min at pH 6.8. A summary of the results of our previous study (in which log-phase cultures were exposed to pH 5.5 and pH 4.5 with no oxalate) and the present study is shown in Fig. 3. In the presence of 1% oxalate at pH 6.8, 16 genes were significantly upregulated (P ≤ 0.05 and a ratio of >2.0) (Table 2), and 315 genes were downregulated (P ≤ 0.05 and a ratio of <0.5). Both the frc and oxc genes were downregulated under these conditions. The most upregulated genes were a cadmium/manganese transport ATPase gene (LBA1234; upregulated 9.6-fold) and the genes encoding two uncharacterized membrane proteins (LBA1119 and LBA1690; upregulated 5.9- and 4.8-fold, respectively). Interestingly, ORFs LBA0038, LBA0039, LBA0040, and LBA0041 were upregulated between 1.4- and 2.4-fold. These four genes appear to form an operon. LBA0041 is predicted to encode a putative adenosylcobalamin-dependent ribonucleoside triphosphate reductase. ORFs LBA0038, LBA0039, and LBA0040 are poorly characterized; however, the LBA0040 product is similar to a putative ATP:cob(I)alamin adenosyltransferase (23), the enzyme responsible for the last step in the activation of vitamin B12 (cyanocobalamin) to coenzyme B12 (adenosylcobalamin). A relationship between altered oxalate metabolism and B vitamin deficiency has been documented (3, 18), which resulted in some interest in why these genes are upregulated in the presence of ammonium oxalate.
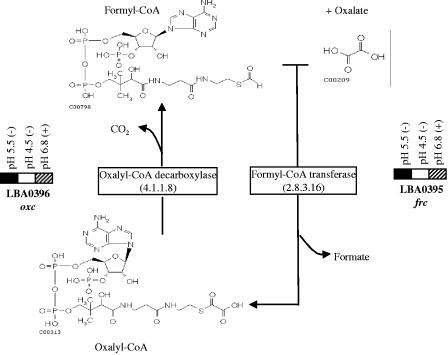
FIG. 3.
Transcriptional response of frc and oxc to pH 5.5, pH 4.5, and 1% (70 mM) ammonium oxalate (pH 6.8) in MRS broth after 30 min. The solid rectangles indicate ≥twofold-higher expression, the cross-hatched rectangles indicate a ≥twofold reduction in expression (P < 0.05), and the open rectangles indicate values of gene expression that are not statistically different from values obtained under the control conditions (L. acidophilus incubated in fresh MRS broth for 30 min). Plus and minus signs indicate that the experiment was carried out in the presence and in the absence of oxalate, respectively. The proposed metabolic pathway of oxalate decarboxylation by L. acidophilus is also shown. The structures of the compounds were obtained from the website http://www.genome.jp/kegg/kegg2.html.
TABLE 2.
Genes upregulated in response to 1% (70 mM) ammonium oxalate at pH 6.8 in L. acidophilus NCFM
| Gene | Description | Expression ratioa | P value |
|---|---|---|---|
| LBA0038 | Hypothetical protein | 2.4 | 3.0E-05 |
| LBA0039 | Hypothetical protein | 2.4 | 7.1E-03 |
| LBA0040 | Hypothetical protein | 2.0 | 1.7E-02 |
| LBA0144 | N-Acetylglucosamine-6-phosphate deacetylase | 3.2 | 5.8E-04 |
| LBA0149 | Hypothetical protein | 2.3 | 1.5E-03 |
| LBA0600 | Xylulose-5-phosphate/fructose phosphoketolase | 3.1 | 4.6E-03 |
| LBA0877 | PTS system IIab | 2.1 | 1.2E-02 |
| LBA1119 | Putative inner membrane protein | 5.9 | 1.5E-05 |
| LBA1234 | Cadmium/manganese transport ATPase | 9.6 | 4.8E-04 |
| LBA1339 | Hypothetical protein | 2.3 | 1.9E-02 |
| LBA1462 | Beta-galactosidase | 2.0 | 3.7E-02 |
| LBA1690 | Putative membrane protein | 4.8 | 3.1E-03 |
| LBA1869 | Beta-phosphoglucomutase | 2.9 | 6.0E-05 |
| LBA1870 | Maltose phosphorylase | 3.1 | 3.4E-03 |
| LBA1877 | Hypothetical protein | 2.6 | 5.4E-03 |
| LBA1948 | Glucosamine-6-phosphate isomerase | 2.3 | 2.0E-02 |
Array ratios obtained from two biological replicates and two technical replicates for each condition were averaged.
PTS, phosphotransferase system.
Transcriptional analysis of the oxc operon by RT-QPCR.
Acid induction of frc and oxc was evaluated in the presence and absence of ammonium oxalate as an inducer of expression of the operon. Primers meeting RT-QPCR criteria were designed for LBA0394, frc, oxc, and LBA0397. L. acidophilus was adapted to oxalate by three consecutive transfers in MRS broth containing 0.05% ammonium oxalate, a noninhibiting concentration. Cells preexposed or not exposed to this compound were then transferred to MRS broth at pH 5.5 (adjusted with lactic acid) containing ammonium oxalate, and samples were taken over time. When L. acidophilus cells were first propagated in the presence of ammonium oxalate and then exposed to pH 5.5 plus 0.5% ammonium oxalate, both frc expression and oxc expression increased dramatically to levels that approached fourfold induction (Fig. 4A). In the absence of oxalate adaptation, exposure to pH 5.5 plus oxalate (Fig. 4B) again resulted in induction of both genes, but oxc was expressed significantly more highly (two- to fourfold) than frc (one- to twofold). It is not clear why the levels of expression of frc and oxc differed under these conditions, particularly since both genes are predicted to be in the same operon and exhibited similar expression levels when they were highly induced (Fig. 4A). Expression of ORFs LBA0394 and LBA0397 remained constant under these conditions.
FIG. 4.
Transcriptional analysis of the oxc operon in L. acidophilus cells at pH 5.5. (A) Cells were first transferred in MRS broth (pH 6.8) containing noninhibitory concentrations of ammonium oxalate (preadapted). Solid bars, frc; cross-hatched bars, oxc. Gene induction was monitored over time after cells were placed in MRS broth containing 0.5% ammonium oxalate at pH 5.5. (B) Gene induction for cells in MRS broth at pH 6.8 (nonadapted). Experiments were carried out in triplicate. The error bars indicate standard deviations.
L. acidophilus cells preexposed or not exposed to 0.05% oxalate were also resuspended in MRS medium containing 0.5% ammonium oxalate at pH 6.8. Under these conditions, none of the genes examined in this study (frc, oxc, bsh1, bsh2, LBA0394, and LBA0397) were induced (data not shown).
LBA0394, the ORF immediately downstream of frc, showed some homology to a bile-inducible protein. The NCFM genome contains genes encoding two bile salt hydrolases, LBA0872 (bsh1) and LBA1078 (bsh2). Therefore, we designed RT-QPCR primers for bsh1 and bsh2 and examined the expression of these genes after exposure of the cells to 0.5% oxalate at pH 5.5 or exposure to oxgall (0.5%). Neither bsh1 nor bsh2 was induced under these conditions, and expression of LBA0394 remained basal and constant.
Inactivation of frc and mutant analysis.
Integrative plasmid pORI28, a pWV01-derived vector (26), was used to replace frc with the deleted version of the same gene by using the protocols described previously (9, 32). PCR and Southern hybridization experiments using an internal fragment of frc as the probe confirmed that the frc gene was replaced with the deleted version in NCK1728 (data not shown).
The survival of log-phase cells (A600, 0.3) of wild-type L. acidophilus NCFM and the survival of the frc mutant were compared at pH 3.0, 3.5, and 4.0 by using hydrochloric acid (HCl), lactic acid, or oxalic acid to acidify MRS broth (Fig. 5). No differences between the parent and the frc mutant were observed when HCl or lactic acid was used to acidify the culture medium. Additionally, no differences in survival were observed in the presence of 5% oxalic acid, at pH 4.0 (>50% survival) or pH 3.0 (<0.01% survival). However, the frc mutant was significantly more sensitive to 5% (wt/vol) oxalic acid after 2 h of exposure at pH 3.5. The Henderson-Hasselbalch equation for oxalic acid predicts that at pH 4.0 most of the oxalate is dissociated (pKa2 = 3.83) and hence unable to enter the cell. At pH 3.5 a larger amount would be undissociated. When combined with a higher concentration of the acid (5%), this would increase the amount of protonated acid available to diffuse into and acidify the cell. At pH 3.0, the combination of a low pH (closer to the pKa1 of oxalate [pKa1 = 1.23]) and the high concentration of acid was lethal for both the wild type and the frc mutant.
FIG. 5.
Survival of log-phase cells of L. acidophilus NCFM and the frc mutant after challenge with MRS broth adjusted to pH 4.0, 3.5, and 3.0 with HCl, lactic acid, or oxalic acid for 2 h. The values are the averages for six separate incubations. The error bars indicate standard deviations.
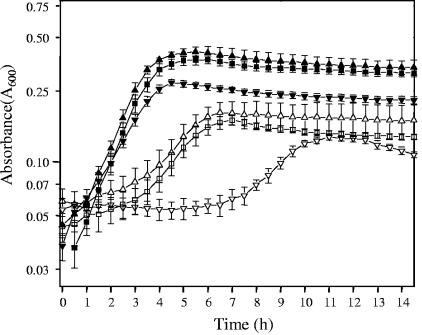
The ability of NCFM to grow in the presence of 0.1% and 0.5% oxalate was also examined (Fig. 6). A semidefined medium (BM with an initial pH of 6.5) was used since addition of the oxalate salt caused formation of a precipitate in MRS broth. The maximum specific growth rate was 0.70 h−1 in BM containing 0.1% glucose, with or without 0.1% oxalate. The maximum specific growth rate decreased somewhat in the presence of 0.5% ammonium oxalate (0.48 h−1), and the maximum cell density was noticeably lower. In the absence of glucose, growth occurred, but the growth rate and maximum cell density were substantially lower. BM is a semidefined medium containing complex sources of nutrients, such as tryptone, yeast extract, and meat extract, which can support limited growth until any residual carbohydrate is exhausted. Interestingly, when 0.5% ammonium oxalate was added to the media without added glucose, a lag phase of 7 h was observed before the cell density of the culture increased to a value similar to that observed in the presence of 0.1% oxalate. The reasons for this delay in growth are unknown. The frc mutant showed similar patterns of growth under these conditions, indicating that initiation of growth at the oxalate concentrations used is not a function of oxalate degradation (data not shown).
FIG. 6.
Growth curves for L. acidophilus NCFM in semidefined BM containing different concentrations of ammonium oxalate. Cell growth was evaluated in BM in the presence of 0.1% glucose (▪), in the presence of glucose plus 0.1% ammonium oxalate (▴) or 0.5% ammonium oxalate (▾), in the absence of glucose (□), or in the absence of glucose plus 0.1% ammonium oxalate (▵) or 0.5% ammonium oxalate (▿). Each point represents the mean of three independent experiments. The error bars indicate standard deviations.
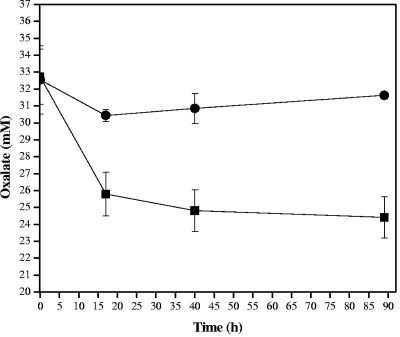
Finally, oxalate utilization was measured for both NCFM and the frc mutant (Fig. 7). The Lactobacillus strains were transferred three times in broth containing a noninhibitory concentration of ammonium oxalate (0.05%) to ensure high levels of expression of the oxalate genes, oxc and frc. For the assay, cells were inoculated into the same medium, propagated to the mid-log phase (A600, ∼0.6), and then centrifuged and resuspended in broth containing 0.1% glucose and 0.5% (35 mM) ammonium oxalate. The concentration of oxalate in the culture supernatant decreased significantly for the control (up to 24%) but not for the frc mutant, for which the oxalate concentration decreased only 6%. Most of the oxalate degradation occurred during the first 16 h. The results indicated that L. acidophilus NCFM was able to degrade oxalate, and Frc participated in this process.
FIG. 7.
Oxalate-degrading activity of L. acidophilus. Strain NCFM (▪) and the frc mutant (•) were consecutively transferred in BMcit containing a noninhibitory concentration of oxalate (0.05%; 3.5 mM) and then exposed to 0.5% (32 mM) oxalate in broth. Samples were taken over time, and the oxalate concentration in the supernatants was measured. Each point represents the mean of three independent experiments. The error bars indicate standard deviations.
DISCUSSION
Studies using a whole-genome microarray of L. acidophilus NCFM (5) showed that there was induction of two ORFs, LBA0395 and LBA0396, at a mildly acidic pH, pH 5.5. Comparative analysis of these genes and the adjacent genes with the available sequences in the GenBank database resulted in identification of genes encoding a formyl-CoA transferase (frc) and an oxalyl-CoA decarboxylase (oxc), which were highly similar to frc and oxc genes which direct oxalate degradation by O. formigenes. In L. acidophilus, frc and oxc appear to form an operon as the two genes are flanked by two predicted terminators. RT-QPCR and microarray experiments showed that oxalate (at a pH above 5.8) did not directly induce the expression of frc and oxc. However, when L. acidophilus was repeatedly transferred in broth containing noninhibitory concentrations of ammonium oxalate and subsequently exposed to pH 5.5 plus oxalate, the expression of oxc and frc was dramatically increased. Moreover, when frc was inactivated and the mutant was exposed to an acidic pH, the strain became more susceptible to oxalic acid specifically at pH 3.5, indicating that frc is involved in the degradation of oxalate by L. acidophilus. In this regard, unlike the wild-type strain, the frc mutant was unable to degrade oxalate.
The concept of autochthonous microorganisms in the GIT has been discussed by several authors (for a review see reference 36). In fact, Tannock proposed a concise definition based on three important characteristics: a long-term association with the host, a stable population in a particular region of the gut, and a demonstrated ecological function. Oxalate occurs widely in nature, and oxalate-rich foods are important sources of oxalate in the diet. Bacteria that specifically degrade oxalate in the GIT can regulate oxalate homeostasis by both preventing absorption and catabolizing free oxalate. Consequently, the ability to detoxify this compound potentially suggests a new ecological function for L. acidophilus.
Other oxalate-degrading bacteria isolated from the human GIT include Eubacterium lentum (22) and Enterococcus faecalis (20). Hokama et al. isolated an oxalate-degrading E. faecalis strain from human stools and identified the formyl-CoA transferase and oxalyl-CoA decarboxylase by Western blotting using antibodies against Frc and Oxc from O. formigenes. Campieri et al. (10) measured oxalate degradation in patients with idiopatic calcium-oxalate urolithiasis that was treated with 8 × 1011 LAB (including L. acidophilus, L. plantarum, Lactobacillus brevis, Streptococcus thermophilus, and Bifidobacterium infantis). They observed a reduction in the excreted oxalate in the patients and showed that L. acidophilus and S. thermophilus could reduce oxalate concentrations in vitro, even when their growth was partially inhibited by this compound. However, the genes responsible for oxalate degradation by these microorganisms were not identified. More recently, an oxalyl-CoA decarboxylase gene was identified in B. lactis, and the oxalate-degrading activity of the enzyme was confirmed by a capillary electrophoresis-based method (12). Therefore, oxalate catabolism in the GIT may be an important property of some commensal and probiotic bacteria.
In other oxalate-degrading organisms, such as O. formigenes, the utilization of oxalate is coupled to energy produced by the antiport of oxalate and formate. By in silico analysis, we were not able to identify a putative permease/antiporter that might incorporate dissociated oxalate into the cell. It is commonly known that the nondissociated forms of organic acids, such as oxalic acid, can freely diffuse through the cytoplasmic membrane. This might explain the apparent absence of a specific transporter for oxalic acid in the genome of NCFM. The concentration of nondissociated oxalate (pKa1 = 1.23; pKa2 = 3.83) entering the cell will increase under acidic conditions, such as those encountered in the digestive tract, where the pH values range from 1 to 7. In the stomach, the pH values range from 1 to 3; in the large intestine, the pH values range from 5 to 7; and in the duodenum, the pH values range from 6 to 6.5. As an alternative hypothesis, an oxalate transporter may be involved, as three genes predicted to encode membrane proteins were strongly upregulated in the presence of ammonium oxalate. Gene expression studies in the presence of oxalate at pH 6.8, which separated the specific effect of the oxalate salt from the effect of the low pH, resulted in identification of a cadmium/manganese transport ATPase gene as the most upregulated gene (9.6-fold) under these conditions. The predicted protein encoded by LBA1234 has two conserved domains, pfam00122 (E1-E2 ATPase) and COG0474 (MgtA, cation transport ATPase). E1-E2 ATPases are primary active transporters that form phospho intermediates during the catalytic cycle. They are classified as P1 to P4 based on the primary structure and potential transmembrane segments (4). E1-E2 ATPases transport divalent cations, and oxalate is a divalent cation. Hence, LBA1234 might be the transporter responsible for the translocation of oxalate into the cell. Two other uncharacterized membrane proteins (LBA1119 and LBA1690) were also upregulated, but they did not have any features that could be used for putative identification.
Since oxalate is normally present in the human GIT, the ability to degrade this compound may provide a selective advantage to certain members of the intestinal microbiota. Additionally, since other microorganisms present in the intestine produce the enzymes for oxalate degradation, we speculate that the ability to decarboxylate oxalyl-CoA was acquired by L. acidophilus via horizontal gene transfer. A number of observations support this hypothesis. The gene upstream of LBA0394 is similar to a gene encoding a transcriptional regulator, and the gene downstream of LBA0397 encodes a putative AT-rich DNA binding protein. The region comprising ORFs LBA0394 to LBA0397, including frc and oxc, is on the complementary strand, and the G+C contents of frc (38.4%) and oxc (40.2%) are notably higher than the average G+C content of the NCFM genome (34.71%). Several studies have reported the occurrence of natural transformation events due to additive integration of DNA, based on two flanking regions with high DNA similarity that initiate the recombination process (for a review see reference 37). It is notable that the region containing ORFs LBA0394 to LBA0397 is flanked by DNA regions that are highly similar to the equivalent segment in the L. johnsonii genome (31), even though oxalate genes are not present in this bacterium.
The efficacy of probiotics as a means to prevent and/or treat urogenital infections and recurrent bladder cancer has been scientifically accepted in the past two decades. More recently, encouraging results were obtained in a clinical trial of O. fomigenes with patients suffering from hyperoxaluria type I, an inherited, life-threatening disease characterized by recurrent oxalate stone formation, nephrocalcinosis, and eventual liver and kidney failure (19). Further characterization of oxalate-degrading probiotic bacteria and efforts to promote the expression, activity, and release of the enzymes involved may lead to a complementary method to manage oxalate-related kidney disease via oral microbial supplements. This is a particularly exciting use of probiotic bacteria, because high levels of these organisms can be safely consumed in food (109 CFU/g) or dietary supplements (1010 CFU/g).
Acknowledgments
This work was partially supported by the Southeast Dairy Foods Research Center, Dairy Management, Inc., the North Carolina Dairy Foundation, and Danisco USA, Inc.
We thank Evelyn Durmaz and B. Logan Buck for helpful discussions and comments.
REFERENCES
- 1.Abe, K., Z.-S. Ruan, and P. C. Maloney. 1996. Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigenes. J. Biol. Chem. 271:6789-6793. [DOI] [PubMed] [Google Scholar]
- 2.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andresson, H., S. Filipsson, and L. Hulten. 1978. Urinary oxalate excretion related to ileocolic surgery in patients with Crohn's disease. Scand. J. Gastroenterol. 13:465-469. [DOI] [PubMed] [Google Scholar]
- 4.Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. [DOI] [PubMed] [Google Scholar]
- 5.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, R. Cano, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barefoot, S. F., and T. R. Klaenhammer. 1983. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourlioux, P., B. Koletzko, F. Guarner, and V. Braesco. 2003. The intestine and its microflora are partners for the protection of the host: report on the Danone Symposium “The Intelligent Intestine,” held in Paris, June 14, 2002. Am. J. Clin. Nutr. 78:675-683. [DOI] [PubMed] [Google Scholar]
- 8.Bruno-Barcena, J. M., J. M. Andrus, S. L. Libby, T. R. Klaenhammer, and H. M. Hassan. 2004. Expression of a heterologous manganese superoxide dismutase gene in intestinal lactobacilli provides protection against the toxicity of hydrogen peroxide. Appl. Environ. Microbiol. 70:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruno-Barcena, J. M., M. A. Azcarate-Peril, T. R. Klaenhammer, and H. M. Hassan. 2005. Marker-free chromosomal integration of the manganese superoxide dismutase gene (sodA) from Streptococcus thermophilus into Lactobacillus gasseri. FEMS Microbiol. Lett. 246:91-101. [DOI] [PubMed] [Google Scholar]
- 10.Campieri, C., M. Campieri, V. Bertuzzi, E. Swennen, D. Matteuzi, S. Stefoni, F. Pirovano, C. Centi, S. Ulisse, G. Famularo, and C. De Simone. 2001. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 60:1097-1105. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, S. H., A. J. Richardson, P. Kaul, R. P. Holmes, M. J. Allison, and C. S. Stewart. 2002. Oxalobacter formigenes and its potential role in human health. Appl. Environ. Microbiol. 68:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federici, F., B. Vitali, R. Gotti, M. R. Pasca, S. Gobbi, A. B. Peck, and P. Brigidi. 2004. Characterization and heterologous expression of the oxalyl coenzyme A decarboxylase gene from Bifidobacterium lactis. Appl. Environ. Microbiol. 70:5066-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill, H. S., and F. Guaner. 2004. Probiotics and human health: a clinical perspective. Postgrad. Med. J. 80:516-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giulietti, A., L. Overbergh, D. Valckx, B. Decallonne, R. Bouillon and C. Mathieu. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386-401. [DOI] [PubMed] [Google Scholar]
- 15.Gold, L. S., T. H. Slone, and B. N. Ames. 2001. Natural and synthetic chemicals in the diet: a critical analysis of possible cancer hazards, p. 95-128. In R. E. Hester and R. M. Harrison (ed.), Issues in environmental science and technology. Food safety and food quality. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 16.Hatch, M., and R. W. Freel. 1995. Alterations in intestinal transport of oxalate in disease states. Scanning Microsc. 9:1121-1126. [PubMed] [Google Scholar]
- 17.Heider, J. 2001. A new family of CoA-transferases. FEBS Lett. 509:345-349. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkinson, A. 1977. Vitamin deficiencies, p. 233-235. In A. Hodgkinson (ed.), Oxalic acid in biology and medicine. Academic Press, Inc., London, United Kingdom.
- 19.Hoesl, C. E., and J. E. Altwein. 2005. The probiotic approach: an alternative treatment option in urology. Eur. Urol. 47:288-296. [DOI] [PubMed] [Google Scholar]
- 20.Hokama, S., Y. Honma, C. Toma, and Y. Ogawa. 2000. Oxalate-degrading Enterococcus faecalis. Microbiol. Immunol. 44:235-240. [DOI] [PubMed] [Google Scholar]
- 21.Holland, B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381-399. [DOI] [PubMed] [Google Scholar]
- 22.Ito, H., N. Miura, M. Masai, K. Yamamoto, and T. Hara. 1996. Reduction of oxalate content of foods by the oxalate degrading bacterium, Eubacterium lentum WYH-1. Int. J. Urol. 3:31-34. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, C. L., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knudsen, S. 2002. A biologist's guide to analysis of DNA microarray data. John Wiley & Sons, Inc., New York, N.Y.
- 26.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lung, H. Y., A. L. Baetz, and A. B. Peck. 1994. Molecular cloning, DNA sequence, and gene expression of the oxalyl-coenzyme A decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J. Bacteriol. 176:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa, Y., T. Miyazato, and T. Hatano. 2000. Oxalate and urinary stones. World J. Surg. 24:1154-1159. [DOI] [PubMed] [Google Scholar]
- 29.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 30.Park, J. S. B., P. M. Wood, M. J. Davies, B. C. Gilbert, and A. C. Whitwood. 1997. A kinetic and ESR investigation of iron(II) oxalate oxidation by hydrogen peroxide and dioxygen as a source of hydroxyl radicals. Free Radic. Res. 27:447-458. [DOI] [PubMed] [Google Scholar]
- 31.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sanders, M. E., and T. R. Klaenhammer. 2001. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84:319-331. [DOI] [PubMed] [Google Scholar]
- 35.Sidhu, H., S. D. Ogden, H. Lung, B. G. Luttge, A. L. Baetz, and A. B. Peck. 1997. DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J. Bacteriol. 179:3378-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, C. H., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Microbiol. Rev. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. J. Eanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Troxel, S. A., H. Sidhu, P. Kaul, and R. K. Low. 2003. Intestinal Oxalobacter formigenes colonization in calcium oxalate stone formers and its relation to urinary oxalate. J. Endourol. 17:173-176. [DOI] [PubMed] [Google Scholar]
- 40.Urzua, U., P. J. Kersten, and R. Vicuna. 1998. Manganese peroxidase-dependent oxidation of glyoxylic and oxalic acids synthesized by Ceriporiopsis subvermispora produces extracellular hydrogen peroxide. Appl. Environ. Microbiol. 64:68-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker, D. C., and T. R. Klaenhammer. 1994. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J. Bacteriol. 176:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, D. C., K. Aoyama, and T. R. Klaenhammer. 1996. Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138:233-237. [DOI] [PubMed] [Google Scholar]