Abstract
During the autumn and winter of 2004 and 2005, an extensive outbreak of waterborne giardiasis occurred in Bergen, Norway. Over 1,500 patients were diagnosed with giardiasis. Analysis of water from the implicated source revealed low numbers of Giardia cysts, but the initial contamination event probably occurred up to 10 weeks previously. While sewage leakage from a residential area is now considered to be the probable source of contamination, during the episode waste from one particular septic tank was thought to be a possible source. Genotyping of cysts from the septic tank demonstrated that they were assemblage A cysts, although the sequences were not identical to any previously published sequences. For the β-giardin gene, the closest published subgenotype was subgenotype A3; for the gdh gene, the closest published subgenotype was subgenotype A2. Genotyping of cysts from 21 patient samples revealed that they were assemblage B cysts; thus, the septic tank was unlikely to be the contamination source. Sequencing of the β-giardin and gdh genes from patient samples and a comparison of the sequences gave complex results. For the β-giardin gene, three isolates had sequences identical to subgenotype B3 sequences. However, other isolates had between one and four single-nucleotide polymorphisms (SNPs). For the gdh gene, none of the sequences were identical to the sequence published for subgenotype B3, and the sequences had between one and three SNPs. One isolate, which was identical to subgenotype B3 at the β-giardin gene, was more similar to subgenotype B2 at the gdh gene. Grouping the isolates on the basis of SNPs resulted in different groups for the two genes. The results are discussed in relation to giardiasis in Norway and to other Giardia genotyping studies.
Human parasitic infections, although probably underdiagnosed, are considered to be rare in Norway. Giardiasis is a reportable infection, and the annual recorded occurrence is 300 to 400 cases (around 8 cases per 100,000 population) for the whole country (6).
During the autumn and winter of 2004 and 2005, an extensive outbreak of waterborne giardiasis occurred in Bergen, Norway (Table 1). Over 1,500 patients were laboratory diagnosed with Giardia infection, although considerably more individuals had symptoms, and Giardia infections considered to be outbreak associated continued to be diagnosed until June 2005.
TABLE 1.
Time line of relevant events related to the waterborne outbreak of giardiasis in Bergena
| Date | Event |
|---|---|
| 20-21 August 2004 | 25-30 mm rainfall recorded each day at Florida Measuring Station in Bergen |
| 28 August 2004 | 45 mm rainfall recorded at Florida Measuring Station in Bergen |
| 30 August-5 September 2004 | Significant increase in Escherichia coli numbers in raw water (to around 65 cells/100 ml, compared to around 5 cells/100 ml at the previous measurement on 16-22 August) |
| 11 September-20 September 2004. | One case of giardiasis recorded at HUS, Bergen; case associated with travel abroad |
| 21 September-30 September 2004. | Three cases of giardiasis recorded at HUS, Bergen; two cases not associated with travel abroad |
| 30 September 2004. | Routine analysis of water from implicated water source due to detection of low concentrations of Clostridium perfringens in treated water; Giardia analysis conducted by commercial laboratory in Stavanger, Norway, and one Giardia cyst per 10 liters raw water documented; at this point no giardiasis outbreak had been recognized |
| 1 October-10 October 2004. | Four cases of giardiasis recorded at HUS, Bergen; three cases not associated with travel abroad |
| 11 October-20 October 2004. | 13 cases of giardiasis recorded at HUS, Bergen; 11 cases not associated with travel abroad |
| 21 October-30 October 2004. | 16 cases of giardiasis recorded at HUS, Bergen; 15 cases not associated with travel abroad |
| 29 October 2004 | Infectious diseases office in Bergen advised of the increase in diagnosis of giardiasis |
| 1 November 2004 | Crisis management team established in Bergen |
| 3 November 2004 | Water samples taken and sent to NVH in Oslo |
| 4 November 2004 | Water samples received at NVH in Oslo and prepared for microscopy by U.S. Environmental Protection Agency Method 1623 |
| 5 November 2004 | Microscopy of water sample concentrates performed at NVH in Oslo, and results (two Giardia cysts in raw water and five Giardia cysts in treated water from implicated water source) reported to Bergen |
| 5-6 November 2004 | Boil water notices announced for the premises served by the implicated water source |
| 24 November 2004 | By this date 900 cases of giardiasis had been registered |
| 9 December 2004 | Septic tank sewage samples collected and sent to NVH, Oslo, for analysis |
| 9 and 13 December 2004 | Soil samples associated with the septic tank collected and sent to NVH, Oslo, for analysis |
| 21 December 2004 | Boil water notice withdrawn |
Much of the information in this time line was obtained from reference 10.
This was the first outbreak of a waterborne parasitic disease recorded in Norway, and various details were published in a report by the local council and members of the Norwegian Food Safety Authority (10). Once the increase in giardiasis cases had been noted, geographic clustering of the cases rapidly made it apparent that the outbreak was probably waterborne, and a particular water source was implicated.
The initial source of contamination of the water supply was obviously of concern and interest, not least to ensure that repeat contamination events did not occur. The final report (10) indicates that sewage leakage from a residential area with drainage toward the water source was the most likely source of contamination, although no definitive evidence was provided and no analyses were conducted. However, during the episode and peak of media and public interest, considerable suspicion was directed toward disposal of waste from a septic tank at a particular tourist installation.
Here we describe how genotyping procedures provided evidence of the lack of likelihood that this installation was the source of the initial contamination event, as well as background information about the water analysis and preliminary information on genotypic analysis of the Giardia cysts isolated from some of the infected patients.
MATERIALS AND METHODS
Fecal samples (purification of Giardia cysts).
Giardiasis was diagnosed at the Unit for Infectious Diseases and Parasitology, Department of Medicine, at Haukeland University Hospital (HUS) in Bergen, either by detection of cysts in fecal samples by microscopy following standard formalin-ether concentration or by a fecal antigen test (ImmunoCard STAT! Cryptosporidium/Giardia rapid assay; Meridian Bioscience, Inc.). Positive samples were forwarded to the Parasitology Laboratory at the Department of Food Safety and Infection Biology at the Norwegian School of Veterinary Science (NVH).
At NVH, samples were refrigerated until further analysis, which was begun as rapidly as possible. Coded samples were washed in detergent solution, and this was followed by flotation on saturated salt (NaCl) solution and washing in distilled water. Each final semipurified suspension (approximately 1.5 ml) was refrigerated.
For the initial analyses described here, 21 stool samples (from 19 different, apparently unrelated individuals) were selected randomly from among the first 50 samples received at NVH. All had been diagnosed at HUS by microscopy, as the fecal antigen test was not in use then. An overview of the samples, patients, and symptoms is shown in Table 2.
TABLE 2.
Summary of patient and sample details and genotyping results
| Patient and sample detailsa
|
Genotyping results
|
||||||
|---|---|---|---|---|---|---|---|
| Patient age (yr) | Patient sex | Record of diarrhea | Designation | Date of sample (day/mo/yr) | Giardia cysts detected (approx % DAPI staining) | β-Giardin gene locus (compared to published subgenotype B3)b | gdh gene locus (compared to published subgenotype B3)c |
| 24 | F | No record | A | 1/11/04 | + (50) | Isolate group BG-ber2 (1 SNP at position 309) | Isolate group gd-ber3 (2 SNPs at positions 243 and 261) |
| 72 | M | 2 to 3 wk | B | 1/11/04 | ++ (5) | Isolate groups BG-ber3 and BG-ber4 (1 SNP at position 60 and 2 SNPs at positions 60 and 333) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
| 29 | M | No record | C | 28/10/04 | ++ (20) | Identical | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
| M | 29/10/04 | +++ (1) | Identical and isolate group BG-ber5 (2 SNPs at positions 519 and 603) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) | |||
| W | 30/10/04 | ++ (5) | Isolate group BG-ber5 (2 SNPs at positions 519 and 603) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) | |||
| 39 | M | No record | H | 1/11/04 | +++ (2) | Isolate group BG-ber2 (1 SNP at position 309) | Isolate group gd-ber3 (2 SNPs at positions 243 and 261) |
| 20 | F | 2 wk | K | 2/11/04 | ++ (50) | Isolate group BG-ber5 (2 SNPs at positions 519 and 603) | Isolate group gd-ber9 (3 SNPs at positions 111, 411, and 426) |
| 48 | F | 3 wk | P | 28/10/04 | +++ (5) | Isolate groups BG-ber7 and BG-ber8 (2 SNPs at positions 60 and 603 and 3 SNPs at positions 60, 519, and 603) | Isolate group gd-ber7 (3 SNPs at positions 111, 174, and 396) |
| 29 | F | 2 mo | R | 1/11/04 | +/++ (20) | Isolate groups BG-ber1 and BG-ber6 (1 SNP at position 183 and 2 SNPs at positions 183 and 309) | Isolate group gd-ber3 (2 SNPs at positions 243 and 261) |
| 22 | F | 2 to 3 wk | Y | 29/10/04 | ++ (10) | Isolate group BG-ber2 (1 SNP at position 309) | Isolate group gd-ber2 (1 SNP at position 354) |
| 42 | M | No record | CA | 3/11/04 | ++ (2) | Isolate group BG-ber9 (3 SNPs at positions 60, 333, and 633) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
| 29 | F | No record | DA | 2/11/04 | ++ (2) | Isolate group BG-ber9 (3 SNPs at positions 60, 333, and 633) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
| 24 | F | Diarrhea recorded, but no time estimate | FA | 3/11/04 | +++ (too much background fluorescence) | Isolate group BG-ber2 (1 SNP at position 309) | Isolate group gd-ber6 (3 SNPs at positions 111, 174, and 396) |
| 25 | M | 5 wk | GA | 3/11/04 | ++ (30) | Isolate group BG-ber5 (2 SNPs at positions 519 and 603) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
| 23 | F | 7 wk | MA | 3/11/04 | + (50) | Isolate group BG-ber2 (1 SNP at position 309) | Isolate group gd-ber3 (2 SNPs at positions 243 and 261) |
| 46 | F | No record | NA | 1/11/04 | ++ (5) | Isolate group BG-ber10 (4 SNPs at positions 165, 240, 333, and 603) | Isolate group gd-ber1 (1 SNP at position 354) |
| 41 | F | 4 wk | TA | 3/11/04 | ++ (5) | Isolate group BG-ber2 (1 SNP at position 309) | Isolate group gd-ber5 (2 SNPs at positions 111 and 396) |
| 38 | M | 4 to 5 wk | VA | 3/11/04 | ++ (2) | Isolate group BG-ber5 (2 SNPs at positions 519 and 603) | Isolate group gd-ber6 (3 SNPs at positions 111, 174, and 396) |
| 35 | F | 10 days | ØA | 2/11/04 | ++ (5) | Sequence identical to B3 sequence | Isolate group gd-ber8 (3 SNPs at positions 243, 261, and 354)d |
| 40 | F | No record | EB | 2/11/04 | ++ (10) | Isolate group BG-ber5 (2 SNPs at positions 519 and 603) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
| 35 | F | Diarrhea recorded, but no time estimate | GB | 4/11/04 | +++ (too much background fluorescence) | Isolate group BG-ber1 (1 SNP at position 183) | Isolate group gd-ber4 (2 SNPs at positions 111 and 174) |
The mean age was 35 years, and the median age was 35 years (n = 19). The female/male ratio was 13:6. The mean duration of diarrhea was 4 weeks (n = 10). The samples were collected from 28 October 2004 to 4 November 2004.
Genotypes were compared to previously described subgenotype B3 (GenBank accession number AY072727). For further details, including division of the samples by subgenotype group, see Table S4 in the supplemental material. The data showed that four samples were identical to B3, two samples were in group BG-ber1, five samples were in group BG-ber2, one sample was in group BG-ber3, one sample was in group BG-ber4, six samples were in group BG-ber5, one sample was in group BG-ber6, one sample was in group BG-ber7, one sample was in group BG-ber8, two samples were in group BG-ber9, and one sample was in group BG-ber10.
Genotypes were compared to previously described subgenotype B3 (GenBank accession number AY178756). For further details, including division of the samples by subgenotype group, see Table S5 in the supplemental material. The data showed that one sample was in group gd-ber1, one sample was in group gd-ber2, four samples were in group gd-ber3, nine samples were in group gd-ber4, one sample was in group gd-ber5, two samples were in group gd-ber6, one sample was in group gd-ber7, one sample was in group gd-ber8, and one sample was in group gd-ber9.
Sample ØA was more similar to previously described subgenotype B2 (GenBank accession no. AY178753), differing by only two SNPs. For further details see Table S5 in the supplemental material.
For each semipurified suspension, 15 μl was spotted on a Spot-On microscope slide (Dynal Biotech ASA, Oslo, Norway), air dried, methanol fixed, and stained with monoclonal antibody against Giardia cysts (Aqua-Glo; Waterborne Inc., New Orleans, La.) and 4′,6′diamidino-2-phenylindole (DAPI) for rapid estimation of the number of cysts and the percentage with nuclei (and therefore DNA for PCR). Cyst quantities were scored with a ×20 objective as follows: no cysts detected, + (1 to 9 cysts per field of view), ++ (10 to 50 cysts per field of view), or +++ (>50 cysts per field of view). The proportion of cysts containing nuclei (stained with DAPI) was estimated from counts of usually around 50 cysts. In some instances background fluorescence was too great for estimation of DAPI staining.
The remainder of the suspension was purified further by immunomagnetic separation (IMS) (GC-Combo; Dynal Biotech ASA, Oslo, Norway) by using a modification of the procedure described by the manufacturer, as briefly described below. To the suspension in each microcentrifuge tube were added 100 μl of each buffer provided and 15 μl of Dynabeads coated with anti-Giardia monoclonal antibody. The suspension was mixed by rotation for 1 h before collection of the beads with a magnet, washing in water, and separation of the beads and cysts by vigorous shaking in 0.1 M acid.
Septic tank samples (purification of Giardia cysts).
Five liters of sewage suspension from the septic tank was concentrated to 10 ml by repeated centrifugation. Giardia cysts were isolated using IMS (GC-Combo) by following the manufacturer's instructions, except that only 50% of the final purified concentrate was dried on a Spot-On slide for fixing, staining (as described above for fecal samples), and microscopy. The remaining 50% was refrigerated for DNA isolation.
Soil samples.
Twelve soil samples from the vicinity of the septic tank were sent to NVH. A series of mixing, settling, decanting, and centrifugation steps were performed, followed either by salt flotation and IMS or only IMS (GC-Combo) following the manufacturer's instructions. Only 50% of the final concentrate was dried on a Spot-On slide for fixing, staining (as described above for fecal samples), and microscopy. The remaining 50% was refrigerated for DNA isolation.
DNA isolation.
Cysts from feces, septic tank sewage, and positive soil samples were resuspended in Tris-EDTA buffer and placed in a heat block set at 100°C overnight. DNA was isolated using a QIAamp DNA mini kit (QIAGEN GmbH, Germany). In the final step, two elutions in distilled water were performed. The isolated DNA was refrigerated until PCR was performed.
Water samples.
The first water samples from the implicated source, raw and treated, were sent to the Parasitology Laboratory at NVH in early November 2004 and were analyzed by using U.S. Environmental Protection Agency Method 1623 (12), with membrane filtration of 10 liters of water as the initial step (11). An additional seven water samples (four treated samples and three raw samples) from other water sources in the area were also collected and analyzed at this time.
Due to limited analytical capacity at NVH, additional water samples were sent from Bergen to the Water and Environmental Microbiology Unit at the Swedish Institute for Infectious Disease Control (SMI) in Solna, Sweden. A few cysts were detected in a number of samples (2 cysts per 10 liters of water) and were largely empty (DAPI negative) (Anette Hansen, SMI, personal communication). DNA was extracted from positive samples at SMI using a QIAamp DNA mini kit. The DNA isolated from 12 positive water samples was forwarded by courier to NVH.
PCR, electrophoresis, purification of PCR product, and sequencing.
Three genes were used for genotyping investigations, the β-giardin gene, the glutamate dehydrogenase (gdh) gene, and the triosphosphate isomerase (tpi) gene, using previously described methods and primers (1, 3, 7), with slight modifications to the protocols described.
For β-giardin and gdh gene investigations, Giardia cysts isolated from all samples of feces, water, and septic tank sewage were used. Only one soil sample (in which a nucleated cyst had been identified) was included, and this sample was used only for analysis of the β-giardin gene.
For the tpi gene, only DNA from one fecal sample (sample DA) and from both elutions of the DNA from the septic tank sewage sample were investigated. The rationale for investigation of this gene is that since two primer sets are used, one for assemblage A and one for assemblage B, the possibility that if both assemblages are present, amplification of the predominant assemblage will mask the occurrence of the other is avoided.
For all genes, the following PCR mixture was used: 10 pmol of each primer, 0.4 μl of bovine serum albumin (20 mg/ml), 17.6 μl of water, 25 μl of HotStartTaqmaster (QIAGEN GmbH, Germany), and 3 μl of template. For each set of reactions, a negative control (water) and a positive control (DNA from Giardia assemblage A trophozoites; Hyperion Research, Medicine Hat, Canada) were included.
The primers and reaction conditions used are described in Table S2 in the supplemental material. PCR products were electrophoresed on 1% agarose gels and stained with ethidium bromide.
Following successful PCR, the products were purified (High Pure PCR product purification kit; Roche Diagnostics GmbH) by using the manufacturer's protocol with minimal modifications. The air-dried purified products were sent, along with appropriate primers, for sequencing of both strands to MWG Biotech in Germany. Chromatograms and sequences were examined using Chromas (http://www.technelysium.com.au/chromas.html) and BioEdit (http://www.mbio.ncsu.edu/BioEdit/page2.html). Sequence searches were conducted using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession numbers.
Sequences described here have been deposited in the GenBank database under accession numbers DQ090522 to DQ090542.
RESULTS
As in the overall infected population during the peak stages of the outbreak, most patients included in the analyses described here were female, and the median age was 35 years (Table 2). For one patient, three samples taken on successive days were included. All samples were positive as determined by microscopy (Table 2), and the predominant cyst score was ++ (10 to 50 cysts per field of view with a ×20 objective).
The septic tank sewage sample was also positive (approximately 20 cysts per field of view with a ×20 objective), but most cysts were broken or distorted; less than 1% were nucleated (DAPI-stained nuclei visibly contained within the cyst wall).
Of the 12 soil samples analyzed, only 4 were positive. Three contained single cysts (nonnucleated), and one contained eight cysts, one of which was nucleated. This sample was analyzed by performing a PCR for the β-giardin gene, but amplification was not successful.
Five Giardia cysts were detected in the treated water from the implicated source, and two cysts were detected in the raw water. None of the cysts were nucleated, and further analysis of these samples was not conducted. Parasites were not detected in six of the seven other water samples. In one other treated water sample, a single cyst, which was nonnucleated and had poor morphology, was detected. PCR amplification of both β-giardin and gdh gene sequences was unsuccessful for the DNA isolated from Giardia-positive water samples from the implicated water sent from SMI in Sweden.
PCR amplification of the β-giardin and gdh gene sequences was successful for all patient samples. The products were the expected size, and sequencing produced clear electropherograms with well-defined peaks. Sequence analysis of the PCR products resulting from β-giardin and gdh gene amplification from all patient samples indicated that the Giardia involved in the outbreak was genotype B and, in general, was most similar to subgenotype B3 or B2. However, not all sequences were identical, nor were they all identical to previously published sequences.
For the β-giardin gene, three samples were identical to Giardia intestinalis assemblage B, genotype B3 (3) (GenBank accession number AY072727), while the other 18 samples could be divided into 10 other groups (for a total of 11 groups) that differed from genotype B3 by between one and four nucleotide substitutions or single-nucleotide polymorphisms (SNPs). At this gene, all samples were more similar to genotype B3 than to genotype B2 (3) (GenBank accession number AY072726). The 11 different groups contained between one and six samples. For three samples, electropherograms indicated that there were 2 nucleotides at the same position, which could be consistent with the presence of genetically different cysts in the same isolate (5). Thus, while 18 samples were each placed into one of the 11 groups, 3 samples were placed into two of the groups. Allelic sequence heterozygosity is an alternative explanation, as previously reported (2), but it was not explored further in this study. A summary of these results is shown in Table 2, and additional information is provided in Table S4 in the supplemental material.
For the gdh gene, no samples were identical to genotype B3 (GenBank accession number AY178756); again, the samples were grouped, this time into 10 groups, on the basis of SNPs. Although for 20 samples the sequences were more similar to genotype B3 (GenBank accession number AY178756), with between one and four SNPs compared with genotype B3, for one sample (sample ØA) the sequence was more similar to genotype B2 (GenBank accession number AY178753), with three differences from genotype B3 but only two differences from genotype B2. The different groups contained between one and nine samples. For one sample, the electropherograms indicated that there were 2 nucleotides at the same position, which could be consistent with the presence of genetically different cysts in the same isolate (5). Thus, while 20 samples were each placed into one of the 10 groups, one sample was placed into two groups. Allelic sequence heterozygosity is an alternative explanation, as previously reported (2), but it was not explored further in this study. A summary of these results is shown in Table 2, and additional information is provided in Table S5 in the supplemental material.
Sequence analysis of the septic tank sewage sample PCR product for β-giardin and gdh gene amplification indicated that the predominant Giardia genotype was genotype A. For the β-giardin gene, the closest previously described subgenotype was subgenotype A3 (3) (GenBank accession number AY072724); however, there were two SNP differences compared with the previously described subgenotype. For the gdh gene, the electropherograms obtained with both forward and reverse primers were truncated, possibly due to the poor quality of DNA extracted from disintegrating cysts. Alignment of forward and reverse sequences resulted in a 168-bp overlap, and the closest previously described genotype for the section of overlap was either genotype A2 (GenBank accession number AY178737) or genotype A1 (GenBank accession number AY178735), with just one SNP. However, in the longer forward sequence there were two additional nucleotide differences which indicated that the genotype was genotype A2. These results are summarized in Table 3.
TABLE 3.
Septic tank sewage sequence analysis for the gdh and β-giardin gene loci; nucleotide substitutions compared with published sequences
| Gene | Genotype or sample | Nucleotide at position:
|
||||
|---|---|---|---|---|---|---|
| 65 | 99 | 143 | 308 | 326 | ||
| β-Giardin | A3 (AY072724)a | G | G | |||
| Septic tank sample | A | A | ||||
| gdh | A1 (AY178735) | C | T | C | ||
| A2 (AY178737) | C | C | T | |||
| Septic tank sample (based on sequence obtained using forward primer) | T | C | T | |||
| Septic tank sample (based on sequence obtained using reverse primer) | T | —b | — | |||
The numbers in parentheses are accession numbers.
—, sequence too short.
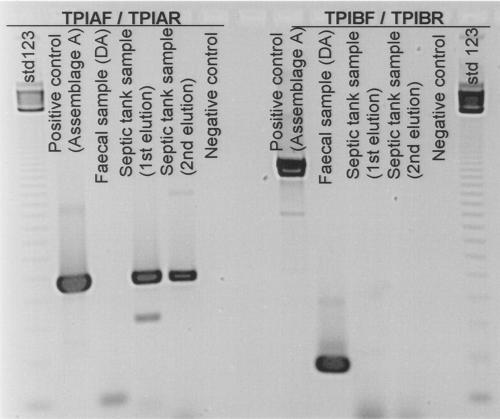
For amplification of the tpi gene, two different primer sets were used, one which should have amplified this sequence (576 bp) from assemblage A cysts and one which should have amplified this sequence (208 bp) from assemblage B cysts. The results (Fig. 1) demonstrated the presence of DNA from assemblage A Giardia cysts but not DNA from assemblage B cysts in the septic tank sewage sample. The patient fecal sample, in contrast, contained DNA from assemblage B Giardia cysts but not DNA from assemblage A cysts.
FIG. 1.
Electrophoretic separation of tpi amplification products using two primer sets. Primers TPIAIF and TPIAR amplify a 576-bp product from assemblage A G. intestinalis, and primers TPIBIF and TPIBR amplify a 208-bp product from assemblage B G. intestinalis.
DISCUSSION
The outbreak of waterborne giardiasis described here is one of the largest outbreaks recorded worldwide (in terms of individuals infected), and it is the first waterborne outbreak of a parasitic infection reported from Norway. Due to the geography and low population density of Norway, drinking water frequently has minimal treatment, unless there is a known problem. The water treatment plant involved in this outbreak is one of the oldest in Norway; the treatment in place during the outbreak was chlorination, although the works were in the process of renovation, and in February 2005 UV treatment was installed. Filtration systems are being installed, and the work should be completed in 2007.
Monitoring of water for parasites in Norway is largely conducted on an ad hoc, mostly very occasional, basis, and there is little infrastructure for such analyses. Nevertheless, the water source involved in this outbreak was monitored regularly, if infrequently, for Giardia parasites, indicating that there was an awareness of the potential for waterborne parasitic infection. When the sharp increase in giardiasis cases in Bergen was first noted during late October 2004, identification of the waterborne route and detection of Giardia cysts in the implicated water occurred relatively rapidly. A “boil water” instruction was implemented quickly by the authorities and was in place for most of November and December 2004.
Retrospective analysis of turbidity and fecal coliform data for the water source, along with weather patterns, suggested that the initial contamination of the water probably occurred in late August 2004 during a period of heavy precipitation. The 7- to 8-week period between contamination and detection of the first cases associated with the outbreak can probably be explained by medical practices in Norway, where exploration for Giardia infection is seldom requested by doctors unless the patient has recently been abroad (6). Additionally, the incubation period for giardiasis is between 1 and 2 weeks, and many patients probably did not immediately seek medical advice. In the weeks preceding identification of the outbreak, the number of fecal specimens sent to the bacteriology laboratory at HUS rose (854 samples were submitted during October 2003, and 1,489 samples were submitted during October 2004 [G. Njølstad, personal communication]).
The data presented here provide evidence that the Giardia that caused the outbreak belonged to assemblage B and was closely related to genotype B3. Interpretation of the differences in sequences between the patient samples for the β-giardin and gdh genes was complex. For the β-giardin gene, three isolates had sequences identical to the previously published sequence designated genotype B3 (3). However, other isolates had between one and four SNPs, based on around 10 polymorphic nucleotide positions. Whether these isolates could or should be classified as members of further genotypes or subgenotypes is uncertain, and we have not done so here. However, other researchers have described new genotypes based on a single SNP at one of these loci (5) (subgenotype B6). For the gdh gene, none of the sequences were identical to the sequence previously described for genotype B3, although genotype B3 was always the most similar (with one exception). Again, between one and three SNPs were found based on around 12 polymorphic nucleotide positions. Also, the possibility that the SNPs were sequencing artifacts cannot be completely excluded.
Grouping of isolates according to SNPs gave different groups for each gene. Although two patient isolates (isolates NA and P) were in their own groups for each gene, other isolates showed completely different grouping for each gene. For example, isolates FA and VA had identical SNPs for the gdh gene, which were not shared with any other isolate; thus, isolates FA and VA were grouped together, with no other isolates. However, with the β-giardin gene, their SNPs were quite different from each other, but these SNPs occurred in other isolates and the isolates were therefore placed in different groups from each other but together with four or five other isolates. One sample (sample ØA) was identical to genotype B3 at the β-giardin gene but was more similar to genotype B2 at the gdh gene. We were unable to find any reports of investigations of nucleotide sequences for both these genes in assemblage B Giardia isolates, and the lack of concurrence between the results for these genes in our analysis calls into question the adequacy of criteria for grouping isolates and indicates that this is an avenue for further research.
As the samples included were selected at random from earlier specimens of the large number of isolates obtained from this outbreak, it was not until these analyses were complete that retrospective study of the documentation showed that three samples (samples C, M, and W) were from the same patient and that the samples were collected on successive days. For the gdh PCR amplification product, all three isolates had the same SNPs, placing them in the same group. However, for the β-giardin gene, the samples fell into two groups. One isolate had two nucleotides at the same position, and the other two isolates resembled each of these.
It is not possible to determine whether the initial contamination involved a range of genetically different assemblage B cysts or whether nucleotide sequences evolved between and within patients. As Giardia cysts and their DNA were isolated from as many patients as possible in this outbreak, future genotyping and sequence studies of these isolates, comparisons between family members, temporal differences, and other factors may provide interesting information.
Determining the source of the initial contamination of the water was obviously a matter of pertinence. A number of suggestions were analyzed for their likelihood (10), including another water source, spreading of manure, contamination from dogs that were being exercised, sewage leakage or discharge from a septic tank, and sewage leakage from three different residential areas in the neighborhood. Ultimately, it was concluded that sewage leakage from a residential area was the most probable source, based largely on geographical criteria and also on evidence of leakage from sewage pipes, although the sewage itself was never analyzed. However, the work described here provides definitive evidence that the septic tank was unlikely to be the contamination source. Although a large number of cysts were detected and cysts were also detected in soil in the vicinity of the septic tank, analyses of three different genes demonstrated that cysts in the septic tank sewage were assemblage A cysts. As sewage is likely to contain cysts from a variety of hosts, it is not surprising that two genotypes (genotypes A2 and A3) were present, as indicated by genotyping of the β-giardin and gdh genes. However, if this is so, it is curious that multiple peaks on the electropherograms did not indicate the presence of more than one assemblage A genotype for either gene. For all three genes there was no evidence that the septic tank contained assemblage B Giardia cysts.
Previous analysis at sewage treatment plants around Norway demonstrated that most Giardia cysts in sewage influent in Norway are assemblage A cysts (8; L. Robertson, B. Gjerde, L. Hermansen, and I. S. Hamnes, Abstr. Int. Giardia Cryptosporidium Conf., abstr. P091, 2004), and similar results have been obtained in studies in Italy (4) and the United States (9). In the United States study, in which a single gene (tpi) was used for genotyping, SNPs were used to divide the isolates into subgenotypes; for the 20 assemblage B isolates, 10 different, distinct subgenotypes were identified, while only 5 subgenotypes were identified for the 111 assemblage A isolates. The authors used this information to suggest that assemblage A parasites may evolve more slowly than assemblage B parasites and that the high genetic diversity in assemblage B isolates indicated that there was more than one source of Giardia infection. While our data may support the suggestion that there is relatively rapid evolution of assemblage B Giardia, our data also suggest that high genetic diversity in assemblage B isolates may occur, even when there is apparently only a single infection source.
Supplementary Material
Acknowledgments
For their various contributions to the data presented in this paper we acknowledge the following individuals: Åshild Våge and Torunn Kleppe of the Parasitology Laboratory at the Norwegian School of Veterinary Science, Oslo, Norway; Rita Kvamsdal, Gro Rasmussen, and Eli Askvik of the Parasitology Laboratory at Haukeland University Hospital, Bergen, Norway; Øystein Strand of Haukeland University Hospital, Bergen, Norway; Ane Gjesdal, Øystein Søbstad, and Ingar Tveit of Bergen kommune, Bergen, Norway; Peter Wallis of Hyperion Research, Medicine Hat, Canada; and Anette Hansen of the Water and Environmental Microbiology Unit at the Swedish Institute for Infectious Disease Control, Solna, Sweden.
Some of this work was supported by the Research Council of Norway.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amar, C. F., P. H. Dear, S. Pedraza-Diaz, N. Looker, E. Linnane, and J. McLauchlin. 2002. Sensitive PCR-restriction fragment length polymorphism assay for detection and genotyping of Giardia duodenalis in human feces. J. Clin. Microbiol. 40:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baruch, A. C., J. Isaac-Renton, and R. D. Adam. 1996. The molecular epidemiology of Giardia lamblia: a sequence-based approach. J. Infect. Dis. 174:233-236. [DOI] [PubMed] [Google Scholar]
- 3.Caccio, S. M., M. De Giacomo, and E. Pozio. 2002. Sequence analysis of the beta-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int. J. Parasitol. 32:1023-1030. [DOI] [PubMed] [Google Scholar]
- 4.Caccio, S. M., M. De Giacomo, F. A. Aulicino, and E. Pozio. 2003. Giardia cysts in wastewater treatment plants in Italy. Appl. Environ. Microbiol. 69:3393-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalle, M., E. Pozio, G. Capelli, F. Bruschi, D. Crotti, and S. M. Caccio. 2005. Genetic heterogeneity at the B-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic genotypes. Int. J. Parasitol. 35:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Nygård, K., L. Vold, L. Robertson, and J. Lassen. 2003. Underdiagnostiseres innenlandssmittede Cryptosporidium- og Giardia-infeksjoner i Norge? Tidsskr. Nor. Laegeforen 123:3406-3409. [PubMed] [Google Scholar]
- 7.Read, C. M., P. T. Monis, and R. C. Thompson. 2004. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect. Genet. Evol. 4:125-130. [DOI] [PubMed] [Google Scholar]
- 8.Robertson, L. 2005. Drikkevann: Giardia-utbruddet i Bergen—en vekker for oss alle. Hva vi vet om parasittene og hvor de stammer fra. Vann 2:193-202.
- 9.Sulaiman, I. M., J. Jiang, A. Singh, and L. Xiao. 2004. Distribution of Giardia duodenalis genotypes and subgenotypes in raw urban wastewater in Milwaukee, Wisconsin. Appl. Environ. Microbiol. 70:3776-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tveit, I., Ø. Søbstad, I. Kalland, A. Seim, R. Ø. Arnesen, and P. Fennell. 2005. Giardia-utbruddet i Bergen. Bergen Kommune and Mattilsynet, Norway. [Online.] http://www.bergen.kommune.no/info_/ekstern/nyheter3/Giardia-rapport_akk_21.02.2005.doc.
- 11.U.S. Environmental Protection Agency. 1997. Method 1622: Cryptosporidium in water by filtration/IMS/FA DRAFT. Publication no. EPA-821-R-97-023. U.S. Environmental Protection Agency, Washington, D.C.
- 12.U.S. Environmental Protection Agency. 1999. Method 1622: Cryptosporidium and Giardia in water by filtration/IMS/FA DRAFT. Publication no. EPA-821-R-99-006. U.S. Environmental Protection Agency, Washington, D.C.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.