Abstract
Bacterial community dynamics of a whole drinking water supply system (DWSS) were studied from source to tap. Raw water for this DWSS is provided by two reservoirs with different water characteristics in the Harz mountains of Northern Germany. Samples were taken after different steps of treatment of raw water (i.e., flocculation, sand filtration, and chlorination) and at different points along the supply system to the tap. RNA and DNA were extracted from the sampled water. The 16S rRNA or its genes were partially amplified by reverse transcription-PCR or PCR and analyzed by single-strand conformation polymorphism community fingerprints. The bacterial community structures of the raw water samples from the two reservoirs were very different, but no major changes of these structures occurred after flocculation and sand filtration. Chlorination of the processed raw water strongly affected bacterial community structure, as reflected by the RNA-based fingerprints. This effect was less pronounced for the DNA-based fingerprints. After chlorination, the bacterial community remained rather constant from the storage containers to the tap. Furthermore, the community structure of the tap water did not change substantially for several months. Community composition was assessed by sequencing of abundant bands and phylogenetic analysis of the sequences obtained. The taxonomic compositions of the bacterial communities from both reservoirs were very different at the species level due to their different limnologies. On the other hand, major taxonomic groups, well known to occur in freshwater, such as Alphaproteobacteria, Betaproteobacteria, and Bacteroidetes, were found in both reservoirs. Significant differences in the detection of the major groups were observed between DNA-based and RNA-based fingerprints irrespective of the reservoir. Chlorination of the drinking water seemed to promote growth of nitrifying bacteria. Detailed analysis of the community dynamics of the whole DWSS revealed a significant influence of both source waters on the overall composition of the drinking water microflora and demonstrated the relevance of the raw water microflora for the drinking water microflora provided to the end user.
Outbreaks of waterborne infectious diseases via the use of contaminated drinking water still pose a serious health threat worldwide, despite the fact that drinking water is one of the most closely monitored and strictly regulated resources. Careful estimates indicate that each year about 350 million people are infected by waterborne pathogens, with 10 to 20 million succumbing to severe cases of infection (45). This phenomenon is far from being restricted to developing countries but also threatens developed countries. In the United States, almost 430,000 cases were reported in 126 outbreaks of waterborne infectious diseases from 1991 to 1999 (1).
Production of drinking water complying with international quality standards does not necessarily ensure good drinking water for the consumer (2). The occurrence of bacterial regrowth may multiply adverse effects in drinking water distribution systems. Composition of the autochthonous microbial community may promote the survival and growth of hygienically relevant and potentially pathogenic bacteria (18). In addition, the autochthonous microflora could sustain the growth of protozoa and metazoa (e.g., crustacean) that are visible (9, 38) or may have adverse effects on the taste of the drinking water (22). Key factors influencing regrowth of heterotrophic bacteria in a drinking water supply system (DWSS) are (i) concentration of organic compounds, (ii) chlorine concentration, (iii) residence time of the water in the distribution system, (iv) water temperature, and (v) physicochemical characteristics of the material lining the distribution pipes (23).
Several studies using cultivation-based approaches helped to characterize some bacteria residing in bulk water (25, 42) or in biofilms at various points in the DWSS (17, 19). In general, heterotrophic plate counts (HPC) are used to assess the overall bacterial quality of drinking water (32). However, the majority of bacterial cells in natural communities are either nonculturable by current cultivation methods or are present in a viable-but-nonculturable state (24, 39). Thus, the real composition and dynamics of bacterial communities in drinking water distribution systems are far from being assessed and understood in detail.
Molecular biology methods now provide tools to elucidate dynamics of microbial communities residing in various aquatic environments (10, 14, 21). These methods have in common that they are based on the molecular analysis of environmental nucleic acids with respect to their rRNA or rRNA genes. These approaches exploit the utility of rRNA as a stable taxonomic marker for microorganisms (47). Although being widely used in many habitats, it was not until recently that these molecular approaches also had been applied to DWSSs (4, 35). The study of community dynamics of the microflora from a DWSS could provide a basic understanding of the effects of processing, such as flocculation and sand filtration, chlorination, and storage plus transportation on the structure and composition of the drinking water microflora.
In the present study, we assessed the structure and composition of microbial communities in a modern European DWSS using single-strand conformation polymorphism (SSCP) fingerprinting of 16S rRNA and its genes (36) based on nucleic acids extracted from a set of water samples. Inclusion of two surface reservoirs with very different drainage basins resulting in different levels of water quality should allow the study of the impact of different source water on the drinking water microflora. New insights into the microbial ecology of DWSSs are provided by a detailed evaluation of bacterial communities at different sampling points throughout the system: i.e., from source to tap. By using DNA- and RNA-based fingerprints, the present and the active members were assessed, respectively. Subsequent sequencing of the electrophoretically separated amplification products allowed phylogenetic identification of the single members in each fingerprint. This approach enabled us to identify the origin of regrowing species: i.e., gave insights into the influences of different source waters, raw water processing methods, and transportation on the composition of microbial communities in tap water intended for human consumption.
MATERIALS AND METHODS
Study site and sampling of bacteria.
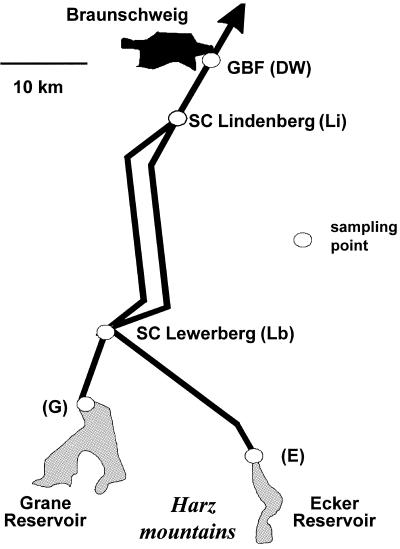
Different samples of raw water and treated water were analyzed from the DWSS of the city of Braunschweig, Germany. The DWSS is operated by the Harzwasserwerke GmbH Corporation, Germany. This water supply company has an average output of about 80 million m3 of drinking water per year and provides drinking water for about two million people. The two surface water reservoirs, the Grane (G) and Ecker (E) reservoirs, which provide raw water for the DWSS of Braunschweig, are located in the northern part of the Harz mountain range 40 km south of Braunschweig (Fig. 1). The oligotrophic Grane reservoir has a maximum retention capacity of 46.4 million m3. The collection of aerobic raw water is located at a depth of 50 m (sampling site G in Fig. 1, sample GR1). The water has an average pH of 7.2. The Ecker reservoir consists of dystrophic water with a mean pH of 5.2 and has a maximum storage capacity of 13.3 million m3. The aerobic raw water is collected at a depth of 58 m (sampling site E, sample ER1). Actual data on the raw water chemistry are summarized in Table 1. At both reservoirs, the obtained raw water is processed using coagulation-flocculation and sand barriers for particle elimination (sampling sites G and E; samples for processed raw water GR2 and ER2, respectively). Depending on the amount of dissolved organic carbon (DOC) of the processed waters, chlorination is performed by addition of 0.2 to 0.3 mg or 0.6 to 0.7 mg chlorine per liter of water originating from the Grane or Ecker reservoir, respectively. After treatment of the raw water by physical and chemical means, pipe systems lead from both reservoirs to the Lewerberg (Lb) storage container, where both waters are mixed. During the first half of 2003, this mixed water consisted of about 77% water from the Grane reservoir and 23% water from the Ecker reservoir, with very little variability (less than 3%). From here, the mixed water is transported to the Lindenberg (Li) storage container, directly located at the southern outskirts of Braunschweig, to which the local drinking water supply net is connected, including the sampled tap water source at the institute. The drinking water (DW) has an average flow time of about 36 to 48 h from the source until it reaches the tap at the institute (GBF).
FIG. 1.
Sampling points for the Harzwasser DWSS near the city of Braunschweig in Germany. SC, storage container. Abbreviations for the single sampling points are given in parentheses.
TABLE 1.
Chemical and biological background data from the raw waters of the drinking water supply system of the Harzwasserwerke Corporation from June 2003
| Parameter | Result fora:
|
||
|---|---|---|---|
| Grane raw water | Ecker raw water | SC Lindenberg | |
| Temp (°C) | 7.3 | 5.9 | 8.2 |
| pH | 7.32 | 4.88 | 9.00 |
| HPC (CFU/ml) | 75 | 30 | 0 |
| Ammonia (mg/liter) | 0.032 | 0.070 | 0.035 |
| Nitrite (mg/liter) | 0.016 | 0.006 | 0.009 |
| Nitrate (mg/liter) | 5.0 | 2.7 | 4.6 |
| Phosphate (mg/liter) | <0.002 | <0.002 | <0.002 |
| DOC (mg/liter) | 2.8 | 6.5 | 2.7 |
| Chlorine (mg/liter) | ND | ND | 0.03 |
All data were from 2 June 2003 except for the HPC results, which were from 11 June 2003. ND, not determined.
Twenty-liter bulk water samples were sampled in autoclaved glass bottles on 11 June 2003 at the various sampling points of the distribution system (Fig. 1). Within less than 4 h after sampling, microorganisms were harvested by filtration of 3 liters of water (three replicates for each sampling site and date) on a filter sandwich consisting of a 0.2-μm-pore-size polycarbonate filter (90-mm diameter; Nuclepore; Whatman, Maidstone, United Kingdom) with a precombusted glass fiber filter on top (90-mm diameter; GF/F; Whatman) (3). After filtration, filter sandwiches were stored at −70°C until further analysis.
Nucleic acid extraction and quantification.
Nucleic acid extraction from frozen filters was performed as described previously by a method that utilizes a pH shift for parallel extraction of RNA and DNA from the same filter sandwich (44). Nucleic acid concentrations were determined by spectrophotometric fluorescence (43) using either RiboGreen (RNA quantification kit; Molecular Probes) or PicoGreen (double-stranded DNA [dsDNA] quantification kit; Molecular Probes) for RNA or DNA quantification, respectively.
Prior to quantification, RNA extracts were purified from contaminating traces of DNA by incubation with DNase I (Roche Diagnostics, Mannheim, Germany) for 60 min at 37°C. For DNA, we included an additional WizardPrep DNA purification step (Promega, Madison, Wis.) with a 50-μl aliquot. Although reducing the yield of total DNA, this additional purification step substantially increased the PCR efficiency for the DNA extracted.
Amplification of 16S rRNA and of 16S rRNA genes from environmental samples.
PCR amplification of 16S rRNA and of its respective genes from the obtained environmental nucleic acid extracts were performed using the previously described primers COM1 (5′-CAGCAGCCGCGGTAATAC-3′) and COM2 (5′-CCGTCAATTCCTTTGAGTTT-3′), amplifying positions 519 to 926 of the Escherichia coli numbering of the 16S rRNA gene (36), with the modification of using a 5′-biotin-labeled forward primer for improved separation (see below). From RNA, reverse transcription-PCR (RT-PCR) amplification of 16S rRNA was achieved using the One Step RT-PCR system (Roche Diagnostics) following the manufacturer's recommendations. Each amplification was carried out using 5 ng RNA template in a final volume of 50 μl, starting with the RT step for 30 min at 60°C and an initial denaturation for 5 min at 95°C. A total of 30 cycles (30s at 95°C, 30s at 55°C, and 1 min at 72°C) was followed by a final extension for 10 min at 72°C. Amplification of 16S rRNA genes from Wizard purified DNA extracts was achieved using HotStarTaq DNA polymerase (QIAGEN, Hilden, Germany) and the same cycling conditions as described above with an increased initial denaturing time (15 min).
16S ribosomal complementary DNA and 16S rRNA gene community fingerprints.
An alternative procedure for preparation of single-stranded DNA (ssDNA) compared to the previously described protocol (36) was used to improve separation and sequencing of the ssDNA. We used a 5′-biotinylated forward primer, COM1, instead of a 3′-phosphorylated reverse primer, COM2. We used magnetic streptavidin beads (Promega, Madison, Wis.) to obtain ssDNA from the amplicons of PCR and RT-PCR. For preparation of the magnetic beads, 47 μl of bead solution per amplification mixture was washed three times and then resuspended in 25 μl of 0.5× SSC (1× SSC consists of 15 mM sodium citrate and 150 mM sodium chloride). Twenty-three microliters of the suspension of prepared beads was added to each completed PCR, and binding of the biotin-labeled amplification products to the streptavidin beads was achieved by incubation at 22°C for 75 min in a thermomixer (350 rpm). After consecutive washing steps for 2 min with 60 μl of 0.1× SSC, the dsDNA was denatured by incubation for 10 min with 0.2 N NaOH at room temperature. These conditions did not affect the biotin-streptavidin binding of the labeled strand band but allowed the nonlabeled strand to dissolve, which was then decanted with the supernatant. After being washed twice with 60 μl of 0.1× SSC, the biotin-labeled ssDNA was eluted from the streptavidin beads by a final incubation for 10 min at 65°C in 22 μl molecular biology-grade water (diethyl pyrocarbonate treated; Bio-Rad) and finally transferred to fresh sterile microtubes. Quantification of the obtained ssDNA was performed on a 1.5% agarose gel by comparison with a low-molecular-weight marker (Invitrogen low-DNA-mass ladder).
For SSCP fingerprinting analysis, 90 ng of the obtained ssDNA was mixed with gel loading buffer (95% formamide, 10 mM NaOH, 0.25% bromphenol blue, 0.25% xylene cyanol) in a final volume of 8 μl. After incubation for 3 min at 95°C, the ssDNA samples were stored on ice, loaded onto a nondenaturing polyacrylamide-like gel (0.6× MDE gel solution; Cambrex BioScience, Rockland, Maine), electrophoretically separated at 20°C at 400 V for 18 h on a Macrophor sequencing apparatus (Pharmacia Biotech, Germany), and finally silver stained according to the method described by Schwieger and Tebbe (36).
Data analysis of SSCP fingerprints.
SSCP fingerprints were analyzed using the GelCompare II software package (Applied Maths, Kortrijk, Belguim) after digitalization of the SSCP gels by an Epson Expression 1600 Pro scanner. Only bands with an intensity of >0.1% of the total lane were considered for further statistical analysis. Similarity coefficients were calculated using Pearson correlation based on the densitometric curves of the lanes. Dendrograms were constructed based on the unweighted pair-group method using arithmetric averages.
Reamplification and sequencing of individual ssDNA bands from SSCP fingerprints.
Reamplification of individual bands excised from SSCP gels using sterile scalpels was performed as described previously (28). After elution of ssDNA from the acrylamide matrix at 95°C for 20 min in Tris buffer (10 mM Tris-HCl, 5 mM MgCl2, 5 mM KCl, 0.1% Triton X-100, pH 9), reamplification was achieved on 5 to 10 μl of the band solution using the same primers and amplification conditions as described above for DNA. To control the reaction products for size and purity, 8 μl was separated on a 1.5% agarose gel. The remaining reaction mixture was purified using the MinElute PCR purification kit, followed by cycle sequencing (ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems, Foster City, Calif.) including the primers applied before. Sequencing products were purified using the BigDye purification kit (QIAGEN) prior to analysis on an ABI Prism 3100 Genetic Analyzer.
Comparative sequence analysis.
The obtained sequences for both forward and reverse reactions were checked for accuracy using the Sequencher software package (www.genecodes.com). Sequences were then included in a local database containing every public 16S rRNA bacterial sequence already aligned and analyzed by phylogeny. Each band sequence was aligned and compared to its most similar sequences. A band sequence was rejected if it contained numerous errors (differences at positions otherwise conserved in every other sequence), if it was shorter than 250 nucleotides, and, finally, if it was suspected to be of chimeric origin (5′ and 3′ parts respectively closest to sequences having a different taxonomic designation). Every alignment was finally checked by eye using SeaView (7). For the similarity tables, each band sequence was blasted (with options no filter and W = 7) first against a database of sequences obtained from validly described species and second against the entire database of sequences. Sequence similarity values between two sequences were calculated as the numbers of identical nucleotides within obtained local alignments divided by the length of the shorter sequence, which therefore corresponds to the most conservative similarity estimate. For the closest described cultured species, we used an 80% limit because otherwise often the closest relatives are in different phyla. For each band sequence, public sequences with a similarity percentage above 95% were retained and the corresponding EMBL files were analyzed in order to identify the presence of various keywords that could indicate the origin of the sequence (such as “soil,” “freshwater,” “drinking water,” etc.).
Phylogenetic analyses were conducted after assigning the single sequences to large taxonomic units such as phyla and classes. For each band sequence, the two most similar public sequences were included as well a number of sequences representative either of well-established species or of some clone sequences known to be frequently isolated from freshwater. Distances were calculated according to the Kimura two-parameter method (Phylogeny Inference Package, version 3.63; distributed by J. Felsenstein, Department of Genetics, University of Washington, Seattle) and using parts of the sequences common to all sequences analyzed. Trees were drawn according to BIONJ (8). A total of 1,000 bootstrap replications were done to test for branch robustness. Bootstrap percentages above 50% only are indicated on the trees. Trees were drawn using NJPLOT (27) or TreeDyn (http://www.treedyn.org/).
Nucleotide sequence accession numbers.
Band sequences determined in this study have been deposited in the GenBank database under accession no. DQ077555 to DQ077627.
RESULTS AND DISCUSSION
Overall community structure of the microflora in the DWSS.
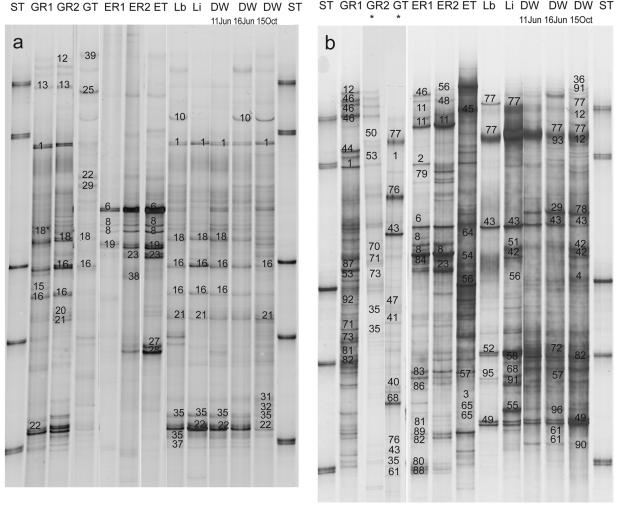
A whole DWSS was studied with raw water coming from two different reservoirs: i.e., the oligotrophic Grane reservoir and the dystrophic Ecker reservoir (Fig. 1). Samples from various sampling points at major steps in the treatment of the raw water and along the supply system were taken and analyzed by SSCP community fingerprinting. Both types of nucleic acids (i.e., DNA and RNA) extracted from the water samples were used to assess the present and active bacterial groups in the single water samples (Fig. 2a and b).
FIG. 2.
(a) Digitized images from the DNA-based SSCP fingerprints of the samples from the Harzwasser DWSS. Sequenced and identified bands are given numbers for the specific phylotype. The phylogenetic information about the identified bands is given in Table 2. Designations of single samples: ST, standards; GR1, raw water from Grane reservoir; GR2, processed raw water from Grane reservoir; GT, chlorinated water from Grane reservoir; ER1, raw water from Ecker reservoir; ER2, processed raw water from Ecker reservoir; ET, chlorinated water from Ecker reservoir; Lb and Li, Lewerberg and Lindenberg storage containers; DW, tap water samples from the respective dates. The asterisk indicates that this phylotype has a slightly different sequence from the phylotype provided under the accession number in Table 2 or 3. (b) Digitized images from the RNA-based SSCP fingerprints of the samples from the Harzwasser DWSS. Sample designations and numbering of sequenced bands are as for panel a. The lanes labeled with asterisks are from a different SSCP gel. The phylogenetic information about the identified bands is given in Table 3.
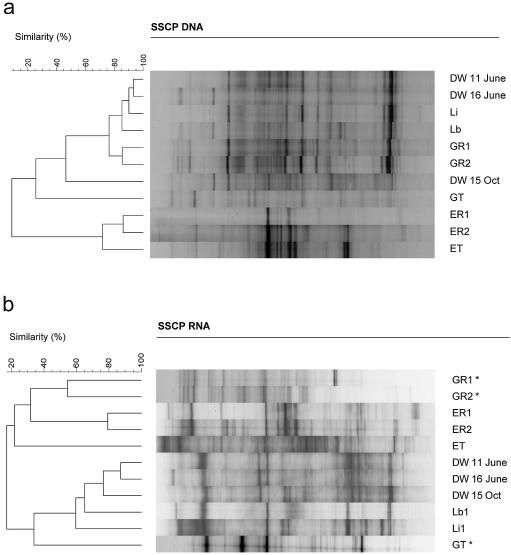
For the DNA-based fingerprints, very significant differences between the bacterial microflora of the two reservoirs could be detected (Fig. 2a). Comparative cluster analyses of the DNA-based fingerprints revealed three major types of communities: (i) the Grane reservoir samples, (ii) the Ecker reservoir samples, and (iii) samples of the supply system after chlorination (Fig. 3a). These three sets of samples clustered very tightly together, with a similarity of more than 80% among each other. The first chlorinated water samples (GT and ET) and the drinking water sample from 15 October 2003 were less related to the other samples (Fig. 3a).
FIG. 3.
Cluster analysis of the two gels given in Fig. 2. (a) DNA-based SSCP fingerprints of the samples from the Harzwasser DWSS. Sample designation as in Fig. 2a. (b) RNA-based SSCP fingerprints of the samples from the Harzwasser DWSS. The lanes labeled with asterisks are from a different SSCP gel.
For the comparison of bacterial communities in the DWSS, the following aspects have to be taken into account: (i) the different steps of the processing in two different waterworks (corresponding to the two different raw waters G and E), (ii) the changes in the drinking water distribution system (DWDS) (i.e., samples Lb, Li, and DW June 11), and (iii) the different time points on the tap side of the system (DW June 11, DW June 16, and DW October 15). The differences between the two reservoir communities were very pronounced and can be understood by the different natures of the aquatic environments. The influence of the treatment of the raw water by flocculation and sand filtration was relatively small. The unspecific removal of particles during these treatments was obviously not affecting the community structure significantly (comparison of the samples GR1/GR2 and ER1/ER2, respectively). Chlorination (samples GT and ET) did show a substantial influence on the community structure of the Grane reservoir water, as can be seen by its low similarity to samples before chlorination. The effect of chlorination was less pronounced in the Ecker reservoir water because the ET sample still clustered with a similarity of about 70%, closest to the other Ecker reservoir samples (Fig. 3a).
After chlorination, the drinking water leaves the waterworks and gets mixed in the storage container at Lewerberg (Lb). All samples of the DWDS (Lb, Li, and DW) showed almost identical fingerprints (similarity greater than 85% in Fig. 3a). In addition, most of the bands derived from fingerprints of the DWDS samples were already found in the Grane raw water, i.e., GR1 and GR2, as indicated by the tight clustering of both groups of samples at a similarity of around 75% in Fig. 3a. The drinking water sample from 15 October clustered apart from the other drinking water samples but still showed almost 50% similarity to the other drinking water samples from June.
The cluster analysis of the RNA-based fingerprints also showed substantial differences between the bacterial community structures of the two reservoirs (Fig. 3b). As for the DNA-based fingerprints, there was not much influence by the processing of the raw water on the RNA-based fingerprints and chlorination showed a strong impact which could be seen for both types of processed raw water (GT and ET). In contrast to the DNA-based observations, the chlorinated Ecker water sample ET clustered with the other raw water samples, whereas the chlorinated Grane water sample GT clustered with the drinking water samples. The DWDS samples all clustered together, but the two storage containers (Lb and Li) were substantially more different from the tap water samples than by the DNA-based fingerprints.
Taxonomic composition of the microbial communities.
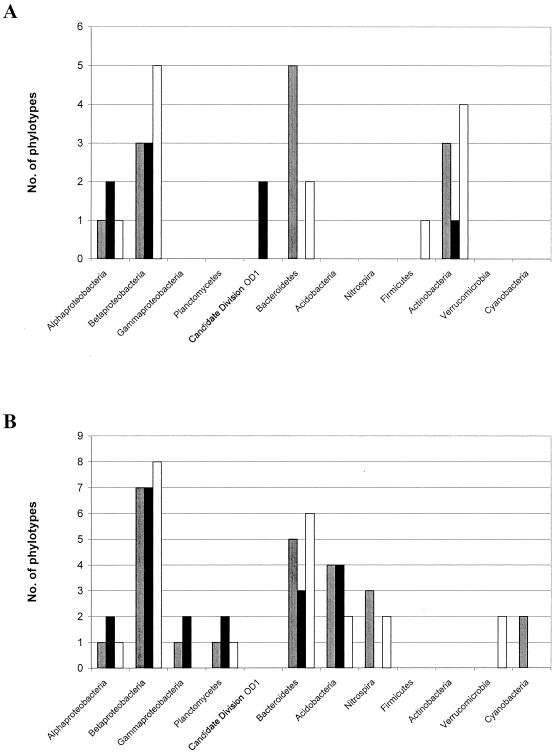
We sequenced a total of 216 bands from the DNA- and RNA-based community fingerprints for the assessment of the taxonomic composition of the single drinking water communities. Among these sequences, 71 unique phylotypes were detected using a limit of ≥98% sequence similarity and phylogenetic uniqueness as discrimination criteria (Tables 2 and 3). Sequences of the single bands were compared with the large data set of 16S rRNA gene sequences available in the GenBank database for identification of the single members of the microflora in this DWSS (Tables 2 and 3). DNA-based fingerprints revealed several members of taxonomic groups typical for freshwater according to Zwart et al. (48), such as Alphaproteobacteria, Betaproteobacteria, Bacteroidetes, and particular freshwater members of the Actinobacteria (Table 2). In addition, one phylotype belonged to the Firmicutes and two phylotypes belonged to candidate division OD1, so far only known from clone libraries of aquatic environments (12). Whereas the three taxonomic groups mentioned first above were also observed in the RNA-based fingerprints, five additional aquatic phyla were detected in the RNA-based fingerprints: i.e., Planctomycetes, Verrucomicrobia, Acidobacteria, Cyanobacteria, and Nitrospira (Table 3).
TABLE 2.
Taxonomic identification of single phylotypes found in the DNA-based SSCP fingerprints shown in Fig. 2a
| Phylotype | Sample(s)a (accession no.) | Taxonomic group | Closest 16S rRNA gene sequence (accession no.) | % Similarity | Closest described relativec (accession no.) | % Similarity |
|---|---|---|---|---|---|---|
| 1b | GR1, GT, Lb, Li, DW11, DW15 (DQ077612) | Betaproteobacteria | Uncultured freshwater bacterium LD28 (Z99999) | 99.8 | Methylophilus methylotrophus (AB193724) | 95.8 |
| 2b | GT, ER1 (DQ077611) | Betaproteobacteria | Uncultured Herbaspirillum CtaxAus-2 clone 2 (AF259596) | 99.5 | “Cenibacterium arsenoxidans” (AY728038) | 98.0 |
| 6b | ER1, ET (DQ077606) | Betaproteobacteria | Uncultured clone MPKCSC9 (AY766004) | 97.3 | Simonsiella crassa (AF328143) | 94.3 |
| 8b | ER1, ER2, ET (DQ077605) | Betaproteobacteria | Uncultured betaproteobacterium clone Stat2-99 (AY562366) | 98.5 | Polynucleobacter necessarius (AJ811014) | 97.8 |
| 10 | Lb, DW16 (DQ077622) | Betaproteobacteria | Uncultured clone G94 (AF407711) | 94.0 | Thiobacillus aquaesulis (U58019) | 92.0 |
| 12b | GR1, GR2, DW15 (DQ077616) | Betaproteobacteria | Uncultured betaproteobacterium clone Stat2-96 (AY562364) | 99.3 | Acidovorax temperans (AF078766) | 99.0 |
| 13 | GR1, GR2 (DQ077603) | Betaproteobacteria | Uncultured lake bacterium P38.4 (AY752086) | 98.3 | Rhodoferax ferrireducens (AF435948) | 97.0 |
| 15 | GR1 (DQ077614) | Actinobacteria | Uncultured lake bacterium P38.2 (AY752084) | 96.3 | Rathayibacter tritici (U96185) | 88.1 |
| 16 | GR1, GR2, GT, Lb, Li, DW11, DW15 (DQ077613) | Actinobacteria | Uncultured lake bacterium P38.2 (AY752084) | 99.5 | Geodermatophilus obscurus (L40621) | 90.3 |
| 18 | GR1, GR2, GT, Lb, DW11 (DQ077609) | Actinobacteria | Uncultured actinobacterium clone Sta2-9 (AY562361) | 99.8 | Geodermatophilus obscurus (L40621) | 90.7 |
| 19 | ER1, ET (DQ077626) | Actinobacteria | Uncultured actinobacterium clone SW9 (AJ575553) | 99.7 | Nostocoida limicola (Y14597) | 90.1 |
| 21 | GR2, Lb, Li, DW15 (DQ077610) | Actinobacteria | Uncultured actinobacterium clone LiUU-3-85.2 (AY497004) | 98.1 | NA | |
| 22 | GR1, Li, DW11, DW15 (DQ077623) | Alphaproteobacteria | Uncultured alphaproteobacterium clone LiUU-14-208 (AY874027) | 99.5 | “Wolbachia pipientis” (AJ548800) | 83.8 |
| 23b | ER1, ER2, ET (DQ077607) | Alphaproteobacteria | Uncultured alphaproteobacterium clone AKYG933 (AY921781) | 97.8 | Blastochloris sulfoviridis (AY117150) | 94.0 |
| 25 | GT (DQ077617) | Firmicutes | Bacillus sp. strain OS-ac-18 (U46747) | 97.7 | Anoxybacillus flavithermus (AY672763) | 97.7 |
| 27 | ER1 (DQ077620) | Candidate division OD1 | Uncultured candidate division OD1 clone DA10 (AY193190) | 90.3 | NA | |
| 28 | ER1 (DQ077619) | Candidate division OD1 | Uncultured candidate division OD1 clone DA10 (AY193190) | 89.4 | NA | |
| 29b | GT, DW16 (DQ077604) | Bacteroidetes | Uncultured Bacteroidetes clone CS17 (AY956649) | 90.5 | Marinicola seohaensis (AY739663) | 84.4 |
| 31 | DW15 (DQ077627) | Bacteriodetes | Uncultured clone KD4-79 (AY218647) | 96.0 | Flavobacterium ferrugineum (M62798) | 94.2 |
| 32 | DW15 (DQ077624) | Bacteriodetes | Uncultured Bacteroidetes clone AKYG1727 (AY921801) | 97.2 | Flavobacterium ferrugineum (M62798) | 94.0 |
| 35b | GR2, GT, Lb, Li, DW11, DW15 (DQ077625) | Bacteriodetes | Uncultured clone TLM09/TLMdgge12a (AF534433) | 99.2 | Flavobacterium ferrugineum (M62798) | 92.2 |
| 37 | Lb (DQ077621) | Bacteriodetes | Uncultured Bacteroidetes clone Sta2-43 (AY562342) | 98.7 | Arcicella aquatica (AJ535729) | 89.3 |
| 38 | ER2 (DQ077608) | Alphaproteobacteria | Uncultured clone DGGE8.2-CcNSW (AY569382) | 99.5 | Ruegeria algicola (X78315) | 97.7 |
| 39 | GT (DQ077618) | Betaproteobacteria | Betaproteobacterium ASRB1 (AY612302) | 100 | Pseudomonas saccharophila (AF368755) | 100 |
TABLE 3.
Taxonomic identification of the single phylotypes only found in RNA-based SSCP fingerprints shown in Fig. 2b
| Phylotype | Sample(s)a (accession no.) | Taxonomic group | Closest 16S rRNA gene sequenceb (accession no.) | % Similarity | Closest described relativeb (accession no.) | % Similarity |
|---|---|---|---|---|---|---|
| 3 | ET, DW16 (DQ077600) | Acidobacteria | Uncultured clone GOUTB8 (AY050595) | 92.9 | NA | |
| 4 | DW15 (DQ077582) | Acidobacteria | Uncultured acidobacterial clone BSR3SF08 (AY690190) | 93.8 | NA | |
| 11 | ER1, ER2 (DQ077562) | Betaproteobacteria | Uncultured Acidovorax sp. clone SK224 (AY882778) | 96.5 | Comamonas terrigena (AJ430343) | 96.5 |
| 36 | DW15 (DQ077555) | Bacteriodetes | New gram-negative bacterium of CFB group (BD175053) | 98.5 | Riemerella anatipestifer (U60104) | 88.2 |
| 40 | GT (DQ077557) | Betaproteobacteria | Uncultured clone D115 (AY274125) | 86.0 | NA | |
| 41 | GT (DQ077592) | Betaproteobacteria | Nitrosospira briensis (AY123800) | 96.4 | Nitrosospira briensis (AJ298741) | 96.4 |
| 42 | Li, DW15 (DQ077580) | Betaproteobacteria | Nitrosomonas sp. isolate Is79A3 (AJ621026) | 99.2 | Nitrosomonas ureae (AF272414) | 97.2 |
| 43 | GT, Lb, Li, DW16, DW15 (DQ077596) | Betaproteobacteria | Nitrosospira briensis (AY123800) | 100 | Nitrosospira briensis (AJ298741) | 100 |
| 44 | GR1 (DQ077590) | Betaproteobacteria | Uncultured betaproteobacterium clone glen99_30 (AY150869) | 98.8 | Methylovorus mays (AY486132) | 93.4 |
| 45 | ET (DQ077595) | Betaproteobacteria | Uncultured clone TM08 (AY838516) | 100 | Burkholderia phenazinium (U96936) | 100 |
| 46 | GR1, ER1 (DQ077577) | Betaproteobacteria | Uncultured clone HB1 (AY996564) | 98.0 | Rhodoferax ferrireducens (AF435948) | 98.0 |
| 47 | GT (DQ077591) | Acidobacteria | Uncultured clone D14305 (AJ617855) | 95.2 | NA | |
| 48 | ER2 (DQ077598) | Gammaproteobacteria | Uncultured bacterium DSSD31 (AY328730) | 97.3 | Thioalkalispira microaerophila (AF481118) | 91.6 |
| 49 | Lb, DW15 (DQ077602) | Gammaproteobacteria | Uncultured gammaproteobacterium clone Sta2-95 (AY562363) | 99.0 | Methylocaldum gracile (U89298) | 92.4 |
| 50 | GR2 (DQ077588) | Verrucomicrobia | Uncultured clone 300G-H06 (AY661974) | 97.5 | Opitutus terrae (AJ229246) | 93.9 |
| 51 | Li (DQ077567) | Cyanobacteria | Uncultured clone B2C70D7 (AY957901) | 99.0 | Glaucocystis nostochinearumc (X82496) | 79.1 |
| 52 | Lb (DQ077587) | Chimera | NA | NA | ||
| 53 | GR1, GR2 (DQ077566) | Acidobacteria | Uncultured clone WD292 (AJ292589) | 95.7 | NA | |
| 54 | ET (DQ077599) | Alphaproteobacteria | Uncultured Caulobacteraceae clone KMD-0-40 (AY323816) | 100 | Phenylobacterium lituiforme (AY534887) | 97.2 |
| 55 | Li (DQ077572) | Alphaproteobacteria | Uncultured bacterium DSSF66 (AY328688) | 99.7 | Sphingomonas oligophenolica (AB018439) | 97.2 |
| 56 | ER2, ET, Li (DQ077581) | Acidobacteria | Uncultured clone LCSA21 (AJ608337) | 94.9 | NA | |
| 58 | Li (DQ077565) | Nitrospira | Uncultured clone LCo23 (AF337199) | 99.7 | “Nitrospira moscoviensis” (AF155155) | 95.5 |
| 61 | GT, DW16 (DQ077576) | Nitrospira | Uncultured Nitrospira-like DGGE gel band ESR BR 14 (AF540049) | 99.5 | “Nitrospira moscoviensis” (AF155155) | 98.4 |
| 64 | ET (DQ077571) | Planctomycetes | Uncultured Planctomycetes clone DSP21 (AJ290180) | 93.9 | Gemmata obscuriglobus (X81957) | 86.8 |
| 65 | ET (DQ077601) | Planctomycetes | Uncultured Planctomycetes clone DSP56 (AJ290192) | 95.5 | Pirellula marina (X62912) | 84.8 |
| 68 | GT, Li (DQ077597) | Planctomycetes | Uncultured Crater Lake bacterium CL500-3 (AF316767) | 96.4 | NA | |
| 70 | GR1, GR2 (DQ077569) | Bacteriodetes | Uncultured clone BIsi7 (AJ318176) | 92.8 | Pedobacter saltans (AJ438173) | 92.0 |
| 73 | GR1, GR2 (DQ077593) | Bacteriodetes | Uncultured clone TLM10/TLMdgge01 (AF534434) | 93.3 | Sejongia jeonii (AY553294) | 81.8 |
| 76 | GT (DQ077589) | Nitrospira | Uncultured ferromanganous bacterium MNF8 (AF293012) | 93.0 | “Nitrospira moscoviensis” (X82558) | 89.5 |
| 77 | Lb, Li, DW16, DW15 (DQ077559) | Betaproteobacteria | Uncultured bacterium DSSD99 (AY328796) | 98.3 | Acidovorax temperans (AF078766) | 97.3 |
| 78 | DW15 (DQ077558) | Bacteriodetes | Uncultured Bacteroidetes clone PS17 (DQ004377) | 88.0 | Flexibacter flexilis (AJ871243) | 86.9 |
| 79 | ER1 (DQ077568) | Bacteriodetes | Flexibacter aggregans (AB078038) | 89.3 | Flexibacter aggregans (AB078038) | 89.3 |
| 80 | ER1 (DQ077573) | Bacteriodetes | Uncultured bacterium FukuN24 (AJ289995) | 99.2 | Flexibacter sancti (AB078068) | 92.7 |
| 81 | GR1 (DQ077594) | Bacteriodetes | Uncultured clone C11-K8 (AJ421121) | 95.6 | Owenweeksia hongkongensis (AB125062) | 91.1 |
| 82 | GR1, DW16, DW15 (DQ077584) | Bacteriodetes | Uncultured gold mine bacterium D28 (AF337883) | 100 | Owenweeksia hongkongensis (AB125062) | 90.1 |
| 83 | ER1 (DQ077570) | Bacteriodetes | Uncultured bacterium DGGE gel band P17 (AY341117) | 98.4 | Pedobacter heparinus (AJ438172) | 95.4 |
| 84 | ER1 (DQ077575) | Betaproteobacteria | Uncultured betaproteobacterium clone BPC1_D08 (AY689932) | 96.1 | Gallionella ferruginea (L07897) | 95.4 |
| 86 | ER1 (DQ077563) | Gammaproteobacteria | Marinomonas primoryensis (AY771708) | 85.3 | Marinomonas primoryensis (AY771708) | 85.6 |
| 87 | GR1 (DQ077578) | Verrucomicrobia | Uncultured lake bacterium P38.38 (AY752101) | 98.3 | “Xiphinematobacter rivesi” (AF217461) | 90.8 |
| 88 | ER1 (DQ077583) | Acidobacteria | Uncultured forest soil bacterium clone DUNssu179 (AY913385) | 97.9 | NA | |
| 89 | ER1 (DQ077564) | Acidobacteria | Uncultured forest soil bacterium clone DUNssu338 (AY913514) | 96.8 | NA | |
| 90 | DW15 (DQ077585) | Acidobacteria | Uncultured Acidobacterium clone EB1129 (AY395448) | 95.3 | NA | |
| 91 | DW15 (DQ077556) | Cyanobacteria | Uncultured bacterium clone B1NR70D4 (AY957889) | 96.5 | Glaucocystis wittrockianac (X82495) | 83.1 |
| 92 | GR1 (DQ077560) | Alphaproteobacteria | Agricultural soil bacterium clone SC-I-27 (AJ252627) | 96.1 | NA | |
| 93 | DW16 (DQ077586) | Betaproteobacteria | Uncultured betaproteobacterium clone SNT3-94 (AY944210) | 92.2 | Rubrivivax gelatinosus (D16214) | 90.1 |
| 95 | Lb (DQ077561) | Betaproteobacteria | Uncultured bacterium FukuN108 (AJ289984) | 98.2 | Ferribacterium limneticum (Y17060) | 91.9 |
| 96 | DW16 (DQ077574) | Nitrospira | Uncultured Nitrospira-like DGGE gel band ESR BR 14 (AF540049) | 94.7 | “Nitrospira moscoviensis” (AF155155) | 92.7 |
Sample nomenclature is given in Fig. 2a.
NA, not applicable (i.e., the closest described relative has a similarity of <80%, as indicated in Materials and Methods).
Plastid of eukaryotic algae.
There was little overlap between the DNA-based and RNA-based phylotypes (Tables 2 and 3). Only eight phylotypes, footnoted in Table 2, occurred in both types of fingerprints. Fifty-five RNA-based phylotypes (Tables 2 and 3) were found in comparison to 24 DNA-based phylotypes (Table 2), indicating a much higher diversity in the RNA-based fingerprints. This finding was supported by the higher number of bands per sample in the RNA-based fingerprints. Some taxonomic groups, like the Actinobacteria, Firmicutes, and candidate division OD1, were only detected by DNA-based fingerprints, whereas the five phyla mentioned above and the Gammaproteobacteria were only detected by RNA-based fingerprints.
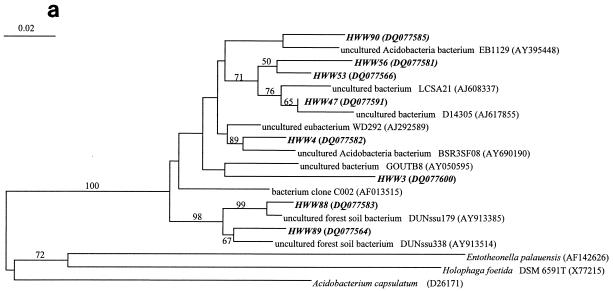
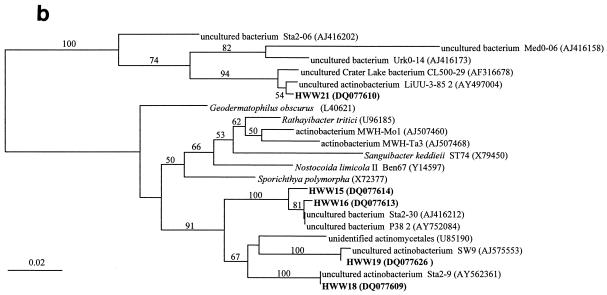
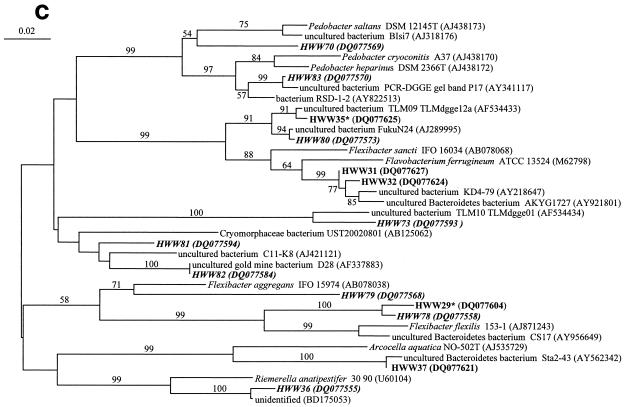
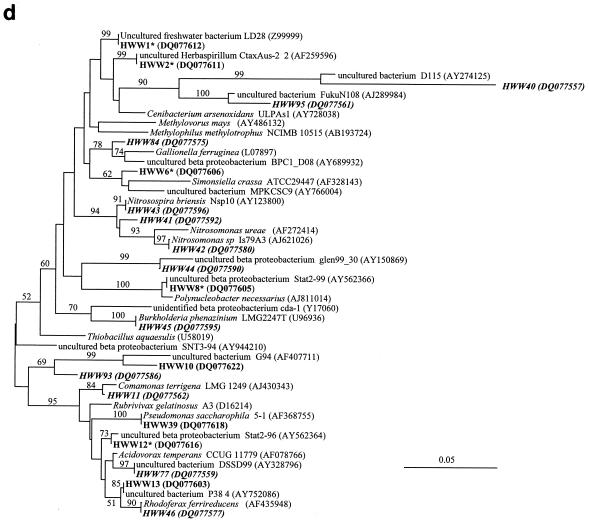
Only 7 of the 71 phylotypes had isolated bacteria or described species as the closest phylogenetic relatives, indicating that the majority of the phylotypes were related to uncultured bacterial taxa (Tables 2 and 3). Detailed phylogenetic analysis demonstrated for the Acidobacteria that all eight phylotypes clustered, despite their broad phylogenetic distribution, among the uncultured acidobacterial sequences very distant from cultured species (Fig. 4a). Similarly, the phylotypes from the Actinobacteria clustered among the uncultured freshwater clusters identified by Zwart et al. (48) away from the freshwater isolates obtained recently by Hahn et al. (11) (Fig. 4b). This distance to cultured species is also indicated by their low 16S rRNA gene sequence similarity, around 90% or below, to cultured species (Table 2). The phylotypes belonging to other major phyla, like Bacteroidetes and Betaproteobacteria, also showed a broad phylogenetic distribution (Fig. 4c and d) but with a closer relationship to cultured species as indicated by their average sequence similarities to cultured bacteria of around 90 and 96%, respectively (Tables 2 and 3). Overall, we could show that more than 90% of the identified phylotypes belonged to uncultured taxa with a broad spectrum of taxonomic groups commonly found in freshwater.
FIG.4.
Comparative sequence analysis of 16S rRNA gene sequences obtained from the bands of the SSCP fingerprints from the samples of the Harzwasser drinking water supply system. They are labeled with HWW (Harzwasserwerke) plus the phylotype number given in Tables 2 and 3. Sequences are coded with different character types according to their origin in terms of nucleic acid type: i.e., DNA-based sequences are shown in bold, RNA-based sequences are shown in bold italic, and sequences occurring in DNA- and RNA-based fingerprints are shown in bold with an asterisk. (a) Phylogenetic tree of the phylum Acidobacteria. (b) Phylogenetic tree of the phylum Actinobacteria. (c) Phylogenetic tree of the phylum Bacteroidetes. (D) Phylogenetic tree of the class Betaproteobacteria.
On the other hand, about 21% of the phylotypes (i.e., 15 phylotypes given in Tables 2 and 3) could be identified to the species level, if we assume the commonly accepted 97% sequence similarity threshold for 16S rRNA gene sequences of the same species. Taxonomic identification is of great relevance for the assessment of human health risk of a specific phylotype detected. For example, most enteropathogenic bacteria, like Salmonella enterica serovar Typhi and Vibrio cholera, belong to a narrow clade within the Gammaproteobacteria. The two gammabacterial phylotypes detected, 48 and 49 (Table 3), were only distantly related (about 92% sequence similarity) to Gammaproteobacteria of genera found only in natural environments. Therefore, a human health threat from these phylotypes is very unlikely. Other phylotypes, belonging to Acidobacteria or freshwater Actinobacteria, are also quite unlikely to be of relevance to human health, because the whole phylum or its environmental part contains no known pathogenic species. One caveat about assessing human health impact from these fingerprint-derived phylotypes is the detection limit of this approach for the single phylotype. It is generally assumed that the detection limit for PCR-based community fingerprints is about 0.1% relative abundance for a single taxon or band. The average total cell count for the drinking water samples was about 3 × 107cells per 100 ml, resulting in a detection limit of 3,000 cells per 100 ml. Considering the current EU requirement for drinking water based on cultivation (i.e., less than 1 E. coli CFU per 100 ml), there is a difference in the sensitivity between both methods of more than 3 orders of magnitude. This renders cultivation-based methods substantially better than community fingerprinting for monitoring the hygienic quality of drinking water.
We compared the occurrence of the major phylogentic groups in the three types of water studied, i.e., Grane reservoir water (samples GR1, GR2, and GT), Ecker reservoir (ER1, ER2, and ET), and drinking water (Li, Lb, DW11, DW12, and DW15) (Fig. 5). The most pronounced difference occurred between the DNA-based and the RNA-based fingerprints (Fig. 5A and B), as detailed above. Mostly all types of water contained the same major groups, except that candidate division OD1 was a marker for Ecker reservoir water and Verrucomicrobia only occurred in the Grane reservoir water. The comparison of the origins of the closest sequences related to the phylotypes (Tables 2 and 3) revealed an additional aspect of the particular contributions of the different types of reservoir waters to the whole microbial community within the distribution system and at the consumer's tap. Taxa belonging to freshwater groups were found in both reservoirs, whereas phylotypes having a closer relationship with soil bacteria originated almost entirely from the dystrophic Ecker reservoir (i.e., phylotypes 23, 45, 83, 88, and 89). These results might indicate a significantly higher introduction of soil material and associated bacteria from the large bog area of the central Harz mountains into the Ecker reservoir compared to the Grane reservoir. These soil-specific phylotypes, especially the two deep-branching acidobacterial phylotypes 88 and 89 (Fig. 4a), could function as taxonomic signatures for the microflora of the Ecker reservoir in RNA-based fingerprints.
FIG. 5.
Comparison of the distribution on the major phylogenetic groups found in the three different types of water studied: i.e., Grane reservoir water (samples GR1, GR2, and GT), Ecker reservoir (ER1, ER2, and ET), and drinking water (Li, Lb, DW11, DW12, and DW15). Open bars, Grane reservoir water; solid bars, Ecker reservoir water; gray bars, drinking water. (A) Phylotypes from the DNA-based SSCP fingerprints. (B) Phylotypes from the RNA-based SSCP fingerprints. Results are based on the phylotypes given in Tables 2 and 3, respectively. There are slightly more phylotypes shown in the bar charts because some of the phylotypes occurred in more than one type of water.
Dynamics of the microbial communities in the DWSS.
The dynamics of the community composition in the DWSS is relatively easy to understand for the DNA-based fingerprints (Fig. 2a). Six phylotypes (1, 16, 18, 21, 22, and 35) from the Grane reservoir water were also observed in the tap water or the DWDS starting with the Lewerberg (Lb) sample. Only two phylotypes, 6, and 8, could have come from the Ecker reservoir water due to corresponding bands in the drinking water fingerprints. The five additional phylotypes found in the drinking water samples (i.e., 10, 26, 31, 32, and 37), were sporadic (26) or could also have come from the Grane water (10, 31, 37) due to their running position on the gel but were not identified by sequencing. Overall, the DNA-based fingerprints of the drinking water at the tap side of the DWSS demonstrated that the drinking water still contained most of the indigenous microflora of the surface reservoirs, with dominance by the Grane reservoir microflora. This finding is in accordance with the cluster analysis of the fingerprints (Fig. 3a), where the Grane raw water samples clustered with about 75% similarity with the drinking water samples. Therefore, structural and compositional dynamics of the microflora showed comparable changes; i.e., the overall community structure and the abundant members showed corresponding dynamics.
The dynamics of the bacterial community in the DWSS was more complex for the RNA-based than for the DNA-based fingerprints due to the higher number of bands per sample and their higher diversity. Only two major phylotypes, 43 and 77, from the chlorinated Grane water sample (GT), were found among the dominating bands of the DWDS (Fig. 2b). Two other phylotypes, 61 and 68, from the GT sample were detected sporadically in the drinking water samples. From the chlorinated Ecker water sample ET, one phylotype, 56, contributed to the drinking water microflora in addition to the two phylotypes 12 and 82 coming from the Grane raw water sample GR1. Five out of the seven phylotypes were detected in chlorinated water and not detected in the raw water before chlorination. Overall, more than half of the phylotypes in the drinking water after chlorination were not detected in the raw water. Thus, the amount of new phylotypes detected after chlorination in the DWDS by RNA-based fingerprints was substantially higher than that detected by the DNA-based fingerprints.
In terms of functional groups, the detection of nitrifying bacteria in the RNA-based fingerprints was important. Betaproteobacterial nitrifiers of the species Nitrosospira briensis (phylotypes 41 and 43) and Nitrosomonas ureae (phylotype 42) and members of the phylum Nitrospira (phylotypes 58, 61, 76, and 96) occurred only after chlorination. More specifically, almost all nitrifiers occurred first in the chlorinated Grane water sample (GT) while no nitrifiers were detected in water from the Ecker reservoir. The ammonia-oxidizing species Nitrosospira briensis (phylotype 43) and the nitrite-oxidizing species “Nitrospira moscoviensis” (phylotypes 61 and 96) were abundant in all drinking water samples and could be responsible for the respective biogeochemical process in the drinking water. Several other studies have reported the occurrence of nitrifying bacteria in drinking water supplies (20, 30, 31).
For an understanding of the dynamics of the community composition based on changes in the banding pattern of SSCP fingerprints, it is necessary to keep in mind that multiple bands of the same phylotype can occur in these fingerprints (34). These multiple bands occur for the following reasons: (i) identical phylotypes can have up to 2% sequence variation according to our threshold value, and these small sequence differences lead to different running distances (see for example phylotypes 18 and 18* in Fig. 2a); and (ii) some amplicons form molecules with a different shape, so-called “conformers,” that have a different running behavior during electrophoresis despite their identical sequences (see, for example, phylotype 16 in Fig. 2a). A special case of multiple bands was the occurrence of very closely related double bands (e.g., phylotypes 1 and 22 in Fig. 2a). These close double bands always had identical sequences and were not labeled twice with a phylotype number.
Insights into bacterial communities by DNA-derived and RNA-derived fingerprints are based on the general assumption that the DNA-derived fingerprints represent the bacterial taxa present in the water sample and the RNA-derived fingerprints represent the active taxa. This general understanding is based on the concept that the number of ribosomes per cell is a good measure of the overall activity of the cell, i.e., its growth rate (33). This concept was exemplified by a variety of bacterial species, but it cannot be assumed for all taxa (6, 15). However, several potential pitfalls have to be taken into account that could skew the assessment of the single phylotypes in the fingerprints: (i) reduced RT-PCR efficiency, (ii) low RNA content of small cells, and (iii) high RNA content of inactive cells. Detection of bacterial groups by DNA-based and not by RNA-based fingerprints could be explained if their cells are known to be very small and contain little RNA. This was reported for freshwater Actinobacteria (37) and could explain our results if we assume, as others have demonstrated (40), that the cDNA bands in the RNA-based fingerprints are a good representation of the RNA content of the respective samples. In addition, a reduced abundance of Actinobacteria has been observed in RNA-derived fingerprints from bacterioplankton of a Finnish lake where Actinobacteria were highly abundant in DNA-derived fingerprints (16). For nitrifying bacteria, it was demonstrated that they contained a high number of ribosomes at a rather low growth rate (41). Therefore, we assume that their relatively high ratio of RNA to DNA let them show up primarily in the RNA-based fingerprints. Most convincing for the assumption that the RNA-derived fingerprints represent an activity indicator was the observation that RNA-derived fingerprints changed strongly after chlorination, including the occurrence of many new phylotypes that had not been detected in the raw waters before. In contrast, the DNA-derived fingerprints behaved rather conservatively and did not show a pronounced community shift. We hypothesize, therefore, that many dead cells present in the drinking water after chlorination are detected by the DNA-based fingerprints. Therefore, RNA-based fingerprints are better suited to assess community shifts in drinking water than DNA-based fingerprints.
Community fingerprints as indicators of stability and functioning of the drinking water microflora.
To our best knowledge, this is the first study comparing the drinking water microflora of the bulk water of a complete DWSS from source to tap using culture-independent molecular techniques. During the last several years, several molecular studies of drinking water biofilms and model systems have been done, including studies done with a metagenome library (4, 35, 46). Our study of the microbial communities in the DWSS of the city of Braunschweig was targeted at bulk water and the understanding of the influence of the two different sources for raw water, the processing of these raw waters to drinking water, and transport along the 40-km-long supply system. The SSCP fingerprints clearly demonstrated that the microflora of the two reservoirs were very different. The first step in the processing of the raw water (i.e., flocculation of the water with aluminum sulfate followed by sand filtration) did not change the structure and composition of the microflora, as indicated by both types of fingerprints. Only the last step of drinking water processing, chlorination, had a significant effect. When the drinking water entered the supply system it was mixed from the waterworks in the first storage container (Lb); therefore, the microflora showed a mixed structure and composition reflecting elements from both raw waters. This mixed structure did not change much during transport to the city of Braunschweig, which takes about 40 h, and remained rather constant for several months.
The origin of the raw water (i.e., the microflora of the two drinking water reservoirs) had great influence on the taxonomic composition of the drinking water microflora, as revealed by the sequencing of the bands from the different reservoir samples. In addition, the large amount of terrestrial taxa in the Ecker reservoir raw water indicated a stronger influence of the drainage basin on the raw water of this reservoir than on the Grane reservoir raw water that was composed of classical freshwater taxa such as Betaproteobacteria, Actinobacteria, and Planctomycetes. A second important effect on the taxonomic composition of the microflora of the DWSS stemmed from the chlorination as detected best by the RNA-based fingerprints. It is well known that chlorination not only kills most bacterial cells but it also produces an increase in assimilable organic carbon (AOC) due to the reaction of free chlorine with the dissolved organic carbon (DOC) present in natural water (29). This AOC consists of a variety of low-molecular-weight compounds that provide substrates for the growth of heterotrophic bacteria (5, 13). Therefore, substantial shifts in the community structure and composition can be expected after chlorination. Overall, the SSCP fingerprints indicated that the source water microflora and the selection process initiated by chlorination shaped the structure and composition of the microflora in the drinking water which is consumed at the tap. We assume that our findings are valid for comparable DWSS in moderate climate and with relative low residence times of the drinking water in the supply system. Elevated temperatures in a hotter climate and compartmented household distribution systems could result in very different microflora in comparison to those in the source water (26).
Key findings of our study were the stability of the drinking water community structure and composition over several months and the increase in activity and diversity of some functional groups like the nitrifiers as indicated by the RNA-based fingerprints. Substantially more phyla and single phylotypes were detected by the RNA-based fingerprints, in addition to the immediate detection of the impact of chlorination on the community structure. Therefore, RNA-based fingerprints could function as universal molecular indicators for the functioning of the drinking water microflora and could help in monitoring the stability of essential steps in the processing of the drinking water such as flocculation and chlorination. Regrowth of undesirable bacteria in the tap water would also be detected by these fingerprints. Such a molecular monitoring of the drinking water microflora could help save maintenance and operational costs for drinking water supply companies. Another advantage of the molecular approach would be that frozen filters with the sampled microflora or extracted nucleic acids can be stored for a long time and are available for later molecular analyses. These samples could provide the basis for a permanent record of the microbial drinking water quality and for later detection and quantification of specific microorganisms or genes of interest (e.g., by using quantitative PCR or DNA chip technology).
Acknowledgments
Support during this study by the Harzwasserwerke Corp. is gratefully acknowledged. The comments and contributions of two anonymous reviewers were greatly appreciated.
This work was supported by funds from the European Commission for the AQUA-CHIP project (QLK4-2000-00764).
The authors are solely responsible for the content of this publication. It does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing herein.
REFERENCES
- 1.Craun, G. F., N. Nwachuku, R. L. Calderon, and M. F. Craun. 2002. Outbreaks in drinking-water systems, 1991-1998. J. Environ. Health 65:16-23. [PubMed] [Google Scholar]
- 2.Dewettinck, T., E. Van Houtte, D. Geenens, K. Van Hege, and W. Verstraete 2001. HACCP (Hazard Analysis and Critical Control Points) to guarantee safe water reuse and drinking water production—a case study. Water Sci. Technol. 43:31-38. [PubMed] [Google Scholar]
- 3.Eichler, S., M. G. Weinbauer, D. Dominik, and M. G. Höfle. 2004. Extraction of total RNA and DNA from bacterioplankton, chapter 1.0.8, p. 103-120. In G. A. Kowalchuk, F. J. D. Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, 2nd ed., Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Emtiazi, F., T. Schwartz, S. M. Marten, P. Krolla-Sidenstein, and U. Obst. 2004. Investigation of natural biofilm formed during the production of drinking water from surface water embankment filtration. Water Res. 38:1197-1206. [DOI] [PubMed] [Google Scholar]
- 5.Escobar, I. C., and A. A. Randall. 2001. Assimilable organic carbon (AOC) and biodegradable dissolved organic carbon (BDOC): complementary measurements. Water Res. 35:4444-4454. [DOI] [PubMed] [Google Scholar]
- 6.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galtier, N., M. Gouy, and C. Gautier. 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 8.Gascuel, O. 1997. BIONJ, an improved version of the NJ algorithm based on a simple method of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 9.Gauthier, V., B. Gerard, J.-M. Portal, J.-C. Block, and D. Gatel. 1999. Organic matter as loose deposits in a drinking water distribution system. Water Res. 33:1014-1026. [Google Scholar]
- 10.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, M. W., H. Lünsdorf, Q. Lu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, J. K., S. T. Kelly, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hem, L. J., and H. Efraimsen. 2001. Assimilable organic carbon in molecular weight fractions of natural organic matter. Water Res. 35:1106-1110. [DOI] [PubMed] [Google Scholar]
- 14.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 16.Kolmonen, E., K. Sivonen, J. Rapala, and K. Haukka. 2004. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in Lake Joutikas, Finland. Aquat. Microb. Ecol. 36:201-211. [Google Scholar]
- 17.LeChevallier, M. W., T. M. Babcock, and R. G. Lee. 1987. Examination and characterization of distribution system biofilms. Appl. Environ. Microbiol. 53:2714-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeChevallier, M. W. 1990. Coliform regrowth in drinking water: a review. J. Am. Water Works Assoc. 82:74-86. [Google Scholar]
- 19.Lee, D.-G., and S.-J. Kim. 2003. Bacterial species in biofilm cultivated from the end of the Seoul water distribution system. J. Appl. Microbiol. 95:317-324. [DOI] [PubMed] [Google Scholar]
- 20.Lipponen, M. T. T., M. H. Suutari, and P. J. Martikainen. 2002. Occurrence of nitrifying bacteria and nitrification in Finnish drinking water distribution systems. Water Res. 36:4319-4329. [DOI] [PubMed] [Google Scholar]
- 21.Liu, W.-T., and D. A. Stahl. 2002. Molecular approaches for the measurement of density, diversity and phylogeny, p. 114-134. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 22.Mallevialle, J., and I. Suffet. 1987. Treatment of tastes and odours in potable water supplies. In AWWA (ed.), Identification and treatment of tastes and odours in drinking water, chapter 7. AWWA Research Foundation Cooperative Research Report no. 211. American Water Works Association, Denver, Colo.
- 23.Niquette, P., P. Servais, and R. Savoir. 2001. Bacterial dynamics in the drinking water distribution system of Brussels. Water Res. 35:675-682. [DOI] [PubMed] [Google Scholar]
- 24.Oliver, J. D. 2000. The public health significance of viable but nonculturable bacteria, p. 277-300. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 25.Olson, B. H., and L. A. Nagy. 1984. Microbiology of potable water. Adv. Appl. Microbiol. 30:73-132. [DOI] [PubMed] [Google Scholar]
- 26.Pepper, I. L., P. Rusin, D. R. Quintanar, C. Haney, K. L. Josephson, and C. P. Gerba. 2004. Tracking the concentration of heterotrophic plate count bacteria from the source to the consumer's tap. Int. J. Food Microbiol. 92:289-295. [DOI] [PubMed] [Google Scholar]
- 27.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 28.Pöhler, I., D. F. Wenderoth, K. Wendt-Potthoff, and M. G. Höfle. 2002. Bacterioplankton community structure and dynamics in enclosures during bioremediation experiments in an acid mining lake. Water, air and soil pollution. Focus 2:111-121. [Google Scholar]
- 29.Polanska, M., K. Huysman, and C. van Keer. 2005. Investigation of assimilable organic carbon (AOC) in Flemish drinking water. Water Res. 39:2259-2266. (First published 31 May 2005; doi: 10.1016/j.watres.2005.04.015.) [DOI] [PubMed] [Google Scholar]
- 30.Regan, J. M., G. W. Harrington, and D. R. Noguera. 2002. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regan, J. M., G. W. Harrington, H. Baribeau, R. D. Leonand, and D. R. Noguera. 2003. Diversity of nitrifying bacteria in full-scale chloraminated distribution systems. Water Res. 37:197-205. [DOI] [PubMed] [Google Scholar]
- 32.Sartory, D. P. 2004. Heterotrophic plate count monitoring of treated drinking water in the UK: a useful operational tool. Int. J. Food Microbiol. 92:297-306. [DOI] [PubMed] [Google Scholar]
- 33.Schaechter, M., O. Maaloe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed] [Google Scholar]
- 34.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effects of primer hybridization to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analysis. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmeiser, C., C. Stöckigt, C. Raasch, J. Wingender, K. N. Timmis, D. F. Wenderoth, H.-C. Flemming, H. Liesegang, R. A. Schmitz, K.-E. Jaeger, and W. R. Streit. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69:7298-7309. [DOI] [PMC free article] [PubMed]
- 36.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Servais, P., P. Laurent, and G. Randon. 1995. Comparison of the bacterial dynamics in various French distribution systems. Aqua 44:10-17. [Google Scholar]
- 39.Szewzyk, U., R. Szewzyk, W. Manz, and K.-H. Schleifer. 2000. Microbiological safety of drinking water. Annu. Rev. Microbiol. 54:81-127. [DOI] [PubMed] [Google Scholar]
- 40.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 42.Ward, N. R., R. L. Wolfe, C. A. Justice, and B. H. Olson. 1986. The identification of gram-negative, nonfermentative bacteria from water: problems and alternative approaches to identification. Adv. Appl. Microbiol. 31:293-365. [DOI] [PubMed] [Google Scholar]
- 43.Weinbauer, M. G., and M. G. Höfle. 2001. Quantification of nucleic acids from aquatic environments by using green fluorescent dyes and microtiter plates, p. 2.1.3/1-2.1.3/10. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 44.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Höfle. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.WHO. 1997. Division of Emerging and Communicable Diseases Surveillance and Control Annual Report—1996. World Health Organization, Geneva, Switzerland.
- 46.Williams, M. M., J. W. S. Domingo, M. C. Meckes, C. A. Kelty, and H. S. Rochon. 2004. Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J. Appl. Microbiol. 96:954-964. [DOI] [PubMed] [Google Scholar]
- 47.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwart G., B. C. Crump, M. P. Kamst-van-Agterveld, F. Hagen and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]