Abstract
Trehalose has many potential applications in biotechnology and the food industry due to its protective effect against environmental stress. Our work explores microbiological production methods based on the capacity of Corynebacterium glutamicum to excrete trehalose. We address here raising trehalose productivity through homologous overexpression of maltooligosyltrehalose synthase and the maltooligosyltrehalose trehalohydrolase genes. In addition, heterologous expression of the UDP-glucose pyrophosphorylase gene from Escherichia coli improved the supply of glycogen. Gene expression effects were tested on enzymatic activities and intracellular glycogen content, as well as on accumulated and excreted trehalose. Overexpression of the treY gene and the treY/treZ synthetic operon significantly increased maltooligosyltrehalose synthase activity, the rate-limiting step, and improved the specific productivity and the final titer of trehalose. Furthermore, a strong decrease was noted in glycogen accumulation. Expression of galU/treY and galU/treYZ synthetic operons showed a partial recovery in the intracellular glycogen levels and a significant improvement in both intra- and extracellular trehalose content.
Trehalose contains two glucose molecules linked by an α-1,1 glycosidic bond. It is a stable, odor-free, and nonreducing disaccharide widely found in nature (27). Its properties as a protein and cell stabilizer make the molecule a promising biotechnological additive for tissue and organ preservation, protein technology, and several food applications.
Trehalose is currently synthesized on an industrial scale by using the method patented by Hayashibara Biochemical Laboratories (17, 18), which is based on direct transformation from maltodextrin using two enzymes obtained from the culture of Rhizobium sp. strain M11 or Arthrobacter sp. strain Q36.
Our approach to production is microbiological and makes use of the fact that Corynebacterium glutamicum a gram-positive bacterium (15), excretes significant amounts of trehalose during batch cultures (29, 32). C. glutamicum has been used extensively for the industrial production of vitamins and numerous l-amino acids for foodstuffs (16).
C. glutamicum uses three pathways in trehalose synthesis (28, 33): the TreS pathway, directly converting maltose into trehalose catalyzed by trehalose synthase (TreS) (5, 21); the OtsAB pathway, wherein trehalose synthesis starts from UDP-glucose and glucose-6-phosphate and is catalyzed in two steps by trehalose-6-phosphate synthase (OtsA), followed by trehalose-6-phosphate phosphatase (OtsB); and the TreYZ pathway, in which enzymatic conversion of the α-glucan polymer into trehalose is catalyzed in two steps by maltooligosyltrehalose synthase (TreY) and maltooligosyltrehalose trehalohydrolase (TreZ), respectively. The regulatory mechanisms of each pathway remain unclear, and just how the three are inter-related is unknown. However, recent studies showed that under hyperosmotic conditions, osmoregulated trehalose synthesis in C. glutamicum is mediated by the TreYZ and not by the OtsAB pathway (28, 33); the physiological role of the OtsAB pathway requires further investigation since it was also shown that it is not essential for mycolic acid synthesis. Finally, TreS apparently compensates for the absence of a classical trehalase by degrading internal trehalose to maltose when present in large amounts.
In previous studies (22, 23), expression of the otsBA operon in C. glutamicum with coexpression of the galU gene-encoded Escherichia coli UDP-glucose pyrophosphorylase produced a significant rise in trehalose synthesis. Increased glycogen supplied during the single expression of galU suggests that the higher trehalose was synthesized through the TreYZ pathway (23). Rather than trying to establish the interrelationship in the two pathways' mechanisms, the aim of the present study was to seek higher trehalose production, making use of the simultaneous expression of C. glutamicum treY and treZ genes in the TreYZ pathway and the E. coli galU gene (11, 23).
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in the present study are listed in Table 1. Luria-Bertani (LB) medium was used as the standard medium for cultivating E. coli DH5α-mcr (9) and the derivative strains. Tryptone soybean broth (TSB) medium was used for the cultivation of C. glutamicum. The culture media defined for C. glutamicum for shake flask experiments (DMCG I) and for batch bioreactor cultivation experiments (DMCG II) were prepared as described previously (7, 22, 23). Antibiotics were added to obtain final concentrations of 50 μg of ampicillin and 20 μg of chloramphenicol/ml, while IPTG (isopropyl-β-d-thiogalactopyranoside) was added to obtain a final concentration of 1.0 mM. IPTG is the inductor that triggers increases in treY, treZ, and galU gene expression that is observable through higher enzymatic activity levels.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genetic characteristicsa | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2,1-TOPO | Ampr | Invitrogen |
| pXMJ-19 | Cfr | 13 |
| pTOPOtreY | treY; Ampr | This study |
| pTOPOtreZ | treZ; Ampr | This study |
| pLPIgalU00 | galU; Ampr | 23 |
| pJCtreY | ptac treY; Cfr | This study |
| pJCtreZ | ptac treZ; Cfr | This study |
| pJCtreYZ | ptac treYZ; Cfr | This study |
| pLPIgalU01 | ptac galU+; Cfr | 23 |
| pJCgalUtreY | ptac galU treY; Cfr | This study |
| pJCgalUtreYZ | ptac galU treYZ; Cfr | This study |
| Strains | ||
| E. coli DH5 α-mcr | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA mcrAΔ(mrr-hsdRMS-mcrBC) | 9 |
| C. glutamicum ATCC 13032 | Wild type | 1 |
| C. glutamicum ΔtreY | ΔtreY derivative of C. glutamicum 13032 | 33 |
| C. glutamicum ΔmluI | ΔmluI derivative of C. glutamicum 13032 | 26 |
| C. glutamicum pJCtreY | treY+; Cfr | This study |
| C. glutamicum pJCtreZ | treZ+; Cfr | This study |
| C. glutamicum pJCtreYZ | treY+treZ+; Cfr | This study |
| C. glutamicum ΔmluI galU01 | ΔmluI galU+; Cfr | 23 |
| C. glutamicum pLPIgalU01 | galU+; Cfr | This study |
| C. glutamicum pJCgalUtreY | galU+treY+; Cfr | This study |
| C. glutamicum pJCgalUtreYZ | galU+treY+ treZ+; Cfr | This study |
Cfr, chloramphenicol resistance; Ampr, ampicillin resistance.
DNA manipulations and bacterial transformation.
DNA cloning steps were performed according to acknowledged procedures (25). E coli DH5α-mcr was transformed by using the method of Inoue et al. (12). C. glutamicum was electroporated according to a protocol published by van der Rest et al. (30).
Plasmid constructs.
The nucleotide sequence accession numbers in the database (http://gib.genes.nig.ac.jp) for C. glutamicum treY and treZ genes are CgI2118 (treY) and CgI2125 (treZ). The following oligodeoxynucleotides for PCR amplification were designed by using Primer Premier Software (Premier Biosoft International, Palo Alto, CA): TREY1, 5′-GTA CCC GGG GAT TTT GCC TAA CTT TTA AGG-3′; TREY2, 5′-TAG CCC GGG TAA CTT TCC TGT GCA G-3′; TREZ1, 5′-CCC GGG TCT CTA GTT CAT GCT CAA AGA C-3′; and TREZ2, 5′-CCC GGG GTA CGA ATA CTT GAC CTA AAC ATA G-3′. Underscores indicate SmaI-cut sites created in both primers for subsequent cloning steps.
The promoterless 2,504-bp fragment that contains the treY gene's coding sequence, harboring the ribosomal binding site, was amplified by PCR from chromosomal C. glutamicum DNA (Promega Corp., Madison, WI). The PCR product was cloned into the pCR2,1-TOPO vector (Invitrogen Corp., Carlsbad, CA). TOPOtreY, the resulting construct, was then transformed into E. coli DH5α-mcr, a DNA methylation mutant (9).
Using the restriction enzyme SmaI, the PCR product was excised from the TOPOtreY plasmid. The excised fragment was filled by using DNA polymerase I (Klenow fragment) and ligated 42 bp downstream of the tac promoter site in the shuttle vector pXMJ19 that had been previously digested with the restriction enzyme SmaI (13). The resulting ligation mixture was used to transform E. coli DH5α-mcr. Transformants were selected according to their chloramphenicol resistance in LB plates. E. coli pJCtreY clones were obtained carrying the treY gene. Plasmid DNA was extracted from these clones and used to electroporate C. glutamicum ATCC 13032 (1). Positive clones were selected on chloramphenicol-LBHIS plates (30), and the construct obtained was tested by restriction analysis. The promoterless 1,992-bp fragment containing the coding sequence of treZ, harboring the ribosomal binding, was amplified by PCR from chromosomal DNA of C. glutamicum. The same ligation procedure was then followed as described above for treY in the pXMJ19 vector to obtain the C. glutamicum strains pJCtreY and pJCtreZ.
Synthetic operons treY/treZ, galU/treY and galU/treYZ were assembled as follows. The treY fragment was excised from the TOPOtreY construct and ligated in the pJCtreZ vector. The latter vector was previously digested with the restriction enzyme XbaI upstream in the treZ gene and filled with DNA polymerase I (Klenow fragment). For the synthetic operon galU/treY, the galU gene was excised from pLPIgalU00 plasmid (22, 23) with the restriction enzyme SmaI and ligated in the pJCtreY vector previously digested with the restriction enzyme XbaI upstream in the treY gene and filled with DNA polymerase I (Klenow fragment). Finally, for the synthetic operon galU/treYZ, the treZ fragment was excised from the TOPOtreZ construct and ligated in the pJCgalUtreY vector—previously digested with the restriction enzyme BsaI downstream in the galU and treY genes and filled with DNA polymerase I (Klenow fragment). Following the procedure detailed above, the resulting ligation mixtures were used to transform E. coli DH5α-mcr. The constructs obtained were tested by restriction and sequence analysis. The resulting strains are summarized in Table 2.
TABLE 2.
Specific activities of maltooligosyltrehalose synthase and UDP-glucose pyrophosphorylase in crude extracts from recombinant C. glutamicuma
| Strain | Induction with 1 mM IPTG | Enzyme | Sp act (pkat mg of protein−1) |
|---|---|---|---|
| C. glutamicum 13032 | + | Maltooligosyltrehalose synthase | 73 |
| C. glutamicum pJCtreY01 | + | Maltooligosyltrehalose synthase | 966 |
| C. glutamicum pJCtreY01 | − | Maltooligosyltrehalose synthase | 80 |
| C. glutamicum pJCtreYZ01 | + | Maltooligosyltrehalose synthase | 950 |
| C. glutamicum pJCtreYZ01 | − | Maltooligosyltrehalose synthase | 95 |
| C. glutamicum pJCgalUtreYZ01 | + | Maltooligosyltrehalose synthase | 655 |
| C. glutamicum pJCgalUtreYZ01 | − | Maltooligosyltrehalose synthase | 101 |
| C. glutamicum 13032 | + | UDP-glucose pyrophosphorylase | 100 |
| C. glutamicum pLPIgalU01 | + | UDP-glucose pyrophosphorylase | 9,000 |
| C. glutamicum pLPIgalU01 | − | UDP-glucose pyrophosphorylase | 200 |
| C. glutamicum pJCgalUtreY01 | + | UDP-glucose pyrophosphorylase | 7,300 |
| C. glutamicum pJCgalUtreY01 | − | UDP-glucose pyrophosphorylase | 200 |
| C. glutamicum pJCgalUtreYZ01 | + | UDP-glucose pyrophosphorylase | 7,000 |
| C. glutamicum pJCgalUtreYZ01 | − | UDP-glucose pyrophosphorylase | 200 |
Each value of tabulated activities represents the mean of three assays, and the standard deviation was always less than 10%. Maltooligosyltrehalose synthase basal activity was corrected on the basis of the value obtained for the extract from C. glutamicum ΔtreY. Extract was prepared from cells grown in shake flasks using DMCG I medium. The cultures were induced with 1 mM IPTG for 6 h after the optical density at 600 nm reached between 0.3 and 0.4.
Cultivation conditions. (i) Shake flasks.
For each induction experiment, 1.5 ml of LB culture grown overnight was inoculated in 30 ml of DMCG I. After 2 h, IPTG was added to a final concentration 1 mM. Samples were taken every 2 h for trehalose, glycogen, and enzymatic analyses.
(ii) Bioreactors.
For batch cultivation, a preinoculum of each strain was made in 100 ml of tryptone soybean broth. After overnight culture in a rotary shaker at 30°C and 250 rpm, the cells were centrifuged and resuspended in 100 ml of DMCG II. The resulting cell suspension was added to a 1-liter Bio-Flo IIc bioreactor (New Brunswick Scientific, Edison, NJ) containing 900 ml of the defined medium. Cultures were run at 30°C and agitated at 700 rpm. The pH was controlled at 7.0 with NaOH 6 M.
Chemical analyses. (i) Extracellular metabolites.
At regular time intervals, 10-ml samples were taken from the culture and immediately centrifuged at 4°C at 1,600 × g for 10 min. The supernatant was collected and subjected to high-pressure liquid chromatography analysis of the extracellular metabolites as described previously (22, 23).
(ii) Intracellular metabolites.
For intracellular metabolite analysis, 2-ml samples were taken from cultures. After collection, the cell pellet was washed twice with 1 ml of 0.25 M Na2CO3 buffer. To determine trehalose, the cell pellet was suspended in 1 ml of methanol and incubated for 20 min at 70°C. Once centrifuged, 500 μl of the supernatant was collected and 10 μl of 1 g of myo-inositol was added/liter as an internal standard. After drying with nitrogen, 35 μl of 20 ng of methoxamine/ml in pyridine was added, and samples were then incubated for 1 h at 30°C. At this point, 65 μl of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) was added, and the resulting mixture was incubated for a further 1.5 h at 65°C. The trehalose concentration in the samples was determined by split injection capillary gas chromatogratography with a GC9000 series gas chromatograph (Fisons Instruments, Mainz, Germany) using a fused-silica capillary column (FS-SE-54-0,25; CS Chromatographie Service, Langerwehe, Germany). Injection and detection devices were kept at 280°C. Separation was achieved by way of a linear temperature gradient from 160°C to 280°C with a heating rate of 12°C/min, starting at 2 min postinjection. The final temperature was kept constant for 3 min for complete elution.
Glycogen was determined by a modified version of the method developed by Parrou and Francois (24), specially adapted for C. glutamicum cells. Cells—obtained from 2 ml of culture medium—were collected by 15 min of centrifugation in a bench centrifuge at 4°C. After the culture medium was carefully drained off, the cell pellet was washed twice with 40 mM potassium acetate (pH 5.2) and resuspended in 0.5 ml of the same buffer in screw-cap Eppendorf tubes. The latter were incubated in an Eppendorf thermomixer for 5 min at 99°C under constant agitation (800 rpm). The suspension was mixed with 300 μl of 0.1-mm glass beads and broken in two 4.5-s cycles in a Mini Bead-beater apparatus (Biospec Products, Inc., Bartlesville, OK). The suspensions were centrifuged at 15,000 rpm for 5 min at 4°C. Once centrifuged, 100 μl of supernatant were incubated with Aspergillus niger amyloglucosidase preparation (1.2 U/ml) in an Eppendorf thermomixer at 57°C with constant agitation. The suspension was centrifuged for 4 min at 4°C in a bench centrifuge, and the glucose level was determined in 30 μl of supernatant by the addition of 1 ml of glucose oxidase mixture (Human, Wiesbaden, Germany), according to supplier instructions.
It is worthy to note that other oligo- and polysaccharides than glycogen might also be included when using this method. However, only glycogen was detected in cell extracts of glucose-grown cells of C. glutamicum after maltooligomer separation by thin-layer chromatography (B. J. Eikmanns, personal communication).
Preparation of cell extracts and enzyme assays.
Harvesting involved centrifuging the cultures 5 h after IPTG addition. Cells were then washed with phosphate-buffered saline (pH 7.4) and resuspended in 1 ml of the same buffer. The suspension was mixed with 300 μl of 0.1-mm glass beads and broken in six 20-s cycles in a Mini Bead-Beater apparatus (Biospec Products). The resulting supernatant was subjected to gel filtration (5) to remove small molecules. The total proteins in the extract were determined by the dye binding assay method (3).
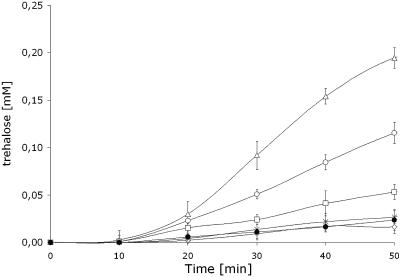
Assay for trehalose biosynthesis by the treYZ pathway has been described elsewhere (5). Briefly, 0.5 mg of protein per ml of the total protein extract (40 μl) was mixed with maltoheptaose (Sigma Chemical Co.), a heptameric oligosaccharide comprising glucose residues linked by α(1→4) glycosidic bonds, at a concentration of 10 mM. The mixture was incubated at 30°C, and samples were taken periodically up to a final time of 50 min (Fig. 1). The reaction was stopped by boiling the samples for 10 min. The resulting supernatant was analyzed by high-pressure liquid chromatography as described above to determine the concentration of trehalose.
FIG. 1.
Trehalose biosynthesis from maltoheptaose in vitro. Trehalose synthesis was monitored by using cell extracts from C. glutamicum pJCtreZ (□), C. glutamicum pJCtreYZ (○), and a mixture of both cell extracts (▵). No significant trehalose synthesis was observed in cell extracts from the wild-type (×), C. glutamicum pJCtreY (•), and C. glutamicum ΔtreY (⋄). Each value represents the mean of three different uptake experiments ± the standard error.
Maltooligosyltrehalose synthase activity (TreY) was determined as follows. A total of 0 to 5 μl of crude extract was incubated in 100 μl of 50 mM sodium phosphate buffer (pH 7.0) containing 45 mM maltopentaose (Sigma Chemical Co.) substrate, a pentameric oligosaccharide comprising glucose residues linked by α(1→4) glycosidic bonds (14). Reaction kinetics were monitored for 10 min at 30°C, and the process was terminated by heating the reaction mixture to 95°C for 5 min. Samples were taken every 2 min. Determination of the substrate consumed was assessed by measuring the decrease in reducing sugars using 3-methyl-2-benzothiazolinonehydrazone (MBTH) (2).
Assay of UDP-glucose pyrophosphorylase (GalU) reaction was carried out according to a previously published protocol (6, 24).
RESULTS
Homologous and heterologous expression of treY, treZ, and galU genes and the synthetic operons treY/treZ, galU/treY, and galU/treYZ.
We first tested in vitro biosynthesis of trehalose from maltoheptaose by using crude extracts prepared from recombinant C. glutamicum cultures (Fig. 1). C. glutamicum pJCtreYZ produced a fivefold increase compared to the control, wild-type strain (C. glutamicum ATCC 13032). When the extract from the treZ-overexpressing strain was mixed with an extract from C. glutamicum pJCtreY, the synthesis of trehalose increased sevenfold, although production was only twice as high as the control strain using the treZ-overexpressing strain extract alone. Trehalose synthesis was not detected in TreY-expressing extracts.
We then analyzed TreY and GalU activity in extracts prepared from induced (1 mM IPTG) and uninduced wild-type and recombinant C. glutamicum strains (Table 2). The treY-overexpressing strain, C. glutamicum pJCtreY, showed a 12-fold increase in TreY activity (966 pkat/mg protein) relative to the uninduced condition (80 pkat/mg protein). TreY activity, in extracts prepared from C. glutamicum pJCtreYZ, increased 10-fold from 95 pkat/mg protein in uninduced cultures to 950 pkat/mg protein after the addition of 1 mM IPTG. Specific activity for TreY in C. glutamicum pJCgalUtreYZ cells was 655 pkat/mg protein.
To confirm that the activity observed in the uninduced strain was indeed caused by endogenous TreY, we also tested the activity in crude extracts of C. glutamicum 13032, which was 73 pkat/mg protein. This result is close to the activity found in the uninduced recombinant strain and demonstrates the functional expression of the encoded treY gene plasmids in the presence of an inductor.
Simultaneously, we measured GalU activity. A cell extract was prepared by using C. glutamicum pLPIgalU01 as a positive control (23). The specific activity of 9,000 pkat/mg protein for the strain pLPIgalU01 was significantly higher than for either its uninduced counterpart or the wild-type strain (specific activity of 200 pkat/mg protein). In induced cultures of recombinant strains, GalU activity was 7,300 pkat/mg protein in crude extracts of C. glutamicum pJCgalUtreY and 7,000 pkat/mg protein from C. glutamicum pJCgalUtreYZ. In uninduced strains, the GalU activity was similar to that of the wild type (Table 2).
Macroscopic response to homologous expression of treY and treZ genes and heterologous expression of galU gene. (i) Effect on internal glycogen and trehalose pools.
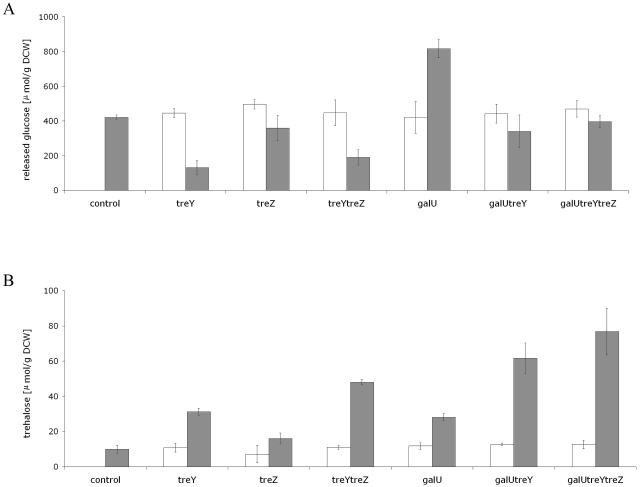
We determined the capacity of the genetically modified strains to accumulate glycogen and trehalose in shake flask experiments (Fig. 2). Measured as released glucose, the glycogen content of all uninduced strains differed little from the wild-type strain. It is noteworthy that the glycogen content in the C. glutamicum strains was much higher than previously reported (23), probably as a consequence of the exhaustive extraction pretreatment used in the present study prior to glycogen determination (see Materials and Methods). The glycogen content decreased in strains induced with the treY gene to a third of the amounts observed in either the uninduced strain or the control. A similar result was noted for the strain overexpressing the synthetic operon treY/treZ, although no significant differences were encountered between the two constructions, treY and treY/treZ (Fig. 2A). Overexpression of the treZ gene resulted in a slight decrease in accumulated internal pools of glycogen. A significant increase in glycogen content was observed from 400 to 840 μmol/g dry cell weight of released glucose in C. glutamicum pLPIgalU01, and this corroborates recent findings (23). When treY or the synthetic operon treY/treZ were coexpressed with the heterologous galU gene, less glycogen was depleted than from the singly expressed strains (Fig. 2A).
FIG. 2.
Effect of treY, treZ, treY/treZ, galU, galU/treY, and galU/treYZ expression on the intracellular content of glycogen (A) and trehalose (B), in the late exponential growth phase in C. glutamicum strains, under IPTG induction (░⃞) and in uninduced states (□). Each uptake value represents the mean for three different uptake experiments ± the standard error.
All the induced recombinant strains produced higher trehalose contents than their uninduced counterparts (Fig. 2B). The results for induced C. glutamicum pJCtreY, C. glutamicum pJCtreYZ, and C. glutamicum pJCtreZ strains are consistent with their concomitant decreases in glycogen levels (Fig. 2A). Intracellular trehalose levels in induced C. glutamicum pJCtreZ strain increased only 1.5-fold in comparison to the control strain, whereas the strains C. glutamicum pJCtreY and C. glutamicum pJCtreYZ accumulated, respectively, 2.5 and 3.5 times more intracellular trehalose than the control strain.
The effect of galU expression on trehalose synthesis through the TreYZ pathway was determined by measuring intracellular trehalose levels in C. glutamicum pJCgalUtreY and C. glutamicum pJCgalUtreYZ (Fig. 2B). The trehalose content of induced cells almost doubled in comparison to the plasmids pJCtreY and pJCtreYZ and exhibited a fourfold increase compared to cells under uninduced conditions and to the wild-type strain. C. glutamicum pJCtreZ was not significantly improved in its capacity to accumulate trehalose and, as expected, the trehalose content of the strain expressing the synthetic operon galU/treZ did not increase significantly (data not shown).
(ii) Effect of treY, treZ, and galU gene expression on kinetic parameters.
Biomass growth rates of galU-induced strains were lower than the other strains. The underlying cause of biomass growth rate inhibition in the presence of the induced galU gene is the focus of ongoing study.
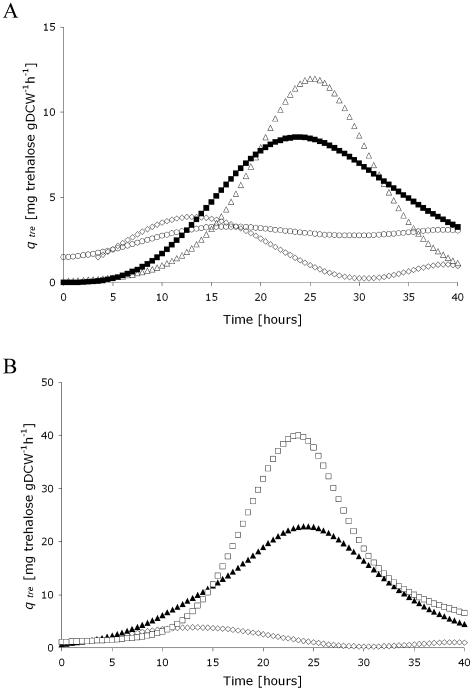
Kinetic profiles for trehalose specific productivities (qtre) were similar for all but one of the recombinant strains: maximum specific productivities were reached during the late exponential growth phase at between 23 and 27 h of cultivation (Fig. 3). Interestingly, the latter coincided with the highest availability of glycogen (data not shown).
FIG. 3.
Fitted curves for the specific productivity of trehalose obtained from batch cultures of C. glutamicum strains in DMCG II supplemented with 1 mM IPTG. (A) ⋄, C. glutamicum 13032; ▪, C. glutamicum pJCtreY; ○, C. glutamicum pJCtreZ; ▵, C. glutamicum pJCtreYZ. (B) ⋄, C. glutamicum 13032; ▴, C. glutamicum pJCgalUtreY; □, C. glutamicum pJCgalUtreYZ.
The two strains harboring the galU gene (Fig. 3B) achieved the highest maximum specific productivities. C. glutamicum pJCgalUtreY reaches a [qtre]max of 24 mg of trehalose g (dry weight)−1 h−1, while the [qtre]max for C. glutamicum pJCgalUtreYZ was even higher at 40 mg of trehalose g (dry weight)−1 h−1, corresponding to a 13-fold increase over the control strain (Table 3). Although the highest specific productivity of trehalose was achieved with C. glutamicum pJCgalUtreYZ, the strain C. glutamicum pJCgalUtreY showed the highest trehalose titer. This difference might result from the reduced growth rate of C. glutamicum pJCgalUtreYZ that was observed after IPTG induction. C. glutamicum pJCtreY and pJCtreYZ strains achieved lower improvements over the control (Fig. 3A).
TABLE 3.
Kinetic parameters of C. glutamicum cultures during exponential and late exponential growth phases
| Strain | Biomass (g liter−1) | μmax (h−1) | Trehalose titer (g liter−1) | qtre (mg gdw−1 h−1)d | Ytre/glu (mmol/mmol of glucose)b | % [Ytre/glu]o |
|---|---|---|---|---|---|---|
| C. glutamicum ATCC 13032 | 27.4 | 0.15 | 1.28 | 3 | 0.007 | 3 |
| C. glutamicum pJCtreY01 | 28.2 | 0.15 | 4.35 | 8 | 0.035 | 14 |
| C. glutamicum pJCtreZ01 | 25.2 | 0.14 | 2.41 | 4 | 0.017 | 7 |
| C. glutamicum pJCtreYZ01 | 26.2 | 0.15 | 5.72 | 12 | 0.043 | 17 |
| C. glutamicum pJCgalUtreY01 | 20.8 | 0.087 | 9.75 | 24 | 0.094 | 38 |
| C. glutamicum pJCgalUtreYZ01 | 18.7 | 0.080 | 7.79 | 40 | 0.080 | 32 |
| C. glutamicum pLPIgalUotsBA01c | 21.0 | 0.096 | 6.8-10a | 22-61a | 0.056 | 22 |
DMCG II medium supplemented with 0.1 mM IPTG.
Experimental trehalose-to-glucose yield, Ytre/glu, over the theoretical yield (with [Ytre/glu ratio]o = 0.25 mol/mol glucose), expressed as a percentage.
Results by Padilla et al. (23) and unpublished data.
gdw, grams dry weight.
All five strains expressing the recombinant genes produced high final trehalose titers (g liter−1). The pJCgalUtreY strain achieved a final trehalose concentration of 9.75 g liter−1, and C. glutamicum pJCgalUtreYZ produced 7.79 g liter−1 (Table 3). Recombinant strains without galU secreted lower amounts of trehalose into the medium, although the levels were still higher than that of the wild type (1.28 g liter−1). Previous results in our laboratory obtained with strains engineered to overexpress the galU/OtsBA genes (23) allowed us to achieve similar final trehalose contents in the extracellular medium, i.e., close to 10 g/liter (Table 3).
The new recombinant strains showed glucose-to-trehalose yields of 0.094 and 0.080 (mol mol−1) for C. glutamicum pJCgalUtreY and C. glutamicum pJCgalUtreYZ, respectively, levels much higher than for the control strain (Table 3). Improvements in trehalose over glucose yields for the recombinant strains, relative to the maximum, theoretical yield, [Ytre/glu]°, were also assessed (Table 3). The latter, i.e., the maximal glucose-to-trehalose conversion yield under the assumption that no substrate is consumed for cell maintenance and that no biomass is produced, is 0.25 mol of trehalose per mol of glucose. Almost 40% of the carbon in the substrate flows toward trehalose biosynthesis in our best-engineered strain, C. glutamicum pJCgalUtreYZ (Table 3).
DISCUSSION
Corynebacterium spp. have three active routes for the biosynthesis of trehalose, i.e., the OtsAB, TreYZ, and TreS pathways. In the present study, we engineered the overexpression of the genes responsible for the TreYZ pathway, in conjunction with the glycogen-related galU gene, to overproduce extracellular trehalose in C. glutamicum.
Glycogen is critical for trehalose synthesis by the TreYZ pathway. This is clearly shown from the significant reduction in glycogen content in the transformants that overexpressed the treY gene, as well as in the one harboring the treY/treZ operon. However, not all of the available glycogen is directed to the TreYZ-mediated pathway for trehalose synthesis; glycogen plays other cellular functions, such as an energy sink, for example, which results in a nonlinear relationship between glycogen and trehalose metabolism. In any case, to maintain a maximal carbon flux toward the TreYZ trehalose biosynthetic pathway, in addition to the overexpression of treY and treZ genes, a permanent and sufficient supply of glycogen is also required. Previous work in our laboratory demonstrated that the heterologous expression of E. coli UDP-glucose pyrophosphorylase, encoded by the galU gene, in C. glutamicum resulted in both higher glycogen levels and higher trehalose levels (23). The significant increase in trehalose production rate and yield obtained here when galU is coexpressed with treY/treZ confirms this result.
The apparent increase in glycogen levels resulting from GalU overexpression can be interpreted either by trehalose stimulation via the OtsAB pathway with concomittant reduction of trehalose formation through the TreYZ pathway, leading to increased glycogen content, or by stimulation not only of the OtsAB pathway but also of the TreYZ pathway via direct stimulation of glycogen synthesis (23). Future work will consider the construction of an ΔotsAB GalU+ transformant of C. glutamicum that might help to define the function of GalU on glycogen synthesis. A third possibility, related to the eventual synthesis of low-molecular-weight malto-oligomers by C. glutamicum cells and their measurement as glycogen, can be excluded, since only glycogen and trehalose are produced in glucose-grown C. glutamicum cells (Eikmanns, personal communication).
Overexpression of heterologous treY gene in C. glutamicum resulted in a 12-fold increase in the TreY enzymatic activity during the exponential growth phase compared to the wild type. On the other hand, the specific enzymatic activity for the GalU-induced strain exhibited 45 times the activity of the uninduced strains or the control. Both constructs were under the same promoter regulation and, accordingly, we expected similar relative increases in the enzymatic activities for both genes; the observed differences may result from differential (i) relative affinities for the specific substrates used in the assays, (ii) enzyme stabilities, and (iii) the crude protein extract's capacity for enzymatic activities of either GalU or TreY. Coexpression of galU and treY, however, resulted in lower specific activities than in the transformants expressing each of the genes individually, which may be due to an imbalance in the translation of the enzymes encoded in the operon (31) or a consequence of a different ribosomal binding efficiency (10).
The enzymatic activity of treZ could not be measured directly in the present study; therefore, the effect of overexpression of the treZ gene alone was evaluated as the in vitro biosynthesis of trehalose from maltoheptaose. The resulting strain prompted a slight increase in trehalose synthesis, although the presence of both overexpressed enzymes in the (treY/treZ) operon were necessary to achieve a substantial rise in the synthesis of trehalose. The fact that treY overexpression alone did not show any significant increase suggests that treZ activity is the rate-limiting step in the in vitro synthesis of trehalose from maltoheptaose. Surprisingly, strains overexpressing treY, treZ, or the treY/treZ operon showed that treY activity was limiting in the synthesis of trehalose in vivo. The differences observed between in vitro and in vivo trehalose synthesis could be attributable to the lack of stability of treY in vitro, since a cofactor may be missing in the extract. An additional cause involves the different specificities of the two substrates used, i.e., maltoheptaose and glycogen.
The increase in carbon flux through the TreYZ pathway in engineered strains significantly increased the glucose-to-trehalose conversion yield, reaching a maximum of 37% of the theoretical Ytre/glu for the C. glutamicum pJCgalUtreYZ strain. It is worth mentioning that similar yields were also obtained when strains carrying the heterologous galUOtsA/OtsB synthetic operon were constructed (22, 23). As a whole, these results suggest that such strategies based on upstream engineering of the trehalose biosynthetic pathways will always reach a maximum. New strategies, relying on strains engineered downstream of the trehalose biosynthetic pathways, particularly those related to trehalose transport and secretion, might lead to improved yields that are closer to the theoretical, maximum, trehalose-over-glucose conversion yields.
Acknowledgments
J.C. received financial support from a doctoral fellowship from the Consejo Nacional de Ciencia y Tecnología de Chile (CONICYT) and the Deutscher Akademischer Austauschdienst (DAAD) of Germany.
We thank Susanne Morbach and Leandro Padilla for critically reading the manuscript and for providing ΔtreY mutants and plasmids pLPIgalU01 and pXMJ19.
REFERENCES
- 1.Abe, S., and K. Takayama. 1972. Amino acid producing microorganisms: variety and classification, p. 3-38. In K. Yamada et al. (ed.), Microbial production of amino acids. Wiley, New York, N.Y.
- 2.Anthon, G. E., and D. M. Barrett. 2002. Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Anal. Biochem. 305:287-289. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cocaign-Bousquet, M., A. Guyonvarach, and N. D. Lindley. 1996. Growth rate-dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose-limited chemostat cultures of Corynebacterium glutamicum. Appl. Environ. Microbiol. 62:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Smet, K. A., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146(Pt. 1):199-208. [DOI] [PubMed] [Google Scholar]
- 6.Degeest, B., and L. de Vuyst. 2000. Correlation of activities of the enzymes alpha-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaunay, S., D. Uy, M. F. Baucher, J. M. Engasser, A. Guyonvarch, and J. L. Goergen. 1999. Importance of phosphoenolpyruvate carboxylase of Corynebacterium glutamicum during the temperature triggered glutamic acid fermentation. Metab. Eng. 1:334-343. [DOI] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershberger, C., E. A. Best, J. Sterner, C. Frye, M. Menke, and E. L. Verderber. 1999. Design and assembly of polycistronic operons in Escherichia coli, p. 539-550. In A. L. Demain and J. E. Davies (ed.), Manual of industrial microbiology, 2nd ed. ASM Press, Washington, D.C.
- 11.Hossain, S. A., K. Tanizawa, Y. Kazuta, and T. Fukui. 1994. Overproduction and characterization of recombinant UDP-glucose pyrophosphorylase from Escherichia coli K-12. J. Biochem. 115:965-972. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Jakoby, M., C.-E. Ngouoto-Nkili, and A. Burkovski. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Techniques 13:437-441. [Google Scholar]
- 14.Kim, Y. H., T. K. Kwon, S. Park, H. S. Seo, J. J. Cheong, C. H. Kim, J. K. Kim, J. S. Lee, and Y. D. Choi. 2000. Trehalose synthesis by sequential reactions of recombinant maltooligosyltrehalose synthase and maltooligosyltrehalose trehalohydrolase from Brevibacterium helvolum. Appl. Environ. Microbiol. 66:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on the amino acid fermentation. I. Production of l-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 16.Leuchtenberger, W. 1996. Amino acids: technical production and use, p. 465-502. In H.-J. Rehm and G. Reed (ed.), Biotechnology: products of primary metabolism 6. VCH-Verlag, Weinheim, Germany.
- 17.Maruta, K., M. Kubota, T. Sugimoto, and T. Miyake. 1995. Enzyme capable of liberating trehalose, its production and use. Hayashibara Biochemical Laboratories, Inc. Japanese patent JP07-213283.
- 18.Maruta, K., M. Kubota, T. Sugimoto, and T. Miyake. 1995. Non-reducing glucide-producing enzyme, its production and use. Hayashibara Biochemical Laboratories, Inc. Japanese patent JP07-143876.
- 19.Reference deleted.
- 20.Reference deleted.
- 21.Nishimoto, T., H. Chaen, T. Sugimoto, and T. Miyake. June 1998. Trehalose and its production and use. U.S. patent 5,759,610.
- 22.Padilla, L., R. Kramer, G. Stephanopoulos, and E. Agosin. 2004. Overproduction of trehalose: heterologous expression of Escherichia coli trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:370-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padilla, L., S. Morbach, R. Kramer, and E. Agosin. 2004. Impact of heterologous expression of Escherichia coli UDP-glucose pyrophosphorylase on trehalose and glycogen synthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:3845-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrou, J. L., and J. Francois. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248:186-188. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Schafer, A., A. Tauch, N. Droste, A. Puhler, and J. Kalinowski. 1997. The Corynebacterium glutamicum cglIM gene encoding a 5-cytosine methyltransferase enzyme confers a specific DNA methylation pattern in an McrBC-deficient Escherichia coli strain. Gene 203:95-101. [DOI] [PubMed] [Google Scholar]
- 27.Schiraldi, C., I. Di Lernia, and M. De Rosa. 2002. Trehalose production: exploiting novel approaches. Trends Biotechnol. 20:420-425. [DOI] [PubMed] [Google Scholar]
- 28.Tzvetkov, M., C. Klopprogge, O. Zelder, and W. Liebl. 2003. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology 149:1659-1673. [DOI] [PubMed] [Google Scholar]
- 29.Vallino, J. J., and G. Stephanopoulos. 2000. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 67:872-885. [Reprint of Biotechnol. Bioeng. 41:633-646, 1993.] [PubMed] [Google Scholar]
- 30.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 31.Watson, J. D., N. H. Hopkins, J. W. Roberts, J. A. Steitz, and A. M. Weiner. 1987. Molecular biology of the gene, 4th ed. Benjamin Cummings Publishing Company, Inc., Menlo Park, Calif.
- 32.Wittmann, C., and E. Heinzle. 2001. MALDI-TOF MS for quantification of substrates and products in cultivations of Corynebacterium glutamicum. Biotechnol. Bioeng. 72:642-647. [PubMed] [Google Scholar]
- 33.Wolf, A., R. Kramer, and S. Morbach. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC 13032 and their significance in response to osmotic stress. Mol. Microbiol. 49:1119-1134. [DOI] [PubMed] [Google Scholar]