Abstract
The successive generation times for single cells of Escherichia coli K-12 were measured as described by A. Elfwing, Y. LeMarc, J. Baranyi, and A. Ballagi (Appl. Environ. Microbiol. 70:675-678, 2004), and the histograms they generated were used as empirical distributions to simulate growth of the population as the result of the multiplication of its single cells. This way, a stochastic birth model in which the underlying distributions were measured experimentally was simulated. To validate the model, analogous bacterial growth curves were generated by the use of different inoculum levels. The agreement with the simulation was very good, proving that the growth of the population can be predicted accurately if the distribution of the first few division times for the single cells within that population is known. Two questions were investigated by the simulation. (i) To what extent can we say that the distribution of the detection time, i.e., the time by which a single-cell-generated subpopulation reaches a detectable level, can be identified with that of the lag time of the original single cell? (ii) For low inocula, how does the inoculum size affect the lag time of the population?
The growth of a population is determined by the multiplication of the cells within that population. In predictive microbiology, the most frequently used mathematical models describe the growth of the population as a whole, without considering the individual cells. Recently, the connection between the generation times of individual cells and the growth of the population as a whole has raised much interest. It is especially important to investigate this connection at low cell concentrations at the early stage of growth, because (due to the exponential nature of bacterial proliferation) exceptionally fast-growing cells and their subsequent subpopulations have a dominant effect on the growth of the total culture. It is understandable, therefore, that the focus of attention has turned to the distribution of single-cell lag times and to the techniques that can measure them (7, 9, 11, 18, 23, 24).
Though the lag time of a growing bacterial population is a commonly used term in predictive microbiology, the relationship between the population lag time and the distribution of single-cell lag times is not straightforward. Even if ideal conditions are assumed, it is practically impossible to identify the distribution of the division times of single cells from the population growth (3) for simple numerical reasons. However, determining population lag from the distribution of the division times of single cells is not a problem. Therefore, models for the growth of single cells allow descriptions of bacterial growth at a deeper level.
Here, we will model the multiplication of cells by a stochastic birth process based on a set of random variables whose realizations are time dependent. Stochastic models are used to describe those processes that are not completely understood or are too complicated to be described deterministically (13). In our case, the first division times for a sufficiently large number of cells could be measured, but the complexity and the lack of knowledge about the intracellular processes during the lag were the main reasons for turning to stochastic models. The question to be answered is how accurately can we predict the growth of the bacterial culture, including the lag and the exponential phase, after we observe the first few division times of some single cells?
It is not easy to observe the division of single cells in such a way that statistically significant distributions can be derived from the observations. Elfwing et al. (7) described a novel automated method that enabled the user to observe the division times of a large number of individual cells during a relatively long interval. Using this technique together with viable-count growth curves measured by traditional plating, single-cell and population growth parameters were measured simultaneously. The measured distributions for the successive division intervals of the single cells were used to model the growth of the population as a stochastic process. The measured growth of the cell culture and that predicted by simulation were compared. The very good agreement justified the use of the simulation to study (i) the relationship between detection times and single-cell lag times and (ii) the effect of the inoculum size on the population lag time.
Detection times (measured by spectrophotometry) of single-cell-generated cultures are commonly used to estimate the lag times of single cells. The use of detection times is based on the assumption that, if the specific growth rate of the population and the detection level are known, the distribution of detection times can be identified with that of the lag times of the original single cells (8, 9, 11, 18, 23). However, this relationship could be seriously compromised by the variability of the first few generation times, when the cells are not necessarily in the exponential phase yet (15, 21). Our simulations, based on data produced by the flow chamber, will make it possible to study this question.
The effect of the inoculum size on the population lag times has also been studied by many microbiologists, who have arrived at mixed conclusions (6, 10, 17, 24). The effect of the inoculum size on the duration of the lag time has, so far, been reported for situations when either the inoculum was very low (1) or, because of stressing conditions, only a small proportion of the population was able to grow (1, 20, 22). The approach developed here could reliably model the population lag time with very low inoculum sizes.
MATERIALS AND METHODS
Bacterial strain and inoculum preparation.
Escherichia coli K-12 was maintained at −80°C. Immediately before the experiments, the cells were revitalized by subculturing twice consecutively in LB medium (10 g of tryptone/liter, 5 g of yeast extract/liter, 10 g of NaCl/liter [pH 7.2]) with 0.2% glucose at 25°C for 48 h. These cells were inoculated simultaneously in the flow chamber, to observe the divisions of single cells, and in broth, to measure population growth.
Single-cell growth measurements.
The flow chamber was set up as described by Elfwing et al. (7). The system was initialized by flushing through sterile phosphate-buffered saline (0.2 g of KCl/liter, 0.2 g of KH2PO4/liter, 1.15 g of Na2PO4/liter, 8 g of NaCl/liter [pH 7.3]) at 0.7 ml/min, generating an average linear velocity of approximately 1.1 cm/s. One hundred microliters of the culture was inoculated onto the system. The flow was stopped for 15 min to allow cells to attach to the slide. Growth was initiated by flushing fresh medium (LB medium plus glucose) though the flow chamber at 0.7 ml/min. This flow was sufficient to feed the bacteria and remove one of the newborn daughter cells without removing significant numbers of cells already attached. The temperature in the flow chamber was ca. 25°C for the experiment with relatively long lag times and ca. 32°C for the experiment with relatively short lag times. The temperature was controlled by immersing both LB medium and phosphate-buffered saline in a water bath and measured by a thermocouple applied to the flow chamber outlet.
The cells were observed by dark-field microscopy (Zeiss Standard 25; Oberkochen, Germany) with a 10× objective (Zeiss, Oberkochen, Germany). A high resolution (1,040- by 1,392-pixel) charge-coupled-device camera (CoolSnap Pro cf; Roper Scientific, Trenton, NJ) was controlled by an image analysis program (Image Pro Plus; Media Cybernetics, Silver Spring, MD) that captured an image every 5 min. From the images, the sizes (in pixels) of the individual cells at the different times were recorded. The time of division was identified by the time a sudden drop in pixel size occurred; this sudden drop was attributed to the fact that, right after cell division, one daughter cell was removed by the flow of the fresh medium, while the other remained. The division times were recorded by using an in-house program written in Visual Basic. The time to the first division was assumed to consist of both the lag time and the first generation time. The subsequent generation times were defined as the time intervals between two successive divisions: ith generation time = time to the ith division − time to the (i − 1)th division (i > 1).
Population growth measurements.
Two batches of seven (0.5-liter) bottles containing 100 ml of LB medium were inoculated with different inoculum sizes (from 0.4 to 106 CFU/ml). One batch was incubated in a shaking water bath at 25°C and the other at 32°C. At each sampling time, bacterial counts were estimated by plating samples onto tryptone soy agar (Oxoid CM131).
Random number generation.
Random numbers had to be generated following the empirical distributions of the single-cell division/generation times measured in the flow chamber, as follows.
First, a histogram of the measured empirical distribution was created. The length of the bins in the histogram was estimated as indicated in reference 12. The length of the bins = L = 1.5 × (Q75 − Q25) × n−1/3, where Q75 is the third quartile or the value with the position 3 × (n + 1)/4 in the sorted vector of measurements (X1 … Xn), Q25 is the first quartile or the value with the position (n + 1)/4 in the sorted vector of measurements, and n is the number of measurements.
Intervals were constructed starting from the median. The relative frequency for each interval is estimated as ni/n, where ni is the number of observations inside the ith interval.
Next, to generate random values from the histograms, two uniformly distributed random variables were generated: u1 between X1 and Xn and u2 between 0 and the greatest relative frequency. The value u1 is accepted as a value randomly generated from the empirical distribution only if 0 < fi < u2, with fi being the associated relative frequency of the interval to which u1 belongs.
Stochastic modeling of the population growth.
The distribution of the first division time of the first k generation times measured by image analysis for a single cells, are used to model the growth of the population. The value of k is usually around 3 or 4.
The assumptions made for this purpose are as follows. (i) The first division is the sum of the lag time and the first generation time. (ii) After the kth division, the daughter cells are in exponential growth phase, having the same distribution for their generation times as for the last (kth) observed one. (iii) The distributions of the second and successive generation times (up to the kth) are those measured by image analysis.
With the usual terminology for stochastic processes, the states of the system are represented as (Y1, Y2, … , Yk), where Y1 is the number of cells that have not divided yet, Y2 is the number of cells at second generation time, and Yk is the number of cells at kth generation time. The initial state of the system, at the starting time, t = t0, is (N, 0, … , 0), meaning that the N initial cells of the population have not started to divide yet.
To the N initial cells, the first division times are assigned by generating randomly N values, (F1… FN), following the empirical distribution of the first division time. The time t1 (the time at which the state of the system first changes) is that of the cell with the shortest division time: t1 = min(F1… FN).
At t = t1, N − 1 cells have not divided yet, while the two daughter cells enter their second generation time. Thus, the state of the system is (N − 1, 2, … , 0). The total number of cells is N − 1 + 2 = N + 1.
Two values, SG1 and SG2, are generated randomly from the empirical distribution of the second generation times for the two daughter cells. The next division time for these cells will be t1 + SG1 and t1 + SG2.
Now there are two possibilities: the time t2 of the second division in the population will be the minimum time to divide among the N − 1 cells that have not divided yet (case 1), or the time t2 will be the division times of the two newborn cells (case 2): t2 = min(F1, … FN − 1, t1 + SG1, t1 + SG2).
In case 1, if t2 is the first division of a cell, then the state of the system will be (N − 2, 4, … , 0). The next division time of each newborn cell is the sum of t2 and a value randomly generated from the second generation time distribution. The time of the third division in the population will be t3 = min(F1, … FN − 2, t1 + SG1, t1 + SG2, t2 + SG3, t2 + SG4).
In case 2, if a cell from the second generation divides first, then the system goes into the state (N − 1, 1, 2, … , 0). The two newborn cells are at the third generation time. Two values will then be obtained following the third generation time distribution (TG1 and TG2). The time to the third division in the population will be t3 = min(F1, … FN − 1, t1 + SG1, t2 + TG1, t2 + TG2).
Continuing with case 2, the third division could happen to a cell that has not divided yet (case 2.1), a cell at second generation time (case 2.2), or a cell at third generation time (case 2.3).
In case 2.1, if the cell divides for its first time, the state of the system will be (N − 2, 3, 2, … , 0). Two values are randomly obtained from the second generation time distribution for the newborn cells, and the time of the fourth division in the population will be t4 = min(F1, … FN − 2, t1 + SG1, t2 + TG1, t2 + TG2, t3 + SG3, t3 + SG3).
In case 2.2, if the cell dividing is in its second generation time, the state of the system will be (N − 1, 0, 4, … , 0). To assign to the newborn cells the time for the next division, two values are randomly obtained from the third generation time distribution. The time of the fourth division in the population will be t4 = min(F1, … FN − 2, t2 + TG1, t2 + TG2, t3 + TG3, t3 + TG3).
In case 2.3, if the cell that divided was in its third generation time, the system goes into the state (N − 1, 1, 1, 2, … , 0). Two values are randomly obtained from the fourth generation time distribution for the newborn cells. If the fourth and/or the successive generation time distributions could not be measured, random values are generated from the third or last measured generation time distribution. The time of the fourth division in the population will be t4 = min(F1, … FN − 1, t1 + SG1, t2 + TG1, t3 + FG1, t3 + FG2).
The iteration can be continued in a similar manner until the population reaches a given number of cells. The system converges to the state (0, … , NT), in which the total number of cells, NT, are in exponential phase and their generation times follow the kth generation time distribution.
Simulations.
The algorithm above was implemented in an in-house Excel add-in program written in Visual Basic. To validate the stochastic model, 100 simulated growth curves were generated for each experimentally measured growth curve.
To compare the distribution of the detection time with the distribution of the first division time, 100 growth curves were generated, starting from a single cell
To study the effect of the inoculum size on the lag time, 100 growth curves were generated for each of the inoculum sizes (1, 2, 4, 8, 16, and 100 cells).
Estimation of the parameters of population growth.
The maximum specific growth rate and the lag time were estimated by fitting the model of Baranyi and Roberts (5) to the growth curves.
Comparison of distributions.
The distributions of division times and the intervals between successive divisions were compared by χ2 tests.
RESULTS
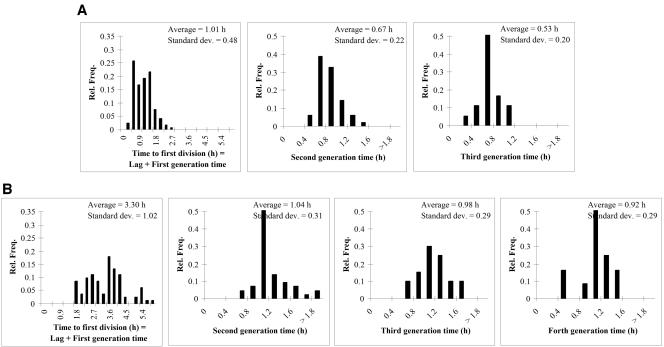
Figure 1 shows the distributions of the first division time (i.e., the sum of the lag time and the first generation time) and the successive generation times of the cells observed in the flow chamber. At 25°C, the division intervals were longer than at 32°C. The generation times decreased gradually in the successive divisions after the lag time. The standard deviations of the division intervals in general decreased in proportion to the averages.
FIG. 1.
Distributions of the single-cell first division intervals measured with a flow chamber (7) by image analysis at 32°C (A) and 25°C (B). Rel. Freq., relative frequency; Standard dev., standard deviation.
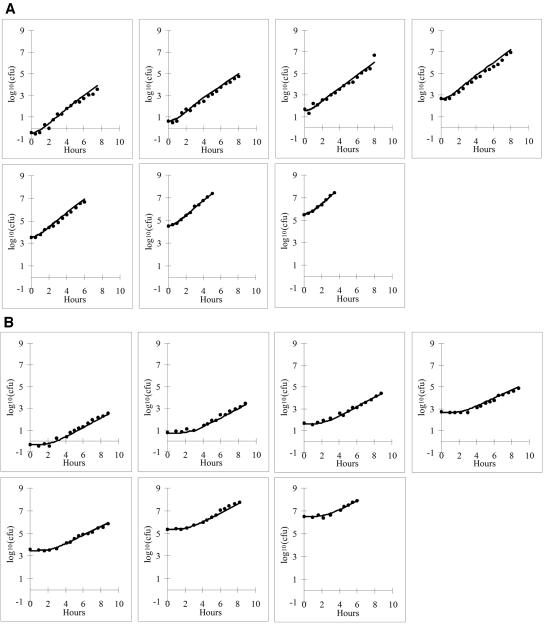
Data in Table 1 show the lag times and growth rates estimated from the measured and simulated growth curves with different inoculum sizes. The growth of the population was simulated by modeling individually the intervals between successive divisions for the cells. These times were assumed to follow the distributions shown by the cells in the flow chamber. The number of cells to start the simulation was the total number of cells inoculated per flask. One hundred growth curves were simulated for each inoculum size. Table 1 shows the averages and standard deviations of the estimated growth parameters from the 100 simulated growth curves together with the growth parameters and their standard errors, estimated by fitting the measured data. Figure 2 shows the average simulated growth curve for each inoculum. The agreement between the simulated and measured growth curves justifies the further use of the simulation approach for questions which are difficult to investigate experimentally.
TABLE 1.
Lag time, maximum specific growth rates, μmax, and doubling times estimated from the measured and simulated growth curves with different inoculum sizes by fitting the model of Baranyi and Roberts (5)
| Temp (°C) | Inoculum cells/100 ml | Result
|
|||||
|---|---|---|---|---|---|---|---|
| Measured
|
Simulatedd
|
||||||
| Lag time ± SE (h)a | μmax ± SE (h−1) | DT (h)b | Lag time ± SD (h)c | μmax ± SD (h−1) | DT (h) | ||
| 32 | 3.98E + 01 | 0.68 ± 0.31 | 1.48 ± 0.11 | 0.47 | 0.66 ± 0.084 | 1.56 ± 0.015 | 0.44 |
| 32 | 3.98E + 02 | 0.44 ± 0.37 | 1.31 ± 0.08 | 0.53 | 0.73 ± 0.06 | 1.61 ± 0.013 | 0.43 |
| 32 | 5.01E + 03 | 0.51 ± 0.46 | 1.37 ± 0.07 | 0.51 | 0.7 ± 0.012 | 1.6 ± 0.016 | 0.43 |
| 32 | 4.47E + 04 | 0.80 ± 0.14 | 1.36 ± 0.03 | 0.51 | 0.64 ± 0.011 | 1.59 ± 0.013 | 0.44 |
| 32 | 3.47E + 05 | 0.58 ± 0.14 | 1.36 ± 0.04 | 0.51 | 0.7 ± 0.011 | 1.6 ± 0.011 | 0.43 |
| 32 | 3.39E + 06 | 0.72 ± 0.10 | 1.51 ± 0.04 | 0.46 | 0.7 ± 0.009 | 1.61 ± 0.0092 | 0.43 |
| 32 | 3.16E + 07 | 0.61 ± 0.11 | 1.51 ± 0.07 | 0.46 | 0.64 ± 0.011 | 1.51 ± 0.0096 | 0.46 |
| 25 | 4.47E + 01 | 2.94 ± 0.51 | 1.03 ± 0.05 | 0.67 | 2.44 ± 0.161 | 1.03 ± 0.010 | 0.67 |
| 25 | 5.62E + 02 | 2.74 ± 0.28 | 1.00 ± 0.06 | 0.69 | 2.7 ± 0.093 | 1.00 ± 0.006 | 0.69 |
| 25 | 4.68E + 03 | 2.18 ± 0.35 | 0.94 ± 0.06 | 0.74 | 2.47 ± 0.027 | 1.02 ± 0.0042 | 0.68 |
| 25 | 5.50E + 04 | 2.86 ± 0.28 | 0.89 ± 0.05 | 0.78 | 2.54 ± 0.0171 | 0.94 ± 0.0014 | 0.74 |
| 25 | 3.80E + 05 | 2.28 ± 0.33 | 0.85 ± 0.05 | 0.82 | 2.56 ± 0.0160 | 0.89 ± 0.0012 | 0.78 |
| 25 | 2.40E + 07 | 2.58 ± 0.26 | 1.02 ± 0.06 | 0.68 | 2.36 ± 0.0133 | 0.96 ± 0.0010 | 0.72 |
| 25 | 3.24E + 08 | 2.67 ± 0.31 | 1.01 ± 0.11 | 0.69 | 2.29 ± 0.0112 | 0.83 ± 0.0011 | 0.84 |
Estimation and standard error of the growth parameters calculated by fitting the growth curves.
DT, doubling time.
Average and standard deviation of the growth parameters fitted to 100 simulated growth curves.
One hundred curves per inoculum.
FIG. 2.
Growth curves measured by viable counts (dots) and generated by simulation (continuous lines). The simulation was based on the distributions of the generation times of single cells measured by the image analysis-aided flow chamber technique. The agreement is very good at both 32 °C (A) and 25°C (B) for different inoculum sizes.
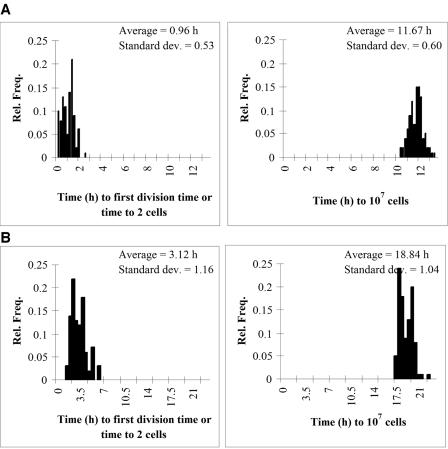
We used this simulation technique to investigate the effect of the variation of the division times on the distribution of the time needed by a single-cell-generated subpopulation to reach a given level (such as 107, representing the lower detection limit of optical density methods). The purpose was to study whether the distribution of those detection times is suitable to identify the distribution of the lag times of the initial single cells. The first division times and the times needed to reach 107 cells were recorded from 100 single-cell-generated simulated growth curves. The simulations were based on single-cell measurements at 25°C and 32°C. Figure 3 shows the distributions for the first division times and for the times needed to reach 107 cells. They were not significantly different from each other or from the distribution of the first division times observed in the flow chamber (P > 0.05).
FIG. 3.
Distributions of the first division times (left panels) and the times to reach 107 cells (right panels) measured from simulated populations growing from one single cell. Simulations are based on the distributions for the single-cell growth parameters measured experimentally in the flow chamber at 32°C (A) and 25°C (B). Rel. Freq., relative frequency; Standard dev., standard deviation.
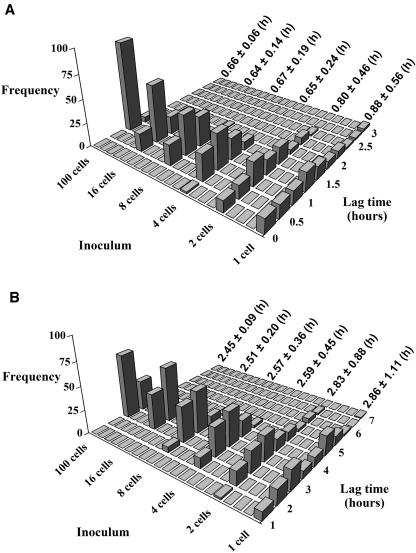
An inoculum effect on the lag time was not apparent from the measured or the simulated growth curve. The reason was that the initial number of cells was >40 cells per total volume (100 ml) (Fig. 2; Table 1). Experimentally, it is very difficult to count levels lower than 0.4 cell/ml by traditional microbiological techniques. However, the above-mentioned agreement between simulation and observation justified the use of simulation, and so we then used the latter technique to study the inoculum effect on the lag time generated by there being less than 40 initial cells. One hundred growth curves were produced for each of the inoculum sizes (1, 2, 4, 8, 16, and 100 cells). The model of Baranyi and Roberts (5) was fitted to the simulated growth curves, and the average growth parameters and their standard deviation were calculated for each inoculum size. These values, together with the histograms of the distributions of the population lag time obtained for each inoculum size, can be seen in Fig. 4. The inoculum size did affect the distribution of the lag time at this low inoculum size. Growth curves initiated with few cells showed longer lag times than those initiated with more cells. Also, the variance of the lag time was greater at small inoculum sizes.
FIG. 4.
Distributions of the lag times estimated from 100 growth curves simulated for each inoculum size. Average values and standard deviations are shown next to each distribution. Growth curves were simulated with the single-cell measurements obtained at 32 °C (A) and 25°C (B) and fitted by the model of Baranyi and Roberts (5).
DISCUSSION
When flow chamber data were used, the average lag time for the single cells (calculated as the difference between the average first division time and the mean generation time) were around 0.5 and 2.4 h at 25°C and 32°C, respectively (Fig. 1). The lag times fitted to the parallel viable-count growth curves were ca. 0.6 and 2.6 h (Table 1). This seems to contradict the results reported in references 2 and 4, which stated that the average single-cell lag time is longer than the lag time of the whole population. However, those theoretical results (2, 4) were based on the assumption that, after the lag, there is no difference between the second, third, and further generations. Recent works suggested that, in fact, the intervals between successive divisions of E. coli decrease gradually and asymptotically after the first division (16, 19, 21). We also observed that successive generation times showed decreasing average values (Fig. 1). By taking into account the measured generation time distributions until the fourth division, our simulation followed the gradual decrease of the generation times; this is why it agrees so well with the viable-count growth curves. The second, third, and maybe even the fourth generation times are still longer than those in the fastest (exponential) phase later, which causes a further delay for the growth of the population. Indeed, when using our simulation approach based only on the distribution of the first division time and the distribution of the last measured generation time, i.e., assuming that cells grow at their maximum potential rate after the first division, the lag parameters derived from the simulated growth curves were ca. 0.45 and 2.1 h. These time values were in good agreement with the results of the approach used in reference 4 when fitting the gamma distribution to the single-cell lag times, 0.40 and 2.0 h, but were shorter than the real population lag times (Table 1). Thus, the population growth curve was accurately predicted only when the model took into account the fact that the cells within that population do not multiply at their maximum potential rate immediately after the first division.
Recently, it has been found that the daughter cells inheriting the preexisting cellular extremes or old poles grow slower than those with newly formed poles (25). Stewart et al. reported that the successive generation times for cells inheriting the old pole increase but that they decrease for cells with newly formed poles. In our work, the division times were measured in cells that always inherited the old pole by which they were attached onto the flow chamber. However, the growth of these cells did not slow down in successive generation times but instead accelerated. This apparent contradiction can be explained by the different growth phases of the cells. While Stewart et al. (25) worked with cells growing exponentially, here the cells were in lag phase. As mentioned above, it has already been demonstrated that, after lag phase, the cells do not divide immediately at their maximum potential rate but reach it progressively (16, 19, 21). Hence, the aging process can be observed only after the cells reach this rate. Another explanation could be that the continuous supply of fresh medium in the flow chamber delayed the aging process described in reference 25.
Single-cell studies are often carried out by means of optical density methods (8, 9, 11, 18, 23) where approximately 1 cell per well is inoculated into a multiwell plate, and the turbidity is measured by an automated plate reader. From the detection time, it is a common practice to derive the single-cell lag time by a simple shift calculated from the population growth rate and the detection level. Some concerns have been raised (15, 21) about the perturbation of the relationship between lag and detection times by the variability of the first generation times, which are not even homogeneous. Here, we demonstrated that in the case of the single cells used in our work, the variance of the detection times would be equal to that of the first division times.
We did not see an inoculum effect on the lag time in the experimental growth curves, where the lowest inoculum used was ca. 40 cells per total volume (100 ml). For inoculum sizes of less than 40 cells, the first division and successive generation times measured by the flow chamber were used to simulate the growth of the population. The results showed that an inoculum effect on the lag time can be noticed only at very low inoculum sizes. However, this conclusion should be treated with caution, because the threshold number of cells to detect an inoculum effect on the duration of lag varies according to the distribution of the single-cell lag times. Indeed, differences in the lag time duration have been reported for greater inoculum sizes, but those cells were severely stressed by starvation (1, 10) or heating (24) or the growth conditions were very close to the growth-no growth boundary (20, 22).
The influence of the inoculum size on the population lag can be derived mainly from the distribution of the single-cell lag time. The population lag is shorter with a greater inoculum size. Any physiological activity during the lag time, such as the secretion of growth inducers (14), could contribute to the inoculum size effect, but that is out of the scope of this paper.
The successive divisions of each cell within a population were randomly generated from the histograms constructed by observing the division times of single cells in the flow chamber. We also tested a method in which gamma distributions were first fitted to those measured histograms, and then the single-cell division times were generated in the simulation to follow the fitted gamma distributions. As expected, the results with the two methods were similar. Hence, the developed stochastic process provides an accurate prediction of the growth of the bacterial population, regardless of whether an empirical or a theoretical distribution function is used to generate the single-cell division times. It is the lag time and the first few generation times of the original single cells which are of major importance for predicting the time by which low numbers of pathogenic bacteria grow above a dangerous level. Our modeling approach can therefore significantly improve predictive tools for quantitative microbial risk assessment purposes.
Acknowledgments
This paper was prepared under the funding of the EU Programme Quality of Life and Management of Living Resources, project no. QLK1-CT-2001-01145 (BACANOVA).
We thank Anders Elfwing and András Ballagi for help with the flow chamber setup and Susie George for help with the experiments.
REFERENCES
- 1.Augustin, J. C., A. Brouillaud-Delattre, L. Rosso, and V. Carlier. 2000. Significance of inoculum size in the lag time of Listeria monocytogenes. Appl. Environ. Microbiol. 66:1706-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranyi, J. 1998. Comparison of stochastic and deterministic concepts of bacterial lag. J. Theor. Biol. 192:403-408. [DOI] [PubMed] [Google Scholar]
- 3.Baranyi, J. 2002. Stochastic modelling of bacterial lag phase. Int. J. Food Microbiol. 73:203-206. [DOI] [PubMed] [Google Scholar]
- 4.Baranyi, J., and C. Pin. 2001. A parallel study on bacterial growth and inactivation. J. Theor. Biol. 210:327-336. [DOI] [PubMed] [Google Scholar]
- 5.Baranyi, J., and T. A. Roberts. 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23:277-294. [DOI] [PubMed] [Google Scholar]
- 6.Duffy, G., J. J. Sheridan, R. L. Buchanan, D. A. McDowell, and I. S. Blair. 1994. The effect of aeration, initial inoculum and meat microflora on the growth kinetics of Listeria monocytogenes in selective enrichment broths. Food Microbiol. 11:429-438. [Google Scholar]
- 7.Elfwing, A., Y. LeMarc, J. Baranyi, and A. Ballagi. 2004. Observing growth and division of large numbers of individual bacteria by image analysis. Appl. Environ. Microbiol. 70:675-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francois, K., F. Devlieghere, K. Smet, A. R. Standaert, A. H. Geeraerd, J. F. Van Impe, and J. Debevere. 2005. Modelling the individual cell lag phase: effect of temperature and pH on the individual cell lag distribution of Listeria monocytogenes. Int. J. Food Microbiol. 100:41-53. [DOI] [PubMed] [Google Scholar]
- 9.Francois, K., F. Devlieghere, A. R. Standaert, A. H. Geeraerd, J. F. Van Impe, and J. Debevere. 2003. Modelling the individual cell lag phase. Isolating single cells: protocol development. Lett. Appl. Microbiol. 37:26-30. [DOI] [PubMed] [Google Scholar]
- 10.Gay, M., O. Cerf, and K. R. Davey. 1996. Significance of pre-incubation temperature and inoculum concentration on subsequent growth of Listeria monocytogenes at 14 degrees C. J. Appl. Bacteriol. 81:433-438. [DOI] [PubMed] [Google Scholar]
- 11.Guillier, L., P. Pardon, and J. C. Augustin. 2005. Influence of stress on individual lag time distributions of Listeria monocytogenes. Appl. Environ. Microbiol. 71:2940-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izenman, A. J. 1991. Recent developments in nonparametric density estimation. J. Am. Statist. Assoc. 86:205-224. [Google Scholar]
- 13.Jaynes, E. T. 2003. Probability theory. The logic of science. Cambridge University Press, Cambridge, United Kingdom.
- 14.Kaprelyants, A. S., and D. B. Kell. 1996. Do bacteria need to communicate with each other for growth? Trends Microbiol. 4:237-242. [DOI] [PubMed] [Google Scholar]
- 15.Kutalik, Z., M. Razaz, and J. Baranyi. 2004. Connection between stochastic and deterministic modelling of microbial growth. J. Theor. Biol. 232:283-297. [DOI] [PubMed] [Google Scholar]
- 16.Kutalik, Z., M. Razaz, A. Elfwing, A. Ballagi, and J. Baranyi. 2005. Stochastic modelling of individual cell growth using flow chamber microscopy images. Int. J. Food Microbiol. 105:177-190. [DOI] [PubMed] [Google Scholar]
- 17.Mackey, B. M., and A. L. Kerridge. 1988. The effect of incubation temperature and inoculum size on growth of salmonellae in minced beef. Int. J. Food Microbiol. 6:57-65. [DOI] [PubMed] [Google Scholar]
- 18.Metris, A., S. M. George, M. W. Peck, and J. Baranyi. 2003. Distribution of turbidity detection times produced by single cell-generated bacterial populations. J. Microbiol. Methods 55:821-827. [DOI] [PubMed] [Google Scholar]
- 19.Metris, A., Y. Le Marc, A. Elfwing, A. Ballagi, and J. Baranyi. 2005. Modelling the variability of lag times and the first generation times of single cells of E. coli. Int. J. Food Microbiol. 100:13-19. [DOI] [PubMed] [Google Scholar]
- 20.Pascual, C., T. P. Robinson, M. J. Ocio, O. O. Aboaba, and B. M. Mackey. 2001. The effect of inoculum size and sublethal injury on the ability of Listeria monocytogenes to initiate growth under suboptimal conditions. Lett. Appl. Microbiol. 33:357-361. [DOI] [PubMed] [Google Scholar]
- 21.Pin, C., and J. Baranyi. 2004. Distribution of the lag times of individual cells as a function of the age of the cells in the inoculum. In P. Raspor, S. Smole Mozina, and A. Cencic (ed.), Food micro 2004: new tools for improving microbial food safety and quality. Conference proceedings. Slovenian Microbiology Society, Ljubljana, Slovenia.
- 22.Robinson, T. P., O. O. Aboaba, A. Kaloti, M. J. Ocio, J. Baranyi, and B. M. Mackey. 2001. The effect of inoculum size on the lag phase of Listeria monocytogenes. Int. J. Food Microbiol. 70:163-173. [DOI] [PubMed] [Google Scholar]
- 23.Standaert, A. R., A. H. Geeraerd, K. Bernaerts, K. Francois, F. Devlieghere, J. Debevere, and J. F. Van Impe. 2005. Obtaining single cells: analysis and evaluation of an experimental protocol by means of a simulation model. Int. J. Food Microbiol. 100:55-66. [DOI] [PubMed] [Google Scholar]
- 24.Stephens, P. J., J. A. Joynson, K. W. Davies, R. Holbrook, H. M. Lappin-Scott, and T. J. Humphrey. 1997. The use of an automated growth analyser to measure recovery times of single heat-injured Salmonella cells. J. Appl. Microbiol. 83:445-455. [DOI] [PubMed] [Google Scholar]
- 25.Stewart, E. J., R. Madden, G. Paul, and F. Taddei. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]