Abstract
Aryl-hydroxylating dioxygenases are of interest for the degradation of persistant aromatic pollutants, such as polychlorobiphenyls (PCBs), or as catalysts for the functionalization of aromatic scaffolds. In order to achieve dioxygenation of technical mixtures of PCBs, enzymes with broadened or altered substrate ranges are essential. To alter the substrate specificity of the biphenyl dioxygenase (BphA) of Burkholderia xenovorans LB400, we applied a directed evolution approach that used structure-function relationship data to target random mutageneses to specific segments of the enzyme. The limitation of random amino acid (AA) substitutions to regions that are critical for substrate binding and the exclusion of AA exchanges from positions that are essential for catalytic activity yielded enzyme variants of interest at comparatively high frequencies. After only a single mutagenic cycle, 10 beneficial variants were detected in a library of fewer than 1,000 active enzymes. Compared to the parental BphA, they showed between 5- and 200-fold increased turnover of chlorinated biphenyls, with substituent patterns that rendered them largely recalcitrant to attack by BphA-LB400. Determination of their sequences identified AAs that prevent the acceptance of specific PCBs by the wild-type enzyme, such as Pro334 and Phe384. The results suggest prime targets for subsequent cycles of BphA modification. Correlations with a three-dimensional model of the enzyme indicated that most of the exchanges with major influence on substrate turnover do not involve pocket-lining residues and had not been predictable through structural modeling.
Aryl-hydroxylating dioxygenases catalyze the addition of two hydroxy groups to vicinal carbons of their substrates, thereby destroying the aromatic system and yielding dihydrodiol compounds of cis,cis stereochemistry (Fig. 1) (4, 6). These enzymes are of increasing interest for the degradation of aromatic pollutants (6, 10) as well as for the regio- and stereospecific functionalization of aromatic scaffolds in general (4, 16).
FIG. 1.
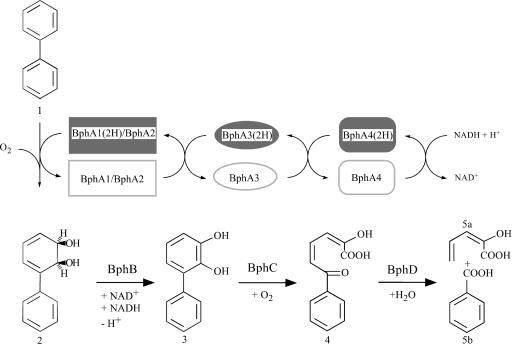
Reaction catalyzed by biphenyl-hydroxylating dioxygenases and the three subsequent steps of aerobic bacterial biphenyl catabolism. Enzymes: BphA1 and BphA2, alpha and beta subunits of biphenyl dioxygenase; BphA3, ferredoxin; BphA4, ferredoxin reductase; BphB, biphenyl-2,3-dihydrodiol 2,3-dehydrogenase; BphC, 2,3-dihydroxybiphenyl 1,2-dioxygenase; and BphD, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase. Compounds: 1, biphenyl; 2, biphenyl-2,3-dihydrodiol; 3, 2,3-dihydroxybiphenyl; 4, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid; 5a, 2-hydroxypenta-2,4-dienoic acid; and 5b, benzoic acid.
The biphenyl dioxygenases (BphAs) are members of this family of enzymes. They have attracted attention as biocatalysts for the removal of polychlorobiphenyls. Their use in this field is hampered by the problem that pollutions typically consists of “technical” polychlorobiphenyls that are mixtures of dozens of compounds, so-called congeners. So far, only a fraction of them can be attacked by known BphAs. Therefore, enzymes with broadened and/or altered substrate ranges are required. These may be obtained either by more extensive and sophisticated screens of the natural resources (17) or by the generation of novel substrate specificities through evolution in the laboratory. In this work, we applied the latter approach.
Given the persisting problem of correctly predicting the properties of designed protein variants, strategies employing random modifications are of increasing importance. Such methods of “directed evolution” have been applied before to improve substrate turnover by BphAs. Most of these approaches were based on some sort of DNA shuffling between the alpha subunit genes of two closely related enzymes (5, 20, 36). In one report (37), random priming recombination was used. Variants accepting novel substrates were detected; however, frequencies at which they were obtained have not been given. After DNA shuffling or related approaches, typically some 104 clones have to be screened to detect variants with novel substrate or product specificities (13, 29).
Obviously, strategies that increase the probability of generating variants of interest are desirable. To this end, we describe an approach that made use of structure-function relationship data to narrow down the overall number of amino acids (AAs) to be exchanged. Previous work of different groups (3, 7, 9, 18, 19, 24-26, 40, 41) has indicated that three particular regions within the alpha subunit play a crucial role in substrate specificity. Therefore, random mutageneses were simultaneously targeted to the respective gene segments. Oligonucleotides were used as mutagenic agents, permitting a precise choice of the positions to be modified as well as of the rate and bias of mutagenesis.
The BphA of Burkholderia xenovorans LB400 (formerly Burkholderia sp.) is one of the most powerful biocatalysts for the dioxygenation of chlorobiphenyls (CBs) (23, 33). However, several di- and trichlorinated biphenyls are poorly transformed by this enzyme. A selection of five of these congeners, namely, 3,3′-, 4,4′-, 2,6,4′-, 3,4,4′-, and 3,5,4′-CB, was used to assess whether, and at which frequencies, beneficial variants of BphA-LB400 could be obtained with the mutagenesis approach outlined above. This not only yielded a number of enzyme derivatives accepting most of these biphenyls but also identified AA substitutions that are crucial for changes in substrate specificity.
MATERIALS AND METHODS
Chemicals including oligonucleotides.
Chemicals were of the highest purity available. CBs of >99% purity were supplied by Promochem (Wesel, Germany). The sequences of all oligonucleotides used are given in Table 1. Primers BPH1883 and PCRD-REV were purchased from Invitrogen Life Technologies (Karlsruhe, Germany). Primers PCRA-REV, PCRB-REV, PCRC-REV, OMS MUT1, OMSMUT2, and OMSMUT3 were obtained from TIBMolBiol (Berlin, Germany). The latter three were degenerate oligonucleotides. During their syntheses, 96.5% of wild-type (WT) and 1.5% of each non-WT nucleotide (NT) precursor were used at each degenerate position. These primers were purified via anion exchange fast-performance liquid chromatography.
TABLE 1.
Sequences of oligonucleotides used
| Oligonucleotide | Sequence (5′ to 3′)a |
|---|---|
| BPH1883 | GGCGACTGCGGCTTTGAC |
| OMSMUT1 | GAAGTGGGTGATTCCGTGCAACTGGAAGTTTGCCGCCGAGCAGTTCTGCAGTGACATGTACCACGCCGGCACCACGACGCACCTGTCCGGCATCCTGGC |
| OMSMUT2 | GGTCGGCCAGCATATGACGATCTTCCCGACCTGTTCATTCCTGCCCACCTTCAACAACATCCGGATCTGGCACCC |
| OMSMUT3 | ATATCGCCGGCACAACATCCGCAACTTCTCCGCAGGCGGCGTGTTTGAGCAGGACGATGGCGAGAACTGGGTGGAGATC |
| PCRA-REV | CACCCACTTCTGCATGCCGCC |
| PCRB-REV | CTGGCCGACCATGCGTCGAAC |
| PCRC-REV | CCGGCGATATTCTTCCTTGA |
| PCRD-REV | CACCATGTTGAATTCTTCCGG |
Degenerate positions are underlined. For details, see the text.
Bacterial strains, plasmids, and culture conditions.
Strains used were Escherichia coli DH10B (12) and BL21(DE3)pLysS (34). Competent cells of these strains were obtained from Invitrogen Life Technologies. Plasmids pAIA6000, harboring genes bphA1A2A3A4BC of Burkholderia xenovorans LB400 (11), and pAIA15, containing gene bphC of B. xenovorans LB400, have been described previously (33, 40). E. coli DH10B harboring pAIA6000 or derivatives (described below) was routinely grown at 37°C in LB medium (28) with 100 μg/ml of ampicillin. In microtiter plates, CTY medium (5) was used. E. coli BL21(DE3)pLysS harboring pAIA6000 or derivatives was routinely grown at 30°C in LB or ZY medium (35) with 25 μg/ml of chloramphenicol and 100 μg/ml of ampicillin.
General DNA techniques.
In vitro DNA modifications, agarose gel electrophoresis, and transformations were carried out according to standard protocols (28) unless described below in detail.
Assembly and cloning of partial mutant bphA1 segments.
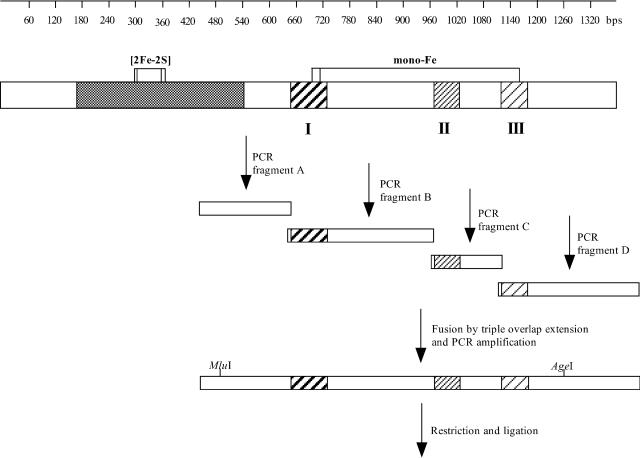
Three libraries of mutant DNA fragments were generated by standard PCRs (27) with primer pairs OMSMUT1 and PCRBREV, OMSMUT2 and PCRCREV, or OMSMUT3 and PCRD-REV using pAIA6000 as a template (Figure 2). A WT DNA fragment was synthesized by PCR with the same template using primers BPH1883 and PCRA-REV (Figure 2). This fragment and the three library fragments overlapped each other. After purification by agarose gel electrophoresis, they were fused in a single overlap extension PCR (14). The resulting fragment library, containing all three mutagenized regions, was purified with a PCR purification kit (QIAGEN, Hilden, Germany), cleaved with AgeI and MluI, purified again with the QIAGEN kit, and ligated with identically cleaved pAIA6000. Transformants of strain DH10B were selected on LB-ampicillin.
FIG. 2.
Scheme of assembly of partial mutant bphA1 genes. A linear representation of the entire bphA1 gene is shown at the top. Segments encoding the catalytic or Rieske domain (7, 18), are indicated by white or patterned backgrounds, respectively. Horizontally connected vertical bars indicate sites encoding AA ligands of the Rieske iron-sulfur cluster ([2Fe-2S]) and of the active-site mononuclear iron (mono-Fe) (7, 18). Hatched areas within the catalytic domain highlight regions (I to III) subjected to random mutagenesis.
Deletion of genes bphB and bphC.
Selected mutagenized plasmids of the library were cleaved with BspEI and recircularized. This deleted almost the entire bphB and bphC genes.
Construction of a plasmid expressing gene bphD.
The multiple cloning site of expression vector pT7-6 (39) was extended by cleavage with SalI and insertion in sense orientation of a chemically synthesized adapter containing restriction sites for NcoI, AflII, KpnI, XbaI, SpeI, and NotI to yield vector pT7-601. This plasmid was linearized with NcoI, and the NcoI fragment of pDD5301 (8) containing the bphD gene of strain LB400 was inserted in sense orientation to yield pAIA51.
DNA sequencing.
About 1 μg of plasmid DNA was subjected to Taq DNA polymerase-catalyzed cycle sequencing as described previously (2).
Screening for enzymatic activity.
BphA activity of clones was assessed by placing solid biphenyl into the lids of petri dishes and incubating them at 30°C for up to 48 h. As the clones also harbored and expressed genes bphB and bphC, appearance of the yellow metabolite 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HOPDA) indicated BphA activity.
Screening for CB turnover.
In 96-well microtiter plates, 250 μl of CTY-ampicillin medium was inoculated with active clones and incubated for 20 h at 37°C. Subsequently, 5 μl of 10 mM stock solutions of 3,3′-, 4,4′-, 2,6,4′-, 3,4,4′-, or 3,5,4′-CB was added, and the incubation was continued at 30°C for up to 48 h. Increases in yellow color were taken as indications of productive CB dioxygenation.
Preparation of resting cells.
E. coli BL21(DE3)pLysS harboring the appropriate plasmid was grown at 30°C in ZY or LB medium with antibiotics to an optical density at 600 nm of about 1.0. Subsequently, IPTG (isopropyl-β-d-galactopyranoside) was added to a 0.4 mM final concentration, and the incubation was continued for 60 min. Cells were harvested, washed with 50 mM sodium phosphate buffer (pH 7.5), and resuspended in the same buffer supplemented with 0.5% (wt/vol) glucose to a final optical density at 600 nm of 2.0.
Protein gel electrophoresis.
Resting cell preparations were concentrated 25-fold, disrupted with a French press (40), and centrifuged for 30 min at 65,000 × g. Of the supernatants, 1.75 μl was mixed with the same volume of 2× cracking buffer (39), and the proteins were separated by 0.1% sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (15). Gels were stained with Coomassie blue (28).
CB incubations with resting cells.
Single CBs were dispensed into Teflon-sealed glass tubes. After evaporation of the solvent, resting cells harboring the appropriate plasmids were added. After vortexing for 20 s, the Teflon-sealed tubes were shaken at 30°C for 20 h. Typically, aliquots of 1 ml were withdrawn, cells were pelleted for 5 min at 13,000 rpm in an Eppendorf centrifuge, and supernatants were subjected to different types of analyses (described below). Cells harboring the cloning vector served as background controls. Experiment-specific details are given in the following sections.
Quantitation of substrate turnover via measurement of HOPDA absorption.
Resting cells harboring pAIA6000 or a mutant derivative were shaken with 50 μM of CB. Electronic spectra of the supernatants were recorded, and absorptions at the maxima were determined. The detection limit was around an absorbance of 0.005.
Quantitation of substrate turnover via high-performance liquid chromatography (HPLC) of dioxygenation products.
Resting cells harboring WT or mutant plasmids devoid of genes bphB and bphC (construction described above) were incubated with 250 μM of CB. Supernatants were analyzed on an RP8 column as previously described (40). The aqueous eluent contained 720 ml of methanol and 1 ml of 85% ortho-phosphoric acid per liter.
Assay of HOPDA formation in the presence or absence of BphB.
Resting cells harboring WT or mutant bphA genes were mixed with equal volumes of cells containing either genes bphBC (pDD372) or only gene bphC (pAIA15) and were incubated with 50 μM of CB. HOPDA formation was monitored as described above.
Assay of chlorobenzoate formation.
Equal volumes of resting cells harboring pAIA6000 or a mutant derivative and of cells harboring pAIA51 (construction described above) were mixed and incubated with 125 μM of CB. Chlorobenzoate (CBA) formation was analyzed in comparison with authentic standards by HPLC of supernatants as previously described (31).
Characterization of dioxygenation products by gas chromatography-mass spectrometry (GC-MS).
Resting cells synthesizing only WT or variant BphA were incubated with 250 μM of CB. Supernatants were extracted with ethyl acetate, and dioxygenation products were converted into butylboronate derivatives (33). The mixtures were then evaporated to dryness under a stream of nitrogen and dissolved in 0.1 volume of n-hexane. Samples (0.01 volume) were injected in the splitless mode (300°C injector temperature) into a Thermo Finnigan GCQ ion trap mass spectrometer (Finnigan MAT Corp., San Jose, CA) running in the positive-ion electron impact mode and equipped with a 30-m DB5 capillary column. The temperature program was as follows: 1 min at 80°C, followed by an increase of 10°C/min to 300°C. Helium served as the carrier gas.
RESULTS
Mutagenesis of the target gene.
Mutations were generated in three segments (I to III) of the BphA alpha subunit gene bphA1 from B. xenovorans LB400 (Fig. 2). Mutagenesis was carried out by the incorporation of degenerate oligonucleotides (Table 1) via normal and overlap extension PCRs (Fig. 2) (14, 27). Within the mutagenic region of the oligonucleotides, specific codons were kept nondegenerate in order to fully conserve certain AAs that are likely to be essential for enzyme activity. Such residues were identified on the basis of their high conservation among homologous sequences and with the aid of a structural model of BphA-LB400 (41). This led to the targeting of random NT changes to 141 positions altogether, representing approximately 10% of the total gene length. At positions to be mutagenized, the oligonucleotides had been designed to contain each mutagenic NT at a frequency of 1.5%. The fused end product of the PCRs was used to replace the respective WT fragment in a plasmid that harbored genes bphA1A2A3A4 of the BphA system and, for screening purposes, also genes bphBC. The latter genes encode the two subsequent enzymes of the pathway for biphenyl utilization (Fig. 1), which convert dioxygenation products into ring-cleaved metabolites, HOPDAs, that absorb light in the visible region. This type of screen implies that the detected dioxygenations are productive in the sense that they are further metabolized through the biphenyl catabolic pathway.
Characterization of the mutant bank by activity screening and DNA sequencing.
The bank of the resulting clones was screened on agar plates for BphA activity by exposure to biphenyl vapor. The percentage of active colonies, identified by conversion of biphenyl into the yellow ring fission product, was about 2%.
Active, inactive, and randomly chosen clones were analyzed by DNA sequencing. This showed that about 75% of the inactive clones contained frameshifts. It also revealed that these were mostly derived from the mutagenic oligonucleotides. Clones that carried frameshifts were categorized as false inactives. Thus, about 23% of all clones were true inactives, and the ratio of true inactives to actives was about 10:1.
The observed average frequency of primer-induced mutations in random clones was 5.45 (Table 2), which represents 86% of the theoretically expected value of 6.35. Of these, 75.2% (4.10 per clone) were nonsilent. This is close to the theoretically expected value of 77.8%. In active clones, the rate of nonsilent mutations was significantly reduced (1.70 per clone), while the rate of silent mutations was virtually unchanged (1.42 per clone). This is consistent with the observation that the rate of mutagenesis applied to the target regions led predominantly to inactive dioxygenases.
TABLE 2.
Frequencies of intentional mutations in different types of clones
| Clone typea | No. of mutations per clone
|
||
|---|---|---|---|
| Nonsilent | Silent | Total | |
| Random | 4.94b | 1.41b | 6.35b |
| Random (20) | 4.10 | 1.35 | 5.45 |
| Active (43) | 1.70 | 1.42 | 3.12 |
| CB selected (10) | 2.00 | 1.50 | 3.50 |
Numbers of sequenced clones are given in parentheses.
As calculated for the specific sequence and degree of degeneracy of the mutagenic oligonucleotides (theoretical value).
Identification of clones with novel substrate specificities and quantitation of CB turnover.
About 660 active clones were screened for improved turnover of five CBs that were not or were inefficiently attacked by the WT BphA, namely, 3,3′-, 4,4′-, 2,6,4′-, 3,4,4′-, and 3,5,4′-CB. The screen identified 11 positive clones. These preliminary results were confirmed in 10 cases. Improved dioxygenation was detected for all congeners except 3,3′-CB.
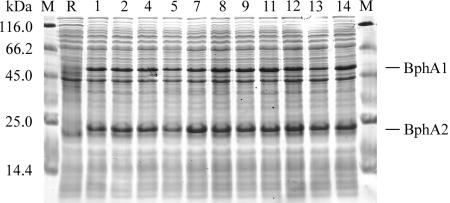
For some of the respective BphA variants, cellular concentrations of dissolved alpha and beta subunits were somewhat reduced, but no major differences were detected (Fig. 3). This rules out that increased turnover of CBs was due to higher concentrations of the variants relative to that of the WT enzyme.
FIG. 3.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of dissolved proteins of E. coli BL21(DE3)pLysS cells synthesizing different BphA1 subunits. Contents of the lanes are indicated at the top. M, marker proteins; R, negative control cells harboring the vector pT7-6; 1, WT BphA; other numbers refer to the respective BphA1-MZ variants (see Fig. 5).
Of the 10 positive variants, 1 showed enhanced turnover of 4,4′-CB, 3 showed enhanced turnover of 4,4′- and 3,4,4′-CB, 4 showed enhanced turnover of 3,5,4′-CB, and 2 showed enhanced turnover of 2,6,4′-CB. Substrate dioxygenation was quantitated at the absorption maxima of the respective HOPDAs. As shown in Table 3, the detected variants increased the dioxygenation of 4,4′-CB up to 40-fold, that of 2,6,4′-CB up to 5-fold, that of 3,4,4′-CB up to 210-fold, and that of 3,5,4′-CB up to 100-fold relative to that of the parental enzyme. After deletion of genes bphBC from the plasmids (see Materials and Methods), these results were confirmed by direct measurements of the dioxygenation products by HPLC. Of the four compounds assayed, only 2,6,4′-CB was measurably attacked by the WT enzyme. It was of interest to see how the dioxygenation of this substrate was affected in the variants that were detected by the screens with the other CBs. We observed a reduced as well as a slightly enhanced or an unchanged turnover of this substrate. This indicates that certain structural changes which enabled improved dioxygenation of one substrate negatively affected the turnover of another.
TABLE 3.
Quantitation of productive CB dioxygenation by WT and variant BphAs
| CB substrate | BphA variant | Amt of HOPDA ± SD (mA)a |
|---|---|---|
| 4,4′ | LB400 | <5 |
| MZ7 | 93 ± 30 | |
| MZ8 | 75 ± 11 | |
| MZ11 | 198 ± 59 | |
| MZ12 | 68 ± 21 | |
| 3,4,4′ | LB400 | <5 |
| MZ7 | 1,060 ± 270 | |
| MZ8 | 110 ± 20 | |
| MZ11 | 822 ± 239 | |
| 3,5,4′ | LB400 | <5 |
| MZ2 | 61 ± 10 | |
| MZ4 | 515 ± 131 | |
| MZ9 | 91 ± 27 | |
| MZ14 | 50 ± 15 | |
| 2,6,4′ | LB400 | 135 ± 39 |
| MZ5 | 640 ± 181 | |
| MZ13 | 509 ± 149 |
Absorbance was recorded at λmax (the wavelength at the absorption maximum) of the respective HOPDA (see Table 4).
Characterization of dioxygenation products.
The conversion of dioxygenation products into HOPDAs implied that the respective dioxygenations were directed against ortho and meta carbons of the substrates (Fig. 1). Thus, oxidations of 4,4′-CB by all BphA variants must have occurred at positions 2 and 3. Consistent with this result, all HOPDAs had the same absorption maximum (Table 4). The absorption maxima of HOPDAs are influenced by, and thus indicative of, their chlorination pattern (30, 31), which is dependent on the site of initial dioxygenation. In order to directly characterize the dioxygenation products, WT and mutant plasmids devoid of genes bphBC were used. GC-MS analysis of the metabolites of BphA-MZ7 and BphA-MZ11, which were the most efficient variants for 4,4′-CB, detected only a single metabolite. Its identification as a dichlorinated biphenyldihydrodiol (BDHD) (Table 5) was in accordance with dioxygenation at carbons 2 and 3.
TABLE 4.
λmaxa values of HOPDAs and BphB dependence of HOPDA formation
| CB substrate | BphA variant | λmax (nm) of HOPDA ± SD formed in the presence of:
|
|
|---|---|---|---|
| BphABC | BphAC | ||
| 4,4′ | LB400 | 432 ± 2 | Not done |
| MZ7 | |||
| MZ8 | |||
| MZ11 | |||
| MZ12 | |||
| 3,4,4′ | MZ7 | 438 ± 2 | Not observed |
| MZ8 | |||
| MZ11 | |||
| 3,5,4′ | MZ2 | 437 ± 2 | Not done |
| MZ4 | |||
| MZ9 | |||
| MZ14 | |||
| 2,6,4′ | LB400 | 402 ± 2 | 402 ± 2 |
| MZ5 | |||
| MZ13 | |||
λmax, wavelength at the absorption maximum.
TABLE 5.
GC-MS characterization of CB dioxygenation products formed by selected BphA variantsa
| CB substrate | BphA variant | tr (min) | M (MS) of derivative, in Da | No. of chlorines | Type of compound |
|---|---|---|---|---|---|
| 4,4′ | MZ7 | 20.9 | 322 | 2 | BDHD |
| MZ11 | 20.9 | 322 | 2 | BDHD | |
| 3,4,4′ | MZ7 | 22.9 | 356 | 3 | BDHD |
| MZ11 | 22.9 | 356 | 3 | BDHD | |
| 3,5,4′ | MZ4 | 22.1 | 356 | 3 | BDHD |
| 2,6,4′ | LB400 | 21.1 | 320 | 2 | DHB |
| MZ5 | 21.1 | 320 | 2 | DHB | |
| MZ13 | 21.1 | 320 | 2 | DHB |
tr, retention time; M (MS), molecular mass determined by MS; DHB, dihydroxybiphenyl.
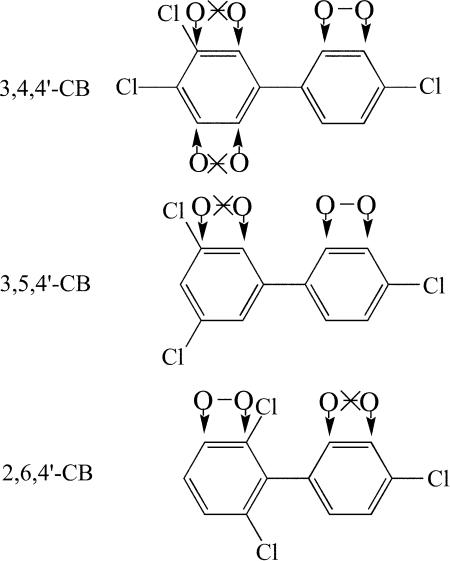
The other CBs attacked by the BphA variants contain two or three different pairs of ortho and meta carbons (Fig. 4). The HOPDAs derived from 3,4,4′-CB by the different BphA variants all showed an absorption maximum ± standard deviation of 438 ± 2 nm (Table 4) and, hence, were probably identical. Available data from structurally related HOPDAs suggest that initial dioxygenations at carbons 2 and 3 or 2′ and 3′ would lead to an absorption maximum above 430 nm, whereas dioxygenations at positions 5 and 6 would yield an absorption maximum around 410 nm (30, 31). Thus, the observed maximum is consistent with the former two possibilities. BphA-MZ7 and BphA-MZ11 were again chosen for further characterization. GC-MS analysis (Table 5) found a trichlorinated BDHD as the only metabolite formed by these variants. This rules out dioxygenation at carbons 2 and 3, which would lead to spontaneous elimination of HCl and, thereby, directly yield the corresponding catechol (32). Attack at positions 2′ and 3′ was eventually confirmed in the presence of cells synthesizing BphD, which converts HOPDAs into benzoates (Fig. 1). Under these conditions, 3,4,4′-CB was transformed into 3,4-CBA (data not shown), which can be formed only via 2′,3′ dioxygenation of this congener (Fig. 1).
FIG. 4.
Regiospecificity of substrate dioxygenation. The different possibilities of dioxygenation of 3,4,4′-, 3,5,4′-, and 2,6,4′-CB at vicinal ortho and meta carbons are shown. The exclusion of attack at specific sites by different analytical data (see the text) is also indicated.
The BphA variants that enabled the conversion of 3,5,4′-CB into ring fission products led to probably identical HOPDAs, as their absorption maxima were invariably at 437 ± 2 nm (Table 4). This is consistent only with dioxygenation at carbons 2′ and 3′ (30, 31). In this case, BphA-MZ4 was selected for further analysis. With the strain synthesizing this dioxygenase variant as the sole biphenyl catabolic enzyme, a trichlorinated BDHD was detected as a dioxygenation product (Table 5). This independently excluded hydroxylations at chlorinated carbons and showed that dioxygenation occurred only at carbons 2′ and 3′ (Fig. 4).
The WT as well as the improved variants converted 2,6,4′-CB into HOPDAs possessing an absorption maximum of 402 ± 2 nm (Table 4). This suggests dioxygenation at carbons 2 and 3 (30, 31). As mentioned above, such an attack would lead directly to the corresponding catechol. Substrate conversion in the presence of WT or variant BphA and of BphC but in the absence of BphB, as described in Materials and Methods, yielded the same HOPDAs (Table 4), thereby confirming hydroxylation at carbons 2 and 3. Furthermore, GC-MS analyses of the dioxygenation products detected that the same dichlorinated dihydroxybiphenyl was formed by WT as well as variant dioxygenases (Table 5). Finally, hydroxylation by BphA-MZ5 in the presence of BphBCD led to the formation of 4-CBA (data not shown), which is consistent only with an initial dioxygenation at positions 2 and 3 (Fig. 1 and 4).
Comparison of mutations in CB-selected and nonselected clones and correlation with substrate acceptance.
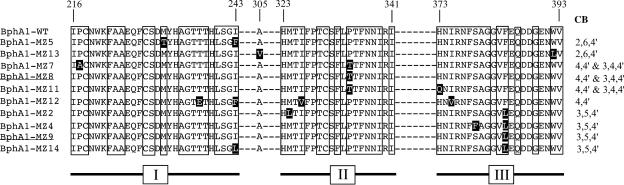
The 10 CB-selected clones contained an average number of 3.50 mutations per clone, of which 2.00 were nonsilent (Table 2). Since one codon harbored two nonsilent mutations, they led to a total of 19 AA exchanges. Additionally, a single mutation not induced by the mutagenic oligonucleotides was found which resulted in an Ala305Val exchange. Individual clones encoded between one and four AA replacements. The AA sequences of the relevant regions and the Ala305Val substitution are shown in Fig. 5.
FIG. 5.
Amino acid exchanges in BphA1 variants exhibiting novel substrate specificities. Segments (I to III) of major importance for interaction with CB substrates are shown. Positions of the first and last residues in each segment are indicated at the top. Subsegments to which AA exchanges were targeted are boxed. The WT sequence is given in the top row. Below, sequences are grouped according to their detection in the CB screen. The respective CBs are given at the right margin. Exchanges of residues are highlighted by inverse colors. The unintended substitution of BphA1-MZ13 at position 305 is also indicated. Designations of subunits harboring only single AA substitutions are underlined.
In Fig. 5, the sequences have been grouped according to the novel properties of the variants. As can be seen, the two variants exhibiting increased turnover of 2,6,4′-CB do not have even one position of AA exchange in common. In striking contrast, the three evolved enzymes showing dioxygenation of 4,4′- and 3,4,4′-CB all share the Pro334Thr substitution. The importance of this replacement for the acceptance of the novel substrates is clearly demonstrated by BphA-MZ8, which harbors only this exchange. Similarly, the four variants that acquired the ability to dioxygenate 3,5,4′-CB all have the Phe384Leu substitution in common. In BphA-MZ9, this is the only difference with the WT, indicating that this exchange alone leads to acceptance of the novel substrate. It can be ruled out that the frequent Pro334Thr and Phe384Leu substitutions were found due to some bias in mutagenesis. Sequencing of 63 variants not selected through the CB screen showed that the majority of mutations in the respective codons encoded other AA replacements.
The variant BphA-MZ12 showed turnover of 4,4′-CB but not of 3,4,4′-CB. It harbors four exchanged residues, none of which is shared by any of the other evolved enzymes attacking this congener.
With some of the double-exchange variants containing the Pro334Thr or Phe384Leu substitutions, turnover of the novel substrates was significantly modulated. It was increased severalfold by the additional AA exchanges (Ser379Phe in BphA1-MZ4, Pro217Ala in BphA1-MZ7, and His373Gln in BphA1-MZ11), as shown in Table 3. This demonstrates a strong amplification of the beneficial effect of the shared substitution by the additional replacement.
DISCUSSION
Due to the limited predictability of the effects of defined AA substitutions on protein structure and function, techniques of artificial or directed evolution have increasingly become methods of choice for the alteration of protein properties such as substrate specificity. The number of possible sequence variants of even medium-sized proteins is far beyond what can be screened with even the most advanced high-throughput methods. Thus, strategies for artificial evolution to introduce mutations less randomly are desirable. To this end, DNA shuffling has been targeted to parts of BphA genes (1). In the present work, we used available sequence-function relationship data to limit mutagenesis to relatively small segments of the BphA alpha subunit gene. This enabled the use of chemically synthesized oligonucleotides as mutagenic agents. The advantages of oligonucleotides include free choices of the rate, bias, and position of mutagenesis at the NT level. When relatively small gene segments are mutagenized, a high rate of NT substitutions can be applied so that AA exchanges are not limited to those requiring only a single base pair substitution per codon.
One problem encountered was the quality of the relatively long oligonucleotides. Sequencing showed that three quarters of all inactive clones were false inactives due to errors in oligonucleotide synthesis. They originated from primers of incorrect length, due typically to erroneous double incorporation of the same NT. Thus, further improvements in oligonucleotide synthesis and/or purification are desirable.
In the current work, we applied a uniform rate of mutagenesis without any bias in the incorporation of non-WT NTs. The chosen rate yielded approximately 90% of true inactive variants. This may appear high; however, it is expected to enrich the library with variants of interest. Theoretical considerations have shown that a rate of mutagenesis that is optimal for the generation of beneficial mutants requiring 1-, 2-, or 3-bp exchanges will yield approximately 63%, 87%, or 95%, respectively, of inactive proteins (22).
The observed frequency in the generation of beneficial mutants was strikingly high. A screen of only some 670 active clones with five potential substrates detected 10 enzyme variants with novel substrate specificities. This result suggests that the limitation of random AA substitutions to regions that are critical for substrate binding and the exclusion of AA exchanges from positions that are essential for catalytic activity are advantageous strategies for the artificial evolution of enzymatic properties such as substrate specificity.
The BphA of strain LB400 has long been known as an enzyme unable to efficiently attack the para-chlorinated ring (21, 23). If the problem for the WT enzyme to dioxygenate this ring were only the para substituent, then a single positive variant would be able to attack it in all CBs containing this moiety. However, discrete solutions were found for its hydroxylation in 3,5,4′-CB on the one hand and in 4,4′- as well as 3,4,4′-CB on the other, and none of the variants was able to attack the para-chlorinated ring in 2,6,4′-CB. These results clearly indicate a crucial role for the substituent pattern of the nonoxidized ring.
The limited number of AA exchanges in the selected variants and the repeated appearance of specific substitutions in variants of similar substrate preference permitted the identification of residues involved in substrate specificity.
BphA1-MZ8 showed strongly enhanced dioxygenation of 4,4′- and 3,4,4′-CB. It harbors only the Pro334Thr substitution. Residue 334 is a direct neighbor of Leu333, which is a constituent of the substrate-binding pocket (SBP), according to a model of the three-dimensional structure of the subunit (41). The Pro334Thr replacement may cause a repositioning of Leu333 via the main chain connection. Recently, a potential influence of Pro334 and several other residues on the structure of the catalytic center has been examined by an approach that determined the influence of directed AA replacements by Ala on the orientation of WT substrates at the active site (41). This strategy indicated that a change at position 334 altered the structure of the SBP. Our present results now demonstrate that an exchange of this residue can lead to the acceptance of novel substrates.
Two additional exchanges, Pro217Ala (BphA1-MZ7) and His373Gln (BphA1-MZ11), further improved the oxidation of 4,4′- and 3,4,4′-CB, respectively. No previous data are available on exchanges at position 373. In the study mentioned above, no significant effect was found for the Pro217Ala replacement. This suggests that a significant structural change through the Pro217Ala replacement is provoked only in concert with another replacement, such as that of Pro334.
BphA1-MZ9 greatly increased the dioxygenation of 3,5,4′-CB. It differs from the WT only by the Phe384Leu exchange. The remarkable effect exerted by this single substitution is consistent with the finding that residue 384 probably belongs to the SBP (41). It is also consistent with previous observations indicating that changes at this position greatly reduce the substrate range of BphA of strain KF707 (38) and influence the regiospecificity of dioxygenation by BphA-LB400 (41) or by the naphthalene dioxygenase of strain NCIB9816 (25).
The additional replacement of Met324 (BphA1-MZ2) or Ile243 (BphA1-MZ14) by Leu only insignificantly modulated the turnover of 3,5,4′-CB. This is in keeping with the result that the Met324Ala substitution had little influence on the structure of the active site, although it is located next to His323, a probable constituent of the SBP (41). The Ile243Ala exchange has previously been shown to affect the regiospecificity of dioxygenation (41). This difference from the present result may be due to the fact that Leu is more similar to Ile than Ala. Furthermore, different CBs were used in this and the previous studies. It has been shown that an effect of a given substitution is not “sensed” by all substrates to the same extent (25, 41).
When the Phe384Leu replacement was accompanied by the Ser379Phe exchange (BphA1-MZ4), the dioxygenation of 3,5,4′-CB was further increased. To our knowledge, this is the first indication that residue 379 can affect substrate specificity. It may exert its influence via a repositioning of the probable SBP residue Phe 378 (41). Substitutions of Ser379Phe have been shown to strongly decrease the substrate range of BphA-KF707 (38) or to alter the regiospecificity of dioxygenation by BphA-LB400 (41).
BphA1-MZ5 showed enhanced dioxygenation of 2,6,4′-CB. It contains two AA exchanges, Met231Thr and Ile243Phe. Residue 231 is probably an SBP constituent (41). Consistent with this, a strong influence of the Met231Ala substitution on the structure of the SBP has experimentally been determined (41). As mentioned above, this has also been shown for the Ile243Ala exchange. We note that residue 243 was also replaced in variants BphA1-MZ12 and BphA1-MZ14. These findings suggest that both exchanges in BphA1-MZ5 contribute to the enhanced dioxygenation of 2,6,4′-CB.
The second variant with significantly improved turnover of this substrate, BphA1-MZ13, also harbors two AA substitutions, Ala305Val and Trp392Leu. The former exchange, which was presumably caused by a PCR error, appears to be quite distant from the active site (41). The replacement of Trp392 by Ala has previously been shown to influence the SBP structure (41). These observations suggest that the improved dioxygenation of 2,6,4′-CB is exclusively or predominantly caused by the latter exchange.
BphA1-MZ12 showed enhanced turnover of 4,4′-CB. It is the only variant with substitutions at four positions. Interestingly, all of these four residues have been replaced separately in previous studies. The Thr237Met and Ile375Ala exchanges showed no significant influence on substrate dioxygenations (24, 41). However, remarkable effects were observed for the substitutions Ile243Ala (as mentioned above) and Ile326Ala (41), suggesting that changes at these two positions are mainly responsible for the better acceptance of 4,4′-CB. None of the four AA exchanges of BphA-MZ12 is shared by any of the other evolved enzymes attacking this substrate. This result and the finding that the two variants that enhance the attack on 2,6,4′-CB have no replacements in common suggest that in many cases, several different solutions exist for the generation of a protein with a particular property.
Taken together, the data for the 10 variants indicate that AAs at the following five positions, 217, 334, 373, 379, and 384, can—alone or in concert with substitutions at other positions—remarkably alter the substrate specificity of BphA-LB400. Moreover, it also appears likely that residues at four additional positions, namely, 231, 243, 326, and 392, can affect the acceptance of novel substrates. According to our model of the three-dimensional structure of BphA-LB400, only two of these nine AAs are SBP constituents. Thus, changes of most of these residues would not have easily been predicted to affect substrate preference. The identification of these positions suggests prime targets for subsequent, more-refined rounds of artificial evolution in order to change the substrate specificity of this and other aryl-hydroxylating dioxygenases.
Acknowledgments
We thank Manfred Nimtz, Esther Surges, and Annette Krüger for help with GC-MS analyses and DNA sequencing.
Support of this work by grants from the Deutsche Forschungsgemeinschaft (Ho 1219/2-1), the Bundesministerium für Bildung und Forschung (WTZ CHL 99/029), CONICYT, FONDECYT (1020221-7020221), and USM (130122) is gratefully acknowledged.
REFERENCES
- 1.Barriault, D., M.-M. Plante, and M. Sylvestre. 2002. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J. Bacteriol. 184:3794-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartels, F., S. Backhaus, E. R. B. Moore, K. N. Timmis, and B. Hofer. 1999. Occurrence and expression of glutathione S-transferase-encoding bphK genes in Burkholderia sp. strain LB400 and other biphenyl-utilizing bacteria. Microbiology 145:2821-2834. [DOI] [PubMed] [Google Scholar]
- 3.Beil, S., J. R. Mason, K. N. Timmis, and D. H. Pieper. 1998. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J. Bacteriol. 180:5520-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D. R., and G. N. Sheldrake. 1998. The dioxygenase-catalyzed formation of vicinal cis-diols. Nat. Prod. Rep. 15:309-325. [Google Scholar]
- 5.Brühlmann, F., and W. Chen. 1999. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol. Bioeng. 63:544-551. [DOI] [PubMed] [Google Scholar]
- 6.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 7.Carredano, E., A. Karlsson, B. Kauppi, D. Choudhury, R. E. Parales, J. V. Parales, K. Lee, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2000. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J. Mol. Biol. 296:701-712. [DOI] [PubMed] [Google Scholar]
- 8.Dowling, D. N., R. Pipke, and D. F. Dwyer. 1993. A DNA module encoding bph genes for the degradation of polychlorinated biphenyls. FEMS Microbiol. Lett. 113:149-154. [DOI] [PubMed] [Google Scholar]
- 9.Erickson, B. D., and F. J. Mondello. 1993. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl. Environ. Microbiol. 59:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 11.Goris, J., P. de Vos, J. Caballero-Mellado, J. Park, E. Falsen, F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400(T) and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 12.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harayama, S. 1998. Artificial evolution by DNA shuffling. Trends Biotechnol. 16:76-82. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofer, B., L. D. Eltis, D. N. Dowling, and K. N. Timmis. 1993. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene 130:47-55. [DOI] [PubMed] [Google Scholar]
- 16.Hudlicky, T., D. Gonzalez, and D. T. Gibson. 1999. Enzymatic hydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodiology. Aldrichim. Acta 32:35-62. [Google Scholar]
- 17.Kahl, S., and B. Hofer. 2003. A genetic system for the rapid isolation of aromatic-ring-hydroxylating dioxygenase activities. Microbiology 149:1475-1481. [DOI] [PubMed] [Google Scholar]
- 18.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, N., A. Nishi, M. Goto, and K. Furukawa. 1997. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J. Bacteriol. 189:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumamaru, T., H. Suenaga, M. Mitsuoka, T. Watanabe, and K. Furukawa. 1998. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol. 17:663-666. [DOI] [PubMed] [Google Scholar]
- 21.McKay, D. B., M. Seeger, M. Zielinski, B. Hofer, and K. N. Timmis. 1997. Heterologous expression of biphenyl dioxygenase-encoding genes from a gram-positive broad-spectrum polychlorinated biphenyl degrader and characterization of chlorobiphenyl oxidation by the gene products. J. Bacteriol. 179:1924-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura, T., and P. Sonigo. 2001. A mathematical model for experimental gene evolution. J. Theor. Biol. 209:497-502. [DOI] [PubMed] [Google Scholar]
- 23.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polyclorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parales, R. E., S. M. Resnick, C.-L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the αsubunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Schmidt-Dannert, C., D. Umeno, and F. H. Arnold. 2000. Molecular breeding of carotenoid biosynthetic pathways. Nat. Biotechnol. 18:750-753. [DOI] [PubMed] [Google Scholar]
- 30.Seah, S. Y., G. Labbé, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 31.Seeger, M., K. N. Timmis, and B. Hofer. 1995. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl. Environ. Microbiol. 61:2654-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeger, M., K. N. Timmis, and B. Hofer. 1995. Degradation of chlorobiphenyls catalyzed by the bph-encoded biphenyl-2,3-dioxygenase and biphenyl-2,3-dihydrodiol-2,3-dehydrogenase of Pseudomonas sp. LB400. FEMS Microbiol. Lett. 133:259-264. [DOI] [PubMed] [Google Scholar]
- 33.Seeger, M., M. Zielinski, K. N. Timmis, and B. Hofer. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 35.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 36.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suenaga, H., M. Goto, and K. Furukawa. 2001. Emergence of multifunctional oxygenase activities by random priming recombination. J. Biol. Chem. 276:22500-22506. [DOI] [PubMed] [Google Scholar]
- 38.Suenaga, H., T. Watanabe, M. Sato, Ngadiman, and K. Furukawa. 2002. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184:3682-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zielinski, M., S. Backhaus, and B. Hofer. 2002. The principal determinants for the structure of the substrate-binding pocket are located within a central core of a biphenyl dioxygenase alpha subunit. Microbiology 148:2439-2448. [DOI] [PubMed] [Google Scholar]
- 41.Zielinski, M., S. Kahl, H.-J. Hecht, and B. Hofer. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]