Abstract
The gene for a novel α-amylase, designated AmyC, from the hyperthermophilic bacterium Thermotoga maritima was cloned and heterologously overexpressed in Escherichia coli. The putative intracellular enzyme had no amino acid sequence similarity to glycoside hydrolase family (GHF) 13 α-amylases, yet the range of substrate hydrolysis and the product profile clearly define the protein as an α-amylase. Based on sequence similarity AmyC belongs to a subgroup within GHF 57. On the basis of amino acid sequence similarity, Glu185 and Asp349 could be identified as the catalytic residues of AmyC. Using a 60-min assay, the maximum hydrolytic activity of the purified enzyme, which was dithiothreitol dependent, was found to be at 90°C. AmyC displayed a remarkably high pH optimum of pH 8.5 and an unusual sensitivity towards both ATP and EDTA.
α-Amylases (EC 3.2.1.1) are glycoside hydrolases which cleave the α-1,4-glycosidic linkages in starch and maltodextrins with an endo mechanism, i.e., in a random fashion within the polysaccharide molecule. Due to the retaining catalytic mechanism the released cleavage products have an α-anomeric configuration. Amylases are of commercial interest, e.g., for starch processing in the food industry. In the amino acid sequence-based classification system for carbohydrate active enzymes (CAZY) the vast majority of α-amylases are sorted into the glycoside hydrolase family (GHF) 13 (6). All enzymes of this family share a (β/α)8-barrel as the common supersecondary structure. Some α-amylases are also classified into GHF 57. This enzyme family is considerably smaller than GHF 13 and less well investigated.
The hyperthermophilic bacterium Thermotoga maritima was first isolated from geothermally heated marine sediments (7). The bacterium grows anaerobically at temperatures up to 90°C with an optimum at 80°C. As an obligate heterotrophic organism it utilizes various polysaccharides and proteinaceous substrates. Sugars are mainly metabolized via the classical, unmodified Embden-Meyerhof pathway and to 15% via the Entner- Doudoroff pathway (7, 23, 24).
T. maritima has the highest number of genes for carbohydrate-degrading and -modifying enzymes in relation to its 1.8-Mbp genome known to date in free-living prokaryotes (19). Among the polysaccharide-degrading enzymes of T. maritima described so far are two α-amylases, one an extracellular putative lipoprotein (AmyA) (15) and one located in the cytoplasm (AmyB) (16).
Here we report the recombinant expression in Escherichia coli and the biochemical characterization of an additional intracellular α-amylase from T. maritima. The enzyme does not belong in the GHF 13 amylase group but has significant similarity to the less well characterized GHF 57.
MATERIALS AND METHODS
Library screening.
A T. maritima MSB8 (type strain) chromosomal gene library, whose construction was described earlier (22), was screened for amylolytic activity. E. coli JM83 transformant colonies were grown overnight at 37°C on Luria-Bertani agar plates containing 0.5% soluble starch (Merck, Darmstadt, Germany). Subsequently, the plates were incubated for 1 h at 65°C and then flooded with Lugol solution (0.3% [wt/vol] I2, 0.6% [wt/vol] KI in H2O). Amylolytic activity underneath and around the colonies resulted in clear halos against a dark violet background.
Construction of the amyC expression vector.
To amplify the complete amyC gene, PCR was carried out using genomic T. maritima DNA as the template. The forward primer (5′-CCCGTTCCATATGAGAGGAAAAATACTGATATT TCTG-3′) was designed to introduce a NdeI restriction site at the start codon of the amyC open reading frame (ORF), and the reverse primer (5′-CGGGAT CCGAGGATAGAGGTGGTGGTGGTG-3′) was used to introduce a BamHI site downstream of the stop codon. The amplified DNA fragment (1.7 kb) was initially cloned into pBluescript (Stratagene, San Diego, Calif.) before it was recloned into pET24c (Novagen, San Diego, Calif.) via the introduced NdeI and BamHI restriction sites, resulting in the plasmid pET24::AmyC. The correct cloning of the amyC ORF was verified by sequencing and restriction enzyme analysis. All recombinant techniques were performed in Escherichia coli strains XL1-Blue and BL21.
Bacterial strains and growth conditions.
E. coli XL1-Blue harboring the pBluescript::AmyC plasmid and E. coli BL21 transformed with the pET24:: AmyC plasmid were cultured in Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl [wt/vol]) containing the appropriate antibiotic, ampicillin (100 μg/ml) or kanamycin (50 μg/ml). For crude extract preparation, the recombinant E. coli BL21 strain carrying pET24::AmyC was grown in 2 liters liquid culture with kanamycin selection in a 5-liter baffled flask. The T7 promoter of the plasmid was induced with 0.1 M isopropyl-1-thio-β-d-galactoside (IPTG) at an optical density (600 nm) of 0.8. After 10 h the cells were harvested by centrifugation (10,400 × g, 15 min, 4°C) and after resuspension in 20 mM Tris buffer (pH 8.0) disrupted by twofold passage through a French pressure cell (American Instruments, Silver Spring, MD). T. maritima was grown anaerobically as previously described (13) in a medium containing 0.5% casein peptone, 0.1% yeast extract, 2.5% Instant Ocean salts, and 0.5% glucose or 0.5% soluble starch (wt/vol).
Purification.
The crude cell extract was centrifuged at 20,000 × g (15 min, 4°C) to sediment the cell debris. The supernatant was incubated at 75°C for 20 min in order to denature the thermolabile host proteins, which were sedimented at 20,000 × g (15 min, 4°C).
The supernatant clarified by the heat treatment was dialyzed against 20 mM Tris (pH 8.0) and subjected to anion-exchange chromatography on a Source 30Q HR10/10 column (GE Healthcare/Amersham, Freiburg, Germany) equilibrated with the same buffer. Elution was done with a 0 to 1,000 mM NaCl gradient in the same buffer in 15 column volumes at a flow rate of 2 ml/min. Amylase activity-containing fractions, which eluted at ∼350 mM NaCl, were pooled and dialyzed against 20 mM Tris, pH 8.0. The purity of the resulting enzyme was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Size-exclusion chromatography.
In order to determine the native molecular masses of the purified protein, analytical size-exclusion chromatography was carried out using a Superdex 200 Prep Grade HiLoad 16/60 column (GE Healthcare/Amersham). An isocratic gradient of 20 mM Tris, pH 8.0, with the addition of 150 mM NaCl was applied for elution. Five proteins with molecular masses from 25 to 450 kDa were used for column calibration under the same conditions. The partition coefficient (KAV) was determined for all proteins with the formula KAV = Ve − V0/Vt − V0 with Ve being the retention volume, V0 the column void volume, and Vt the total bed volume. The molecular mass of AmyC was calculated from the regression curve in a KAV:log Mr plot.
Gel electrophoresis.
SDS-PAGE according to the method of Laemmli (14) was routinely performed to test the purity of the samples during purification and to estimate the molecular mass of proteins under denaturing conditions. SDS-PAGE was carried out with 12% gels in Mini-Protean II electrophoresis units (Bio-Rad, München, Germany), and the gels were stained with Coomassie brilliant blue. Sizes of the analyzed proteins were estimated by comparison with the standard protein marker mixture SDS-6H (Sigma-Aldrich, Taufkirchen, Germany) separated on the same gel.
Western blotting.
For Western blot analysis according to the method of Towbin et al. (27) polyclonal anti-AmyC antiserum from rabbit was used as the primary antibody and alkaline phosphatase-coupled anti-rabbit immunoglobulin G (Sigma- Aldrich) was used as secondary antibody.
Activity assay.
Amylolytic activity with various substrates was assayed with the 3,5-dinitrosalicylic acid method (17). The reaction mixture contained 250 μl substrate (1% [wt/vol] soluble starch [Sigma-Aldrich] or other polysaccharides as specified in the text), 100 μl 250 mM Tris buffer adjusted to pH 8.0 at 90°C, 5 μl dithiothreitol (DTT; 1 M), 145 μl H2O, and 50 μg appropriately diluted enzyme. After incubation for 1 h at 90°C, 750 μl of 3,5-dinitrosalicylic acid solution (1% [wt/vol] 3,5-dinitrosalicylic acid, 0.2% [vol/vol] phenol, 0.05% [wt/vol] Na2SO4, 20% KNa-tartrate, 1% [wt/vol] NaOH in H2O) was added, and the mixture was incubated for 15 min at 98°C. The absorption was measured at 575 nm, and the specific activity was calculated using maltose as a standard. One unit of α-amylase was defined as the amount of enzyme that liberates 1 μmol of reducing groups per min.
Influence of pH and temperature on activity.
The influence of the pH value on AmyC activity was measured at 90°C in Bis-Tris (pH 5.5 to 6.5) and Tris buffer (pH 7.0 to 9.0), adjusted at this temperature to various pH values, using the standard assay described above. The influence of the temperature on the enzyme activity was determined at pH 8.5 with Tris buffer using standard assay reaction mixtures incubated at temperatures ranging from 45 to 100°C.
Thermoinactivation.
The recombinant enzyme was diluted to a protein concentration of about 25 μg ml−1 in 250 mM Tris buffer adjusted to pH 8.5 at the selected temperatures. Thermoinactivation kinetics were determined through preincubation of the diluted enzyme at 80°C, 90°C, and 100°C for various periods of time followed by determination of the residual activity with the standard enzyme assay after allowing the samples to cool.
Influence of chelating agents.
The effect of EDTA and ATP on AmyC activity was analyzed in 50 mM Tris buffer, pH 8.0. The enzyme was incubated with various additives (ATP, ADP, or EDTA at 0.1 or 1 mM) in the reaction mixture described above without substrate at 90°C. In some experiments, MgCl2 or CaCl2 (1 or 3 mM) was added after 5 min. The enzyme reaction was started through the addition of prewarmed substrate solution and carried on for 1 h at 90°C.
Analysis of sugars by TLC.
Thin-layer chromatography (TLC) of mono- and oligosaccharides was done on 0.2-mm silica gel-coated aluminum sheets (type 60; Merck, Darmstadt, Germany) with a solvent system composed of 1-propanol-ethyl acetate-H2O at a volume ratio of 6:1:3 or 1-propanol-nitromethane-H2O at a volume ratio of 5:3:2. The chromatograms were sprayed with aniline-diphenylamine reagent (1% [wt/vol] diphenylamine, 1% [vol/vol] aniline in acetone mixed with 0.1 volume 87% [vol/vol] phosphoric acid prior to use) and baked for 10 min at 160°C to visualize the carbohydrate spots.
Quantitative determination of iron.
The amount of nonheme iron bound to the protein was determined according to the colorimetric method of Fish (4).
RESULTS
Identification of the T. maritima amyC gene.
A gene library of T. maritima MSB8 was screened for amylase genes by plating E. coli transformants on 0.5% starch plates. The amylolytic activity of one of the positive clones was attributed to ORF tm1438 (annotation according to the work of Nelson et al. [20]) after sequence analysis of the approximately 3-kb genomic DNA insert of its recombinant plasmid, pTAA31 (data not shown). ORF tm1438 was renamed amyC.
Database search.
A homology search carried out with the BlastP algorithm (1) with the deduced amino acid sequence of AmyC revealed no similarity to the sequences of GHF 13 proteins, a family to which most known α-amylases belong. Instead, high similarities to family 57 glycoside hydrolases were found and the five conserved regions of this protein family are present in AmyC (Fig. 1). GHF 57 consists of amylases, pullulanases, and transferases, few of them biochemically characterized. Apart from the conserved regions a significant overall amino acid sequence similarity was found only to parts of the protein family, specifically to a cluster of orthologous group (COG) 1543-like proteins (25), which seem to form a distinct subfamily within GHF 57. The only two characterized amylases, AmyA from Dictyoglomus thermophilum and AmyA (=PF0272) from Pyrococcus furiosus, have a very low overall sequence similarity (E > 10) to AmyC (5, 13). Among the proteins of GHF 57 is a 4-α-glucanotransferase from Thermococcus litoralis, which has been examined and whose crystal structure is known (8-11). By amino acid sequence comparison to the T. litoralis enzyme it was possible to predict the catalytically essential amino acid residues of AmyC. Glu185 of AmyC is presumably the nucleophilic residue, and Asp349 acts as the acid/base catalyst. These catalytic residues are conserved within GHF 57 (Fig. 1) (28). In a P. furiosus amylopullulanase, also a GHF 57 protein, Kang et al. have identified a glutamate as a third putative catalytic residue (12). The corresponding residue, Glu351, can also be identified in AmyC.
FIG. 1.
Partial alignment of the amino acid sequences of AmyC from T. maritima, the 4-α-glucanotransferase (Gtra) from Thermococcus litoralis, and the amylopullulanase (Apul) from Thermococcus hydrothermalis (27). Catalytic residues of GHF 57 are marked with a star; the additional catalytic glutamate residue identified in a P. furiosus amylopullulanase is marked with a star in parentheses (12).
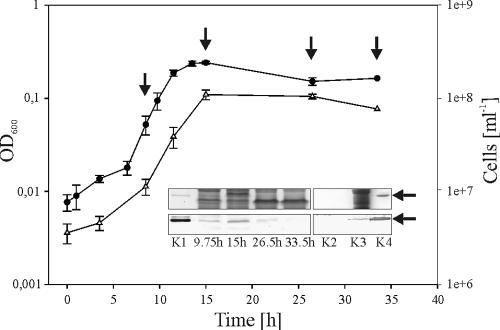
AmyC apparently is an intracellular enzyme. No signal peptide can be found in the amino acid sequence, and AmyC was detected in T. maritima crude extracts during the exponential and the early stationary growth phases by Western blot analysis, whereas it was no longer found in the late stationary phase. No AmyC protein could be detected in the culture supernatant (Fig. 2).
FIG. 2.
Growth curve of T. maritima and immunoassay of AmyC. Growth in a medium containing 0.5% glucose was measured by determination of optical density at 600 nm (OD600) (black circles) and cell count (white triangles). In various growth phases samples were drawn from the culture and the cytoplasmic crude extract was tested for the presence of AmyC via SDS-PAGE (top inset) and Western blotting with anti-AmyC antibody (bottom inset). K1 and K4, purified recombinant AmyC (0.4 μg); K2, 10-fold-concentrated culture supernatant (0.75 μg) of a culture grown on 0.5% starch; K3, cytoplasmic crude extract (30 μg) of the same culture. The arrows at the right side of the SDS-polyacrylamide gel and the Western blot indicate the mobility of AmyC (62 kDa).
Cloning, expression, and purification of AmyC.
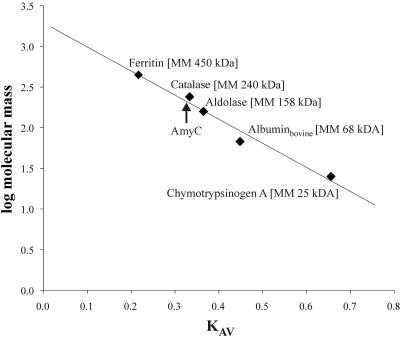
The original publication of the T. maritima genome claims tm1438 (=amyC) to contain an authentic frameshift (19). Upon closer inspection by sequencing no stop codon or such a frameshift could be detected within the amyC ORF. The 1,587-bp full-length ORF codes for a polypeptide with a calculated molecular mass of 62,859 Da. This is in agreement with the apparent molecular mass of 62 kDa of AmyC as calculated from protein migration in gel electrophoresis under denaturing conditions (Fig. 3). Under nondenaturing conditions the recombinant protein has a molecular mass of 241 kDa as determined by gel filtration chromatography with a Superdex 200 column, which was calibrated with five standard proteins (Fig. 4). These data suggest a homotetramer as the quaternary form. The crystallization data of AmyC also point towards a tetramer, confirming the result of the gel filtration (3).
FIG. 3.
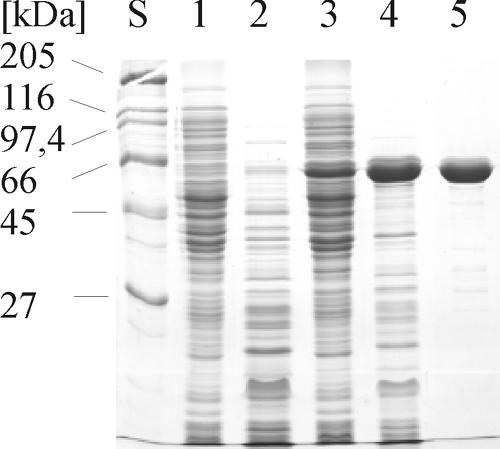
Purification of recombinant AmyC. Samples from all purification steps were separated with SDS-PAGE, and the proteins were stained with Coomassie brilliant blue. Lane S, size standard; lane 1, E. coli BL21 crude extract; lane 2, E. coli BL21 heat-treated crude extract; lane 3, E. coli BL21(pET24c::AmyC) crude extract; lane 4, E. coli BL21(pET24c::AmyC) heat-treated crude extract; lane 5, Source 30Q column pool.
FIG. 4.
Analytical gel filtration. AmyC had a retention volume, Ve, of 63.0 ml on a Superdex 200 column (Amersham Biosciences). From the regression curve in the KAV:log Mr plot a molecular mass of 241 kDa (arrow) was calculated for the nondenatured enzyme.
During the overexpression in E. coli a large portion of the recombinant protein remained insoluble despite various efforts to increase the yield of soluble enzyme (data not shown). Since the specific activity of different purified enzyme preparations varied from 300 to 1,600 mU mg−1, it cannot be excluded that a changing fraction of the purified soluble protein was inactive. Figure 3 shows the overexpression and the purification steps of AmyC.
Physicochemical and biochemical characterization.
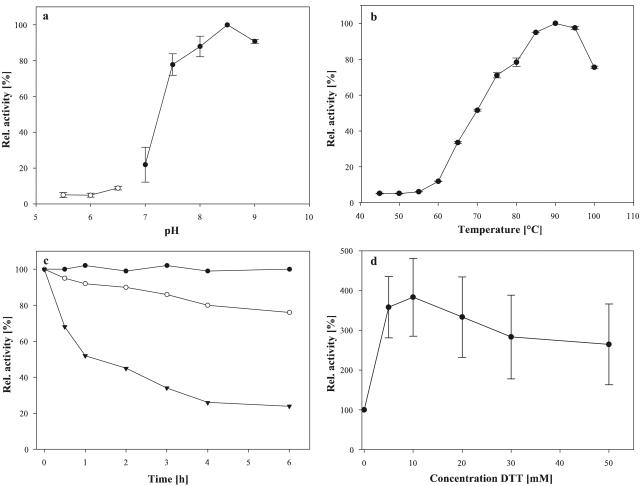
With a 1-h assay using starch as the substrate the maximum enzymatic activity of AmyC was measured at 90°C and pH 8.5 (Fig. 5a and b). Thermoinactivation experiments demonstrated that, after 6 h of incubation at 90°C, the upper temperature limit of T. maritima growth (7), AmyC retained about 80% of its initial activity. At 80°C, the optimal growth temperature of T. maritima, no significant loss of activity could be detected (Fig. 5c). The protein's thermostability at 90°C was slightly decreased by commonly used stabilizing additives such as betaine (15%), Ca2+ (10 mM), bovine serum albumin (40 mg/ml), or Triton X-100 (0.01%) (data not shown).
FIG. 5.
Effects of pH, measured with Bis-Tris (white circles) and Tris buffer (black circles) (a); temperature (b); long-term incubation at elevated temperatures of 80°C (black circles), 90°C (white circles), and 100°C (black inverted triangles) (c); and DTT (d) on AmyC activity.
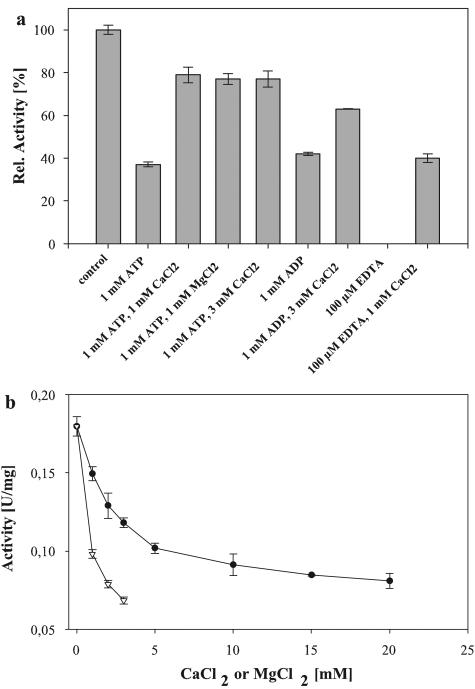
The enzyme's activity was greatly increased in the presence of reducing agents. DTT (5 to 10 mM) led to a threefold-enhanced activity (Fig. 5d). Alternatively, l-cysteine and 2-mercaptoethanol had a slightly smaller stimulating effect (data not shown). The enzyme was unusually sensitive to chelating agents. The addition of 0.1 mM EDTA or EGTA completely abolished activity. Unexpectedly, nucleotides also had a negative effect on the activity. ATP at 5 mM caused an approximately 90% drop of activity (data not shown). The inhibitory effect of EDTA and nucleotides was reversed by the addition of Ca2+ or Mg2+ (Fig. 6a). Surprisingly, Ca2+ and Mg2+, if added alone, led to a decrease in AmyC activity, as did most of the metal ions tested (Fig. 6b). The enzyme activity was also decreased by 50% in the presence of 50 mM NaCl.
FIG. 6.
Effect of EDTA, ATP, and divalent cations on AmyC activity. (a) Effect of EDTA and ATP on AmyC. In reaction mixtures with divalent cations the enzyme was preincubated with ATP or EDTA at 90°C for 5 min before Ca2+ or Mg2+ was added. (b) Effect of MgCl2 (white inverted triangles) and CaCl2 (black circles) on the activity of AmyC. Measurements with MgCl2 were stopped at a concentration of 3 mM because activity dropped below the assay's detection limit.
Remarkably, the purified AmyC protein was found to contain iron (0.4 to 1.7 mol/mol protein in different enzyme preparations), varying with each purification batch. The amount of iron, however, did not correlate with the varying specific activity of the purified protein, and the addition of FeCl3 (1 mM) did not improve the enzyme activity. Iron was also detected in AmyC protein crystals prepared with the sitting-drop method by X-ray fluorescence spectroscopy (data not shown).
Substrate specificity.
AmyC was hydrolytically active on a variety of α-1,4-linked carbohydrates. Apart from amylose (relative activity 100%), the enzyme also cleaved soluble starch (68%) and β-limit-dextrin (28%) while glycogen (4%) and β-cyclodextrin (6%) were poor substrates. Pullulan was not significantly cleaved by AmyC (data not shown).
AmyC action resulted in glucose, maltose, maltotriose, and a small amount of longer maltodextrins as final products. While maltotriose was hydrolyzed, maltose was no substrate for AmyC. The enzyme hydrolyzed amylose by apparently random cleavage of the polysaccharide chain (endo mechanism), which was determined by thin-layer chromatography with samples drawn at different time points during amylose degradation. First, maltooligosaccharides of various lengths were generated and later glucose, maltose, and maltotriose predominated as the products of hydrolysis (Fig. 7). On the basis of its substrate specificity and characteristics of product formation by cleavage of α-1,4-glycosidic bonds in polysaccharides, AmyC from T. maritima was classified as an α-amylase (EC 3.2.1.1).
FIG. 7.
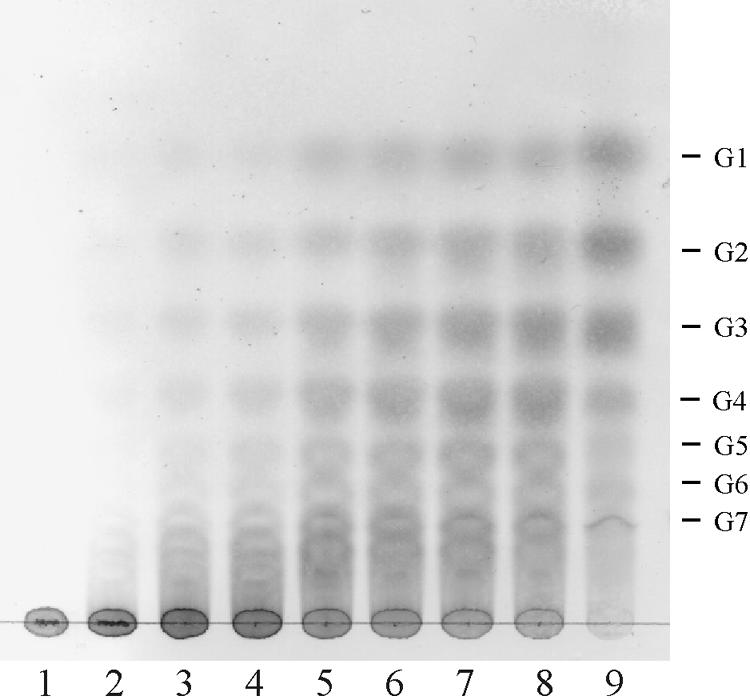
TLC analysis of product formation during degradation of amylose. Amylose (0.4%) was digested with 50 μg recombinant AmyC at 90°C in 20 mM Tris buffer, pH 8.5. The products were separated on a silica gel TLC plate in 1-propanol-nitromethane-H2O (5:3:2). G1 to G7, oligosaccharides glucose to maltoheptaose; lane 1, after 0 min; lane 2, after 15 min; lane 3, after 30 min; lane 4, after 1 h; lane 5, after 2 h; lane 6, after 3 h; lane 7, after 4 h; lane 8, after 5 h; lane 9, after 16 h.
DISCUSSION
The putative intracellular α-amylase AmyC from the hyperthermophilic bacterium T. maritima was heterologously overexpressed in E. coli and biochemically characterized. The maximum activity under the standard assay conditions used in this study was measured at pH 8.5 and 90°C, which is the upper growth limit of T. maritima. AmyC is one of two presumably cytoplasmic α-amylases of T. maritima MSB8. The other enzyme (AmyB) displays maximum activity below neutral pH (pH 6.0) and at a temperature of 70°C, 20°C below the temperature of the maximum rate of hydrolysis by AmyC (16). The pH range of AmyC activity (pH 7.5 to 9.0) is unusually basic for an intracellular enzyme. Other α-amylases known to be localized in the cytoplasm are reported to have a pH optimum at neutral pH or below, e.g., AmyA from E. coli has a pH optimum of pH 7.2, PFIA from Pyrococcus furiosus has a pH optimum of pH 7.0, and AmyC from the thermoalkaliphilic Anaerobranca gottschalkii and TVAII from Thermoactinomyces vulgaris display maximum activity at pH 6.0 (2, 13, 21, 26).
The activity of AmyC is reduced by nucleotide triphosphates. To our knowledge only one glycoside hydrolase is described as being regulated by ATP, a debranching enzyme from bovine brain (18). It does not seem unreasonable for an intracellular enzyme such as AmyC, which helps to break down α-glucan polysaccharides or maltooligosaccharides to glucose, to be allosterically regulated through nucleotides, which reflect the energy status of the cell. However, the inhibitory effect of the nucleotides could be reversed by the addition of divalent cations. Therefore, it may be that the observed decrease in activity is merely caused by the chelating effect of the nucleotides and not by their allosteric interaction with the enzyme. Nevertheless, it remains peculiar that the activity of this cytoplasmic α-amylase is significantly inhibited at millimolar concentrations of ATP which are thought to be physiological concentrations in microbial cells. To our knowledge, the inhibition of amylases by ATP has not been described yet.
A strong inhibitory effect was also observed with EDTA. The relatively low concentration of 100 μM EDTA abolished AmyC activity completely, but the activity could, at least partly, be restored by addition of metal ions. It seems likely that either the conformational stability or the catalytic reaction of AmyC requires metal ions. In this context, the presence of iron ions in the AmyC protein, which was detected both biochemically and with X-ray fluorescence spectroscopy, is intriguing. Yet no direct correlation between the presence of iron and the AmyC activity was found. Considering the stimulatory effect of reducing agents on AmyC it seems possible that not the only the presence of iron but also its redox state is important for the protein's activity.
Although Ca2+ counteracts the negative effect of EDTA and ATP on AmyC, neither the enzyme's activity nor the thermostability was increased by the addition of Ca2+. Adding Ca2+ or Mg2+ even decreased the activity. During long-term incubation (6 h) at 90°C Ca2+ even had a destabilizing effect on the AmyC protein. In contrast, many other α-amylases, especially from GHF 13, are known to depend on Ca2+, e.g., the other two known α-amylases from T. maritima. The activity of the extracellular amylase AmyA is greatly increased by Ca2+, and the thermostability of the second intracellular α-amylase of T. maritima is enhanced by Ca2+ (15, 16).
The AmyC amino acid sequence shows no similarity to sequences of members of the classical α-amylase family GHF 13. Instead, the five characteristic conserved regions invariably present in GHF 57 proteins (28) are found in AmyC. The enzyme has significant overall sequence similarities to GHF 57 proteins, especially to the distinct subgroup formed by the uncharacterized and mostly not annotated enzymes with high amino acid sequence similarity to COG1543. By identifying AmyC as an α-amylase an activity is assigned to the COG1543 enzyme group for the first time.
The atypical, for an α-amylase, amino acid sequence of AmyC is also reflected in its three-dimensional structure, which was recently elucidated by X-ray crystallography. While GHF 13 amylases are based on a (β/α)8-barrel structure motif (TIM barrel), AmyC has a distorted (β/α)7-barrel as a supersecondary structure. Despite the differences in structure and amino acid sequence the catalytic residues of AmyC, Glu185 and Asp349, are the same as in most amylases and the crucial amino acid side chains in the catalytic center of the enzyme are arranged in the same sterical orientation as in typical α-amylases and related enzymes (3).
The intracellular location of the novel α-amylase AmyC and its presence during the exponential growth phase point towards a physiological role in either maltodextrin utilization or storage polysaccharide metabolism. In T. maritima further details of both pathways remain to be elucidated.
Acknowledgments
We thank A. Dickmanns and R. Ficner for performing X-ray fluorescence spectroscopy and for helpful discussions.
Financial support by the Deutsche Forschungsgemeinschaft (Li 398/6) is gratefully acknowledged.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballschmiter, M., M. Armbrecht, K. Ivanova, G. Antranikian, and W. Liebl. 2005. AmyA, an alpha-amylase with beta-cyclodextrin-forming activity, and AmyB from the thermoalkaliphilic Anaerobranca gottschalkii: two alpha-amylases adapted to their different cellular localization. Appl. Environ. Microbiol. 71:3709-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickmanns, A., M. Ballschmiter, W. Liebl, and R. Ficner. Crystal structure of the novel α-amylase AmyC from Thermotoga maritima. Acta Crystallogr. D Biol. Crystallogr., in press. [DOI] [PubMed]
- 4.Fish, W. W. 1988. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158:357-364. [DOI] [PubMed] [Google Scholar]
- 5.Fukusumi, S., A. Kamizono, S. Horinouchi, and T. Beppu. 1988. Cloning and nucleotide sequence of a heat stable amylase gene from an anaerobic thermophile, Dictyoglomus thermophilum. Eur. J. Biochem. 174:15-21. [DOI] [PubMed] [Google Scholar]
- 6.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huber, R., T. A. Langworthy, H. Köning, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 8.Imamura, H., S. Fushinobu, B. S. Jeon, T. Wakagi, and H. Matsuzawa. 2001. Identification of the catalytic residue of Thermococcus litoralis 4-alpha- glucanotransferase through mechanism-based labeling. Biochemistry 40:12400- 12406. [DOI] [PubMed] [Google Scholar]
- 9.Imamura, H., S. Fushinobu, M. Yamamoto, T. Kumasaka, B. S. Jeon, T. Wakagi, and H. Matsuzawa. 2003. Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor. J. Biol. Chem. 278:19378-19386. [DOI] [PubMed] [Google Scholar]
- 10.Imamura, H., B. Jeon, T. Wakagi, and H. Matsuzawa. 1999. High level expression of Thermococcus litoralis 4-alpha-glucanotransferase in a soluble form in Escherichia coli with a novel expression system involving minor arginine tRNAs and GroELS. FEBS Lett. 457:393-396. [DOI] [PubMed] [Google Scholar]
- 11.Jeon, B. S., H. Taguchi, H. Sakai, T. Ohshima, T. Wakagi, and H. Matsuzawa. 1997. 4-Alpha-glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis—enzyme purification and characterization, and gene cloning, sequencing and expression in Escherichia coli. Eur. J. Biochem. 248:171-178. [DOI] [PubMed] [Google Scholar]
- 12.Kang, S., C. Vieille, and J. G. Zeikus. 2005. Identification of Pyrococcus furiosus amylopullulanase catalytic residues. Appl. Microbiol. Biotechnol. 66:408-413. [DOI] [PubMed] [Google Scholar]
- 13.Laderman, K. A., B. R. Davis, H. C. Krutzsch, M. S. Lewis, Y. V. Griko, P. L. Privalov, and C. B. Anfinsen. 1993. The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Biol. Chem. 268:24394-24401. [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Liebl, W., I. Stemplinger, and P. Ruile. 1997. Properties and gene structure of the Thermotoga maritima alpha-amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. J. Bacteriol. 179:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, W. J., S. R. Park, C. L. An, J. Y. Lee, S. Y. Hong, E. C. Shin, E. J. Kim, J. O. Kim, H. Kim, and H. D. Yun. 2003. Cloning and characterization of a thermostable intracellular alpha-amylase gene from the hyperthermophilic bacterium Thermotoga maritima MSB8. Res. Microbiol. 154:681-687. [DOI] [PubMed] [Google Scholar]
- 17.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 32:426-428. [Google Scholar]
- 18.Narahara, E., Y. Makino, and K. Omichi. 2001. Glycogen debranching enzyme in bovine brain. J. Biochem. (Tokyo) 130:465-470. [DOI] [PubMed] [Google Scholar]
- 19.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, C. M. Fraser, et al. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 20.Nelson, K. E., J. A. Eisen, and C. M. Fraser. 2001. Genome of Thermotoga maritima MSB8. Methods Enzymol. 330:169-180. [DOI] [PubMed] [Google Scholar]
- 21.Raha, M., I. Kawagishi, V. Muller, M. Kihara, and R. M. Macnab. 1992. Escherichia coli produces a cytoplasmic alpha-amylase, AmyA. J. Bacteriol. 174:6644-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruile, P., C. Winterhalter, and W. Liebl. 1997. Isolation and analysis of a gene encoding alpha-glucuronidase, an enzyme with a novel primary structure involved in the breakdown of xylan. Mol. Microbiol. 23:267-279. [DOI] [PubMed] [Google Scholar]
- 23.Schröder, C., M. Selig, and P. Schönheit. 1994. Glucose fermentation to acetate, CO2 and H2 in the anaerobic hyperthermophilic eubacterium Thermotoga maritima: involvement of the Embden-Meyerhof pathway. Arch. Microbiol. 161:460-470. [Google Scholar]
- 24.Selig, M., K. B. Xavier, H. Santos, and P. Schönheit. 1997. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch. Microbiol. 167:217-232. [DOI] [PubMed] [Google Scholar]
- 25.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonozuka, T., S. Mogi, Y. Shimura, A. Ibuka, H. Sakai, H. Matsuzawa, Y. Sakano, and T. Ohta. 1995. Comparison of primary structures and substrate specificities of two pullulan-hydrolyzing alpha-amylases, TVA I and TVA II, from Thermoactinomyces vulgaris R-47. Biochim. Biophys. Acta 1252:35-42. [DOI] [PubMed] [Google Scholar]
- 27.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zona, R., F. Chang-Pi-Hin, M. J. O'Donohue, and S. Janecek. 2004. Bioinformatics of the glycoside hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus hydrothermalis. Eur. J. Biochem. 271:2863-2872. [DOI] [PubMed] [Google Scholar]