Abstract
The availability of a diverse set of 23S rRNA gene sequences enabled evaluation of the specificity of 39 previously published and 4 newly designed primers specific for bacteria. An extensive clone library constructed using an optimized primer pair resulted in similar gene richness but slightly differing coverage of some phylogenetic groups, compared to a 16S rRNA gene library from the same environmental sample.
There has been renewed interest in the use of the 23S rRNA gene with the decrease in sequencing costs and the growing popularity of techniques such as microarrays (3, 13), analysis of the 16S-23S intergenetic region (7, 9), fluorescence in situ hybridization, and quantitative PCR. The 23S rRNA gene offers the same advantages as the 16S rRNA gene (e.g., universal distribution, conserved function, and invariant and variable regions), yet it includes additional diagnostic sequence stretches due to a greater length, characteristic insertions and/or deletions (12), and possibly better phylogenetic resolution because of greater sequence variation (4, 10-12, 20). However, use of the 23S rRNA gene for bacterial community analysis is hampered by the lack of established broad-range bacterial PCR amplification and sequencing primers.
This study incorporates data from large-scale sequencing efforts to develop new and evaluate existing bacterium-specific 23S rRNA PCR amplification primers. Additionally, this study includes the first well-sampled environmental clone library of 23S rRNA sequences, greatly increasing the number of 23S rRNA gene sequences.
Evaluation of primers.
To check the specificity of PCR primers, an alignment of 23S rRNA gene sequences was developed using the ARB software package (http://www.arb-home.de). Bacterial 23S rRNA sequences were obtained from published sources: the European rRNA database (22), National Center for Biotechnology Information complete bacterial genomes (as of 6 February 2005), the ARB LSU database, and environmental bacterial artificial chromosome clones (16, 18). To ensure broad environmental representation of these primers, sequences were also retrieved using BLAST from the Sargasso Sea assembled database (21) with full-length query 23S rRNA sequences from the genomes of representative organisms (Shigella flexneri 2a strain 301, Pirellula strain 1, Prochlorococcus marinus CCMP 1986, Streptomyces coelicolor A3, Bradyrhozobium japonicum USDA110, and Bacteriodes fragilis YCH46). Using this method, 1,415 nonredundant 23S rRNA sequences of >400 bp each were retrieved from the Sargasso Sea data set. Initial alignments of a total of 2,176 sequences were constructed using the ARB Fast Aligner with manual editing based on secondary structure and the existing ARB alignment. This data set was not corrected for skewing, due to overrepresentation of common laboratory organisms, pathogens, and organisms abundant in the Sargasso Sea.
The primers developed in this study (129f, 189r, 457r, and 2490r, with numbering based on Escherichia coli position) (6) show excellent correspondence to sequences in the aligned database (Table 1); additionally, some mismatches may be the result of PCR or sequencing error. Although some previously published “universal” bacterial primers display broad range, this extensive database indicates that other suggested target regions are not sufficiently conserved to serve as bacterial PCR primers (Table 2). Primers for ITS amplification show various degrees of specificity: the region corresponding to the position of primer 129f is highly conserved (8, 15), but other primers are less conserved and exclude a large fraction of bacterial diversity (7, 19).
TABLE 1.
Percentage mismatches to the 23S rRNA gene dataset for primers designed in this studya
| Primerb | Nucleotide and % mismatch | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 129f | C | Y | G | A | A | T | G | G | G | G | V | A | A | C | C | ||||
| 0.3 | 0.1 | 2.4 | 0.2 | 0.5 | 0.6 | 2.4 | 2.1 | 1.9 | 9.0 | 4.0 | 0.5 | 1.1 | 2.2 | 2.4 | |||||
| 189r | T | A | C | T | D | A | G | A | T | G | T | T | T | C | A | S | T | T | C |
| 0.1 | 0.2 | 2.3 | 0.1 | 0.0 | 0.0 | 0.3 | 0.1 | 0.3 | 4.7 | 1.6 | 0.6 | 0.0 | 0.3 | 0.3 | 4.8 | 0.0 | 0.0 | 0.2 | |
| 457r | C | C | T | T | T | C | C | C | - | T | C | A | C | G | G | T | A | C | T |
| 3.0 | 0.1 | 4.4 | 0.1 | 0.0 | 1.3 | 0.7 | 5.5 | 0.5 | 0.2 | 1.3 | 3.7 | 0.1 | 5.2 | 0.6 | 0.0 | 0.0 | 0.2 | 0.1 | |
| 2490r | C | G | A | C | A | T | C | G | A | G | G | T | G | C | C | A | A | A | C |
| 0.3 | 0.3 | 0.2 | 0.1 | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 | 0.2 | 0.9 | 0.9 | 0.1 | 0.9 | 0.1 | 0.7 | 0.2 | |
Degenerate positions in the sequences were assumed to equally contribute to all possible nucleotides. Boldface type indicates that >5% of database sequences do not match the primer. A hyphen indicates insertions in more than two sequences.
Primer 129f is modified from 130f (9b), 189r is modified from 11A (20a), and 457r is modified from 473r (10).
TABLE 2.
Percentage mismatches to the 23S rRNA gene dataset at each position for previously described universal primersa
| Primer (reference) | Nucleotide and % mismatch | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITSReub (7) | G | C | C | A | A | - | G | G | C | A | T | C | C | A | C | C | |||||||
| 7.9 | 6.2 | 2.9 | 16.4 | 10.8 | 0.4 | 2.9 | 4.2 | 5.8 | 2.1 | 0.5 | 10.6 | 2.8 | 12.8 | 5.3 | 4.0 | ||||||||
| 66r (19) | C | A | C | G | T | C | T | T | T | C | A | T | C | G | S | C | T | ||||||
| 24.4 | 17.6 | 3.1 | 0.9 | 7.7 | 0.1 | 94.4 | 0.0 | 2.6 | 0.4 | 17.0 | 0.4 | 2.9 | 4.2 | 6.2 | 6.0 | 1.3 | |||||||
| fprimer6 (3a) | G | C | G | A | T | T | T | C | Y | G | A | A | Y | G | G | G | R | A | A | A | C | C | C |
| 50.6 | 80.3 | 7.0 | 11.7 | 0.1 | 46.9 | 5.1 | 0.2 | 0.1 | 2.4 | 0.2 | 0.5 | 0.3 | 2.4 | 2.1 | 2.0 | 0.9 | 39.3 | 0.5 | 1.1 | 2.2 | 2.4 | 7.5 | |
| 130f (9b) | C | C | G | A | A | T | G | G | G | G | V | A | A | G* | G* | G* | |||||||
| 0.3 | 8.4 | 2.4 | 0.2 | 0.5 | 0.6 | 2.4 | 2.1 | 2.0 | 9.0 | 4.0 | 0.5 | 1.1 | 100 | 99.9 | 100 | ||||||||
| 130r (9b) | C* | C* | T | T | G | C | C | C | C | A | T | T | C | G | G | ||||||||
| 99.9 | 100 | 1.1 | 0.5 | 84.5 | 9.0 | 2.0 | 2.1 | 2.4 | 0.6 | 0.5 | 0.2 | 2.4 | 8.4 | 0.3 | |||||||||
| 11A (20a) | G | G | A | A | C | T | G | A | A | A | C | A | T | C | T | A | A | G | T | A | |||
| 47.0 | 0.2 | 0.0 | 0.0 | 17.1 | 0.3 | 0.3 | 0.0 | 0.6 | 1.6 | 4.7 | 0.3 | 0.1 | 0.3 | 0.0 | 34.0 | 0.1 | 2.3 | 0.2 | 0.1 | ||||
| 242r (9b) | K | T | T | C | G | C | - | T | C | G | C | C | R | C | T | A | C | ||||||
| 2.9 | 0.2 | 0.6 | 0.8 | 0.5 | 1.7 | 0.6 | 0.0 | 0.0 | 2.2 | 5.5 | 0.0 | 0.3 | 0.2 | 0.3 | 0.0 | 0.1 | |||||||
| 256f (9b) | A | G | T | A | G | Y | G | G | C | G | A | - | G | C | G | A | A | ||||||
| 3.1 | 0.1 | 0.0 | 0.3 | 0.2 | 0.3 | 0.0 | 5.5 | 2.2 | 0.0 | 0.0 | 0.6 | 1.7 | 0.5 | 0.8 | 0.6 | 0.2 | |||||||
| 23ar (20a) | C | G | G | T | A | C | T | - | G | G | T | T | C | A | C | T | A | T | C | G | G | ||
| 0.1 | 5.2 | 0.6 | 0.0 | 0.0 | 0.2 | 0.1 | 0.4 | 38.4 | 10.6 | 0.2 | 13.0 | 0.2 | 17.2 | 0.1 | 0.2 | 0.0 | 0.1 | 1.3 | 0.0 | 2.4 | |||
| rprimer10 (3) | T | T | C | G | C | C | T | T | T | C | C | C | - | T | C | A | C | G | G | T | A | C | T |
| 0.5 | 0.3 | 0.9 | 66.5 | 3.1 | 0.1 | 4.4 | 0.1 | 0.0 | 1.3 | 0.7 | 5.5 | 0.5 | 0.2 | 1.3 | 3.7 | 0.1 | 5.2 | 0.6 | 0.0 | 0.0 | 0.2 | 0.1 | |
| 473f (10) | A | G | T | A | C | C | G | Y | G | A | - | G | G | G | A | A | A | G | |||||
| 0.1 | 0.2 | 0.0 | 0.0 | 0.6 | 5.2 | 0.1 | 0.1 | 1.3 | 0.2 | 0.5 | 5.5 | 0.7 | 1.3 | 0.0 | 0.1 | 4.4 | 0.1 | ||||||
| 559r (9b) | C | A | T | T | M | T | A | C | A | A | A | A | G | G | Y | A | C | G | C | ||||
| 0.3 | 2.5 | 0.1 | 6.2 | 0.3 | 0.7 | 25.6 | 0.2 | 0.3 | 0.1 | 4.9 | 0.3 | 4.3 | 0.1 | 0.3 | 0.1 | 6.7 | 6.6 | 4.9 | |||||
| 559r (21a) | C | A | T | T | M | T | R | C | A | A | A | A | G | G | Y | A | C | G | C | ||||
| 0.3 | 2.5 | 0.1 | 6.2 | 0.3 | 0.7 | 11.4 | 0.2 | 0.3 | 0.1 | 4.9 | 0.3 | 4.3 | 0.1 | 0.3 | 0.1 | 6.7 | 6.6 | 4.9 | |||||
| 803r (21a) | T | T | C | G | G | R | G | A | G | A | A | C | S | A | G | M | T | A | |||||
| 0.3 | 0.5 | 0.5 | 0.2 | 45.0 | 2.8 | 0.8 | 0.5 | 15.8 | 3.7 | 0.3 | 0.2 | 4.9 | 0.3 | 0.2 | 0.4 | 0.1 | 0.5 | ||||||
| 820f (20a) | T | A | G | C | T | G | G | T | T | C | T | C | Y | Y | C | G | A | A | |||||
| 0.5 | 0.1 | 10.5 | 0.2 | 0.3 | 9.5 | 0.2 | 0.3 | 3.7 | 15.8 | 0.5 | 0.8 | 2.8 | 26.7 | 0.2 | 0.5 | 0.5 | 0.3 | ||||||
| 975r (10) | T | C | T | - | G | G | G | Y | T | G | T | T | Y | C | C | C | - | T | |||||
| 0.2 | 1.6 | 6.3 | 0.3 | 0.8 | 0.2 | 2.2 | 0.1 | 0.4 | 21.0 | 0.5 | 0.5 | 1.1 | 2.3 | 0.2 | 0.4 | 0.4 | 0.7 | ||||||
| 43a (20a) | G | G | A | T | G | T | T | G | G | C | T | T | A | G | A | A | G | C | A | G | |||
| 2.0 | 4.5 | 0.4 | 55.2 | 0.2 | 0.0 | 2.4 | 5.2 | 0.0 | 0.1 | 1.6 | 2.2 | 2.7 | 0.2 | 0.0 | 2.6 | 0.0 | 0.0 | 0.0 | 0.1 | ||||
| 1075f (10) | G | T | T | G | G | C | T | T | R | G | A | R | G | C | A | G | C | ||||||
| 0.2 | 0.0 | 2.4 | 5.2 | 0.0 | 0.1 | 1.6 | 2.2 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.3 | |||||||
| 1091r (9b) | R | G | T | G | A | G | C | T | R | T | T | A | C | G | C | ||||||||
| 0.0 | 1.9 | 0.0 | 1.1 | 0.3 | 0.2 | 0.1 | 0.3 | 0.0 | 0.1 | 0.0 | 0.1 | 0.2 | 0.2 | 0.1 | |||||||||
| 1104f (9b) | W | G | C | G | T | A | A | Y | A | G | C | T | C | A | C | ||||||||
| 0.0 | 0.0 | 0.2 | 0.2 | 0.1 | 0.0 | 0.1 | 0.0 | 0.3 | 0.1 | 0.2 | 0.3 | 1.1 | 0.0 | 1.9 | |||||||||
| 1200f (10) | G | G | T | A | G | R | R | G | A | - | G | C | G | T | T | - | C | ||||||
| 0.6 | 2.3 | 0.6 | 0.6 | 0.3 | 25.8 | 2.6 | 1.0 | 0.8 | 0.4 | 16.7 | 1.5 | 4.6 | 0.8 | 10.2 | 0.3 | 15.0 | |||||||
| 1363f (10) | G | A | G | G | C | C | G | A | N | - | - | A | R | G | C | G | - | T | A | ||||
| 6.3 | 2.4 | 4.5 | 8.6 | 22.4 | 24.3 | 2.9 | 1.9 | 0.0 | 1.1 | 0.4 | 5.4 | 11.4 | 22.9 | 11.4 | 0.5 | 0.4 | 2.9 | 1.8 | |||||
| 53a (20a) | G | G | A | C | - | A | A | C | A | G | G | T | T | A | A | - | T | A | T | T | C | C | |
| 0.0 | 0.6 | 53.3 | 80.0 | 3.8 | 0.2 | 29.9 | 23.9 | 28.6 | 3.0 | 7.7 | 0.9 | 11.2 | 26.4 | 0.1 | 2.8 | 8.6 | 0.5 | 1.3 | 0.1 | 8.4 | 3.3 | ||
| 1623f (9b) | A | A | A | C | C | G | W | C | A | C | A | G | G | T | R | G | |||||||
| 3.1 | 0.0 | 1.7 | 0.1 | 5.5 | 3.4 | 0.1 | 0.1 | 3.0 | 0.1 | 2.5 | 4.9 | 0.0 | 0.4 | 0.6 | 5.7 | ||||||||
| 62ar (20a) | G | G | G | G | C | C | A | T | T | T | T | G | C | C | G | A | G | T | T | C | |||
| 10.8 | 8.4 | 54.9 | 42.1 | 55.9 | 27.8 | 0.2 | 26.6 | 2.9 | 0.1 | 0.0 | 0.3 | 0.2 | 8.7 | 34.4 | 0.0 | 0.2 | 0.1 | 0.0 | 0.2 | ||||
| 1685r (21a) | C | C | T | T | M | T | C | S | C | - | G | A | A | S | T | T | A | C | G | G | |||
| 8.5 | 6.6 | 0.0 | 0.0 | 0.9 | 0.1 | 21.6 | 0.1 | 2.7 | 0.9 | 4.9 | 0.0 | 0.4 | 0.1 | 5.6 | 0.1 | 0.1 | 0.0 | 10.8 | 8.4 | ||||
| 69ar (20a) | C | T | T | A | G | G | A | C | C | G | T | T | A | T | A | G | T | T | A | C | |||
| 0.3 | 0.3 | 0.3 | 1.5 | 0.8 | 2.0 | 0.3 | 5.8 | 5.9 | 0.7 | 0.1 | 3.0 | 0.0 | 0.8 | 0.6 | 0.5 | 0.3 | 0.1 | 0.0 | 0.2 | ||||
| 1930r (9b) | C | G | A | C | A | A | G | G | A | A | - | T | T | T | C | G | C | T | A | C | |||
| 3.7 | 0.0 | 0.7 | 0.5 | 0.9 | 0.1 | 0.4 | 0.3 | 1.1 | 0.0 | 0.4 | 1.1 | 0.4 | 0.2 | 0.3 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | ||||
| 2069f (9b) | G | A | C | G | Y* | A | A | A | G | A | C | C | C | C | R | T | G | ||||||
| 0.1 | 0.0 | 0.2 | 3.1 | 99.8 | 0.5 | 0.5 | 0.3 | 0.1 | 0.1 | 0.3 | 0.0 | 0.0 | 3.5 | 0.0 | 5.8 | 1.6 | |||||||
| 2241r (9b) | A | C | C | G | C | C | C | C | A | G | T | H | A | A | A | C | T | ||||||
| 0.4 | 0.4 | 0.7 | 2.6 | 0.4 | 0.4 | 0.1 | 0.0 | 0.0 | 0.3 | 0.9 | 1.0 | 0.1 | 0.1 | 0.2 | 0.2 | 0.6 | |||||||
| 2436f (10) | T | C | - | G | C | T | C | A | A | C | G | G | A | T | A | A | A | A | G | ||||
| 0.9 | 2.7 | 0.3 | 3.0 | 1.5 | 0.3 | 4.1 | 0.3 | 3.0 | 6.1 | 0.7 | 3.5 | 1.9 | 0.8 | 0.4 | 0.2 | 0.0 | 0.3 | 0.2 | |||||
| 2498r (9b) | G | A | G | Y | C | G | A | C | A | T | C | G | A | G | G | ||||||||
| 0.3 | 0.1 | 0.3 | 0.0 | 0.3 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 | |||||||||
| 93ar (20a) | C | G | A | C | G | - | T | T | C | T | G | A | A | C | C | C | A | G | C | T | C | ||
| 0.2 | 0.2 | 0.1 | 0.2 | 0.2 | 0.2 | 2.0 | 0.1 | 10.7 | 0.2 | 38.6 | 0.2 | 0.2 | 0.2 | 0.2 | 0.1 | 0.1 | 2.9 | 0.2 | 0.2 | 0.2 | |||
| 2603f (21a) | A | R | A | M | - | C | G | T | C | G | T | G | A | G | A | C | A | G | |||||
| 0.2 | 0.1 | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 | 0.1 | 0.2 | 0.2 | 2.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 | ||||||
| 2669f (9b) | A | G | T | A | C | G | A | G | - | A | G | G | A | C | C | G | G | ||||||
| 0.5 | 0.2 | 0.1 | 0.2 | 0.3 | 0.2 | 0.2 | 0.6 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.6 | 0.3 | 3.5 | 8.0 | |||||||
| 2744r (10) | C | T | T | - | A | G | A | T | G | C | Y | T | T | C | A | G | C | ||||||
| 2.2 | 0.1 | 0.2 | 0.4 | 0.1 | 5.7 | 0.1 | 0.2 | 0.2 | 0.4 | 1.5 | 0.1 | 0.0 | 0.2 | 0.2 | 2.3 | 3.2 | |||||||
| 2747r (9b) | G | Y | T | T | - | A | G | A | T | G | C | Y | T | T | C | ||||||||
| 17.4 | 0.0 | 0.1 | 0.2 | 0.4 | 0.1 | 5.7 | 0.1 | 0.2 | 0.2 | 0.4 | 1.5 | 0.1 | 0.0 | 0.2 | |||||||||
| 97ar (20a) | C | C | C | G | C | T | T | - | A | G | A | T | G | C | T | T | T | C | A | G | C | ||
| 2.3 | 35.9 | 19.0 | 17.4 | 2.2 | 0.1 | 0.2 | 0.4 | 0.1 | 5.7 | 0.1 | 0.2 | 0.2 | 0.4 | 5.6 | 0.1 | 0.0 | 0.2 | 0.2 | 2.3 | 3.2 | |||
| 2758f (9b) | Y | T | G | A | A | R | G | C | A | T | C | T | - | A | A | ||||||||
| 0.0 | 0.2 | 0.2 | 0.0 | 0.1 | 1.5 | 0.4 | 0.2 | 0.2 | 0.1 | 5.7 | 0.1 | 0.4 | 0.2 | 0.1 | |||||||||
Primers follow the naming convention of the original publication but are ordered according to their position along the 23S rRNA sequence. Positions shown in boldface indicate that >5% of sequences do not match the primer at that position. Degenerate positions in the sequences were assumed to equally contribute to all possible nucleotides. Hyphens indicate insertions in more than two sequences. *, probable typographic error in the published primer sequence.
On the basis of their broad specificity, length of amplified sequence, and good amplification properties, we propose using the primers 129f (modified in this study) and 2241r for studies of bacterial 23S rRNA diversity. These primers amplify a large portion of the 23S rRNA, consistently produce only a single band of PCR product, and are highly conserved across the bacterial sequences currently available (Tables 1 and 2). Positive amplification was achieved with a diverse set of isolates under the following conditions: 3 min at 94°C; then 30 cycles, each consisting of 1 min at 94°C, 1 min at 57°C, and 2 min at 72°C; and a final 5-min extension at 72°C. All isolates used to test the primers produced PCR product of the correct size; the phyla of bacteria are listed and the number of isolates tested is given in parentheses: α-Proteobacteria (7), β-Proteobacteria (2), δ-Proteobacteria (1), ɛ-Proteobacteria (1), γ-Proteobacteria (22), Firmicutes (7), Bacteroidetes (8), and Cyanobacteria (2).
Analysis of 23S rRNA clone library.
The 129f-2241r primer set was subsequently used to construct a clone library to evaluate coverage and relative distribution of phyla in comparison with a 16S rRNA clone library constructed from a parallel sample (1). A surface seawater sample from the marine end of Plum Island Sound estuary (northeastern Massachusetts) was collected as previously described (1). Cells were lysed using bead beading (5), and DNA was purified using phenol:chloroform:isoamyl alcohol extraction, sodium acetate and ethanol precipitation, and RNase I treatment (17). DNA was amplified in 10 replicate 20-μl PCRs, each reaction mixture containing 50 ng of purified DNA template. PCR conditions were as follows: 3 min at 94°C; then 15 cycles, each consisting of 1 min at 94°C, 1 min at 57°C, and 2 min at 72°C; and a final 5-min extension at 72°C. PCR products were pooled, precipitated with ethanol, and gel extracted (QIAGEN gel extraction kit). Amplicons were cloned using the TOPO-TA kit (Invitrogen).
A total of 535 operational taxonomic units (OTUs) were identified, based on sequential digests with restriction enzymes HhaI and MspI of cloned inserts amplified using internal plasmid primers (M13). Inserts with restriction patterns adding up to >2,500 nucleotides were excluded, as they were assumed to originate from more than one cloned 23S rRNA gene insert. To determine the phylogenetic coverage, at least one member of each OTU was sequenced and grouped into higher taxonomic groups (subphylum or phylum). Both 129f and 457r were used as sequencing primers on plasmids extracted using a QIAprep Spin Miniprep kit (QIAGEN) and M13-amplified PCR products, respectively. A total of 614 clone library sequences were edited using Sequencher, and phylum-level identification of the OTUs was made using discontinuous megaBLAST with a scoring metric (match = 4; mismatch = −5) to allow identification of sequences highly divergent from those present in the database. The cutoff for categorization of a sequence was a sequence length of 300 bp of at least 85% similarity to an organism of known phylogeny.
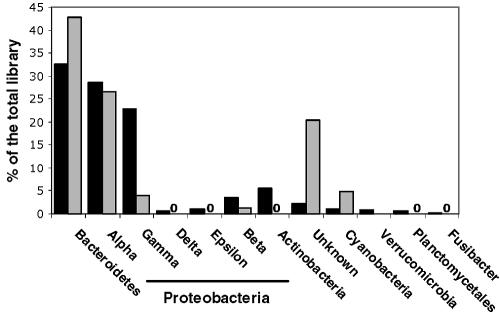
A comparison of 23S and 16S rRNA (1) gene clone libraries constructed from replicate water samples yielded gross similarities but also some important differences (Fig. 1). The observed levels of richness in the two libraries were comparable when the digestion-defined OTUs in the 23S library were approximated by 99% sequence identity clusters (1, 14) in the 16S rRNA library (535 versus 520 for the 23S and 16S rRNA gene libraries, respectively). In both libraries, Bacteroidetes and α-Proteobacteria were the most abundant groups (2). However, the 23S library displayed a higher percentage of Bacteriodetes (42.8% versus 32.5%) and lower percentages of γ-Proteobacteria (3.9% versus 22.8%), Actinobacteria, and minor groups. This comparison is of interest because it may reflect the primer bias of either 23S or 16S rRNA primers, a shallower depth of sequence coverage in the 23S library masking rare variants, or a limited 23S rRNA database preventing identification of certain groups. Planctomycetales were probably excluded by these 23S rRNA primers because the forward primer targets a region not present in their 23S rRNA gene. Additionally, >5% mismatches were observed at position 10 of primer 129f to the set of aligned sequences (Table 1); these mismatches occurred primarily in environmental sequences rather than in cultured isolates, confirming the value of incorporating environmental shotgun sequences in the alignment. This mismatch may explain the low level of abundance of γ-Proteobacteria in the clone library, as this alternate sequence is present in the SAR-86 group (16, 18) and other γ-Proteobacteria in the database, as well as members of other phyla. This problem can be remedied by adding an additional degeneracy to primer 129f with the final sequence as CYGAATGGGRVAACC; this modified primer paired with 2241r positively amplified a subset of 14 isolates from diverse phyla. The large number of unknowns is due to the difficulty of 23S rRNA sequence identification because of the poor depth of sequence coverage, especially for less-well-studied phyla (i.e., β-, δ-, and ɛ-Proteobacteria).
FIG. 1.
Relative frequency distribution of major phylogenetic groups detected among the environmental sequences from a 16S rRNA library (black) and a 23S rRNA library (gray).
Applications using 23S primers, especially techniques such as automated rRNA intergenic spacer analysis that are highly sensitive to the primers chosen (7), should be reevaluated and perhaps modified in light of this data. Nonetheless, this comparison of 16S and 23S rRNA gene sequences shows that reasonable coverage and agreement between broad-range primer pairs can be achieved.
The alignment used to check the primers is available online (see the supplemental material and the ARB database, available for download at http://web.mit.edu/polz/seq=align.html).
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank with accession numbers DQ312516 to DQ313129.
Supplementary Material
Acknowledgments
This research was supported by grants from NSF, NIEHS, and DOE (Genomes to Life).
Thanks to Ivan Ceraj for programming assistance and Patricia McAndrew for help with initial clone library screening.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Acinas, S. G., R. Sarma-Rupavtarm, V. Klepac-Ceraj, and M. F. Polz. 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71:8966-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony, R. M., J. T. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Anthony, R. M., J. T. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anton, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1999. Intraspecific diversity of the 23S rRNA gene and the spacer region downstream in Escherichia coli. J. Bacteriol. 181:2703-2709.10217757 [Google Scholar]
- 5.Bertilsson, S., C. M. Cavanaugh, and M. F. Polz. 2002. Sequencing-independent method to generate oligonucleotide probes targeting a variable region in bacterial 16S rRNA by PCR with detachable primers. Appl. Environ. Microbiol. 68:6077-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 7.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 9a.Ibrahim, A., J. Hofman-Bang, and B. K. Ahring. 2001. Amplification and direct sequence analysis of the 23S rRNA gene from thermophilic bacteria. BioTechniques 30:414-416. [DOI] [PubMed] [Google Scholar]
- 9b.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons Ltd., New York, N.Y.
- 10.Ludwig, W., G. Kirchhof, N. Klugbauer, M. Weizenegger, D. Betzl, M. Ehrmann, C. Hertel, S. Jilg, R. Tatzel, H. Zitzelsberger, S. Liebl, M. Hochberger, J. Shah, D. Lane, and P. R. Wallnoef. 1992. Complete 23S ribosomal RNA sequences of gram-positive bacteria with a low DNA G+C content. Syst. Appl. Microbiol. 15:487-501. [Google Scholar]
- 11.Ludwig, W., R. Rossellomora, R. Aznar, S. Klugbauer, S. Spring, K. Reetz, C. Beimfohr, E. Brockmann, G. Kirchhof, S. Dorn, M. Bachleitner, N. Klugbauer, N. Springer, D. Lane, R. Nietupsky, M. Weizenegger, and K. H. Schleifer. 1995. Comparative sequence analysis of 23S rRNA from Proteobacteria. Syst. Appl. Microbiol. 18:164-188. [Google Scholar]
- 12.Ludwig, W., and K. H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev 15:155-173. [DOI] [PubMed] [Google Scholar]
- 13.Mitterer, G., M. Huber, E. Leidinger, C. Kirisits, W. Lubitz, M. W. Mueller, and W. M. Schmidt. 2004. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J. Clin. Microbiol. 42:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyer, C. L., J. M. Tiedje, F. C. Dobbs, and D. M. Karl. 1996. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature? Appl. Environ. Microbiol. 62:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjard, L., F. Poly, J. C. Lata, C. Mougel, J. Thioulouse, and S. Nazaret. 2001. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabehi, G., O. Beja, M. T. Suzuki, C. M. Preston, and E. F. DeLong. 2004. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6:903-910. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Suzuki, M. T., O. Beja, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki, M. T., C. M. Preston, O. Beja, J. R. de la Torre, G. F. Steward, and E. F. DeLong. 2004. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb. Ecol. 48:473-488. [DOI] [PubMed] [Google Scholar]
- 20.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Van Camp, G., S. Chapelle, and R. De Wachter. 1993. Amplification and sequencing of variable regions in bacterial 23S ribosomal-RNA genes with conserved primer sequences. Curr. Microbiol. 27:147-151. [DOI] [PubMed] [Google Scholar]
- 21.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 21a.Ward, N. L., F. A. Rainey, B. P. Hedlund, J. T. Staley, W. Ludwig, and F. Stackebrandt. 2000. Comparative phylogenetic analyses of members of the order Planctomycetales and the division Verrucomicrobia: 23S rRNA gene sequence analysis supports the 16S rRNA gene sequence-derived phylogeny. Int. J. Syst. Evol. Microbiol. 50:1965-1972. [DOI] [PubMed] [Google Scholar]
- 22.Wuyts, J., G. Perriere, and Y. V. de Peer. 2004. The European ribosomal RNA database. Nucleic Acids Res. 32:D101-D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.