Abstract
Nitrobenzene 1,2-dioxygenase from Comamonas sp. strain JS765 catalyzes the initial reaction in nitrobenzene degradation, forming catechol and nitrite. The enzyme also oxidizes the aromatic rings of mono- and dinitrotoluenes at the nitro-substituted carbon, but the basis for this specificity is not understood. In this study, site-directed mutagenesis was used to modify the active site of nitrobenzene dioxygenase, and the contribution of specific residues in controlling substrate specificity and enzyme performance was evaluated. The activities of six mutant enzymes indicated that the residues at positions 258, 293, and 350 in the α subunit are important for determining regiospecificity with nitroarene substrates and enantiospecificity with naphthalene. The results provide an explanation for the characteristic specificity with nitroarene substrates. Based on the structure of nitrobenzene dioxygenase, substitution of valine for the asparagine at position 258 should eliminate a hydrogen bond between the substrate nitro group and the amino group of asparagine. Up to 99% of the mononitrotoluene oxidation products formed by the N258V mutant were nitrobenzyl alcohols rather than catechols, supporting the importance of this hydrogen bond in positioning substrates in the active site for ring oxidation. Similar results were obtained with an I350F mutant, where the formation of the hydrogen bond appeared to be prevented by steric interference. The specificity of enzymes with substitutions at position 293 varied depending on the residue present. Compared to the wild type, the F293Q mutant was 2.5 times faster at oxidizing 2,6-dinitrotoluene while retaining a similar Km for the substrate based on product formation rates and whole-cell kinetics.
Nitroaromatic compounds are commonly used for the production of pesticides, dyes, and polymers (34). Widespread application and improper disposal of these chemicals have resulted in their release into the environment. Past synthesis of the explosive 2,4,6-trinitrotoluene at military facilities in the United States has contaminated soil and groundwater with mixtures of mono-, di-, and trinitrotoluene isomers (34, 39). Nitrotoluenes pose a significant risk to human health and the environment due to their acute toxicity and suspected carcinogenicity, and several of them are listed on the U.S. Environmental Protection Agency's list of priority pollutants (25, 34, 44). Additionally, nitroaromatic compounds are rapidly reduced to aromatic amines in the environment; these chemicals are established human carcinogens and piscicides (34). In general, nitroaromatic compounds are recalcitrant to biodegradation; however, bacterial strains that grow on and completely mineralize nitrobenzene, 2-nitrotoluene (2NT), 2,4-dinitrotoluene, or 2,6-dinitrotoluene have been isolated and characterized (8, 13, 23, 24, 40). In Comamonas sp. strain JS765, the key enzyme allowing growth on nitrobenzene is nitrobenzene 1,2-dioxygenase (NBDO), which catalyzes the oxidation of nitrobenzene to catechol and nitrite (24, 29). The reaction requires molecular oxygen as a substrate and involves an iron-sulfur flavoprotein reductase (NbzAa) that transfers electrons from NAD(P)H through a ferredoxin (NbzAb), to the oxygenase (NbzAcAd) (29). Catechol is subsequently transformed to tricarboxylic acid cycle intermediates through a meta-ring cleavage pathway (24). Genes encoding NBDO are clustered in a catabolic operon (nbzAaAbAcAd) under the control of a divergently transcribed LysR-type transcriptional regulator, designated NbzR (20, 21).
Numerous similarities between nitrobenzene and naphthalene degradation gene clusters suggest that the two enzyme systems are evolutionarily related (20). Comparisons of the deduced amino acid sequences of the NBDO proteins from JS765 and the naphthalene dioxygenase proteins from Ralstonia sp. strain U2 (NDOU2) showed ≥88% sequence identity (20). In addition, NBDO from JS765 retains the ability to oxidize naphthalene to naphthalene cis-dihydrodiol, a reaction necessary for naphthalene metabolism, although JS765 is unable to grow on naphthalene or any of its degradation intermediates (20). In contrast, dioxygenation of nitrobenzene appears to be a novel activity acquired by NBDO that NDOU2 is unable to catalyze. However, mutant forms of NDOU2 that can oxidize dinitrotoluenes have been engineered (15).
There is significant interest in the development and application of Rieske nonheme iron dioxygenases as biocatalysts to produce compounds for use in the synthesis of industrial chemicals and pharmaceuticals (2, 3, 10, 36). However, in contrast to naphthalene dioxygenase, much less is known about nitroarene dioxygenase substrate specificity. More importantly, we wished to understand the unique ability of nitroarene dioxygenases to specifically attack the nitro-substituted carbons of benzene rings. To address these issues, we initiated an analysis of NBDO by rational mutagenesis. By using the available crystal structure of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 (NDO9816-4) and amino acid alignments of NBDO from Comamonas sp. strain JS765 (20), 2-nitrotoluene dioxygenase (2NTDO) from Acidovorax sp. strain JS42 (26), 2,4-dinitrotoluene dioxygenase (DNTDO) from Burkholderia sp. strain DNT (DNTDODNT) and Burkholderia cepacia R34 (13, 43), and NDOs from Pseudomonas sp. strain NCIB 9816-4 (26) and Ralstonia sp. strain U2 (7) together with previous detailed analyses of NDO9816-4 (30, 31, 46), three amino acid positions were predicted to be critical for substrate positioning and were selected for analysis. These predicted active-site residues were quite variable in the closely related enzymes that were compared (Table 1), suggesting that they might play a role in the very different substrate specificities exhibited by these enzymes. Six NBDO mutants with single amino acid substitutions at the active site were tested for their activities with a series of aromatic substrates. Based on the results, we were able to provide experimental evidence for predictions made from the recently solved crystal structure of NBDO (6).
TABLE 1.
Amino acid differences at the active sites of nitroarene and naphthalene dioxygenases
| Enzyme | Strain | α subunit amino acid sequence identity (%)a | Amino acid residue at positionb:
|
||
|---|---|---|---|---|---|
| 258 | 293 | 350 | |||
| NBDO | Comamonas sp. strain JS765 | 100 | Asn | Phe | Ile |
| 2NTDO | Acidovorax sp. strain JS42 | 95 | Asn | Ile | Ile |
| DNTDODNT | Burkholderia sp. strain DNT | 84 | Val | Gln | Thr |
| DNTDOR34 | Burkholderia cepacia R34 | 84 | Ile | Gln | Val |
| NDO9816-4 | Pseudomonas sp. strain NCIB 9816-4 | 82 | Val | His | Phe |
| NDOU2 | Ralstonia sp. strain U2 | 88 | Val | His | Phe |
Relative to NBDO.
NBDO numbering.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain DH5α (Life Technologies, Gaithersburg, Md.) was used for cloning and expression of recombinant dioxygenase constructs. Plasmid pKSJ4 was generated by subcloning a 4.6-kb SacI-EcoRI DNA fragment containing the nbzAaAbAcAd genes from pDTG925 (20) into SacI-EcoRI-digested pDTG850 (28), replacing the ntdAaAbAcAd genes. E. coli strains ES1301 mutS (Promega Corp., Madison, Wis.) and JM109 (45) were used for site-directed mutagenesis. DH5α(pUC18) (45) was used as a vector control in biotransformation reactions.
Media and growth conditions.
E. coli strains were grown at 37°C in Luria-Bertani (LB) medium (5) with ampicillin and tetracycline supplemented to 200 μg/ml and 12.5 μg/ml, respectively, as appropriate. For whole-cell biotransformations and kinetic experiments, DH5α strains expressing wild-type and mutant dioxygenase genes were grown in minimal medium with glucose as the sole carbon source to ensure balanced growth and the absence of competing indigo formation. DH5α strains carrying plasmids of interest were cultured aerobically at 37°C in 500-ml flasks containing 100 ml of a modified minimal medium (MSB-EDTA [nitrilotriacetic acid substituted with an equimolar concentration of EDTA]) (41) supplemented with 20 mM glucose, 1 mM thiamine, and ampicillin (200 μg/ml). Exponentially growing cultures were harvested when they reached an optical density of 1.6 to 1.8 (A660) for biotransformation experiments as described below. A previous study showed that the dioxygenase activity of DH5α cells expressing nitroarene dioxygenase genes from pUC-derived plasmids decreased upon the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (27), and therefore, IPTG induction was not used in this study. For plates, MSB-EDTA was solidified with 1.8% (wt/vol) Noble agar (Difco), and LB medium was solidified with 1.5% Bacto agar (Difco).
DNA purification and manipulations.
Standard methods were used to manipulate plasmids and DNA fragments (35). Restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs (Beverly, Mass.). Plasmids were purified as previously described (19) or with a QIAprep Miniprep kit (QIAGEN, Valencia, Calif.). DNA fragments were purified from gel slices using a QIAquick gel extraction kit (QIAGEN). Transformation of E. coli strains was carried out by standard procedures (9). Nucleotide and amino acid sequence analyses were performed using the Lasergene sequence analysis package (DNAStar, Inc., Madison, Wis.).
Site-directed mutagenesis.
Mutagenesis of nbzAc, which encodes the α subunit of NBDO, was carried out using the Altered Sites II in vitro mutagenesis system (Promega Corp.). Plasmid pKSJ2, the master template for mutagenesis, was constructed by subcloning a 949-bp PstI-KpnI DNA fragment that encodes the C-terminal portion of the α subunit of NBDO and the N-terminal part of the β subunit of NBDO into PstI-KpnI-digested pAlter-1 (Promega Corp.). The following mutagenic oligonucleotides were designed with silent mutations that introduced new restriction sites to allow easy screening for desired clones (underlined bases indicate the position of the introduced restriction site; base changes are in boldface type): N258V (5′-CTACTACTCCGGTGTCTTCAGCGCTGA-3′) (BbsI), F293H (5′-ACGGATTTACCGGAGCCATCTGAACGGCAC-3′) (NlaIV), F293I (5′-GGATTTACCGCAGTATACTGAACGGCACGA-3′) (AccI), F293Q (5′-GATTTACCGCAGCCAGCTGAACGGCACG-3′) (PvuII), I350F (5′-CGCGGTTCAGCGAAGCTTCGGACCAGCAGG-3′) (HindIII), and I350T (5′-GTTCAGCGCAGTACTGGACCAGCAGGAT-3′) (ScaI). Phosphorylated oligonucleotides were synthesized by MWG-Biotech (Greensboro, N.C.). After restriction analysis and nucleotide sequencing of both strands of the insertion in pKSJ2 to verify the desired mutations, the 949-bp PstI-KpnI DNA fragments were individually cloned into pKSJ4. The presence of each mutation was reconfirmed by restriction analysis and nucleotide sequencing. Fluorescent automated DNA sequencing was carried out by the University of California, Davis, DNA sequencing facility with an Applied Biosystems 3730 automated DNA sequencer. The resulting derivatives of pKSJ4 were used to transform DH5α for expression and substrate specificity studies.
Indigo formation.
DH5α strains carrying pKSJ4 derivatives with the various mutations were grown overnight at 37°C on nitrocellulose filters placed onto MSB-EDTA agar plates containing 20 mM glucose, 1 mM thiamine, and 200 μg/ml ampicillin. Dried Whatman no. 1 filters soaked in a 10% solution of indole in acetone were placed into petri dish covers after colony formation. Plates were incubated at 30°C, and production of indigo from indole was observed after 24 h.
Biotransformation of aromatic substrates.
E. coli cultures expressing recombinant wild-type and mutant dioxygenase enzymes were prepared as described above, and 25-ml volumes were dispensed into 125-ml Erlenmeyer flasks. DH5α(pUC18) served as a negative control for biotransformation reactions. Cultures were supplemented with 20 mM glucose and 40 mM phosphate buffer (pH 7.3). Reaction mixtures were incubated at 30°C with shaking (200 rpm) for 6 h with 0.1% (wt/vol) naphthalene or 4-nitrotoluene (4NT), 0.1% (vol/vol) nitrobenzene, 2NT, or 3-nitrotoluene (3NT) or for 16 h with 0.05% (wt/vol) 2,4-dinitrotoluene (24DNT) or 2,6-dinitrotoluene (26DNT).
Analytical methods.
Nitrite released from biotransformation reactions was determined by a modification of a previously described method (37). Specifically, 50 μl of clarified sample supernatants was transferred into 96-well microtiter dishes and mixed with 50 μl of 1% (wt/vol) sulfanilamide in 1.5 N HCl. After 5 min, 50 μl of 0.02% (wt/vol) N-(1-napthyl)ethylenediamine in 1.5 N HCl was added. After an additional 5 min, formation of the pink-colored azo dye complex was quantified on a Victor3 multilabel counter (Perkin-Elmer, Boston, Mass.) by measuring the absorbance at 550 nm compared with a sodium nitrite standard curve.
Clarified supernatants from whole-cell biotransformations were extracted with sodium hydroxide-washed ethyl acetate and concentrated by rotary evaporation under reduced pressure. Biotransformation products were dissolved in 0.5 ml of acetonitrile and analyzed by thin-layer chromatography (TLC) as previously described (33). Trimethylsilyl derivatives were prepared by derivatizing an equal volume of dissolved product with N,O-bis(trimethysilyl)trifluoroacetamide according to instructions supplied by the distributor (Supelco, Bellefonte, Pa.). Gas chromatography-mass spectrometry (GC-MS) analyses were performed with an Agilent 6890N gas chromatograph equipped with a Supleco Equity-1 capillary column (30 m by 250 μm; 25-μm film thickness), a 7683 series autoinjector, and an Agilent 5973 network mass selective detector (Agilent Technologies, Palo Alto, Calif.). Helium was used as the carrier gas with a constant flow rate of 0.5 ml min−1. The injector and transfer lines were 220 and 300°C, respectively. The chromatography program was as follows: an initial column temperature of 70°C, a temperature increase of 10°C min−1 to 240°C. The ionization voltage and electron multiplier settings were 70 eV and 1,294 V, respectively. Product identities were confirmed by a comparison of retention times and MS fragmentation profiles to authentic chemical standards.
Naphthalene cis-dihydrodiol was purified by preparative-layer liquid chromatography using 0.1-mm silica gel 60F254 plates (EMD Chemicals, Gibbstown, N.J.) as previously described (28). Enantiomeric composition of naphthalene cis-dihydrodiol was determined using a Beckman Coulter System Gold 125 high-performance liquid chromatography system with a model 168 diode array multiwavelength detector (Beckman Coulter, Inc., Fullerton, Calif.) and a Chiracel OJ column (Chiral Technologies, Inc., Exton, Pa.) as previously described (33).
Measurement of nitrobenzene and 26DNT oxidation rates.
E. coli cultures were grown as described above, harvested by centrifugation, and resuspended in 40 mM phosphate buffer (pH 7.3) with 20 mM glucose to an optical density of 1.6 to 1.8 (A660), and 25-ml volumes were dispensed into 125-ml Erlenmeyer flasks. The maximum solubilities of nitrobenzene and 26DNT in water were assumed to be approximately 15 and 1 mM, respectively. Nitrobenzene was added from 0.2 and 1 M methanol stock solutions to final concentrations ranging from 12.5 μM to 15 mM. 26DNT was added from a 50 mM methanol-acetone (50:50) stock solution to final concentrations ranging from 25 to 1,000 μM. Cultures were incubated at 30°C with shaking (200 rpm), and 1-ml volumes were periodically withdrawn and immediately frozen on dry ice during the initial 0 to 3 h. Samples were stored at −20°C until assayed. Nitrite in clarified sample supernatants was determined as described above. Total protein was determined by the Bradford assay (4) after resuspending cell pellets in 100 mM NaOH and boiling them for 10 min. Bovine serum albumin was used as the standard. Kinetic coefficients of nitrite formation resulting from nitrobenzene and 26DNT oxidation were estimated by nonlinear regression with the Michaelis-Menten equation using Microsoft Excel Solver (38).
SDS-PAGE and Western analysis.
Cell pellets (from 1-ml culture suspensions) were resuspended in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, boiled for 10 min, and separated by SDS-PAGE (12% polyacrylamide) (1). Gels were subjected to Western blotting with a polyclonal antibody for NDO9816-4, as described previously (28), and crude cell extract from JM109(DE3)(pDTG141) (42) expressing NDO9816-4 and purified NBDO (29) as controls. Antigens were visualized using alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Pierce, Rockford, Ill.).
Chemicals.
1,2-Dihydronaphthalene (98%), catechol (>99.5%), 3-methylcatechol (98%), 4-methylcatechol (>95%), 2-nitrobenzyl alcohol (97%), 3-nitrobenzyl alcohol (98%), 4-nitrobenzyl alcohol (99%), 4NT (99%), and 26DNT (98%) were purchased from Aldrich (Milwaukee, Wis.). Naphthalene (99%) and nitrobenzene were obtained from Acros Organics (Morris Plains, N.J.), and 2NT (99%), 3NT (99%), and 24DNT (97%) were from Avocado (Heysham, Lancashire, United Kingdom). 4-Methyl-5-nitrocatechol, 4-methyl-3-nitrocatechol, 3-methyl-4-nitrocatechol, 3-amino-4-methyl-5-nitrocatechol, and 3-amino-6-methyl-5-nitrocatechol were generously provided by Jim C. Spain (Georgia Institute of Technology). (+)-(1R, 2S)-cis-1,2-Dihydroxy-1,2-dihydronaphthalene was prepared as previously described by Jeffrey et al. (11) using Pseudomonas sp. strain 9816/11 (17), a naphthalene cis-dihydrodiol dehydrogenase mutant of the naphthalene-degrading strain Pseudomonas sp. strain NCIB 9816-4 that accumulates the product when incubated with naphthalene.
RESULTS
Construction and preliminary analysis of NBDO variants.
Amino acid residues near the active site of NBDO were modeled based on the crystal structure of NDO9816-4 (14). Asn-258, Phe-293, and Ile-350 were predicted to line one face of the active site of NBDO, and the corresponding residues in NDO9816-4 (Val-260, His-295, and Phe-352) were previously shown to mediate regioselectivity and enantioselectivity (30, 31, 46). To evaluate the contribution of these amino acids to substrate specificity and catalytic efficiency in these five enzymes, missense mutations were introduced into the gene encoding the α subunit of NBDO, generating mutant enzymes with amino acid substitutions at positions 258, 293, and 350. Specific amino acid substitutions were chosen based on sequence comparisons primarily between NBDO, 2NTDO, DNTDODNT, and NDO9816-4 (Table 1), because more is known about the substrate specificities of these enzymes. The following amino acids were mutated in NBDO: Asn-258 was changed to valine; Phe-293 was changed to isoleucine, histidine, and glutamine; and Ile-350 was changed to phenylalanine and threonine.
Indigo formation was used as an initial measurement of NBDO activity. DH5α strains carrying pDTG850 (2NTDO), pKSJ4 (wild-type NBDO), and mutant derivatives of pKSJ4 were grown on plates and then incubated in the presence of indole vapor. All of the strains formed blue colonies except for the strain expressing the F293I mutant. Wild-type NBDO and 2NTDO formed dark blue and pale blue colonies, respectively, in the presence of indole. Substitution of phenylalanine at position 293 with histidine or isoleucine appeared to reduce or abolish the ability of NBDO to attack indole, but substitution of Asn-258 with valine resulted in increased indigo formation. Substitution of Ile-350 with phenylalanine or threonine did not have a significant effect on indole oxidation. In contrast, mutant NDO9816-4 enzymes with amino acid substitutions at the positions corresponding to position 258, 293, or 350 all produced less indigo than wild-type NDO (46).
To ensure that recombinant strains were expressing similar levels of NBDO and NBDO variants, total protein was resolved by 12% SDS-PAGE, and Western analysis was carried out using polyclonal serum that reacts with the α and β subunits of NDO9816-4 (28). The polyclonal antibody reacted positively with both subunits of purified NBDO and showed similar amounts of dioxygenase enzyme present in the recombinant cultures (data not shown). More importantly, these results indicate that differences in the catalytic activities of the NBDO mutants were not due to differences in enzyme production.
Biotransformation reactions with naphthalene.
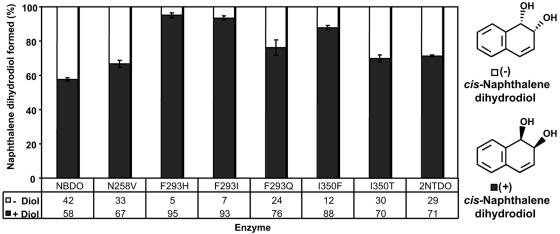
Biotransformations with naphthalene as the substrate yielded cis-1,2-dihydroxy-1,2-dihydronaphthalene (naphthalene cis-dihydrodiol) from all of the NBDO variants. The stereochemistry of the cis-naphthalene dihydrodiols was analyzed by chiral stationary-phase high-performance liquid chromatography. NBDO and 2NTDO showed characteristic ratios of (+)- and (−)-naphthalene cis-dihydrodiols (Fig. 1) (20, 27). The mutant forms of NBDO showed slight to moderate increases in enantioselectivity; each formed a higher ratio of (+)- to (−)-naphthalene cis-dihydrodiol compared to wild-type NBDO. No product was detected in biotransformations with the vector control strain DH5α(pUC18).
FIG. 1.
Enantiomeric composition of naphthalene cis-dihydrodiol produced from naphthalene in biotransformation reactions (n = 3) with wild-type NBDO, NBDO variants, and 2NTDO. (−)-Naphthalene cis-dihydrodiol (− Diol) is represented by white bars, and (+)-naphthalene cis-dihydrodiol (+ Diol) is represented by dark gray bars.
Biotransformation reactions with nitrobenzene.
2NTDO and the NBDO enzymes all oxidized nitrobenzene to catechol and nitrite. To determine the effects of amino acid substitutions on the catalytic efficiency of the dioxygenases, nitrite was measured at the end of 6-h biotransformation reactions (Table 2). Wild-type NBDO released 923 ± 55 nmol NO2−/mg total protein; 2NTDO produced twice as much product from nitrobenzene as NBDO. While similar amounts of nitrite were produced by wild-type NBDO and enzymes carrying F293Q and I350T substitutions, 7- to 23-fold-less nitrite was detected in nitrobenzene biotransformation reactions with the N258V, F293H, F293I, and I350F variants. The amounts of nitrite detected were consistent with the amounts of catechol formed based on TLC and GC-MS analyses that compared the intensity of fluorescence quenching and peak area of total ion chromatograms, respectively (data not shown); increased nitrite correlated with a higher abundance of catechol. Only trace amounts of catechol were detected in extracted culture supernatants from N258V, F293I, and I350F biotransformation reactions. Biotransformation of nitrobenzene by DH5α(pUC18) did not produce any detectable catechol based on TLC and GC-MS analyses of extracted culture supernatants. These results show that the introduction of the N258V, F293H, F293I, and I350F mutations in the active site reduced the ability of the enzyme to oxidize its native substrate (Table 2).
TABLE 2.
Nitrobenzene and 26DNT as substrates: nitrite formation and whole cell biotransformation kinetics
| Enzyme | Mean nitrite releaseda ± SD from:
|
Estimated kinetic coefficientsb
|
||||
|---|---|---|---|---|---|---|
| Nitrobenzene | 26DNT | Nitrobenzene
|
26DNT
|
|||
| Km (μM) | Vmax | Km (μM) | Vmax | |||
| NBDO | 923 ± 55 | 563 ± 77 | 7.7 ± 0.2 | 3.6 ± 0.1 | 851 ± 131 | 5.48 ± 1.25 |
| N258V | 81.6 ± 3.1 | 384 ± 43 | 1780 ± 70 | 0.23 ± 0.12 | 808 ± 69 | 3.20 ± 2.74 |
| F293H | 127 ± 44 | 281 ± 128 | 23.9 ± 1.4 | 2.0 ± 1.4 | 12.0 ± 4.2 | 0.51 ± 0.21 |
| F293I | 39.8 ± 16.3 | 25.8 ± 17.5 | NDc | BLDd | 288 ± 55 | 1.74 ± 1.29 |
| F293Q | 1,080 ± 180 | 2,040 ± 320 | 60.7 ± 12.8 | 4.5 ± 2.0 | 717 ± 44 | 13.5 ± 2.0 |
| I350F | 41.9 ± 5.6 | 7.7 ± 3.9 | ND | BLD | ND | BLD |
| I350T | 1,380 ± 290 | 43.1 ± 23.8 | 159 ± 35 | 16 ± 1 | ND | 2.83 ± 0.42e |
| 2NTDO | 1,870 ± 110 | 7.2 ± 3.0 | 21.0 ± 4.1 | 12.1 ± 1.7 | ND | 0.56 ± 0.25e |
Expressed as nanomoles of NO2− per milligram of protein. Nitrobenzene and 26DNT products were assayed after 6 and 16 h, respectively.
Kinetic coefficients were estimated for whole cells expressing dioxygenase variants by nonlinear regression with the Michaelis-Menten equation (38). Vmax is reported as nanomoles per minute per milligram of protein.
ND, could not be determined. These reactions were very slow and did not follow Michaelis-Menten kinetics.
BLD, below the limit of detection.
Reactions did not follow Michaelis-Menten kinetics. Reported values reflect maximum observed oxidation rates under substrate-saturating conditions (1 mM).
Biotransformation reactions with mononitrotoluenes.
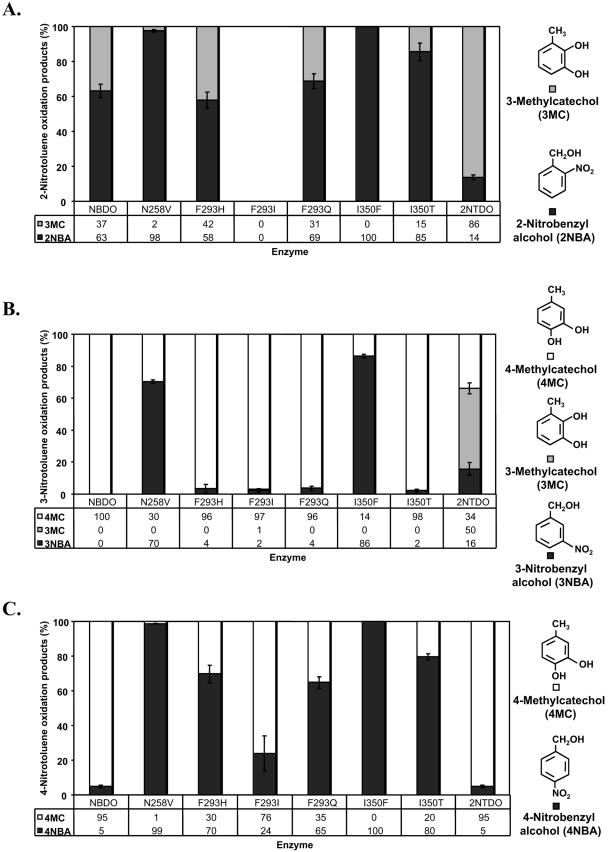
The ability of the mutant enzymes to oxidize 2NT, 3NT, and 4NT is presented in Fig. 2A to C. Replacement of Asn-258 with valine resulted in an overall decreased specificity for attack at nitro-substituted positions of the benzene ring, with 2-nitrobenzyl alcohol, 3-nitrobenzyl alcohol, and 4-nitrobenzyl alcohol formed as the dominant products from 2NT, 3NT, and 4NT, respectively. In general, mutant enzymes with changes at position 293 had slight differences in regiospecificity with 2NT and 3NT (Fig. 2A and B) and more significant differences with 4NT as the substrate (Fig. 2C). The F293I mutant completely lost the ability to oxidize 2NT but gained the ability to oxidize 3NT at the 2,3 positions to form a small amount of 3-methylcatechol (3MC). Both substitutions at position 350 reduced or eliminated the ability of NBDO to oxidize the aromatic ring of mononitrotoluenes (Fig. 2). No catechols or nitrobenzyl alcohols were detected in biotransformations with the vector control strain DH5α(pUC18). These results provide experimental evidence for recent predictions suggesting that the amino acids at positions 258, 293, and 350 in NBDO are critical for directing nitroarene substrates for oxidative attack on the aromatic ring (6).
FIG. 2.
Oxidation products from mononitrotoluene biotransformation reactions with wild-type NBDO, NBDO variants, and 2NTDO. (A) 2NT as the substrate (n = 3). 3-Methylcatechol (3MC) is represented by light gray bars, and 2-nitrobenzyl alcohol (2NBA) is represented by dark gray bars. (B) 3NT as the substrate (n = 3). 4-Methylcatechol (4MC) is represented by white bars, 3MC is represented by light gray bars, and 3-nitrobenzyl alcohol (3NBA) is represented by dark gray bars. (C) 4NT as the substrate (n = 3). 4-Methylcatechol is represented by white bars, and 4-nitrobenzyl alcohol (4NBA) is represented by dark gray bars.
Biotransformation reactions with 24DNT.
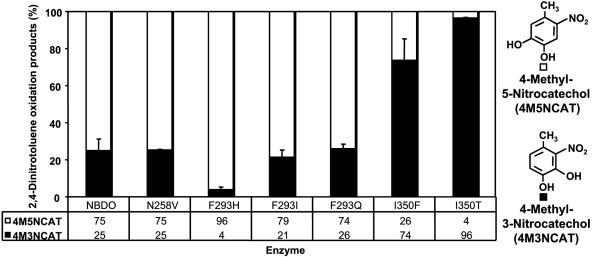
Wild-type NBDO oxidized 24DNT to 4-methyl-5-nitrocatechol (4M5NCAT) (75%) and 4-methyl-3-nitrocatechol (4M3NCAT) (25%) (Fig. 3). 2,4-Dinitrobenzyl alcohol was previously reported to be formed from 24DNT by NBDO (20) but was not detected in any reactions with wild-type or mutant enzymes. Our observation that 2NTDO did not form any detectable oxidation products from 24DNT is consistent with findings of previous studies that examined the substrate specificity of that enzyme (27). The N258V, F293I, or F293Q mutations did not alter the ratios of methylnitrocatechols relative to wild-type NBDO. The greatest change in product ratios was observed with F293H, I350F, and I350T mutant enzymes, which were improved in their regioselectivities for attack at the 4,5 positions or 3,4 positions (Fig. 3). The F293H enzyme greatly favored attack at the 4,5 positions, forming 96% 4M5NCAT. In contrast, enzymes with substitutions at position 350 preferentially attacked at the 3,4 positions, forming 74% and 96% 4M3NCAT, respectively. These results demonstrate that the residues at positions 293 and 350 play important roles in positioning 24DNT in the active site.
FIG. 3.
Oxidation products from 24DNT biotransformation reactions (n = 3) with wild-type NBDO, NBDO variants, and 2NTDO. 4M5NCAT is represented by white bars, and 4M3NCAT is represented by dark gray bars.
Biotransformation reactions with 26DNT.
2NTDO and the NBDO enzymes all oxidized 26DNT to 3-methyl-4-nitrocatechol (3M4NCAT) and nitrite. Nitrite was measured at the end of the 16-h biotransformation reactions with 26DNT to examine how changes in the active site affected the catalytic activity with dinitro-substituted compounds (Table 2). In contrast to nitrobenzene oxidation, NBDO (563 ± 77 nmol NO2−/mg total protein) was more active towards 26DNT than 2NTDO, which released 78-fold-less nitrite. The F293I, I350F, and I350T variants all produced less nitrite than wild-type NBDO (13- to 73-fold-reduced activity). In contrast, the N258V and F293H substitutions did not cause a major change in the amount of nitrite released. Interestingly, the F293Q variant released four times more nitrite than NBDO. These results are consistent with the amounts of 3M4NCAT formed as determined by TLC and GC-MS analyses (data not shown); increased nitrite correlated with a higher abundance of 3M4NCAT. Biotransformation of 26DNT by DH5α(pUC18) did not produce any detectable 3M4NCAT as judged by TLC and GC-MS analyses of extracted culture supernatants. These results support the hypothesis that amino acids at positions 293 and 350 are important for the positioning of 26DNT in the active site to favor oxidative attack on the nitro-substituted benzene ring, while the residue at position 258 may not play a direct role.
Nitrobenzene and 26DNT oxidation kinetics.
The endpoint nitrite assays provided an initial measurement of enzyme activity towards nitrobenzene and 26DNT. In order to explore the role of amino acids at positions 258, 293, and 350 in mediating activity towards nitroarene compounds and to clarify the effects of the amino acid substitutions on enzyme efficiency, the kinetic coefficients for nitrobenzene and 26DNT oxidation were determined with whole cells expressing wild-type and mutant dioxygenase enzymes (Table 2).
The half-saturation constant (Km) and maximum observed oxidation rate (Vmax) were 7.7 ± 0.2 μM and 3.6 ± 0.1 nmol NO2−/mg protein/min, respectively. 2NTDO had a threefold-higher Vmax but also had a threefold-higher Km, suggesting that the enzyme is less specific for nitrobenzene than NBDO. All of the substitutions in the NBDO active site resulted in enzymes with a decreased affinity for nitrobenzene (higher Km values). F293H and F293Q enzymes had Vmax values comparable to those of NBDO (Table 2). Although the I350T enzyme had an increased Km for nitrobenzene, it had the highest oxidation rate for nitrobenzene of all of the tested enzymes, exhibiting a 4.5-fold-higher maximum rate with nitrobenzene than the wild-type enzyme. Nitrobenzene was a very poor substrate for enzymes with the N258V, F293I, and I350F substitutions. The enzyme with N258V not only was 200 times less specific for nitrobenzene but also had a dramatically reduced maximum oxidation rate. During the 3-h assay, the F293I and I350F variants did not produce nitrite above the minimum level of detection (1 μM). In general, whole-cell kinetic results are consistent with the results of the endpoint nitrite assays (Table 2).
Wild-type NBDO had an apparent Km of 851 ± 131 μM and a Vmax of 5.5 ± 1.2 nmol NO2−/mg protein/min for 26DNT. The N258V enzyme had comparable Km and Vmax values for 26DNT which are consistent with endpoint nitrite assay data (Table 2). Mutations at position 293 generated variants with increased affinities (lower Km values) and altered Vmax constants. The F293H and F293I enzymes, respectively, showed 71- and 3-fold improvement in 26DNT affinity but were 10- and 3-fold decreased in their Vmax values. F293Q had a comparable affinity for 26DNT compared to wild-type NBDO but had a 2.5-fold increase in Vmax (Table 2). 26DNT was a very poor substrate for 2NTDO and for NBDO variants with I350F and I350T substitutions. The I350F-containing enzyme did not produce nitrite above the minimum level of detection during the kinetic assay. 26DNT oxidation rates by NBDO-I350T and 2NTDO did not exhibit Michaelis-Menten kinetics. Despite increasing substrate concentrations to the solubility limit of 1 mM and extended monitoring, rates reflecting substrate saturation were never achieved. The reported values reflect the maximum observed oxidation rate over 3 h (Table 2).
DISCUSSION
Amino acid sequences of the catalytic (α) subunits from NBDO, 2NTDO, DNTDO, NDO9816-4, and NDOU2 show greater than 80% sequence identity, but different amino acids are present at positions 258, 293, and 350 among the enzymes (Table 1). As one might predict, a substitution of single active-site residues at positions 258, 293, or 350 in NBDO with the corresponding amino acids present in 2NTDO, DNTDO, and NDO9816-4 (Table 1) increased the ratio of (+)-naphthalene cis-dihydrodiol formed (Fig. 1), as these dioxygenases oxidize naphthalene with greater enantioselectivity than NBDO (11, 12, 20, 27, 43).
The residue at position 258 in NBDO and 2NTDO is an asparagine, while in most of the other sequenced dioxygenases, a valine or other small hydrophobic residue is present. This residue appears to be significant in both NBDO and 2NTDO. The amino group of Asn-258 forms a hydrogen bond with the nitro groups of nitrobenzene and 3NT based on NBDO crystal structure data (6) and is required for efficient attack at the nitro-substituted carbon of nitroarene substrates in NBDO (Fig. 2 and Table 2) and 2NTDO (18). Interestingly, DNTDOs do not have an asparagine at position 258 (Table 1). They are able to oxidize the aromatic rings of dinitrotoluenes with the release of nitrite but do not catalyze efficient oxidation of the aromatic rings of mononitrotoluenes or nitrobenzene (22, 43). Consistent with this information, the N258V substitution in NBDO had a major effect on mononitrotoluene and nitrobenzene oxidation (Table 2 and Fig. 2) but had little effect on 24DNT regioselectivity (Fig. 3) or 26DNT oxidation (Table 2), indicating that the presence of an asparagine at this position is important only for the oxidation of mononitroarene compounds.
The residue at position 350 in many ring-hydroxylating dioxygenases is phenylalanine; however, all four of the sequenced nitroarene dioxygenases have a smaller residue at this position (Table 1). This residue has been shown to be important for determining substrate specificity in several different dioxygenases (15, 16, 18, 30-32, 47). Changes in product ratios formed by the I350F mutant can be attributed to the increased size of the residue. Based on the wild-type NBDO structure, a phenylalanine would likely prevent binding of nitrobenzene and mononitrotoluenes in the correct position to allow hydrogen bonding to Asn-258 (see above) and subsequent oxidation of the aromatic ring. As a result, these substrates would probably bind in alternative positions, which, as shown here for the nitrotoluenes, results in preferential oxidation of the methyl group. Similarly, a phenylalanine at position 350 might push 24DNT out of position for efficient oxidation at the 4,5 position of the 24DNT molecule, resulting in increased oxidation at the 3,4 position and the production of more 4M3NC.
To our knowledge, the residue at position 293 has been shown to play a role in substrate specificity only in NDO9816-4 and only in conjunction with other amino acid substitutions at the active site (46). A single replacement of the residue at the position corresponding to position 293 in NDO9816-4 did not affect the regiospecificity or enantioselectivity of NDO9816-4 with any of the tested substrates (46). However, this residue plays an important role in determining substrate specificity in NBDO based on the results presented here. Amino acid substitutions at position 293 in NBDO resulted in changes in regiospecificity with 4NT (Fig. 2C) and 24DNT (Fig. 3), a change in the enantioselectivity with naphthalene (Fig. 1), and improved product formation rates with 26DNT (Table 2). In the case of the F293Q mutant, this change in substrate specificity may result from the formation of a favorable hydrogen bond between the glutamine and 24DNT or 26DNT in the active site to promote oxidation of the ring. In contrast, the ability to oxidize 2NT was lost by the F293I variant of NBDO, and nitrobenzene oxidation was severely reduced (Fig. 2A and Table 2). Therefore, the increased size of the active-site pocket resulting from the reduced size of the hydrophobic residue at position 293 appears to be unfavorable for the appropriate positioning of small substrates for ring oxidation but not as deleterious for larger substrates such as 26DNT (Table 2).
In order to assess the relative efficiencies of the NBDO variants, we carried out whole-cell kinetic analyses with nitrobenzene and 26DNT as substrates. These results gave a measure of the efficiency of substrate oxidation while eliminating the complexity associated with the in vitro analysis of three-component enzyme systems where the ratios of individual proteins affect activity. As a biocatalyst, E. coli expressing NBDO-F293Q had the highest activity reported to date with 26DNT. The F293Q mutant had a similar Km for 26DNT but a 2.5-fold-higher Vmax than wild-type NBDO (Table 2). Recombinant E. coli carrying wild-type NBDO was 8- and 14-fold faster at 26DNT oxidation than recombinant strains expressing DNTDO from either Burkholderia cepacia R34 or Burkholderia sp. strain DNT, respectively (22). Recombinant E. coli expressing NBDO-F293Q formed nitrite from 26DNT 12 to 38 times faster than E. coli strains expressing mutant forms of NDOU2 (F350T and F350T/G407S) or DNTDODNT (I204L and I204Y), which were generated by saturation mutagenesis (15, 22).
We are currently purifying several variants of NBDO in order to obtain in vitro kinetic data and to pursue high-resolution crystal structures. These studies will allow us to confirm predictions made based on the wild-type structure of NBDO.
Acknowledgments
We thank Jim C. Spain for generously providing methylnitrocatechols and aminomethylnitrocatechols, Chi-Li Yu for providing purified NBDO, Juan Parales for assistance with GC/MS analysis, Kyung-Seon Lee for carrying out Western blot analysis, and Rosie Friemann for modeling studies and for helpful discussions.
This work was supported by grant CU1212 awarded to R.E.P. from the U.S. Department of Defense Strategic Environmental Research and Development Program (SERDP). K.-S.J. was supported by an NIH Traineeship in Molecular and Cellular Biology (NIH TM32 GM070377).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Boyd, D. R., N. D. Sharma, and C. C. R. Allen. 2001. Aromatic dioxygenases: molecular biocatalysis and applications. Curr. Opin. Biotechnol. 12:564-573. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D. R., and G. N. Sheldrake. 1998. The dioxygenase-catalysed formation of vicinal cis-diols. Nat. Prod. Rep. 15:309-324. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 6.Friemann, R., M. M. Ivkovic-Jensen, D. J. Lessner, C.-L. Yu, D. T. Gibson, R. E. Parales, H. Eklund, and S. Ramaswamy. 2005. Structural insights into the dioxygenation of nitroarene compounds: the crystal structure of the nitrobenzene dioxygenase. J. Mol. Biol. 348:1139-1151. [DOI] [PubMed] [Google Scholar]
- 7.Fuenmayor, S. L., M. Wild, A. L. Boyles, and P. A. Williams. 1998. A gene cluster encoding steps in the conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigler, B. E., W. H. Wallace, and J. C. Spain. 1994. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol. 60:3466-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Hudlicky, T., D. Gonzalez, and D. T. Gibson. 1999. Enzymatic dihydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodology. Aldrichim. Acta 32:35-62. [Google Scholar]
- 11.Jeffrey, A. M., H. J. C. Yeh, D. M. Jerina, T. R. Patel, J. F. Davey, and D. T. Gibson. 1975. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 14:575-583. [DOI] [PubMed] [Google Scholar]
- 12.Jerina, D. M., J. W. Daly, A. M. Jeffrey, and D. T. Gibson. 1971. cis-1,2-Dihydroxy-1,2-dihydronaphthalene: a bacterial metabolite from naphthalene. Arch. Biochem. Biophys. 142:394-396. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 15.Keenan, B. G., T. Leungsakul, B. F. Smets, M. A. Mori, D. E. Henderson, and T. K. Wood. 2005. Protein engineering of the archetypal nitroarene dioxygenase of Ralstonia sp. strain U2 for activity on aminonitrotoluenes and dinitrotoluenes through alpha-subunit residues leucine 225, phenylalanine 350, and glycine 407. J. Bacteriol. 187:3302-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keenan, B. G., T. Leungsakul, B. F. Smets, and T. K. Wood. 2004. Saturation mutagenesis of Burkholderia cepacia R34 2,4-dinitrotoluene dioxygenase at DntAc valine 350 for synthesizing nitrohydroquinone, methylhydroquinone, and methoxyhydroquinone. Appl. Environ. Microbiol. 70:3222-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klecka, G. M., and D. T. Gibson. 1979. Metabolism of dibenzo[1,4]dioxan by a Pseudomonas species. Biochem. J. 180:639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, K.-S., J. V. Parales, R. Friemann, and R. E. Parales. 2005. Active site residues controlling substrate specificity in 2-nitrotoluene dioxygenase from Acidovorax sp. strain JS42. J. Ind. Microbiol. Biotechnol. 32:465-473. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S.-Y., and S. Rasheed. 1990. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques 9:676-679. [PubMed] [Google Scholar]
- 20.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessner, D. J., R. E. Parales, S. Narayan, and D. T. Gibson. 2003. Expression of nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds. J. Bacteriol. 185:3895-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leungsakul, T., B. G. Keenan, H. Yin, B. F. Smets, and T. K. Wood. 2005. Saturation mutagenesis of 2,4-DNT dioxygenase of Burkholderia sp. strain DNT for enhanced dinitrotoluene degradation. Biotechnol. Bioeng. 92:416-420. [DOI] [PubMed] [Google Scholar]
- 23.Nishino, S. F., G. C. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and the pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishino, S. F., and J. C. Spain. 1995. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl. Environ. Microbiol. 61:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padda, R. S., C. Wang, J. B. Hughes, R. Kutty, and G. N. Bennett. 2003. Mutagenicity of nitroaromatic degradation compounds. Environ. Toxicol. Chem. 22:2293-2297. [DOI] [PubMed] [Google Scholar]
- 26.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 27.Parales, J. V., R. E. Parales, S. M. Resnick, and D. T. Gibson. 1998. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the α subunit of the oxygenase component. J. Bacteriol. 180:1194-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parales, R. E., M. D. Emig, N. A. Lynch, and D. T. Gibson. 1998. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol. 180:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parales, R. E., R. Huang, C.-L. Yu, J. V. Parales, F. K. N. Lee, M. M. Ivkovic-Jensen, W. Liu, D. J. Lessner, R. Friemann, S. Ramaswamy, and D. T. Gibson. 2005. Purification, characterization, and crystallization of the components of the nitrobenzene and 2-nitrotoluene dioxygenase enzyme systems. Appl. Environ. Microbiol. 71:3806-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parales, R. E., S. M. Resnick, C. L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the α subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollmann, K., V. Wray, H.-J. Hecht, and D. H. Pieper. 2003. Rational engineering of the regioselectivity of TecA tetrachlorobenzene dioxygenase for the transformation of chlorinated toluenes. Microbiology 149:903-913. [DOI] [PubMed] [Google Scholar]
- 33.Resnick, S. M., D. S. Torok, K. Lee, J. M. Brand, and D. T. Gibson. 1994. Regiospecific and stereoselective hydroxylation of 1-indanone and 2-indanone by naphthalene dioxygenase and toluene dioxygenase. Appl. Environ. Microbiol. 60:3323-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rieger, P.-G., and H.-J. Knackmuss. 1995. Basic knowledge and perspectives on biodegradation of 2,4,6-trinitrotoluene and related nitroaromatic compounds in contaminated soil, p. 1-18. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds, vol. 49. Plenum Press, New York, N.Y. [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sheldrake, G. N. 1992. Biologically derived arene cis-dihydrodiols as synthetic building blocks, p. 127-166. In A. N. Collins, G. N. Sheldrake, and J. Crosby (ed.), Chirality in industry: the commercial manufacture and application of optically active compounds. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 37.Smibert, R. M., and N. R. Krieg. 1981. General characterization, p. 409-443. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 38.Smith, L. H., P. L. McCarty, and P. K. Kitanidis. 1998. Spreadsheet method for evaluation of biochemical reaction rate coefficients and their uncertainties by weighted nonlinear least-squares analysis of the integrated monod equation. Appl. Environ. Microbiol. 64:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spain, J. C., J. B. Hughes, and H.-J. Knackmuss. 2000. Perspectives of bioelimination of polynitroaromatic compounds, p. 91-126. In H. Lenke, C. Achnich, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromaic compounds and explosives. CRC Press, Boca Raton, Fla.
- 40.Spanggord, R. J., J. C. Spain, S. F. Nishino, and K. E. Mortelmans. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 42.Suen, W.-C. 1991. Gene expression of naphthalene dioxygenase from Pseudomonas sp. NCIB 9816-4 in Escherichia coli. Ph.D. thesis. University of Iowa, Iowa City, Iowa.
- 43.Suen, W.-C., B. E. Haigler, and J. C. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U.S. Environmental Protection Agency. 20 January 2006, posting date. Water Quality Standards Database. [Online.] http://oaspub.epa.gov/wqsdatabase/wqsi_epa_criteria.rep_parameter.
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 46.Yu, C.-L., R. E. Parales, and D. T. Gibson. 2001. Multiple mutations at the active site of naphthalene dioxygenase affect regioselectivity and enantioselectivity. J. Ind. Microbiol. Biotechnol. 27:94-103. [DOI] [PubMed] [Google Scholar]
- 47.Zielinski, M., S. Kahl, H. J. Hecht, and B. Hofer. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]