Abstract
We characterized the arsenate-reducing, sulfide-oxidizing population of Mono Lake, California, by analyzing the distribution and diversity of rrnA, cbbL, and dissimilatory arsenate reductase (arrA) genes in environmental DNA, arsenate-plus sulfide-amended lake water, mixed cultures, and isolates. The arsenate-reducing community was diverse. An organism represented by an rrnA sequence previously retrieved from Mono Lake and affiliated with the Desulfobulbaceae (Deltaproteobacteria) appears to be an important member of the arsenate-reducing, sulfide-oxidizing community. Sulfide oxidation coupled with arsenate reduction appears to proceed via a two-electron transfer, resulting in the production of arsenite and an intermediate S compound that is subsequently disproportionated. A realgar-like As/S mineral was formed in some experiments.
Soda lakes and similar alkaline, hypersaline environments are widespread today and have been important features of the hydrosphere since the earliest stages of the formation of the Earth's oceans (15). Because of their constrained watersheds, local geology strongly influences the composition of dissolved salts in these endorheic (closed-basin) lakes. Hydrothermal processes associated with volcanism in the watershed of Mono Lake, an endorheic lake in eastern California, have led to the accumulation of arsenic compounds (12, 17). These compounds play an active role in the lake's biogeochemical cycles (26), serving both as electron acceptors for the oxidation of organic matter (23) and as electron donors (24). Arsenate reduction in Mono Lake appeared to be relatively insensitive to organic-carbon additions (11, 26), suggesting that another electron donor, possibly sulfide, might be significant in this environment, a conclusion that was supported by geochemical evidence (12).
Arsenate reduction coupled with sulfide oxidation was first demonstrated with an archived sample of Mono Lake bottom water (10) and was subsequently repeated with slurries of sediments from an arsenic-enriched playa basin south of Mono Lake (25). An organism (deltaproteobacterium strain MLMS-1) capable of chemoautotrophic growth by oxidizing sulfide with arsenate was isolated from the Mono Lake sample (10). While Hoeft et al. (10) noted the relatively close alignment (∼97% rrnA sequence similarity) of strain MLMS-1 with ribotypes previously detected in Mono Lake (13), extensive characterizations of Mono Lake microbial assemblages using culture-independent techniques (13; J. T. Hollibaugh, unpublished data) failed to retrieve ribotypes more closely related to strain MLMS-1. This prompted us to examine arsenate reduction coupled with sulfide oxidation in enrichments of freshly collected water samples and to compare arsenate-reducing, sulfide-oxidizing populations with nitrate-reducing, sulfide-oxidizing populations in similar enrichments. We also compared the populations we obtained in this way with diagnostic sequences retrieved directly from DNA extracted from lake water samples.
MATERIALS AND METHODS
Sample collection.
Water was collected at station 6 in Mono Lake's central basin (37°57.822′N, 119°01.305′W) using a Niskin water sampler as described previously (12). Care was taken to avoid exposing the samples to sunlight and to keep them anoxic. Water samples for enrichment experiments were collected in 2003 and were chosen to cover a range of microaerophilic to anoxic conditions and the seasonal progression from a stratified water column to fall overturn: 29 May (experiment 1; 16 m), 4 September (experiment 2; 26 m), 14 November (experiment 3; 28 m), and 23 November (experiment 4; 35 m). Archived DNA samples collected on 22 May 2002 (23 and 30 m) and 6 August 2002 (5, 23, 26. and 35 m) were used for comparison with enrichment experiments.
The water column was stably stratified with the base of the thermo- and oxyclines at 13 to 15 m during summer (May to September). The lake was stratified and oxygenated to 17 m on 22 October 2003. Fall cooling initiated complete overturn of the lake's water column, ending an 8-year period of meromixis and mixing anoxic bottom water containing high concentrations of reduced substrates with oxygenated surface waters (12, 17, 19). The lake was hypoxic ([O2], <1 mg/liter) or anoxic when sampled in November 2003. Profiles of water column properties are included in the supplemental material, and additional information is available at http://www.monolake.uga.edu.
Enrichment experiments.
Filtered (0.22-μm nominal pore size) lake water was dispensed (125 to 300 ml) into replicate sterile bottles. These were wrapped in several layers of aluminum foil to exclude light and then placed, loosely capped, in an anaerobic chamber for at least 48 h to achieve anoxia before being amended with combinations of arsenate and sulfide. The bottles were then inoculated with unfiltered water (∼10% [vol/vol]). Controls were treatments with arsenate only and with sulfide only, with no additions, and with no inoculum. The bottles were tightly capped and then incubated at room temperature (∼25°C) in an anaerobic chamber and were sampled periodically. Arsenate and sulfide concentrations were monitored to track microbial activity; some treatments received additional arsenate and sulfide once the original amendment was depleted. A similar set of nitrate amendments was run for comparison with arsenate during experiments 1 and 2 (data not shown). Incorporation of 13C-bicarbonate into organic matter was determined at the beginning, middle, and end of experiment 2. Samples were also taken for cell enumeration or were filtered for subsequent extraction of DNA from retained particulate material.
Chemical analyses.
Vertical profiles of water properties were taken with a SeaBird conductivity-temperature-depth sensor package equipped with a fluorometer and transmissometer as reported previously (12). Oxygen distributions were determined with a YSI oxygen meter equipped with a Clark-type electrode. Arsenate was determined colorimetrically (14) after dilution in distilled water. Sulfide was measured using an ion-specific electrode as described previously (12). 13C was determined in samples collected on GF/F glass fiber filters and analyzed with a CHN analyzer coupled with an isotope ratio mass spectrometer.
Molecular analysis.
DNA was extracted from bacteria retained on filters and purified using a MoBio Soil Extraction kit (Bio 101, Solana Beach, CA). PCR amplification and denaturing gradient gel electrophoretic (DGGE) analysis of rrnA amplicons were performed as described previously (3, 20). PCR amplification and cloning and sequencing of rrnA and cbbL gene fragments were performed as described previously (8, 13).
PCR amplification of arsenate reductase (arrA) gene fragments used primers reported previously (18). Genomic DNA from Sulfurospirillum barnesii served as a positive control. The amplicons were then cloned using the TOPO-TA Cloning kit (Invitrogen) and sequenced using the cloning vector primer M13, as reported previously (8, 16). A total of 425 clones from 17 libraries were sequenced. We discarded 33 poor-quality sequences and 18 that were shorter and 3 that were much longer (>246 bp) than expected. The inferred amino acid sequences were aligned and examined for conserved residues, indels, and other structural features. A total of 340 nucleotide sequences of 112 bp were then used in a phylogenetic analysis, and 31 additional, longer sequences were included in a second phylogenetic analysis based on inferred amino acid residues.
Reference sequences.
An organism identified as an Ectothiorhodospira strain by its rrnA gene sequence was isolated from Mono Lake water into pure culture (strain MLCB). Ectothiorhodospira sp. strain “Bogoria Red” was provided by M. Madigan; Sulfurospirillum barnesii, Alkalispirillum mobile, Alkalilimnicola halodurans, strain MLHE-1, and strain MLMS-1 were obtained from R. S. Oremland. DNA fragments presumed to represent partial sequences of arrA genes were amplified from genomic DNA extracted from these cultures and then cloned and sequenced as described above. Additional reference sequences were retrieved from GenBank.
Isolation media and growth conditions.
The base mineral medium for isolation, purification, and characterization of bacteria consisted of 650 mM NaCl, 10 mM Na2CO3, 5 mM NH4Cl, 0.15 mM Na2HPO4, 0.25 mM MgSO4, and added vitamins and trace metals; the pH was adjusted to 9.3 (4). Anoxia was achieved by allowing the medium to sit in an anaerobic chamber for 48 h. The medium was dispensed into 125-ml serum bottles with butyl rubber stoppers. Reduced sulfur compounds, organic carbon, arsenate, arsenite, or nitrate was added separately as required. The bottles were incubated at ∼25°C. Agar plates were prepared with purified agar, poured aerobically, and placed in an anaerobic chamber for 48 h before use. Enrichment cultures A, B, and C were inoculated from arsenate-plus-sulfide (As+S) treatments in experiments 2 and 3 and carried through at least two transfers into fresh mineral medium before use.
Enumeration.
Bacteria were counted by epifluorescence microscopy using a modification of the DAPI (4′,6′-diamidino-2-phenylindole) technique (27). Briefly, samples (0.1 ml) were diluted in 2 ml of 10% acetic acid containing 1 μg/ml of DAPI, stained for 5 min, and then counted. We have found that stained cells fluoresce more brightly and that background fluorescence is lower with this modification than if they were stained in Mono Lake water.
Nucleotide sequence accession numbers.
All sequences determined in this study have been deposited in GenBank under accession numbers DQ155316 to DQ155373, DQ206427 to DQ206430 (arrA), and DQ206405 to DQ206426 (rrnA).
RESULTS AND DISCUSSION
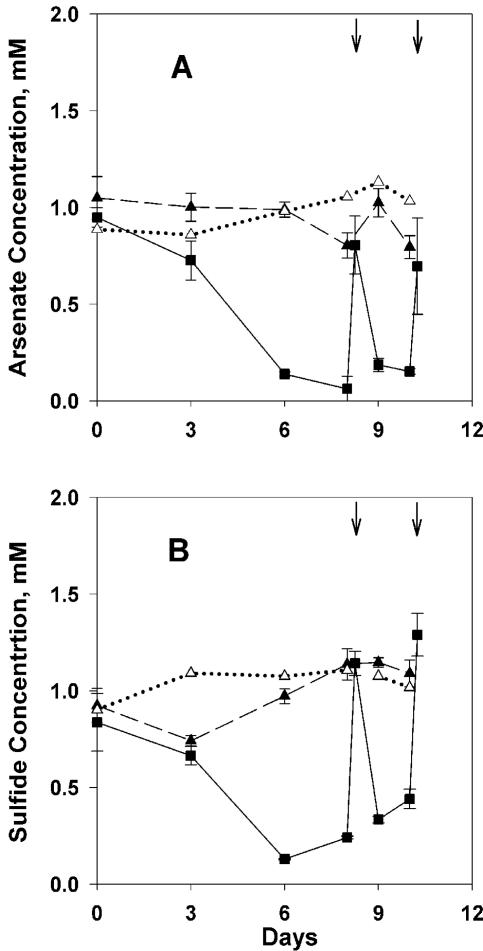
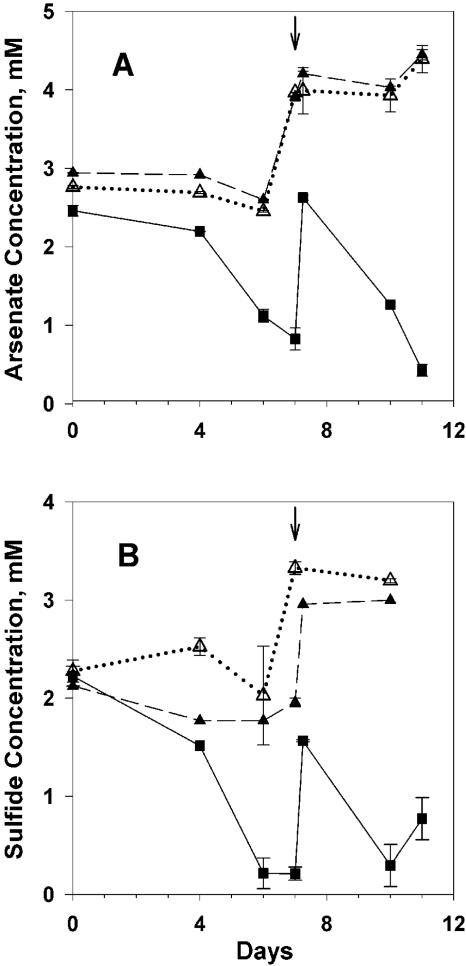
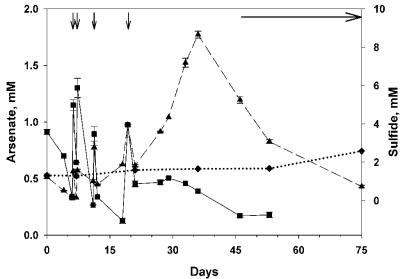
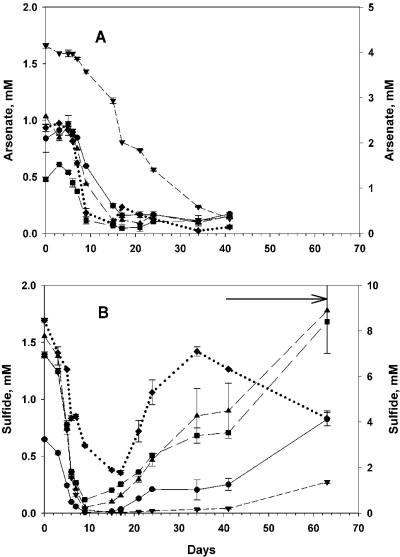
Arsenate and sulfide concentrations declined rapidly in As+S amendments (Fig. 1, 2, 3, and 4) and decreased more rapidly when the As+S treatments were amended again following depletion of the substrate (Fig. 1, 2, and 3). Arsenate concentrations decreased slightly in the arsenate-alone treatment (As), while sulfide concentrations increased slightly in the sulfide-alone (S) treatment of experiment 1, presumably due to slow heterotrophic growth using arsenate or sulfate as an electron acceptor (Fig. 1). Arsenate concentrations in filtered As+S controls did not change except when amended (Fig. 1A and 2A), indicating that the decrease in arsenate in the As+S treatment was not due to an abiotic reaction. The results for sulfide in filtered controls (Fig. 1B, 2B, and 3) were similar, except that sulfide measurements were more variable in experiment 2 (Fig. 2B).
FIG. 1.
Time courses of arsenate reduction and sulfide oxidation during an experiment conducted with Mono Lake water collected on 29 May 2003 (experiment 1). (A) Arsenate concentration. (B) Sulfide concentration. Additional substrate was added to As+S treatments at the times indicated by the vertical arrows at the top of each panel. The error bars (standard deviation; n = 3 except filtered control, where n = 1) are smaller than the symbol when not shown. Solid line and filled box, As+S; dashed line and filled triangle, arsenate or sulfide alone; dotted line and open triangle, As+S filtered.
FIG. 2.
Time course of arsenate reduction and sulfide oxidation during an experiment conducted with Mono Lake water collected on 4 September 2003 (experiment 2). (A) Arsenate concentration. (B) Sulfide concentration. Solid line and filled box, As+S; dashed line and filled triangle, arsenate or sulfide alone; dotted line and open triangle, As+S filtered. Additional substrate was added to all treatments at the time indicated by the vertical arrow at the top of each panel. The error bars (range; n = 2) are smaller than the symbol when not shown.
FIG. 3.
Time course of arsenate reduction and sulfide oxidation during an experiment conducted with Mono Lake water collected on 14 November 2003 (experiment 3). Additional substrate was added to As+S treatments at the times indicated by vertical arrows. The horizontal arrow indicates when precipitate was observed. The error bars (range; n = 2) are smaller than the symbol when not shown. Solid line and filled squares, arsenate in As+S treatment; dashed line and filled triangles, sulfide in As+S treatment; dotted line and filled diamonds, sulfide in As+S filtered control.
FIG. 4.
Time course of arsenate reduction and sulfide oxidation during an experiment conducted with a Mono Lake water sample collected on 23 November 2003 (experiment 4). (A) Arsenate concentration. (B) Sulfide concentration. All treatments were As+S amendments; the nominal amendments were as follows: filled circle and solid line, 1 mM arsenate-0.5 mM sulfide; filled square and long dashed line, 0.5 mM arsenate-1 mM sulfide; filled triangle and medium dashed line, 1 mM arsenate-1 mM sulfide; filled inverted triangle and short dashed line, 5 mM arsenate-1 mM sulfide; filled diamond and dotted line,1 mM arsenate-5 mM sulfide. The right ordinates apply only to the 5 mM amendments. The solid horizontal arrow along the top of panel B indicates when precipitate was observed. The error bars (range; n = 2) are smaller than the symbol when not shown.
There was no growth in the S-alone and no-additions controls of experiment 1; however, cell numbers increased in the As-alone control (data not shown), presumably due to growth of heterotrophs. DAPI counts increased 2- to 10-fold in As+S compared to As or S treatments; (data not shown). As+S amended treatments incorporated inorganic C into particulate organic material, confirming the inferred autotrophic growth (Table 1). The estimated net efficiency for this process (assuming 2 mol e− available per mol of arsenate reduced and 4 mol e− required per mol of CO2 fixed) was ∼8%.
TABLE 1.
Carbon fixed into bacterial biomass during experiment 2a
| Treatment | μM 13C incorporated |
|---|---|
| Arsenate + sulfide | 200 ± 2 |
| Arsenate alone | 50 ± 4 |
| Sulfide alone | 68 ± 3 |
| Filtered arsenate + sulfide | 5 ± 0.5 |
| No added electron acceptors or donors | 46 ± 4 |
| Blank | 15 |
The amount of carbon fixed after 11 days of incubation (∼5 mM of arsenate reduced) was measured by incorporation of 13C supplied as bicarbonate. The values are means ± standard errors; n = 2.
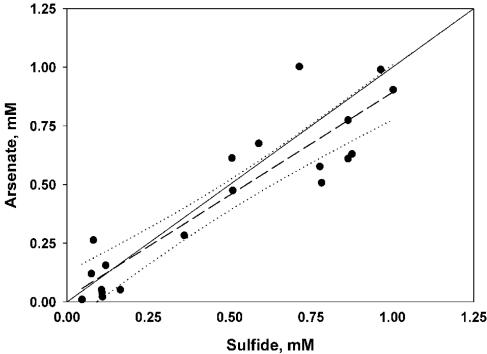
Data taken prior to the second amendment of experiment 1 (Fig. 1) supported two e− exchanges between arsenate and sulfide (Fig. 5), implying the formation of arsenite and S0. The apparent ratio of arsenate reduced to sulfide oxidized (lost) varied in other experiments. We assume that this stoichiometric variability was due to the formation of arsenic-thiol complexes (12) and polysulfides (5-7) that are not detected by the electrode used to measure sulfide (Hollibaugh, unpublished). This is demonstrated in experiment 4 (Fig. 4), where excess sulfide was apparently consumed in some treatments before all of the added arsenate was consumed, also suggesting that these complexed forms of reduced S are not directly available to arsenate reducers. Sulfide concentrations eventually increased again (Fig. 3 and 4), presumably as a result of the disproportionation of elemental sulfur or polysulfides formed during the initial oxidation of sulfide (2, 6, 22), rather than from sulfate reduction, as sulfide production varied with the initial sulfide concentration (Fig. 4) and only small increases in sulfide concentration were seen in controls (Fig. 1 to 3). Eventually, sulfide concentrations declined in As+S enrichments, and a fine reddish or yellowish precipitate was observed. The precipitate appeared to be crystalline, and its X-ray fluorescence spectrum indicated an As-S stoichiometry of 1:1.06, consistent with the 1:1 stoichiometry expected of realgar and distinct from the arsenic trisulfide produced by pure cultures of a sulfate-reducing bacterium (21). The precipitate formed only when sulfide concentrations exceeded ∼6 mM (Fig. 3 and 4), suggesting abiotic precipitation. Precipitation of As minerals may explain the loss of arsenic from Mono Lake's sulfidic bottom water during meromixis (12) or during arsenate reduction in Searles Lake sediment slurries (25). The sulfide concentrations at which the precipitate formed were greater than those observed in Mono Lake water (∼3 mM) (12); however, they can be exceeded in Mono Lake sediments (S. B. Joye, unpublished data).
FIG. 5.
Scatter plot of arsenate versus sulfide concentration during the first 10 days of experiment 1 (29 May 2003). Also shown are least squares linear regression lines (short dashes) and 95% confidence limit belts (curved dotted lines) for the regression lines. A 1:1 line (solid) is shown for comparison.
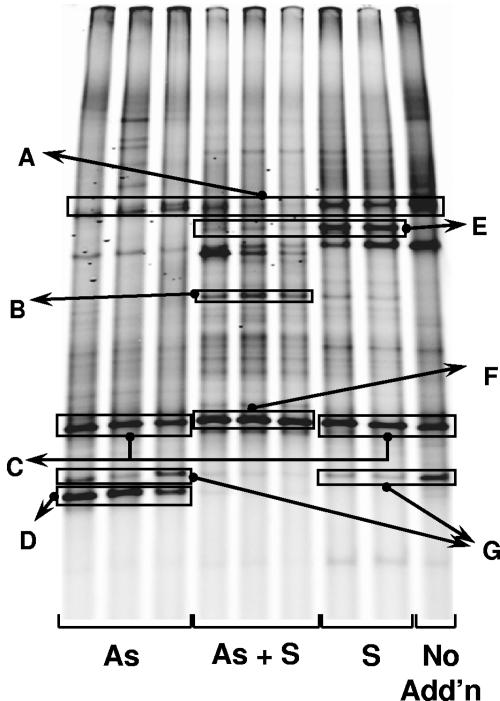
Analysis of rrnA sequences in clone libraries derived from As+S enrichments revealed that a sequence >99% similar to that of environmental clone ML623J-57 (a deltaproteobacterium affiliated with the Desulfobulbaceae and ∼97% similar to MLMS-1) was dominant (7 of 18 clones). The ML623J-57-like sequence was also found in an intense band in DGGE analyses of As+S enrichments in experiment 1 (Fig. 6) and in other experiments (not shown). The ML623J-57 sequence was also recovered in libraries derived from As (2/20) and S (3/18) treatments. Amplicons similar to Marinospirillum minutulum, which was the dominant sequence found in libraries from As and S treatments, had the same mobility as ML623J-57 in DGGE gels (Fig. 6); however, this sequence was not found in As+S treatments. Comparison of DGGE banding patterns from the enrichments, Mono Lake isolates, and samples, supported by sequencing DNA excised from DGGE bands, indicated that the organism containing the ML623J-57 rrnA gene is a consistent member of the Mono Lake bacterial assemblage. It has been recovered consistently in samples from the water column between the summer thermocline and the chemocline (13; Hollibaugh, unpublished), a seasonally anoxic environment in which the organism carrying this sequence would encounter both arsenate and sulfide or other reduced S compounds (12). Four of the 18 sequences recovered from the As+S treatment were 94% similar to the Fusobacteria-like sequence GalB35. The remaining sequences from this library were distributed among a variety of Firmicutes.
FIG. 6.
DGGE analysis of rrnA amplicons retrieved from various treatments in experiment 1, sampled after 12 days. No Add'n, no additions. DNAs in bands enclosed in boxes were identified either by sequencing or by comparison with the mobilities of cloned fragments. The most similar sequences and similarities were as follows: (A) Picocystis salinarum chloroplast AF454327, >99%; (B) Fusobacteria clone GalB35 AY193168, 94%; (C) mixture of Marinospirillum minutulum AF275713, 95%, and clone ML623J-57, AF507840, >99%; (D) Firmicutes clone un-c23, AJ431345, 90%; (E) Paenibacillus lentimorbus AB110988, 87%; (F) Deltaproteobacteria clone ML623J-57, AF507840, >99%; (G) Desulfuromusa succinoxidans X79415, 95%.
Ribulose bisphosphate carboxylase (RuBisCo) sequences retrieved from the As+S enrichments (10/10) were >99% similar to cbbL sequences previously retrieved from Mono Lake (8) (clade C, ML35J-3R and AY291528). They were also >99% similar to cbbL sequences obtained from mixed culture A, derived from experiment 3. The rrnA sequences retrieved from this culture indicated that it contained ML623J-57 and a sequence most similar (87%) to Paenibacillus lentimorbus. Four of five cbbL sequences retrieved from N+S amendments were >99% similar to sequences in clade F1 (ML35J2R and AY291531) previously retrieved from Mono Lake (8); the fifth was 95% similar to Rhodobacter blasticus.
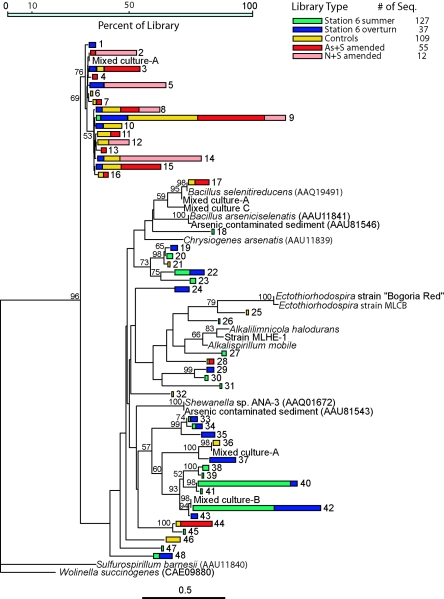
We retrieved partial arrA sequences from Mono Lake samples collected under two different hydrographic conditions: summer stratification (summer) and during fall overturn (overturn), which in this case coincided with the breakdown of 8 years of meromixis. With the exception of six sequences that were identical to those of arrA genes from the Mono Lake isolate Bacillus selenitireducens (1, 4), none of the sequences we retrieved was very similar to any currently in the databases (Fig. 7). The sequences in clades 1 to 16 (Fig. 7) were 115 bp long, while those in clades 17 to 48 were 112 bp long. We retrieved 31 longer sequences that were not included in Fig. 7. These fell into three clades: 145 bp (12 sequences, all >99% similar), 175 bp (2 identical sequences), and 178 bp (17 sequences, 97 to 99% similar). Phylogenetic analysis of their inferred amino acid sequences (see the supplemental material) indicated that the 145-bp clade was most similar to sequences from a cluster of Firmicutes, while the 175- and 178-bp sequences were most similar to sequences from the Ectothiorhodospiraceae. The alignments of these sequences (see the supplemental material) contained two indels separated by two partially conserved residues. Our search did not identify any matches to motifs currently in the Prosite (http://www.expasy.ch/prosite/) database.
FIG. 7.
Neighbor-joining maximum likelihood tree of arrA gene sequences (112 bp) retrieved from Mono Lake, enrichment experiments, mixed cultures, and isolates. The numbered (1 to 48) clades represent groups of sequences that are >99% similar. The stacked bars represent the percentages of arrA sequences from each treatment type in that clade; the total number of sequences in each treatment type is given. The scale bar indicates distance. Bootstrap values of >50% (100 iterations) are shown. The tree is unrooted; the molybdopterin binding subunit of molybdopterin oxidoreductase from Wolinella succinogenes strain DSMZ 1740 is the outgroup.
We retrieved presumptive arrA sequences from a number of Ectothiorhodospiraceae that have not yet been shown to respire arsenate (Ectothiorhodospira, Alkalilimnicola, and Alkalispirillum) (Fig. 7). One of these (strain MLHE-1) is able to oxidize arsenite (24) but is unable to respire arsenate (Hollibaugh, unpublished), and the Mono Lake isolate Ectothiorhodospira sp. strain MLCB cannot grow on arsenate and acetate (C. Budinoff, unpublished data). These discrepancies suggest that the Ectothiorhodospiraceae genes are paralogs of arrA; however, the sequences we obtained were too short to explore this relationship further.
We obtained three different arrA sequences from mixed culture A: 28% percent were identical to sequences in clade 2 (Fig. 7). Most (57%) of the arrA sequences from mixed culture A were identical to sequences in clade 36, while 14% were 97% similar to those from Bacillus selenitireducens. The arrA sequence from mixed culture C was 83% similar to arrA from Bacillus selenitireducens (the rrnA sequence retrieved from this culture was 95% similar to Bacillus sp. strain SFB; AY669375). The arrA sequences retrieved from mixed culture B fell into clade 42. Two rrnA sequences were retrieved from this culture: one was 96% similar to Tindallia californiensis, AF373919, and a second was 87% similar to Paenibacillus lentimorbus, AB110988.
The arrA sequences we retrieved from the different types of samples appeared to represent distinct populations (Fig. 7). The following pairs of libraries were tested (9, 28) and found to be statistically significantly different at P < 0.05: controls (As alone, N alone, S alone, and no additions) versus As+S, all enrichments versus summer, all enrichments versus overturn, and overturn versus summer. Only 2% of the sequences retrieved from summer samples were found in clades (1 to 16) dominated by sequences from enrichments; most of them (72%) fell into clades 40 to 42. Only 23% of all sequences retrieved from the enrichments fell into clades (17 to 48) dominated by summer samples, and only 1 of 55 sequences retrieved from As+S enrichments fell into clades 40 to 42. In contrast to the summer samples, arrA sequences retrieved from the overturn sample were more widely distributed among the clades, with 32% falling into clades 1 to 16 and only 22% into clades 40 to 42.
We also noted that arrA sequences retrieved from N+S amendments fell into the same group of clades (1 to 16), and in many cases (2, 5, 8, and 9) the same clades, as arrA sequences retrieved from As+S enrichments. This suggests that at least some of the arsenate-reducing sulfide oxidizers can also use nitrate as an electron acceptor for the oxidation of sulfide. We retrieved arrA sequences from controls that should not have favored growth by arsenate reduction (S alone and no additions) that were identical to those retrieved from As+S treatments. Since there was little growth in the controls, these likely represent arrA genes that were present in the inoculum.
We recovered presumptive arrA sequences from organisms that are not known to reduce arsenate. We also recovered presumptive arrA sequences that diverge significantly from arrA genes reported thus far (Fig. 7) (see the supplemental material). Initially, we hypothesized two guilds of arsenate reducers: sulfide oxidizers and heterotrophs. It is likely that additional guilds are present, for example, organisms oxidizing other reduced S compounds, H2, and Fe(II). Other arsenate reducers, such as strain MLMS-1, which apparently oxidizes sulfide to sulfate (10) rather than to S0 or another intermediate, may use different pathways or contain divergent arrA genes, since we were unable to amplify arrA genes from it. We retrieved sequences presumed to be partial sequences of arrA genes from organisms (the Ectothiorhodospiraceae) that are not known to respire arsenate. These may represent paralogous genes, and some of the clades shown in Fig. 7 may correspond to guilds with a completely different physiology: arsenite oxidation rather than arsenate reduction.
Supplementary Material
Acknowledgments
We thank R. Jellison for help with sample collection; S. B. Joye, M. A. Moran, R. S. Oremland, J. Stolz, W. Whitman, and J. Zehr for valuable discussions; two anonymous reviewers for comments on the manuscript; and M. Madigan and R. S. Oremland for cultures.
This work was supported by NSF grant MCB 99-77886 to J.T.H.
Footnotes
Supplemental material for this article can be found at http://aem.asm.org/.
REFERENCES
- 1.Afkar, E., J. Lisak, C. Saltikov, P. Basu, R. S. Oremland, and J. F. Stolz. 2003. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 226:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Bak, F., and H. Cypionka. 1987. A novel type of energy conservation involving fermentation of inorganic sulphur compounds. Nature 326:891-892. [DOI] [PubMed] [Google Scholar]
- 3.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum, J. S., A. B. Bindi, J. Buzzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 5.Chen, K. Y., and S. K. Gupta. 1973. Formation of polysulfides in aqueous solution. Environ. Lett. 4:187-200. [DOI] [PubMed] [Google Scholar]
- 6.Fuseler, K., and H. Cypionka. 1995. Elemental sulfur as an intermediate of sulfide oxidation with oxygen by Desulfobulbus propionicus. Arch. Microbiol. 164:104-109. [Google Scholar]
- 7.Giggenbach, W. F. 1974. Equilibria involving polysulfide ions in aqueous sulfide solutions up to 240°. Inorg. Chem. 13:1724-1730. [Google Scholar]
- 8.Giri, B. J., N. Bano, and J. T. Hollibaugh. 2004. Distribution of RuBisCO genotypes along a redox gradient in Mono Lake, California. Appl. Environ. Microbiol. 70:3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henriksen, J. R. 2004. webLIBSHUFF. http://libshuff.mib.uga.edu.
- 10.Hoeft, S. E., T. R. Kulp, J. F. Stolz, J. T. Hollibaugh, and R. S. Oremland. 2004. Dissimilatory arsenate reduction with sulfide as electron donor: experiments with Mono Lake water and isolation of Strain MLMS-1, a chemoautotrophic arsenate respirer. Appl. Environ. Microbiol. 70:2741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoeft, S. E., F. Lucas, J. T. Hollibaugh, and R. S. Oremland. 2002. Characterization of bacterial arsenate reduction in the anoxic bottom waters of Mono Lake, California. Geomicrob. J. 19:1-19. [Google Scholar]
- 12.Hollibaugh, J. T., S. Carini, H. Gürleyük, R. Jellison, S. B. Joye, G. Lecleir, C. Meile, L. Vasquez, and D. Wallschläger. 2005. Distribution of arsenic species in alkaline, hypersaline, Mono Lake, California and response to seasonal stratification and anoxia. Geochim. Cosmochim. Acta 69:1925-1937. [Google Scholar]
- 13.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, D. L., and M. E. Q. Pilson. 1972. Spectrophotometric determination of arsenite, arsenate, and phosphate in natural waters. Anal. Chim. Acta 58:289-299. [Google Scholar]
- 15.Kempe, S., and E. T. Degens. 1985. An early soda ocean? Chem. Geol. 53:95-108. [Google Scholar]
- 16.LeCleir, G. R., A. Buchan, and J. T. Hollibaugh. 2005. Chitinase gene sequences from diverse aquatic habitats reveal complex patterns of diversity. Appl. Environ. Microbiol. 69:6977-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maest, A. S., S. P. Pasilis, L. G. Miller, and D. K. Nordstrom. 1992. Redox geochemistry of arsenic and iron in Mono Lake, California, p. 507-511. In Y. K. Kharaka and A. S. Maest (ed.), Water-rock interaction. A. A. Balkema, Rotterdam, The Netherlands.
- 18.Malasarn, D., C. W. Saltikov, K. M. Campbell, J. M. Santini, J. G. Hering, and D. K. Newman. 2004. arrA is a reliable marker for As(V) respiration. Science 306:455. [DOI] [PubMed] [Google Scholar]
- 19.Miller, L. G., R. Jellison, R. S. Oremland, and C. W. Culbertson. 1993. Meromixis in hypersaline Mono Lake, California. 3. Biogeochemical response to stratification and overturn. Limnol. Oceanogr. 38:1040-1051. [Google Scholar]
- 20.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis of polymerase chain reaction-amplified genes coding for 16S rDNA fragments. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman, D. K., T. J. Beveridge, and F. M. M. Morel. 1997. Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 63:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obraztsova, A. Y., C. A. Francis, and B. M. Tebo. 2002. Sulfur disproportionation by the facultative anaerobe Pantoea agglomerans sp1 as a mechanism for chromium(VI) reduction. Geomicrob. J. 19:121-132. [Google Scholar]
- 23.Oremland, R. S., P. R. Dowdle, S. Hoeft, J. O. Sharp, J. K. Schaefer, L. G. Miller, J. S. Blum, R. L. Smith, N. S. Bloom, and D. Wallschläger. 2000. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim. Cosmochim. Acta 64:3073-3084. [Google Scholar]
- 24.Oremland, R. S., S. E. Hoeft, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oremland, R. S., T. R. Kulp, J. S. Blum, S. E. Hoeft, S. Baesman, L. G. Miller, and J. F. Stolz. 2005. A microbial arsenic cycle in a salt-saturated, extreme environment. Science 308:1305-1308. [DOI] [PubMed] [Google Scholar]
- 26.Oremland, R. S., J. F. Stolz, and J. T. Hollibaugh. 2004. The microbial arsenic cycle in Mono Lake, California. FEMS Microbiol. Ecol. 48:15-27. [DOI] [PubMed] [Google Scholar]
- 27.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 28.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.