Abstract
The Pho regulon integrates the sensing of environmental inorganic phosphate (Pi) availability with coregulation of gene expression, mediating an adaptive response to Pi limitation. Many aspects of the Pho regulon have been addressed in studies of Escherichia coli; however, it is unclear how transferable this knowledge is to other bacterial systems. Here, we report work to discern the conservation of the Pho regulon in Pseudomonas fluorescens Pf0-1. We demonstrate by mutational studies that PhoB/PhoR and the Pst system have conserved functions in the regulation of Pi-induced phosphatase activities, as well as expression of other Pi-regulated genes. A genetic screen was carried out to isolate factors that affect Pho-regulated phosphatase activity. We identified the Pho-regulated phosphatases PhoX and PhoD and present evidence that these enzymes are exported via the Tat system. The phoX and phoD genes were shown to be members of the Pho regulon by reverse transcription-PCR, as well as by functional assessment of putative PhoB binding sites (Pho boxes). Our data also suggested that at least one other non-Tat-secreted Pho-regulated phosphatase exists. From the genetic screen, numerous siderophore mutants that displayed severe defects in Pho-activated phosphatase activity were isolated. Subsequently, iron was shown to be important for modulating the activity of Pho-regulated phosphatases, but it does not regulate this activity at the level of transcription. We also identify and demonstrate a novel role in siderophore production and Pho-regulated phosphatase activity for ApaH, the hydrolase for the nucleotide-signaling molecule AppppA. Finally, numerous mutations in multiple cellular pathways were recovered that may be required for maximal induction of the Pho regulon under Pi-limiting conditions.
Bacteria are remarkable in their capacity to monitor and respond to fluctuations in their microenvironment, adapting their metabolic and physiological potential to better suit their immediate requirements. This is particularly true of Pseudomonas spp., which inhabit a broad range of environmental habitats, from soil to animal hosts (48, 51).
Inorganic phosphate (Pi) is the preferred source of phosphate for bacteria, and thus, adaptation to fluctuations in its extracellular concentration is an important biological trait (57). One system that has evolved to perform this function is the Pho regulon, which can be defined as the set of genes whose transcription is modulated by the response regulator PhoB when concentrations of exogenous Pi become limiting (42). In response to low-Pi environments, PhoB is phosphorylated (PhoB∼P) by the histidine kinase PhoR, allowing it to bind within the promoters of specific genes, modulating their transcription. In Escherichia coli, PhoB∼P is known to bind a specific sequence termed a Pho box, which is comprised of two 7-bp direct repeats separated by a 4-bp linker sequence. One monomer of PhoB binds to each 7-bp direct repeat, facilitating recruitment of the σ70 subunit of RNA polymerase and initiation of transcription. Under Pi-sufficient conditions, PhoR is prevented from phosphorylating PhoB via an interaction with the Pst system (57). The Pst system is comprised of five proteins, PstS, -C, -A, and -B and PhoU, which are expressed from a single operon. In addition to its role as a negative regulator of Pho, the Pst system is a high-affinity Pi transport system that is itself expressed as part of the Pho regulon. The exact mechanism by which Pst inhibits PhoR-mediated phosphorylation of PhoB is not known; however, Pst is thought to form a repression complex with PhoR in the inner membrane when Pi levels are not limiting (57).
Many genes within the Pho regulon have roles in Pi transport and/or assimilation that facilitate adaptation to limiting Pi environments (57). For example, alkaline phosphatase (APase) liberates inorganic phosphate from a range of organic molecules and it is a well-conserved member of the Pho regulon (55, 57). The canonical Pho system links the detection of limiting Pi environments to regulatory mechanisms directing the adjustment of gene expression to specifically facilitate growth/survival in such an environment.
Most of our knowledge of the Pho regulon has been derived from experiments with E. coli, so it would be interesting to know at what level this information is transferable to other bacterial systems. Various degrees of evidence supporting conservation of the Pho regulon have been published for a number of bacteria, mostly within the alpha- and gammaproteobacteria (14, 31, 35, 50, 53, 56) and gram-positive bacteria (19, 36, 54). Among the alphaproteobacteria, the Pho regulon of Sinorhizobium meliloti has been best characterized. As in E. coli, PhoB is required for activation of gene expression in response to limiting phosphate and is likely to bind a similar Pho box motif (4, 53). However the roles of PhoR and the Pst system in regulating PhoB activity remain unclear (25).
Studies of several pseudomonads, including Pseudomonas aeruginosa, Pseudomonas putida, and Pseudomonas aureofaciens, have demonstrated conservation of the major regulators of Pho regulon expression. The PhoB-PhoR two-component system is conserved in all three species and is required for sensing and modulation of gene expression under Pi-limiting conditions (3, 11, 31, 35, 63). PhoB binding sites have been identified using the E. coli consensus, a limited number of which have been functionally assessed (35, 47). The Pst system has also been shown to be functionally conserved in all three species for both Pi transport and negative regulation of Pho induction (31, 35, 63). The genomic organization of the pst operons in Pseudomonas fluorescens Pf0-1 (http://genome.ornl.gov/microbial/pflu/) and P. putida KT2440 (34) is identical to that of E. coli, with the pstSCAB-phoU genes likely forming an operon. In P. aeruginosa PAO1, the pstS gene is situated at a locus distinct from the pstCAB-phoU genes (35) but remains under PhoB control (62). In pseudomonads, the major regulatory elements of the Pho regulon appear to be functionally conserved; however, very little is known about the composition of the Pho regulon and the nuances of its regulation.
We were interested in knowing to what extent the Pho regulon, as described in E. coli, is conserved in Pseudomonas fluorescens Pf0-1. We first demonstrated the conserved functions of both PhoB/PhoR and the Pst system in the regulation of Pho expression in response to Pi. Secondly, we performed a genetic screen to isolate mutants defective in induction of the Pho-regulated phosphatase activity(s). This screen led to the identification of enzymes responsible for Pho-regulated phosphatase activity, factors required for their transport to the periplasm, and cellular factors required for maximal phosphatase activity. The isolation of genes that may generally affect Pho regulon induction are also reported and briefly discussed.
MATERIALS AND METHODS
Strains and media.
The strains and plasmids used in this study are listed in Table 1. P. fluorescens and E. coli were routinely cultured in lysogeny broth (LB) unless stated otherwise and were grown at 30°C and 37°C, respectively (5). K10Tπ was used for low-phosphate conditions and consisted of 50 mM Tris-HCl (pH 7.4), 0.2% (wt/vol) Bacto tryptone, 0.15% (vol/vol) glycerol, and 0.61 mM MgSO4. K10Tπ contains 0.14 mM Pi, measured as described below. For Pi-sufficient conditions, K10Tπ medium was amended with 1 mM K2HPO4, yielding K10T-1. All K10T media were made from concentrated stock solutions and sterilized by passing them through a 0.22-μm filter. TSP salts consisted of 2 g/liter (NH2)SO4, 2 g/liter NaCl, and 1.65 g/liter KCl. TSP medium was made by amending TSP salts with 50 mM Tris HCl (pH 7.4), 0.2% (vol/vol) glycerol, 1 mM MgSO4, and the desired phosphorus source. For chelation of iron, K10Tπ medium was supplemented with 2,2′-dipyridyl (Sigma-Aldrich, St. Louis, MO) at concentrations ranging from 0.1 to 0.3 mM. Kings B medium (KB) consisted of 20 g/liter Bacto tryptone, 1.5 g/liter MgCl2, 1.5 g/liter K2HPO4, and 1.5% (vol/vol) glycerol (23).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 thi-1 relA1 recA1 | 15 |
| S17-1(λpir) | thi pro hsdR hsdM+ ΔrecA RP4-2::TcMu-Km::Tn7 | 49 |
| Pseudomonas fluorescens | ||
| Pf0-1 | Wild type | |
| Δpst | Pf0-1 with deletion of pstSCAB-phoU; Gmr | This study |
| ΔphoB | Pf0-1 with deletion of phoB; Gmr | This study |
| Δpst phoB | Δpst with unmarked deletion of phoB; Gmr | This study |
| ΔphoD | Pf0-1 with deletion of phoD; Gmr | This study |
| PF-B6136 | Pf0-1; tatA::mini-Tn5lacZ1Km; Kmr | This study |
| PF-E280 | Pf0-1; tatC::mini-TnM; Gmr | This study |
| PF-E313 | Pf0-1; phoX::mini-Tn5lacZ1Km; Kmr | This study |
| PF-E313 ΔphoD | PF-E313 with deletion of phoD; Kmr Gmr | This study |
| PF-H1190 | Pf0-1; phoR::mini-Tn5lacZ1Km; Kmr | This study |
| PF-B337 | Pf0-1; apaH::mini-Tn5lacZ1Km; Kmr | This study |
| PF-B582 | Pf0-1; pvdL::mini-TnM; Gmr | This study |
| PF-apaH | Pf0-1::pKO-apaH; Tetr | This study |
| Plasmids | ||
| pAC-Ω-GM | Source of Gm cassette | 45 |
| pBBRMCS-5 | Broad-host range cloning vector; Gmr | 24 |
| pBB-apaGH | pBBRMCS-5 expressing apaGH | This study |
| pBT20 | Mini-TnM delivery vector; Apr Gmr | 26 |
| pEX18Tc | Positive selection knockout vector; sacB Tetr | 16 |
| pEX-phoB-Gm | pEX18Tc-derived vector for deletion of phoB; Tetr Gmr | This study |
| pEX-phoB | pEX18Tc-derived vector for unmarked deletion of phoB; Tetr | This study |
| pEX-pst-Gm | pEX18Tc-derived vector for deletion of pstSCABphoU; Tetr Gmr | This study |
| pEX-phoD-Gm | pEX18Tc-derived vector for deletion of phoD; Tetr Gmr | This study |
| pMini-CTX-lacZ | Site-specific integration vector; Tetr | 17 |
| pKO3 | Single-crossover knockout vector derived from pMini-CTX-lacZ | This study |
| pKO-apaH | pKO3 containing internal apaH fragment; Tetr | This study |
| pJB785TTKm1 | RK2 origin vector for luciferase transcriptional fusions; Kmr | 44 |
| pME3280a | Tn7 cloning vector; Apr Gmr | 65 |
| pTn7-PphoD-luc | Tn7 delivery vector for PphoD luciferase fusion; Apr Kmr | This study |
| pTn7-PphoD-Δbox | Tn7 delivery vector for PphoD-Δbox luciferase fusion; Apr Kmr | This study |
| pTn7-Ppst-luc | Tn7 delivery vector for Ppst luciferase fusion; Apr Kmr | This study |
| pUC-lucK | Derivative of pJB785TTKm with colE1 origin; Kmr | This study |
| pUC-Ppst-luc | pUC-lucK with Ppst luciferase fusion; Kmr | This study |
| pUC-PphoD-luc | pUC-lucK with PphoD luciferase fusion; Kmr | This study |
| pUC-PphoD-Δbox | pUC-lucK with PphoD luciferase fusion with mutated Pho box; Kmr | This study |
| pUC-PphoX-luc | pUC-lucK with PphoX luciferase fusion; Kmr | This study |
| pUC19 | colE1-based cloning vector; Apr | 64 |
| pUT-miniTn5-lacZ1-Km | Tn5 delivery vector; Apr Kmr | 7 |
| pUX-BF13 | Tn7 helper plasmid encoding Tn7 transposition functions; Apr | 65 |
Antibiotics were used at the following concentrations, unless otherwise stated: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 50 μg/ml; tetracycline (Tet), 10 to 15 μg/ml (E. coli) and 30 μg/ml (P. fluorescens); gentamicin (Gm), 30 μg/ml; and chloramphenicol (Cm), 20 μg/ml.
Quantification of the inorganic-phosphate concentration.
The methodology for the quantification of the inorganic-phosphate concentration was as described previously with minor modifications (20). Briefly, 500 μl of medium was added to 500 μl of freshly prepared reagent containing 0.5% sodium molybdate and 2% ascorbic acid. Samples were mixed and incubated for 20 min at 50°C. After the samples cooled to room temperature, their absorbances were read at 820 nM. The concentrations of Pi in samples were determined by the construction of a standard curve using K2HPO4.
Construction of phoB, pstSCAB-phoU, and phoD gene deletions.
Fragments (800 bp to 1 kb) flanking both sides of the gene(s) to be deleted were amplified by PCR and ligated together before being cloned into the sacB-based suicide vector pEX18Tc. The constructs were further modified for positive selection by cloning a Gm omega cassette in the junction between flanking regions to create pEX-phoB-Gm, pEX-pst-Gm, and pEX-phoD-Gm. The Gm omega cassette was amplified from pAC-Ω-GM. To construct the pst phoB double mutant, pEX-phoB was used to make an unmarked phoB deletion in the pst deletion background. Plasmids were mobilized into host strains by conjugation, and single crossovers were selected with either Tet or Gm. Subsequently, strains that had undergone a second single-crossover event at this locus were selected for by plating them on LB plus 5% (wt/vol) sucrose or LB plus 5% (wt/vol) sucrose plus Gm (16). Genuine knockouts were assessed for Tet sensitivity and validated by PCR analysis utilizing at least one primer located outside the area of recombination.
Fluorescent-phosphatase assay.
Stationary-phase cultures grown in LB were subcultured to 5 ml of K10Tπ in a 1:50 dilution. After 6 h of aerated growth, 1 ml of culture was pelleted and resuspended in 1 ml of TSP salts. For measurement of optical density, 500-μl aliquots were removed, diluted twofold, and read in a Spectra Max M2 microplate reader (Molecular Devices, Sunnyvale, CA) at 600 nM. To measure phosphatase activity, 500 μl of washed cells was lysed by adding 10 μl 0.1% (wt/vol) sodium dodecyl sulfate and 20 μl chloroform, mixed, and incubated at 30°C for 10 min. Cell debris was pelleted by centrifugation at 13,000 rpm for 10 min. The phosphatase reaction mixture consisted of 400 μl cell extract, 100 μl 1 M Tris-HCl, pH 8.0, and 10 μl of 10 mM 4-methylumbelliferyl phosphate (MUP) (Sigma-Aldrich, St. Louis, MO). Reaction mixtures were incubated at 30°C for 30 min before termination by the addition of 20 μl of 3 N NaOH. MUP is a substrate for alkaline phosphatase that when cleaved produces the fluorescent product 4-methylumbelliferone (13). The relative fluorescences of samples were measured using a Spectra Max M2 microplate reader set at 360-nM excitation and 449-nM emission. Relative fluorescence units (RFU) were corrected for background fluorescence and then normalized to readings of the optical density at 600 nm (OD600).
In development of this assay, we determined that (i) the concentration of MUP substrate is not limiting over the course of the assay, (ii) the phosphatase reaction is linear over time up to at least 1 h, (iii) NaOH added at the specified concentration effectively prevents further cleavage of substrate, and (iv) all fluorescence readings were within the linear range of the fluorometer (data not shown).
Qualitative phosphatase assay.
Strains were picked onto K10Tπ agar medium supplemented with 5-bromo-4-chloro-3-indolylphosphate (BCIP) (40 μg/ml; Sigma-Aldrich, St. Louis, MO) and incubated for 24 to 48 h at 30°C. The colonies were then scored for levels of blue coloration due to cleavage of the phosphatase substrate BCIP (31).
Tn mutagenesis.
Transposon (Tn) mutagenesis was performed with both mini-Tn5 and mariner transposons. E. coli S17-1(λpir) harboring either pBT20 or pUT-miniTn5-KmlacZ1 was washed twice with LB and mixed with P. fluorescens in a 1:4 donor/host ratio, spotted on LB plates, and incubated at 30°C for 45 min to 1 h. Cells were then recovered and plated on selective media. The mini-Tn5 transformants were selected with Km and Cm. Mariner transformants were selected with Gm and Cm. P. fluorescens Pf0-1 is naturally resistant to Cm at concentrations below 30 μg/ml. Individual transformants were then picked onto BCIP test plates and screened for Pho induction using the qualitative phosphatase assay.
Arbitrary primed PCR and sequencing of Tn insertions.
Arbitrary primed PCR and sequencing were performed as reported previously (39) with the modifications outlined below.
(i) First-round PCR.
For mini-Tn5-lacZ1-Km insertions, the Tn-specific primer used was lacZ-1 (5′ GGC GAT TAA GTT GGG TAA CG), and for TnM insertions, the specific primer was TnM-3 (5′ GAC ATC ATA ACG GTT CTG GCA AAT). The appropriate Tn-specific primer was used in conjunction with two degenerate primers, ARB1 (5′ GGC CAC GCG TCG ACT AGT ACN NNN NNN NNN GAT AT) and ARB6 (5′ GGC CAC GCG TCG ACT AGT ACN NNN NNN NNN ACG CC). The PCR conditions were 200 μM deoxynucleoside triphosphates (Roche, Indianapolis, IN), 5% dimethyl sulfoxide, 0.8 μM Tn-specific primer, 2 μM of both ARB1 and ARB6, and 1 unit of Taq polymerase (QIAGEN, Valencia, CA). The PCR cycling conditions were 94°C for 5 min and then 6 cycles of 94°C for 30 s, 30°C for 30 s, and 72°C for 1 min, followed by 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 2 min, ending with a single-step extension for 7 min at 72°C.
(ii) Second-round PCR.
One microliter of the first-round reaction product was used as a template for the second-round reaction as follows. For mini-Tn5-lacZ1-Km insertions, the nested Tn-specific primer used was lacZ-2 (5′ TTT TCC CAG TCA CGA CGT T), and for TnM insertions, the nested specific primer was TnM-2 (5′ TGA GCT GTT GAC AAT TAA TCA TCG G). The appropriate Tn-specific primer was used in conjunction with ARB2 (GGC CAC GCG TCG ACT AGT AC), using the same standard PCR conditions described above. The PCR cycling conditions were 94°C for 5 min and then 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 1 min, followed by a single-step extension for 7 min at 72°C. The reaction products yielded were purified with a QIAquick PCR purification kit (QIAGEN, Valencia, CA) and sequenced using either the lacZ-2 primer for Tn5-derived products or TnM-3 for TnM-derived products.
Reverse transcription (RT)-PCR.
Stationary-phase cultures grown in LB were subcultured to 5 ml of K10T-1 at a 1:50 dilution. The cells were grown to mid-log phase (OD600 = 0.6) before 1.5 ml was removed and mixed with 2 volumes of RNAprotect reagent (QIAGEN, Valencia, CA). RNA was extracted using an RNeasy kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions, except that a postcolumn DNase treatment was performed using 15 U of RQ1 DNase (Promega, Madison, WI). The RNA was subsequently purified using the RNeasy kit. RNA integrity was assessed by gel electrophoresis, and genomic DNA contamination was assessed by PCR. The RNA concentration was measured using an Eppendorf Biophotometer.
First-strand cDNA synthesis was performed using Superscript III (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Random hexamers were used to prime cDNA synthesis, using 1.5 μg of total RNA for each reaction.
PCR was performed on cDNA samples using primers designed to amplify 200- to 300-bp fragments internal to the phoX, phoD, and rplU coding regions. rplU served as a control for cDNA synthesis. Luciferase transcriptional fusions indicated that rplU expression does not change between the strains and conditions used in this experiment (data not shown).
Construction of luciferase transcriptional fusions.
To allow construction of merodiploids, the RK2 origin of pJB785TTKm1 was replaced with the colE1 origin from pUC19, thereby creating a Pseudomonas suicide vector. The RK2 origin was excised from pJB785TTKm1 by digestion with XmnI and MslI, followed by gel extraction of the 5-kb band. The colE1 origin was amplified from pUC19 using the primers pUC-ori-F (5′ TGG TTT GAT ATC GCG TTT TTC CAT AGG CTC C) and pUC-ori-R (5′ TGG TTT GAT ATC TCA AAG GAT CTT CTT GAG ATC C). The approximately 600-bp product was digested with EcoRV and blunt-end ligated to the XmnI/MslI pJB785TTKm1 fragment. The resultant plasmid is referred to as pUC-lucK.
Luciferase fusions were constructed by individually cloning the promoter regions of phoX, phoD, and pst as 400- to 500-bp NcoI/BglII fragments into pUC-lucK. All promoters were inclusive of the predicted Pho box. The NcoI site of pUC-lucK overlaps the ATG start codon of luciferase so that fusions to luciferase included the native ribosomal binding site for each gene. phoD, phoX, and pst fusions are referred to as pUC-PphoD-luc, pUC-PphoX-luc, and pUC-Ppst-luc, respectively.
To construct a phoD promoter fusion with a mutated Pho box, two sets of primers were designed to amplify regions on each side of the 5′ Pho box. A PacI restriction site was engineered into the ends of both amplicons so that restriction with PacI and ligation of the two fragments would ultimately replace the 5′ Pho box repeat with a PacI restriction site. This ligation was then used as a template for PCR with the same primers used to construct the wild-type (WT) phoD promoter fusion. The resultant product was purified and then cloned into pUC-lucK, using NcoI and BglII to create the plasmid pUC-PphoD-Δbox.
phoD, phoD-Δbox, and pst luciferase fusions were shifted into a Tn7 vector for mobilization onto the chromosome. Internal regions of Tn7 from pME3280a were removed by digestion with XhoI, followed by end filling of the 5′ overhang with DNA polymerase I, large (Klenow) fragment. Subsequently, the approximately 5-kb band was extracted and is referred to as pTn7-Blunt. pUC-PphoD-luc, pUC-PphoD-Δbox, and pUC-Ppst-luc were digested with XmnI and MslI and shotgun cloned into pTn7-Blunt, selecting for the Km resistance determinant present on the fragment containing the luciferase fusion. The resultant plasmids are referred to as pTn7-PphoD-luc, pTn7-PphoD-Δbox, and pTn7-Ppst-luc.
Luciferase assays.
Luciferase activity was assessed using the luciferase assay system (Promega, Madison, WI). Assays were carried out essentially as described by the manufacturer. Briefly, 86 μl of culture was combined with 4 μl of 0.5 M EDTA and 10 μl of 1 M K2HPO4, pH 7.8; mixed; and crash frozen in an ethanol-dry-ice bath. Samples were then thawed at room temperature in a water bath before addition of cell culture lysis reagent and mixing. Concurrent with the preparation of cell lysates, optical-density readings (OD600) for cultures were measured with a Spectra Max M2 microplate reader, using either the microplate or cuvette reader. The luciferase activities of cell lysates were measured on a Lumimark luminometer (Bio-Rad, Hercules, CA) following the manufacturer's instructions. Luciferase activity was expressed as relative light units/OD600.
Transformation with Tn7.
The recipient strain, S17-1(λpir); Tn7 donors; and the Tn7 helper strain S17-1(λpir)(pUX-BF13) were washed twice in LB before being mixed in a 4:1:1 (recipient/donor/helper) ratio. Conjugations were incubated at 30°C for 8 h before transformants were selected for by plating them on LB plus Km plus Cm. Tet was used in place of Cm where appropriate. Tn7 is known to insert at a specific location between the coding regions for Pfl5726 (glmS) and Pfl5725 (27). The correct insertion of Tn7 was verified by PCR using a primer that anneals to the right-hand arm of Tn7 (Tn7-F [5′ CAG CAT AAC TGG ACT GAT TTC AG]) and another primer that anneals inside the glmS gene (glmS-R [5′ TGC TCA AGG GCA CTG ACG]). Correct insertion allows the amplification of an approximately 100-bp band.
Construction of single-crossover knockouts.
To construct the single-crossover knockout vector pKO3, pMini-CTX-lacZ was digested with PflFI (an isoschizmer of Tth111L) and MscI. Cohesive 5′ overhangs were then end filled with DNA polymerase I, large (Klenow) fragment, before blunt-end religation and recovery. This effectively deleted the integrase gene, thus preventing site-specific integration and converting pMini-CTX-lacZ into a suicide vector for Pseudomonas and other bacterial species unable to maintain colE1 replicons.
To disrupt apaH, a 310-bp fragment from the N terminus of the apaH open reading frame was amplified with the primers apaH-KO-F-EcoRI (5′ TGA TGG GAA TTC CCT CGA ACC GCT CAA GTG CC) and apaH-KO-R-BamHI (5′ CTG TTT GGA TCC GAA GGT TGT CGT CAC GCA GC). The restriction enzymes EcoRI and BamHI were then used to clone the apaH PCR fragment into pKO3, creating the vector pKO-apaH. Conjugation was used to transform P. fluorescens Pf0-1 with pKO-apaH. Strains that had integrated pKO-apaH onto their chromosomes were selected by resistance to Tet. Homologous recombination at the apaH locus was confirmed with the primers lacZ-pKO-F (5′ GCC AGT GAA TCC GTA ATC ATG GTC) and apaH-KO-verify (5′ ATG GCG ACG TAT GCC GTC). The apaH single-crossover knockout is referred to as PF-apaH.
Complementation of the apaH mutant.
For complementation studies, apaGH inclusive of the putative promoter region was amplified by PCR with the primers apaGH-F-BamHI (5′ AAG GCT GGA TCC GGG TCG AGC ACC TGT TCA AC) and apaGH-R-EcoRI (5′ ACG TTT GAA TTC GCT CAT GGC). The resultant 1.75-kb fragment was digested with BamHI and EcoRI and cloned into pBBRMCS-5, generating the plasmid pBB-apaGH. The apaGH fragment was cloned so that it was under the control of its native promoter rather than the lacZα promoter.
Siderophore fluorescence assays.
Strains were picked onto a low-iron agar medium, either KB or K10T-1. After incubation for 24 h, fluorescence due to excretion of siderophores was visualized by UV transillumination on a Kodak image station 2000R (Eastman Kodak Company, New Haven, CT).
Pyoverdine quantification.
The relative quantification of pyoverdine levels was performed essentially as described previously (1). Briefly, overnight cultures grown in KB were adjusted to the same optical density before the isolation of the supernatant by centrifugation at 13,000 rpm for 3 min. Absorbance spectra from 350 nM to 450 nM were obtained for individual supernantants using a Spectra Max M2 microplate reader. Pyoverdine has maximal absorbance at 403 nM.
RESULTS
Conservation of PhoBR and Pst as regulators of the Pho regulon.
Initially, we assessed the involvement of the PhoBR-Pst system in regulating expression of phosphatase activity in response to Pi-limiting environments. Alkaline phosphatase is known to be a well-conserved member of the Pho regulon (55, 57). P. fluorescens Pf0-1 was also shown to activate expression of an alkaline phosphatase activity when grown in Pi-limiting media (Fig. 1A).
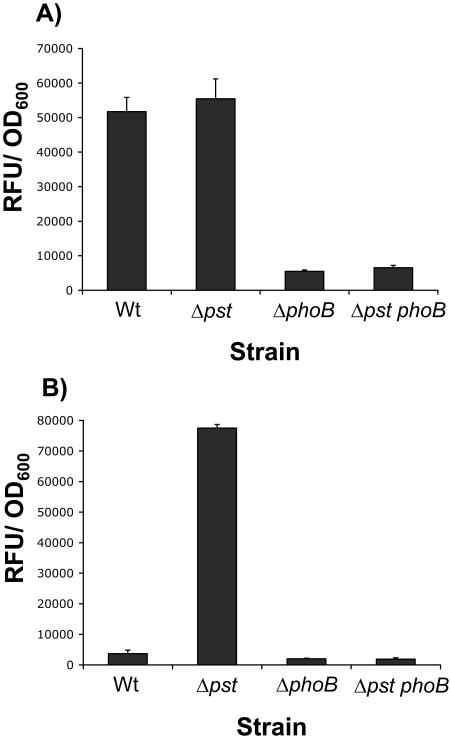
FIG. 1.
Assessment of PhoB and the Pst system as regulators of Pho regulon expression. Phosphatase activities expressed as RFU normalized to the OD600 are shown for the WT and ΔphoB, Δpst, and Δpst phoB mutants grown under (A) Pi-limiting conditions (K10Tπ) and (B) Pi-sufficient conditions (K10T-1) for 6 h. Standard errors based on four replicates are shown.
In E. coli, PhoB is required to activate expression of Pho regulon genes, including the alkaline phosphatase phoA gene. Similarly, deletion of phoB in P. fluorescens prevented expression of alkaline phosphatase activity in limiting Pi environments (Fig. 1A). Conversely, disruption of the Pst system was predicted to prevent repression of PhoB phosphorylation under Pi-sufficient conditions, thereby leading to a constitutive Pho system. Consistent with this prediction, deletion of the entire pstSCAB-phoU operon led to expression of phosphatase activity even in Pi-sufficient environments (Fig. 1B). To demonstrate that the phenotype of the pst mutant is dependent on Pho regulon expression, we mutated pst in the ΔphoB background and assessed the expression of alkaline phosphatase in Pi-sufficient environments relative to the pst mutant. In this case, phoB was shown to be epistatic to pst; the double mutant was unable to activate phosphatase expression under either Pi-sufficient or limiting conditions (Fig. 1A and B).
Interaction of the Pho system with alternative phosphorus sources.
E. coli is able to utilize various alternative sources of phosphorus as the sole source for growth. These include organophosphates, phosphonates, and phosphites. However, only Pi is thought to act as the sensory cue for control of Pho regulon induction via its interaction with the Pst-PhoR complex (57). We tested the abilities of organophosphates (rac-glycerol-3-phosphate [G3P], glucose-6-phosphate [G6P], and β-glycerophosphate [βGP]), phosphonates (2-aminoethylphosphonate [2AEP] and methylphosphonate [MP]), and sodium phosphite to support the growth of P. fluorescens Pf0-1 as the sole phosphorus source and to suppress Pho regulon induction.
We quantified the growth of the WT and ΔphoB in TSP minimal medium amended with the appropriate phosphate source to a final concentration of 1 mM. The OD600 was measured after 24 h of growth and normalized to growth in TSP plus 1 mM Pi. All organophosphates, specifically, βGP (18%), G3P (123%), and G6P (54%), supported various levels of growth of the WT when supplied as the sole phosphorus source. Interestingly, only growth on G6P was phoB dependent; ΔphoB showed no detectable growth even after 48 h. Although all organophosphates supported growth, only G3P could suppress activation of Pho regulon induction when added to K10Tπ (BCIP) at a concentration of 1 mM (data not shown). P. fluorescens was capable of growth only on the phosphonate 2AEP (124%), but not MP, even after 48 h. Similarly only 2AEP was able to inhibit induction of the Pho regulon. Growth on 2AEP was not phoB dependent. Sodium phosphite as the sole phosphorus source could not support the growth of P. fluorescens and was also unable to suppress Pho regulon induction under Pi-limiting conditions.
Identification of factors required for Pho-regulated phosphatase activity.
We were interested in extending our understanding of the P. fluorescens Pho system in several areas, including identification of (i) the Pho-regulated phosphatases, (ii) cellular factors required for phosphatase activity, and (iii) cellular factors required for Pho regulon induction in general. As a means to address these three questions, we performed a genetic screen for mutants of Pf0-1 that were unable to activate expression of alkaline phosphatase expression to the same degree as the wild type when grown in Pi-limiting environments.
From approximately 10,000 insertional mutants, generated with a combination of both Tn5 and mariner transposons, we recovered 96 mutants showing various levels of defects in the ability to cleave BCIP, a colorimetric organic substrate for phosphatase activity. Of these mutants, 75 were sequenced using arbitrary primed PCR. Table 2 shows the major categories of mutants. As expected, insertions within phoB and phoR were recovered from this screen. Furthermore, multiple independent mutant alleles were recovered for a number of loci, providing good genetic evidence for causal genotype-phenotype relationships.
TABLE 2.
Mutants with reduced Pho-regulated phosphatase activty
| Category | Pfl number | Gene | No. of hits | Predicted function |
|---|---|---|---|---|
| Siderophore synthesis | Pfl3940 | pvdL | 8 | Nonribosomal peptide synthetase |
| Pfl1849 | pvdD | 2 | Nonribosomal peptide synthetase | |
| Pfl1845 | 2 | Nonribosomal peptide synthetase | ||
| Pfl1844 | 1 | Nonribosomal peptide synthetase | ||
| Pfl1846 | 6 | Nonribosomal peptide synthetase | ||
| Pfl1847 | 1 | Nonribosomal peptide synthetase | ||
| Pfl1859 | pvdA | 1 | l-Ornithine N5-oxygenase | |
| Pfl3942 | 2 | Acetyltransferase | ||
| Phosphatase | Pfl5179 | phoX | 4 | Alkaline phosphatase |
| Regulation | Pfl5606 | phoB | 1 | Pho regulon response regulator |
| Pfl5607 | phoR | 2 | Pho regulon histidine kinase | |
| Pfl3471 | 1 | Transcriptional regulator, Asr-like | ||
| Pfl5020 | 1 | Transcriptional regulator | ||
| Pfl3848 | 1 | Anti-anti-sigma factor | ||
| Flagellar | Pfl1535 | fliE | 2 | Flagellar-hook basal body |
| Pfl1536 | fliF | 1 | Flagellar M ring protein | |
| Pfl1537 | fliG | 1 | Flagellar motor switch | |
| Pfl1507 | flgL | 1 | Flagellar P ring protein precursor | |
| Pfl1504 | flgI | 1 | Flagellar hook associated protein | |
| Pfl1508 | 1 | Glycosyl transferase | ||
| Membranes and transport | Pfl0838-40 | mreBCD | 2 | Rod shape determination |
| Pfl4402 | tolB | 1 | Outer membrane transport | |
| Pfl1816 | 2 | Cation transport | ||
| Pfl4531 | glpF | 1 | Glycerol uptake channel | |
| Pfl4062 | wecE | 1 | Pyridoxal-phosphate-dependent aminotransferase | |
| Pfl0382 | tatA | 1 | Twin arginine translocase | |
| Pfl0380 | tatC | 2 | Twin arginine translocase | |
| Metabolism | Pfl5512 | pps | 1 | Phosphoenolpyruvate synthase |
| Pfl0463 | aceF | 2 | Pyruvate dehydrogenase | |
| Pfl1766 | acnA | 1 | Aconitate hydratase | |
| Pfl4532 | glpK | 2 | Glycerol kinase | |
| Pfl1764 | prpB | 1 | PEP phosphomutase | |
| Pfl3612 | nouK | 1 | NADH dehydrogenase I | |
| Nucleotide biosynthesis | Pfl4728 | upp | 1 | Phosphoribosyltransferase |
| Pfl1902 | purF | 1 | Amidophosphoribosyltransferase | |
| Pfl5387 | serA | 1 | Phosphoglycerate dehydrogenase | |
| Aminoacid biosynthesis | Pfl4517 | argG | 5 | Argininosuccinate synthase |
| Pfl4527 | argF | 1 | Ornithine carbamoyltransferase | |
| Pfl5115 | trpG | 1 | Anthrinilate synthase component II | |
| Pfl0411 | aroB | 1 | 3-Dehydroquinate synthase | |
| Defense and stress | Pfl5309 | gshB | 1 | Glutathione synthetase |
| Pfl1901 | cvpA | 1 | Colicin production | |
| Pfl2563 | 2 | Penicillin amidase | ||
| Pfl5137 | apaH | 1 | Bis-(5′ nucleosyl) tetraphosphotase | |
| Hypothetical | Pfl4948 | 1 | Hypothetical protein | |
| Total | 75 |
Identification of the Pho-regulated phosphatases.
As a consequence of our screen, we expected to find mutations that directly affected the enzymatic activities of the Pho-regulated phosphatases. Intriguingly, sequence analysis had indicated that P. fluorescens Pf0-1 does not have a phoA homologue (http://genome.ornl.gov/microbial/pflu/). PhoA is the Pho-regulated alkaline phosphatase of E. coli, which is conserved in Pseudomonas aeruginosa. Four independent mutants had insertions that mapped to Pfl5179, a gene encoding a predicted phosphatase. Based on the cluster of orthologous groups (COGs) comparison match reported in the genome annotation, Pfl5179 is subsequently referred to as phoX (http://genome.ornl.gov/microbial/pflu/). After 24 h on BCIP plates, no phosphatase activity could be detected in a phoX mutant; however, at later time points, a low level of phosphatase activity was observed that was not present in the phoB mutant (Fig. 2A). This result was corroborated by quantitative assays, which showed the phoX mutant to have an activity higher than that of the phoB mutant (Fig. 2B). These phenotypes were consistent across all four phoX alleles. Together, these results indicate that PhoX is likely to be the major Pho-regulated phosphatase activity detected by BCIP and MUP substrates but that it is not sufficient to account completely for Pho-regulated phosphatase activity.
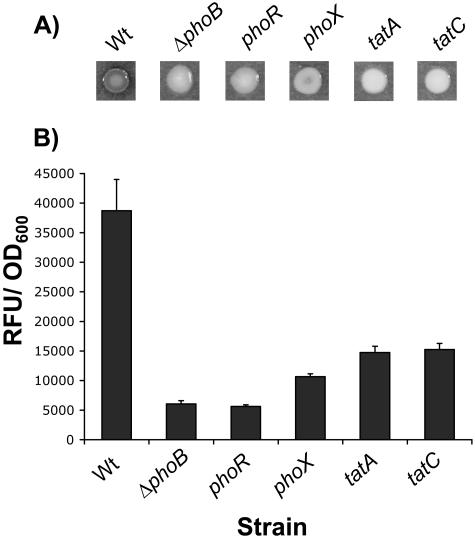
FIG. 2.
Requirement for PhoX and the Tat system in Pho-regulated phosphatase activity. (A) Qualitative phosphatase activities are shown for strains grown on Pi-limiting medium supplemented with BCIP. Higher degrees of darkness indicate higher levels of phosphatase activity. (B) Quantitative phosphatase assays for strains grown, with shaking, in Pi-limiting medium for 6 h. Activity is expressed as RFU normalized to the OD600. Standard errors based on four replicates are shown.
PhoX is secreted by the twin arginine transport system.
PhoX is predicted to have a twin arginine transport (Tat) signal sequence (http://genome.ornl.gov/microbial/pflu/). Tat provides an alternative pathway for the translocation of proteins across the inner membrane of gram-negative bacteria (40). For E. coli, the consensus leader peptide motif has been reported as SRRXFLK, with the twin arginine residues (underlined) essentially invariant (52). PhoX contains a predicted N-terminal leader sequence with the Tat-motif SRRGFIS. Consistent with PhoX being Tat transported, we recovered transposon insertions in structural genes of the Tat export machinery, tatC and tatA, which prevented detectable cleavage of BCIP under Pi-limiting conditions; tat mutants essentially had null phosphatase phenotypes on plates, comparable to those of phoB deletion strains (Fig. 2A). Quantitative phosphatase assays indicated that the tat mutants had slightly higher activities than the phoB mutant (Fig. 2B). This is consistent with the fact that tat mutants are likely to produce all of the phosphatases but cannot export them to the periplasm. Our quantitative assay utilizes chloroform to permeabilize cells, circumventing the need for transport. The fact that tat mutants do not exhibit normal levels of phosphatase activity suggests that, similar to E. coli PhoA, the majority of phosphatases are not active until they have been properly exported to the periplasm.
Further supporting evidence for Tat export of PhoX was provided by demonstrating that phoX transcription is not reduced in a tatC mutant relative to the wild type. For this experiment, we constructed a phoX merodiploid with the luciferase gene under the transcriptional control of the native phoX promoter. When grown under phosphate-limiting conditions, the WT phoX fusion was recorded at 4,579 ± 213 relative light units/OD600, whereas the ΔtatC phoX-fusion was recorded at 4,990 ± 127 RFU/OD600. Therefore, the Tat system does not impinge on expression of phoX at the level of transcription.
Identification of other Pho-regulated phosphatases.
Based on the residual phosphatase activities of phoX mutants, we predicted that other Pho-regulated phosphatases existed. Furthermore, the apparent null phosphatase phenotype of the tat mutants suggested that other phosphatase activities would also be Tat exported and would therefore have a Tat transport leader sequence. A search of the Pf0-1 genome annotation for other putative Tat-transported proteins with similarity to known phosphatases identified Pfl0796. Pfl0796 has a predicted N-terminal leader sequence containing the degenerate twin arginine motif RRRVMQA and shows 26% identity to PhoD, one of three characterized alkaline phosphatases in Bacillus subtilis (8).
Characterization of PhoD and PhoX as Pho-regulated phosphatases.
Given that a phoB mutant expresses no phosphatase activity, we predicted that both phoD and phoX should be expressed under Pho regulon control. In E. coli, PhoB∼P is known to bind to a consensus sequence referred to as a Pho box. Analysis of promoter regions for the well-conserved Pho genes pst (Fig. 3A) and phoB indicated the presence of putative Pho boxes with good matches to the E. coli consensus. Furthermore, transcriptional fusions and RT-PCR indicated that pstS was regulated by levels of Pi and PhoB (data not shown), supporting its membership in the Pho regulon. Together, these data suggested that the PhoB binding consensus was likely to be conserved in P. fluorescens. With this information, we searched for and located putative Pho boxes upstream of phoD and phoX with good matches to the E. coli consensus (Fig. 3A). RT-PCR was carried out to assess the transcript levels of phoD and phoX for WT and Δpst strains grown under Pi-sufficient conditions (Fig. 3B). Both phoX and phoD showed much higher levels of transcript in the Δpst strain than in the wild type, which is consistent with membership in the Pho regulon, as the pst mutants expressed Pho regulon genes even in Pi-sufficient environments (Fig. 1B). The rplU gene served as a Pho-independent control and demonstrated constant levels of transcript irrespective of Pho induction.
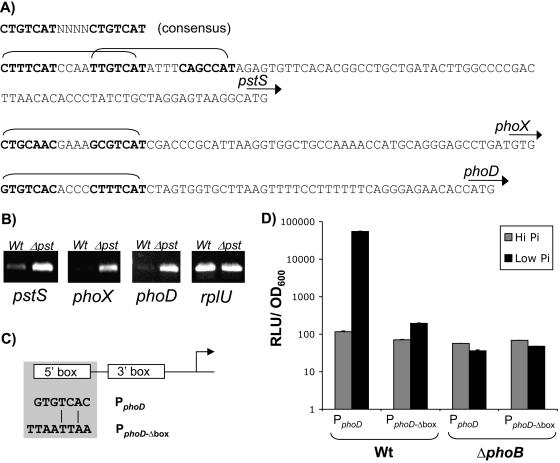
FIG. 3.
Pho regulon activation of phoD and phoX transcription. (A) Identification of putative Pho boxes upstream of pstS, phoX, and phoD based on the E. coli consensus. Direct repeats are in boldface, the complete Pho box is bracketed, and the start codon is indicated by an arrow for each gene; pstS has two overlapping Pho boxes. (B) RT-PCR detection of transcripts for the pstS, phoX, and phoD genes in WT and Δpst genetic backgrounds grown in Pi-sufficient medium (K10T-1). RT primers for pstS anneal within the N-terminal region of pstS that is still expressed in the pst mutant. The rplU gene expression levels serve as a Pho-independent control for cDNA synthesis. (C) Schematic of the site-directed mutational strategy employed to assess the functionality of the putative Pho box for phoD. The 5′ repeat of the predicted Pho box was replaced with a PacI restriction site (shaded box); conservation of bases between the 5′ repeat of Pho boxes for promoters PphoD and PphoDΔbox is also indicated. (D) Requirement for the putative Pho box in mediating activation of phoD transcription. Wild-type phoD promoter fusions to luciferase were compared to a promoter fusion with a mutated Pho box during growth in both Pi-sufficient (K10T-1) and Pi-limiting (K10Tπ) media. This analysis was done in both WT and ΔphoB genetic backgrounds. Standard errors based on three replicates are shown.
We conducted a further experiment to test whether the Pho box of phoD was indeed the sequence motif required for activation. For this experiment, two versions of the phoD promoter were cloned upstream of the luciferase transcriptional reporter gene, a wild-type version and one with the 5′ arm of the Pho box replaced with a PacI restriction site (Fig. 3C). Tn7 was then used to integrate the fusions at a neutral site on the chromosome of both the WT and ΔphoB strains. Transcriptional activation was assessed for both promoters under Pi-limiting conditions relative to Pi-sufficient conditions. Pf0-1 harboring the WT phoD promoter fusion showed 471-fold activation under limiting Pi conditions relative to expression under Pi-sufficient conditions (Fig. 3D). Activation was reduced to only threefold for the phoD promoter fusion with a mutated Pho box. As expected, no activation was seen for either promoter in the phoB mutant background, indicating that activation is PhoB dependent. Overall, these results provide strong evidence for Pho regulon control of phoD and phoX expression, as well as conservation of the Pho box binding sequence in Pseudomonas fluorescens.
Assessment of PhoD's contribution to Pho-regulated phosphatase activity.
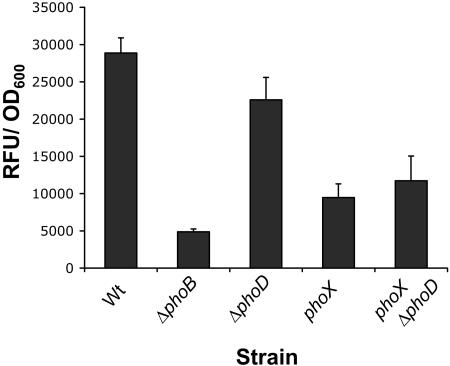
We assessed the contribution of PhoD to Pho-regulated phosphatase activity by comparing the phosphatase activities of ΔphoD and the phoX phoD double mutant relative to the wild type and the phoX mutant (Fig. 4). We could detect a small but significant (Student's t test, P < 0.05) reduction in phosphatase activity for ΔphoD versus the WT. However, we were unable to detect any further reduction of phosphatase activity in the phoD phoX double mutant compared to the phoX mutant. The contribution of PhoD to Pho-regulated phosphatase activity could not be visually detected by the qualitative assay, which explains why it was not recovered from the mutagenesis screen.
FIG. 4.
The contribution of PhoD to Pho-regulated phosphatase activity. Shown are phosphatase activities expressed as RFU/OD600 unit for the ΔphoD mutant and the phoX phoD double mutant compared to the WT and the ΔphoB and phoX mutants. The strains were grown in Pi-limiting media (K10Tπ) for 6 h. The standard error was calculated based on four replicates.
Iron is required for Pho-regulated phosphatase activity.
In addition to phoX and tat mutants, many other categories of mutant were recovered from the screen showing reduced expression of phosphatase activity (Table 2). Of these, siderophore-biosynthetic mutants were the largest class recovered. The majority of siderophore mutants (20) had insertions that mapped within genes encoding nonribosomal peptide synthetases (NRPS). NRPS direct the synthesis of short peptides in the absence of an RNA template, and their requirement for biosynthesis of many pseudomonad-derived siderophores has been well documented (32). Two other genes were recovered that had reported roles in siderophore synthesis, an acetyltransferase (http://genome.ornl.gov/microbial/pflu/) and an l-ornithine N5-oxygenase (2). In support of their roles in siderophore synthesis, all strains within this group failed to exhibit fluorescence characteristic of excreted pyoverdines when cultured on low-iron media (data not shown). All siderophore mutants showed very low levels of phosphatase activity when grown on Pi-limiting media supplemented with BCIP. Due to the similarities of phenotypes among the NRPS mutants, we selected one strain, PF-B582, for further characterization. PF-B582 has a TnM insertion within the coding region of Pfl3940, an NRPS homologous to the NRPS pvdL from Pseudomonas aeruginosa. pvdL was shown to be required for siderophore production by mutational analysis in P. aeruginosa (32).
As a first step to better understanding the nature of the phosphatase defect caused by disruptions of siderophore synthesis, we quantified the phosphatase defect of PF-B582 (pvdL) when grown in Pi-limiting media. We were able to detect a 30% decrease in phosphatase activity for PF-B582 (pvdL) compared to that of the wild type (Fig. 5A, bars marked 0 mM dipyridyl). However, this reduction did not correlate to the severity of the defect seen for siderophore mutants when grown on low-Pi agar media. It seemed likely that this disparity between quantitative and qualitative assays was the consequence of differences in methodology. The phosphatase test medium, K10Tπ, is a low-iron medium, which suggested, combined with the fact that siderophore mutants are compromised in the ability to scavenge iron, that iron limitation could be the biological basis for the effect on phosphatase activity.
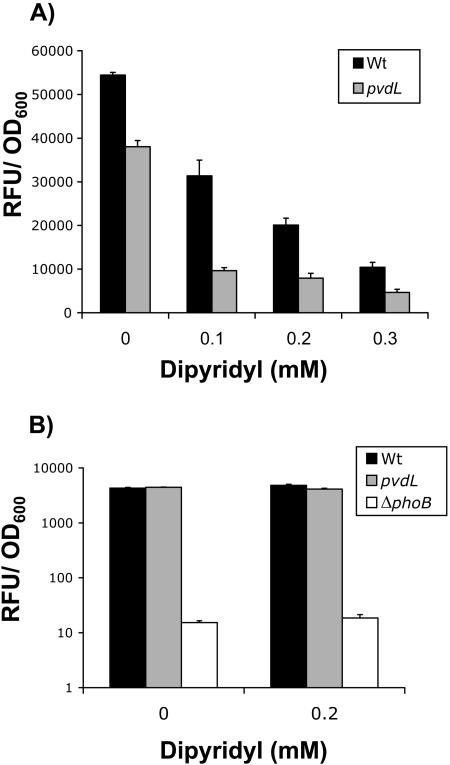
FIG. 5.
Effect of iron limitation on phosphatase activity. (A) Phosphatase activities were measured for the WT and the pvdL mutant (PF-B582) after supplementation of cultures with different amounts of the iron chelator 2,2′dipyridyl. The strains were grown in Pi-limiting medium (K10Tπ) for 6 h, with dipyridyl added after 3 h. Growth of the strains was not affected until the addition of 0.3 mM dipyridyl (data not shown). Standard errors calculated from three replicates are shown. (B) phoX transcription in response to iron limitation. The luciferase fusion to phoX was measured for the WT and the PF-B582 (pvdL) and ΔphoB mutants with and without the addition of 0.2 mM 2,2′dipyridyl. Cells were cultured in Pi-limiting medium (K10Tπ) for 6 h. Standard errors were calculated from three replicates.
Two components of the quantitative assay were likely to aid in reducing iron limitation: (i) the quantitative phosphatase assay is done in shaking liquid cultures instead of on agar medium and (ii) cultures are back diluted 1:50 from LB overnight cultures into test media, thereby potentially carrying over iron. To test these ideas, we measured the effects of iron chelation on the phosphatase activities of the WT and PF-B582 (pvdL). After 3 hours of growth in Pi-limiting medium, cultures were amended with different concentrations of the iron-specific chelator 2,2′dipyridyl and assessed for phosphatase activity after 3 h of further growth (Fig. 5A). Strikingly, addition of increasing amounts of 2,2′dipyridyl led to increasing reductions in phosphatase activity for both WT and PF-B582. The siderophore mutant was more sensitive to the addition of chelator, which supports the idea that chelation of free iron is the cause of the effect on phosphatase activity. It is important to note that the addition of dipyridyl did not markedly reduce growth in the assay until 0.3 mM for both WT and PF-B582 (pvdL). Overall, these results indicate that iron limitation reduces phosphatase activity in P. fluorescens. The question remained as to whether this effect was the result of a requirement for iron in Pho regulon induction.
PhoX was determined to be the major enzyme responsible for the phosphatase activity in both quantitative and qualitative assays. We assessed whether iron limitation had an impact on phoX transcription by utilizing a transcriptional fusion to the firefly luciferase gene. Based on previous experiments, we chose to assess phoX transcription in Pi-limiting media, with and without the addition of 0.2 mM 2,2′dipyridyl. We detected no difference in phoX transcription for either the wild type or PF-B582 (pvdL), irrespective of the dipyridyl concentration (Fig. 5B). The very low levels of transcription seen in a phoB mutant indicated that the phoX fusion was under Pho regulon control. The fact that iron limitation does not reduce phoX transcription suggests that low levels of intracellular iron affect the activity of the phosphatase(s) rather than transcription of phoX or other Pho regulon genes. We further showed that addition of exogenous iron during growth could stimulate phosphatase activity but that this could not be recapitulated by adding iron back to extracts in enzyme assays (data not shown).
Tat mutants are partially rescued by iron supplementation.
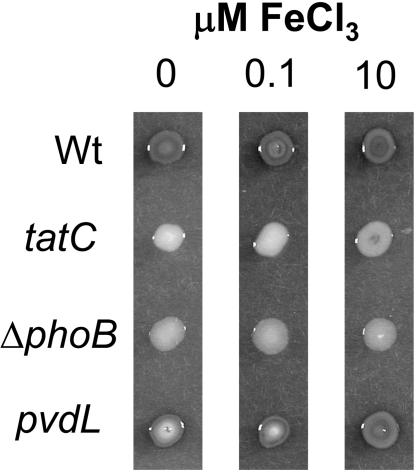
tat mutants showing a null phosphatase phenotype comparable to that of the phoB mutant were isolated. Subsequent analysis indicated that, similar to other siderophore mutants, tat mutants did not produce the fluorescent siderophore pyoverdine (data not shown). This has also been reported for tat mutants of Pseudomonas aeruginosa (37). The pleiotropy associated with loss of tat made it necessary to distinguish between the effects of Tat on modulation of phosphatase activity due to the transport of phosphatases and iron scavenging. To do this, we assessed the ability of iron to rescue the qualitative phosphatase defect of the tatC mutant PF-E280. Partial recovery of phosphatase activity was observed for PF-E280 (tatC) at 10 μM iron but not 0.1 μM iron (Fig. 6). Similar rescue of phosphatase activity was seen for the tatA mutant, as well as the other, tatC, allele (data not shown). Importantly, phosphatase recovery of the tat mutants in response to iron supplementation was much less than that seen for the siderophore mutant, PF-B582 (pvdL). These results suggest that both defects in transport of phosphatases and defects in iron scavenging have an impact on the phosphatase activities of tat mutants. Furthermore, in conjunction with the nonresponsiveness of the phoB mutant to iron supplementation, it seems likely that not all Pho-regulated phosphatases are exported by the Tat system in P. fluorescens. The ability of iron to promote the phosphatase activity of tat mutants is another factor likely to have contributed to the increased phosphatase activity seen for the tat mutants in the fluorescent-phosphatase assay versus the qualitative plate assay (Fig. 2B).
FIG. 6.
Partial rescue of the tatC phosphatase defect with iron supplementation. Phosphatase activity was assessed qualitatively for strains grown on Pi-limiting medium plus BCIP unsupplemented or supplemented with either 0.1 μM or 10 μM FeCl3. The assay was developed for 48 h. Higher degrees of darkness indicate higher levels of phosphatase activity.
Identification of additional factors involved in siderophore synthesis.
Based on the number of mutations mapping to known siderophore-biosynthetic genes, it was possible that other mutations affecting siderophore pathways were recovered in our Pho screen. To investigate this idea, we screened the entire Pho mutant collection for defects in excretion of fluorescent siderophores when grown on low-iron agar media. As a result, we identified two additional loci with putative roles in siderophore biosynthesis and phosphatase expression.
PF-B337 has a transposon insertion that maps to the coding region of Pfl5137 and did not detectably fluoresce when grown on low-iron agar medium. The predicted translation product for Pfl5137 has 51% identity to E. coli ApaH, a bis-(5′nucleosyl) tetraphosphatase known to hydrolyze the nucleotide signaling molecule AppppA (9). Further evidence of Pfl5137's homology to apaH is provided by the conservation of genomic context. As in E. coli, the apaH gene is upstream of pdxA, ksgA, and apaG (43).
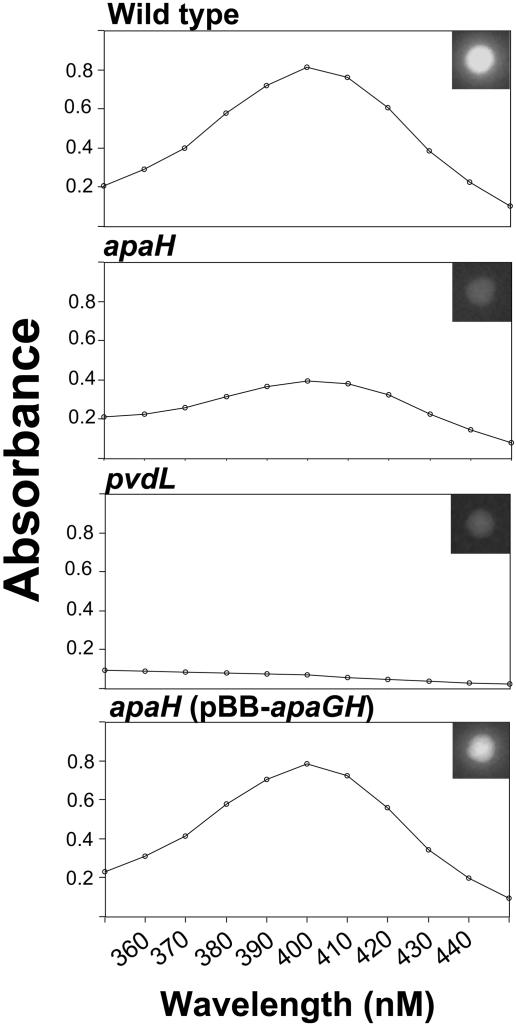
To confirm that loss of apaH is necessary and sufficient to confer defects in phosphatase activity and siderophore production, we reconstructed the apaH mutation and performed complementation studies. We utilized a single-crossover knockout strategy to disrupt apaH (yielding strain PF-apaH) and compared this to the transposon mutant PF-B337 (apaH). The reconstructed strain, PF-apaH, showed defects in phosphatase activity and siderophore-mediated fluorescence similar to those of PF-B337 (apaH) (data not shown). Quantification of pyoverdine levels indicated that PF-B337 (apaH) produced reduced levels of pyoverdine but was not completely defective, as is the case for the pvdL mutant, PF-B582 (Fig. 7). For complementation studies, the apaGH operon was cloned into pBBRMCS-5 under the transcriptional control of its native promoter, creating plasmid pBB-apaGH. PF-B337 (apaH) harboring plasmid pBB-apaGH had phosphatase activity (data not shown) and siderophore-mediated fluorescence (Fig. 7, insets) restored. Quantitative analysis indicated that pBB-apaGH restored pyoverdine excretion to wild-type levels (Fig. 7). The vector alone did not confer any restoration of siderophore-mediated fluorescence on PF-B337 (data not shown).
FIG. 7.
Analysis of siderophore production. Absorbance spectra (350 to 450 nM) were obtained for supernatants from overnight cultures of strains grown in Kings B medium. The spectra were corrected for background absorbance. Pyoverdine absorbs maximally at 403 nM. The apaH mutant produces less pyoverdine than the wild type but significantly more than the pvdL mutant. (Insets) Siderophore-mediated fluorescence is shown for each strain when grown on KB agar medium.
It should be noted that disruption of apaH conferred a lower growth rate on P. fluorescens, which was rescued by trans complementation (data not shown). Pyoverdine quantification was normalized to the optical density, and colony sizes were similar for PF-B337 (apaH) and the WT for fluorescence assessment of pyoverdine excretion (Fig. 7). However, it is still formally possible that the slower growth of apaH mutant strains indirectly exacerbates the defect in production and/or secretion of pyoverdine.
Our screen of the Pho mutants for siderophore-defective strains also recovered five strains with mutations mapping to argininosuccinate synthase (argG), as well as one mutation mapping to ornithine carbamoyltransferase (argF) (Table 2). As predicted by disruptions to the arginine biosynthesis pathway, all mutants were arginine auxotrophs (data not shown). Defects in siderophore fluorescence were conditional in that loss of fluorescence was observed only on K10T-based media, not KB medium (data not shown).
A broad range of cellular processes affect Pho-induced APase activity.
Through the course of our genetic screen we recovered numerous other classes of mutants showing reduced activity of APase under low-Pi conditions. In addition to the major phosphate regulators, phoB and phoR, we recovered mutations in genes encoding three other potential regulators, including two putative transcriptional regulators and a predicted anti-anti-sigma factor. Seven mutants with insertions in putative flagellar biosynthetic genes were recovered. These mutants mapped to three distinct flagellar operons and potentially disrupted basal-body formation (flgI, fliE, and fliF), motor switch complex formation (fliG), and junction between the hook and filament (flgL). In addition to the Tat system, several other transport-related mutations were recovered. These included mutations to a tolB homologue (outer membrane transport), glpF (glycerol uptake), and a putative cation transporter. A large number of mutants had insertions in genes involved in general metabolism, nucleotide biosynthesis, or amino acid biosynthesis. Genes involved in diverse areas of general metabolism were recovered, including the tricarboxylic acid cycle (aceF and acnA), electron transport (nouK), gluconeogenesis (pps), phosphonate biosynthesis (prpB), and glycerol assimilation (glpK). Nucleotide biosynthesis genes were implicated in the modulation of phosphatase activity by the recovery of mutations to pathways involved in uracil scavenging (upp) and de novo purine biosynthesis (purF). Disruptions to arginine-biosynthetic pathways have already been introduced within the context of siderophore regulation; however, several other amino acid-biosynthetic mutations were recovered, disrupting genes involved in serine biosynthesis (serA) and aromatic amino acids (trpG and aroB). We categorized four mutations under the general theme of defense and stress. These included glutathione synthesis (gshB), colicin production (cvpA), and penicillin amidase and nucleotide-alarmone synthesis (apaH; see above).
DISCUSSION
We performed a set of experiments to assess the conservation of the Pho regulon in Pseudomonas fluorescens Pf0-1. The major regulatory networks, defined by PhoBR and the Pst system, were conserved relative to E. coli; however, a genetic screen revealed differences in the Pho-regulated phosphatase complement of P. fluorescens and the transport apparatus utilized for their export, and also identified iron as a requirement for phosphatase activity. We also identified potential cellular factors required for maximal expression of Pho regulon genes.
Based on work with E. coli, the Pho regulon is defined as the group of genes coregulated by the concentration of extracellular Pi and the response regulator PhoB (42, 57). The Pst system is a member of the Pho regulon and, in addition to being a high-affinity Pi transporter, is required for inhibition of PhoB phosphorylation under Pi-sufficient conditions. As in E. coli, deletion of phoB in P. fluorescens prevented activation of the Pho regulon in Pi-limiting environments. Deletion of the entire pstSCAB-phoU operon led to Pho regulon expression in Pi concentrations that repress Pho in the WT. Furthermore, this expression was dependent on the presence of PhoB. These data agree with the results of research performed on other Pseudomonas species showing that PhoBR and Pst are functionally conserved as the major regulators of the adaptive response to low-Pi environments (31, 35, 63).
For E. coli, Pi serves as the primary source of phosphorus; however, other sources of phosphorus can be utilized, such as organophosphates, phosphonates, and phosphites (30). We tested a selection of organophosphates, phosphonates, and phosphites known to support the growth of E. coli for the ability to support the growth of P. fluorescens as the sole phosphorus source and to suppress induction of Pho under Pi-limiting conditions. All organophosphates tested could support the growth of P. fluorescens; however, only G3P was able to suppress Pho induction. Genes for G3P utilization do not appear to be members of the Pho regulon, as phoB was not required for growth on G3P. This suggests either that G3P is specifically transported into the cell for integration into phosphate metabolism or that other non-Pho phosphatases are capable of breaking down G3P and releasing Pi into the periplasm. Distinguishing between these two mechanisms will be important for understanding the basis of G3P repression of Pho induction. Of the phosphonates, 2AEP could support growth whereas MP could not. In agreement with this result, P. fluorescens has homologues of the phnWX genes, which are known to be required for the biodegradation and synthesis of 2AEP in Salmonella (22). In contrast to Salmonella, growth on 2AEP was not phoB dependent, suggesting that the genes for 2AEP utilization are not under Pho regulon control. Sodium phosphite could not support the growth of P. fluorescens and could not repress Pho induction. Pseudomonas stutzeri has been shown to require the phnCDEFGHIJKLMNP operon for phosphonate and phosphite utilization (61). In accordance with these results, only the transport modules of the phn operon (phnCDE) are present in P. fluorescens Pf0-1. Lack of an intact phn operon may also be the reason that methylphosphonate was unable to be utilized. It is interesting that the abilities of phosphonates to repress Pho induction have not been previously reported in other bacterial systems. Phosphonates have a C—P bond, making them very stable relative to phosphate monoesters (57). As a result, the ability of 2AEP to repress Pho induction is not likely due to the release of Pi in the periplasm and its subsequent interaction with the Pst-PhoR system. Presumably, the biodegradation of 2AEP within the cell can suppress PhoB activation by an as-yet-unknown mechanism. In any case, P. fluorescens appears to process alternate phosphorus sources in a manner distinct from that of E. coli and closer relatives, such as P. stutzeri, offering an interesting avenue for investigation.
A genetic screen for reduction in the activity of Pho-regulated phosphatases allowed the potential recovery of at least three classes of mutants with (i) mutations in genes encoding the phosphatase enzymes, (ii) mutations affecting factors required for phosphatase activity or expression, or (iii) mutations affecting general expression of the Pho regulon.
Four mutants were recovered with severe defects in phosphatase activity that had independent insertions mapping to a gene encoding a predicted phosphatase, which we refer to as phoX, based on the Pf0-1 genome annotation. RT-PCR was used to confirm that phoX is a member of the Pho regulon and that its conserved Pho box is likely to be functional. No publication referring to PhoX could be found; however, a BLAST search identified a putative homologue in Vibrio cholerae with an associated reference. Majumdar and colleagues (28) reported the identification and purification of an alkaline phosphatase, referred to as phoAvc. PhoX has 57% identity to PhoAvc, providing good support for its classification as homologous. PhoAvc is known to be a monomeric phosphatase that differs from other known phosphatases, such as PhoA, which are dimeric enzymes. Beyond what is reported and discussed here, very little is known about this class of phosphatase, making the members interesting candidates for more structural and functional investigation.
The twin arginine transport system (Tat) provides a pathway alternative to Sec for translocation of proteins across the inner membrane. Three proteins make up the minimal translocon, TatA, -B, and -C, which, in contrast to the Sec system, can mediate the translocation of fully folded proteins across the inner membrane (33, 40). PhoX has a predicted Tat leader sequence that matches well with the consensus from E. coli. Support for Tat-mediated export of PhoX to the periplasm was gained through isolation of mutations to tatA and tatC that prevented detectable phosphatase activity on Pi-limiting agar medium. We showed that phoX transcription is not reduced in a tatC mutant, making defects in transport the parsimonious explanation for loss of phosphatase activity in tat mutants. To our knowledge, this is the first experimental evidence for Tat secretion and Pho regulon membership of PhoX-type phosphatases in gram-negative bacteria. In contrast to PhoX, PhoA is known to be secreted by the Sec system. The easily measured activity of PhoA has provided an invaluable tool to dissect the mechanisms of Sec protein translocation. PhoX activity is also easily assayed and could serve as a tractable intrinsic marker of functional Tat-mediated secretion.
The residual phosphatase activity of the phoX mutants, combined with the null phosphatase phenotype of phoB and tat mutants, suggested the existence of another phosphatase activity(s) that was Pho regulated and Tat exported. With this information, we identified phoD in the Pf0-1 genome. PhoD is a predicted phosphatase with 26% identity to PhoD from the gram-positive bacterium Bacillus subtilis. PhoD has a predicted Tat leader sequence and was shown to have a functional Pho box required for Pho-regulated expression. Deletion of phoD in Pf0-1 resulted in a small but significant decrease in phosphatase activity, but no further decrease could be detected for the phoX phoD double mutant. Our inability to resolve the contribution of PhoD to the ΔphoX strain's phosphatase activity is likely due to noise associated with readings closer to background for the assay. The only information available on PhoD comes from studies of B. subtilis (8). As was the case with Pf0-1, PhoD was shown to be Tat secreted in B. subtilis (21). Furthermore, PhoD did not contribute significantly to the overall phosphatase activity of B. subtilis; 98% of the activity was accounted for by the phosphatases PhoA and PhoB (8). Our analysis of PhoD also suggested it was not a major contributor to the Pho-regulated phosphatase activity of P. fluorescens. This was true for both substrates used, MUP and BCIP. The natural substrate(s) of PhoD is not known, so substrate specificity, rather than low expression levels, could explain the apparent low phosphatase activity. Our results indicate that phoD is transcribed at relatively high levels under Pho-activating conditions (Fig. 3D).
The genus Pseudomonas represents one of the most diverse groups of bacteria on the planet (51). This diversity seemingly underpins their ability to inhabit an extremely broad range of environments. A survey of the sequenced genomes for P. aeruginosa PAO1, P. putida KT2440, and Pseudomonas syringae DC3000 indicated different profiles for the phosphatases PhoX, PhoD, and PhoA. Putative PhoX homologues could be located in P. putida (33% identity), P. aeruginosa (44%), and P. syringe (78%). Putative PhoD homologues could be located in P. aeruginosa (71%), and P. syringae (79%), but not P. putida. The only species to have a phoA homologue was P. aeruginosa. True to its reputation, the genus Pseudomonas shows substantial diversity in phosphatase profiles between species, which may reflect specialist roles and evolutionary selection forces for different classes of phosphatase.
Based on the fact that the phoX phoD double mutant demonstrates visually detectable phosphatase activity and that a phoB mutant is completely null, it is likely that P. fluorescens has at least one other unidentified Pho-regulated phosphatase. P. fluorescens does not have a phoA homologue; however, similar to PhoA, the unidentified phosphatase is likely to be transported by the Sec system rather than Tat. We infer this from the ability of iron to partially rescue the phosphatase activity of a tatC mutant, but not the phoB mutant. Addition of iron was shown to increase phosphatase activity in general, such that it likely allowed the detection of small amounts of phosphatase excreted by the tat mutant.
Numerous researchers have identified putative Pho boxes upstream of putative Pho genes (35, 60, 63). In many cases, the gene has been shown to be a member of the Pho regulon, but the functionality of the putative Pho box has not been directly assessed. In the course of our characterization of phoD as a Pho regulon gene, we mutated the putative Pho box and assessed its ability to activate expression of phoD. When the 5′ arm of the putative Pho box was replaced with a PacI restriction site, induction of phoD dropped from 471-fold to 3-fold. Residual activation of the mutated phoD promoter suggests that some productive PhoB binding is still occurring. Further analysis of the phoD promoter identified a low-consensus (three of seven) PhoB binding site downstream of the previously identified Pho box, which in the absence of the high-affinity PhoB binding site may be responsible for low-level activation of phoD expression. Overall, these results provide good evidence that the Pho box sequence from E. coli is functionally conserved in Pseudomonas fluorescens. This is not to say that the Pho box consensus sequence of P. fluorescens is identical to that of E. coli. Only comparison of multiple confirmed Pho boxes will provide this information.
The largest class of mutants recovered from the screen had insertions within genes involved in siderophore-biosynthetic pathways. All mutants in this class had strong reductions in phosphatase activity when grown on K10Tπ agar, which is a low-iron medium. We investigated the link between iron limitation and Pho-regulated phosphatase activity by assessing the effect of iron chelation on the phosphatase activity of the wild type and a siderophore mutant. Iron chelation effectively reduced the phosphatase activity of the wild type in a concentration-dependent fashion, mimicking the effect of a siderophore mutation. Importantly, the siderophore mutant was more severely affected by lower dipyridyl concentrations than the wild type. The effect of dipyridyl is not mediated by reduction in growth rates, as levels of dipyridyl that did not affect growth still led to significant reductions in phosphatase activity (Fig. 5A). These results suggest that the concentration of intracellular iron can have an impact on the extent of phosphatase activity under Pi-limiting conditions.
The link between iron-scavenging and phosphate-scavenging pathways was intriguing, such that we wanted to discern whether iron modulated Pho regulon expression in general. We addressed this point by assessing the effect of iron sequestration on phoX transcription. No reduction in phoX transcription could be detected in either the wild type or the siderophore mutant, irrespective of the dipyridyl concentration (Fig. 5B). This result suggests that the iron concentration is important for the activity of Pho-regulated phosphatases, rather than transcription of phosphatase genes or Pho regulon genes in general. It is possible that iron acts as an enzymatic cofactor for PhoX and/or PhoD. Zinc and magnesium are known metal cofactors for the E. coli alkaline phosphatase PhoA (6, 18), but to our knowledge, no similar role for iron has been reported for PhoA or other bacterial alkaline phosphatases. In eukaryotes, iron is a known cofactor of purple acid phosphatases, suggesting that the potential exists for iron to facilitate the cleavage of phosphate monoesters (38). We observed that iron must be present during the growth of P. fluorescens to promote alkaline phosphatase activity and could not simply be added back to crude extracts. This may indicate that iron must be incorporated into the phosphatase during formation of the tertiary structure. This hypothesis is supported by the fact that the Tat system is known to export fully folded proteins, many of which are complexed to metal cofactors (40). More focused structural and enzymological studies will be required to determine the role of iron in PhoX and/or PhoD enzymatic activity.
The link between siderophore synthesis and phosphatase activity prompted us to screen the entire Pho mutant collection for other factors involved in siderophore production. PF-B337 has a Tn5 insertion in apaH and did not show detectable fluorescence on low-iron agar media. Loss of apaH was also shown to confer a defect in pyoverdine excretion during growth in shaking culture. We confirmed the role of apaH in siderophore production by strain reconstruction and trans-complementation of the siderophore defects of PF-B337 (apaH). From studies of E. coli, ApaH is known to hydrolyze the nucleotide-signaling molecule AppppA (9). High intracellular levels of AppppA, due to loss of apaH, have been correlated with multiple phenotypes, such as reduction in expression of catabolite-repressible genes (9), increased sensitivity to heat and UV (12), and decreased expression of flagellar operons (9). A role for ApaH in siderophore biosynthesis has not been reported. At present, the mechanism by which ApaH interacts with siderophore pathways is not understood.
We also isolated mutations in the arginine-biosynthetic pathway that conferred a conditional defect in siderophore-mediated fluorescence. l-Arginine is a structural component of pyoverdine (1), so limitation of arginine on K10T-based agar medium may be the reason for its siderophore defect; however, further studies need to be done to confirm this hypothesis.
The genetic screen we performed recovered mutants with defects in Pho-regulated phosphatase activity that cover a wide range of cellular processes and biology (Table 2). The question remains as to how disruptions to such a wide range of cellular processes and factors could affect APase activity. At this time, several competing explanations exist: the process/factor either directly or indirectly affects (i) phosphatase enzymatic activity, (ii) phosphatase gene expression, (iii) Pho regulon expression, or (iv) gene expression in general. Distinguishing between these alternatives is important, because many genes are involved in core cellular metabolic processes, making it formally possible that their effects on APase activity are nonspecific. Clearly, too much conjecture at this point is unwarranted; however, several groups of mutants are worthy of limited elaboration.
Multiple insertions in flagellar operons led to reductions in APase activity. At first this seems an obscure relationship; however, there is precedent in the literature for flagella regulating the expression of nonflagellar phenotypes. For instance, disruption to formation of the flagellum in Vibrio cholerae O139 has been shown to act as a signal for increased exopolysaccharide production (59). In another example, inhibition of polar-flagellum function in Vibrio parahaemolyticus induces expression of swarmer cell genes (laf) (29). In this case, it is thought that the flagellum is sensing external forces, essentially acting as a dynamometer. Based on these examples, it is possible that loss of flagellar function acts as a signal to down regulate phosphatase expression and/or Pho regulon expression in general.
A large number of mutants were recovered in metabolic pathways, including those for carbon metabolism, nucleotide metabolism, amino acid biosynthesis, and energy metabolism (Table 2). Interestingly, several genes recovered from this screen are involved in the transfer of phosphoryl groups to their substrates. For example, glycerol kinase utilizes ATP to phosphorylate glycerol that is transported into the cell (46). This is the first committed step in glycerol metabolism. Uracil phosphoribosyltransferase is required to phosphorylate uracil, activating it for integration into the nucleotide metabolism of the cell (10). In this case, GTP serves as the phosphate donor. Finally, phosphoenolpyruvate (PEP) synthase utilizes ATP to drive the formation of PEP from pyruvate in the first step of gluconeogenesis (41). Potentially, the flux of ATP and GTP in the cell serves as an internal signal to modulate Pho expression.
The potential involvement of nonphosphate metabolic pathways in regulating Pho induction is not unprecedented. Experiments carried out with E. coli have shown that a two-component system, CreBC (57), and the levels of acetylphosphate can regulate Pho induction (58). Furthermore, over 90 mutants that affect Pi-independent control of the Pho regulon have been isolated (57). Similar to the screen we report in this study, mutants of genes involved in a wide range of metabolic activities were recovered, including the tricarboxylic acid cycle, nucleotide synthesis, and catabolite control. Our screen did not recover the creBC genes or genes involved in direct modulation of acetyl phosphate concentrations; however, in a broader sense, both E. coli and P. fluorescens may monitor global fluxes in carbon and energy metabolism to fine tune activation of the Pho regulon in any given Pi environment. The challenge for researchers is to demonstrate that such effects are specific rather than an indirect cause of perturbations to core cellular processes. We are trying to identify which mutations result in defects in the activation of multiple Pho regulon promoters, so that we can begin to address the role of central metabolism in the regulation of Pho expression.
Acknowledgments
We thank Svein Valla, Michael Kovak, and Dieter Haas for plasmids. We also thank Robin Hulbert and an anonymous reviewer for their critical comments, which were of much help in revision of the manuscript.
This work was supported by funding from the NSF (CAREER 9984521).
REFERENCES
- 1.Ackerley, D. F., and I. L. Lamont. 2004. Characterization and genetic manipulation of peptide synthetases in Pseudomonas aeruginosa PAO1 in order to generate novel pyoverdines. Chem. Biol. 11:971-980. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosi, C., L. Leoni, L. Putignani, N. Orsi, and P. Visca. 2000. Pseudobactin biogenesis in the plant growth-promoting rhizobacterium Pseudomonas strain B10: identification and functional analysis of the l-ornithine N5-oxygenase (psbA) gene. J. Bacteriol. 182:6233-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anba, J., M. Bidaud, M. L. Vasil, and A. Lazdunski. 1990. Nucleotide sequence of the Pseudomonas aeruginosa phoB gene, the regulatory gene for the phosphate regulon. J. Bacteriol. 172:4685-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardin, S. D., and T. M. Finan. 1689. Regulation of phosphate assimilation in Rhizobium (Sinorhizobium) meliloti. Genetics 148:1689-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, J. E. 1992. Structure and mechanism of alkaline phosphatase. Annu. Rev. Biophys. Biomol. Struct. 21:441-483. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eder, S., L. Shi, K. Jensen, K. Yamane, and F. M. Hulett. 1996. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology 142:2041-2047. [DOI] [PubMed] [Google Scholar]
- 9.Farr, S. B., D. N. Arnosti, M. J. Chamberlin, and B. N. Ames. 1989. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. USA 86:5010-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fast, R. 1978. Isolation of Escherichia coli mutants with changed regulation of uracil uptake. J. Bacteriol. 136:839-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filloux, A., M. Bally, C. Soscia, M. Murgier, and A. Lazdunski. 1988. Phosphate regulation in Pseudomonas aeruginosa: cloning of the alkaline phosphatase gene and identification of phoB- and phoR-like genes. Mol. Gen. Genet. 212:510-513. [DOI] [PubMed] [Google Scholar]
- 12.Fuge, E. K., and S. B. Farr. 1993. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J. Bacteriol. 175:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee, K. R., W. C. Sun, M. K. Bhalgat, R. H. Upson, D. H. Klaubert, K. A. Latham, and R. P. Haugland. 1999. Fluorogenic substrates based on fluorinated umbelliferones for continuous assays of phosphatases and beta-galactosidases. Anal. Biochem. 273:41-48. [DOI] [PubMed] [Google Scholar]
- 14.Gonin, M., E. M. Quardokus, D. O'Donnol, J. Maddock, and Y. V. Brun. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 182:337-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 17.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 18.Holtz, K. M., and E. R. Kantrowitz. 1999. The mechanism of the alkaline phosphatase reaction: insights from NMR, crystallography and site-specific mutagenesis. FEBS Lett. 462:7-11. [DOI] [PubMed] [Google Scholar]
- 19.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 20.Izco, J. M., M. Tormo, A. Harris, P. S. Tong, and R. Jimenez-Flores. 2003. Optimization and validation of a rapid method to determine citrate and inorganic phosphate in milk by capillary electrophoresis. J. Dairy Sci. 86:86-95. [DOI] [PubMed] [Google Scholar]
- 21.Jongbloed, J. D., U. Martin, H. Antelmann, M. Hecker, H. Tjalsma, G. Venema, S. Bron, J. M. van Dijl, and J. Muller. 2000. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275:41350-41357. [DOI] [PubMed] [Google Scholar]
- 22.Kim, A. D., A. S. Baker, D. Dunaway-Mariano, W. W. Metcalf, B. L. Wanner, and B. M. Martin. 2002. The 2-aminoethylphosphonate-specific transaminase of the 2-aminoethylphosphonate degradation pathway. J. Bacteriol. 184:4134-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 24.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 25.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genom. 272:1-17. [DOI] [PubMed] [Google Scholar]
- 26.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368-380. [DOI] [PubMed] [Google Scholar]
- 27.Lambertsen, L., C. Sternberg, and S. Molin. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726-732. [DOI] [PubMed] [Google Scholar]
- 28.Majumdar, A., A. Ghatak, and R. K. Ghosh. 2005. Identification of the gene for the monomeric alkaline phosphatase of Vibrio cholerae serogroup O1 strain. Gene 344:251-258. [DOI] [PubMed] [Google Scholar]
- 29.McCarter, L., M. Hilmen, and M. Silverman. 1988. Flagellar dynamometer controls swarmer cell differentiation of Vibrio parahaemolyticus. Cell 54:345-351. [DOI] [PubMed] [Google Scholar]
- 30.Metcalf, W. W., and B. L. Wanner. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173:587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monds, R. D., M. W. Silby, and H. K. Mahanty. 2001. Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 42:415-426. [DOI] [PubMed] [Google Scholar]
- 32.Mossialos, D., U. Ochsner, C. Baysse, P. Chablain, J. P. Pirnay, N. Koedam, H. Budzikiewicz, D. U. Fernandez, M. Schafer, J. Ravel, and P. Cornelis. 1995. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45:1673-1685. [DOI] [PubMed] [Google Scholar]
- 33.Muller, M. 2005. Twin-arginine-specific protein export in Escherichia coli. Res. Microbiol. 156:131-136. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 35.Nikata, T., Y. Sakai, K. Shibat, J. Kato, A. Kuroda, and H. Ohtake. 1996. Molecular analysis of the phosphate-specific transport (pst) operon of Pseudomonas aeruginosa. Mol. Gen. Genet. 250:692-698. [DOI] [PubMed] [Google Scholar]
- 36.Novak, R., A. Cauwels, E. Charpentier, and E. Tuomanen. 1999. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J. Bacteriol. 181:1126-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olczak, M., B. Morawiecka, and W. Watorek. 2003. Plant purple acid phosphatases—genes, structures and biological function. Acta Biochim. Pol. 50:1245-1256. [PubMed] [Google Scholar]
- 39.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 40.Palmer, T., F. Sargent, and B. C. Berks. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13:175-180. [DOI] [PubMed] [Google Scholar]
- 41.Patnaik, R., W. D. Roof, R. F. Young, and J. C. Liao. 1992. Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. J. Bacteriol. 174:7527-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao, N. N., and A. Torriani. 1990. Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4:1083-1090. [DOI] [PubMed] [Google Scholar]
- 43.Roa, B. B., D. M. Connolly, and M. E. Winkler. 1989. Overlap between pdxA and ksgA in the complex pdxA-ksgA-apaG-apaH operon of Escherichia coli K-12. J. Bacteriol. 171:4767-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos, P. M., I. Di Bartolo, J. M. Blatny, E. Zennaro, and S. Valla. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 45.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 46.Schweizer, H. P., R. Jump, and C. Po. 1997. Structure and gene-polypeptide relationships of the region encoding glycerol diffusion facilitator (glpF) and glycerol kinase (glpK) of Pseudomonas aeruginosa. Microbiology 143:1287-1297. [DOI] [PubMed] [Google Scholar]
- 47.Siehnel, R. J., E. A. Worobec, and R. E. Hancock. 1988. Regulation of components of the Pseudomonas aeruginosa phosphate-starvation-inducible regulon in Escherichia coli. Mol. Microbiol. 2:347-352. [DOI] [PubMed] [Google Scholar]
- 48.Silby, M. W., and S. B. Levy. 2004. Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization. J. Bacteriol. 186:7411-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 50.Slater, H., M. Crow, L. Everson, and G. P. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 51.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 52.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275:11591-11596. [DOI] [PubMed] [Google Scholar]
- 53.Summers, M. L., J. G. Elkins, B. A. Elliott, and T. R. McDermott. 1994. Expression and regulation of phosphate stress inducible genes in Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 11:1094-1101. [DOI] [PubMed] [Google Scholar]
- 54.Torres, A., M. D. Juarez, R. Cervantes, and C. Espitia. 2001. Molecular analysis of Mycobacterium tuberculosis phosphate specific transport system in Mycobacterium smegmatis. Characterization of recombinant 38 kDa (PstS-1). Microb. Pathog. 30:289-297. [DOI] [PubMed] [Google Scholar]
- 55.Torriani, A. 1959. Influence of inorganic phosphate in the formation of phosphatases by Escherichia coli. Biochim. Biophys. Acta 38:460-469. [DOI] [PubMed] [Google Scholar]
- 56.von Kruger, W. M., S. Humphreys, and J. M. Ketley. 1999. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145:2463-2475. [DOI] [PubMed] [Google Scholar]
- 57.Wanner, B. L. 1996. Phosphorous assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 58.Wanner, B. L., and M. R. Wilmes Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White, A. K., and W. W. Metcalf. 2004. The htx and ptx operons of Pseudomonas stutzeri WM88 are new members of the Pho regulon. J. Bacteriol. 186:5876-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, A. K., and W. W. Metcalf. 2004. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 186:4730-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Worobec, E. A., R. J. Siehnel, P. Gladman, and R. E. Hancock. 1988. Gene cloning and expression of the Pseudomonas aeruginosa periplasmic phosphate-binding protein. FEMS Microbiol. Lett. 52:235-238. [Google Scholar]
- 63.Wu, H., H. Kosaka, J. Kato, A. Kuroda, T. Ikeda, N. Takiguchi, and H. Ohtake. 1999. Cloning and characterization of Pseudomonas putida genes encoding the phosphate-specific transport system. J. Biosci. Bioeng. 87:273-279. [DOI] [PubMed] [Google Scholar]
- 64.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 65.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]