Abstract
The distinction between viable and dead bacterial cells poses a major challenge in microbial diagnostics. Due to the persistence of DNA in the environment after cells have lost viability, DNA-based quantification methods overestimate the number of viable cells in mixed populations or even lead to false-positive results in the absence of viable cells. On the other hand, RNA-based diagnostic methods, which circumvent this problem, are technically demanding and suffer from some drawbacks. A promising and easy-to-use alternative utilizing the DNA-intercalating dye ethidium monoazide bromide (EMA) was published recently. This chemical is known to penetrate only into “dead” cells with compromised cell membrane integrity. Subsequent photoinduced cross-linking was reported to inhibit PCR amplification of DNA from dead cells. We provide evidence here that in addition to inhibition of amplification, most of the DNA from dead cells is actually lost during the DNA extraction procedure, probably together with cell debris which goes into the pellet fraction. Exposure of bacteria to increasing stress and higher proportions of dead cells in defined populations led to increasing loss of genomic DNA. Experiments were performed using Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium as model pathogens and using real-time PCR for their quantification. Results showed that EMA treatment of mixed populations of these two species provides a valuable tool for selective removal of DNA of nonviable cells by using conventional extraction protocols. Furthermore, we provide evidence that prior to denaturing gradient gel electrophoresis, EMA treatment of a mature mixed-population drinking-water biofilm containing a substantial proportion of dead cells can result in community fingerprints dramatically different from those for an untreated biofilm. The interpretation of such fingerprints can have important implications in the field of microbial ecology.
Whether bacterial cells are dead or alive is an important question with many implications for monitoring of food and water safety and for analysis of the sterility of pharmaceutical drugs. Due to the relatively long persistence of DNA after cell death, in the range of several days to 3 weeks (10, 14), quantitative analysis of total DNA can lead to a substantial overestimation of the presence of living microorganisms and the accompanying pathogenic threats. The lack of differentiation between DNA from viable and dead bacterial cells is therefore a major obstacle to broad-range application of DNA-based molecular diagnostics (10, 12). The most commonly used strategy to overcome this difficulty is to focus on the presence of the rapidly degrading RNA instead of the stable DNA (16, 27, 28, 31). Due to its rapid turnover, the detection of RNA is far more indicative of the presence of viable cells (1, 2). Nevertheless, working with RNA is technically demanding, and RNA is prone to contamination with RNA-degrading enzymes, resulting in problems of reproducibility. Moreover, the RNA expression level depends on the physiological status of the cell, making accurate measurements of bacterial numbers difficult. Another strategy is the use of dyes for microscopic differentiation between viable and dead cells (15, 13, 20, 29). Although these methods are well established, quantification by extrapolating counts of a limited number of bacteria is problematic and cannot be applied on a routine basis. Moreover, due to the lack of an amplification step, the sensitivity of microscope-based techniques is a limiting factor, as is the inability to specify individual species. Culture-based techniques, on the other hand, are slow, providing results only after a few days, and are dependent on the medium and the incubation temperature.
A promising alternative has recently been described by Nogva et al. (18) and Rudi et al. (24, 25), who have introduced ethidium monoazide bromide (EMA)-PCR as a diagnostic DNA-based method combining the use of a live-dead staining dye with the speed and sensitivity of real-time PCR. EMA has been used as a dye for microscopic differentiation between viable and dead cells (4, 17, 19, 21, 22). Like ethidium bromide, the structurally similar EMA is a phenanthridinium DNA/RNA-intercalating agent (30) with a binding constant of 2 × 105 to 3 × 105 M (3) that can enter only bacterial cells with compromised cell walls and cell membranes (24). Following intercalation into the DNA of those cells, EMA can be covalently linked by photoactivation with high yields, up to 75% (3). Photolysis of EMA using visible light (maximum absorbance at 460 nm) produces a nitrene that can form a covalent linkage to DNA and other molecules (5, 6, 9). The unbound EMA, which remains free in solution, is simultaneously inactivated by reacting with water molecules (6). The resulting hydroxylamine is no longer capable of covalently binding to DNA (11). DNA from viable cells, protected from reactive EMA before light exposure by an intact cell membrane/cell wall, should thus not be affected by the inactivated EMA after cell lysis during the DNA extraction procedure.
The amplification of DNA with bound EMA has been shown to be inhibited, leading to a strong signal reduction in subsequent real-time PCR (24). However, the previous studies (18, 24, 25), where this was shown, did not provide information about the amount of genomic DNA added to the diagnostic PCR mixtures as a template following DNA extraction. In order to be able to attribute the viable-dead discrimination solely to PCR inhibition, one needs to ensure that equal amounts of genomic DNA are used as a template for quantitative PCR or that the template DNA is quantified. This point has remained unaddressed.
Surprisingly, we found a strong effect of EMA on the genomic DNA yield when we started with the same bacterial biomass for DNA extraction. The effect of EMA is selective in that it reduces the DNA yield of stressed cultures significantly more than the DNA yield of unstressed cultures. As a consequence, equal volumes of genomic DNA preparations from stressed and unstressed cells used as templates for quantitative PCR result in different amplification thresholds.
We share the view that EMA-PCR might be very useful for viable/dead discrimination of bacterial communities but suggest that the selection occurs mainly during DNA extraction and only to a lesser extent on the level of PCR. Signal reduction may be due in part to PCR inhibition but is also due, more importantly, to a selective loss of genomic DNA from dead cells during the extraction procedure. Understanding the underlying principle might allow the combination of EMA treatment not only with real-time PCR but also with other downstream analysis tools. Furthermore, we provide evidence that EMA can affect PCR-generated community fingerprints. Denaturing gradient gel electrophoresis (DGGE) of a mature mixed-population microbial drinking-water biofilm produced dramatically different community structures when EMA-treated and untreated aliquots of the same biofilm were compared.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and biofilm sampling.
Escherichia coli O157:H7 (strain 932) and Salmonella enterica serovar Typhimurium (environmental isolate; Department of Microbiology, Montana State University) were used as model organisms. Single colonies from Luria-Bertani (LB) agar streak plates were transferred to 50-ml culture tubes containing approximately 10 ml of LB medium. The cultures were incubated on a shaker at 280 rpm and 37°C and were grown to log phase. Cultures were adjusted to an optical density at 600 nm of 1 by dilution with LB medium. Loss of viability was examined by plating 50 to 100 μl of serial dilutions (10−2 to 10−6 in LB medium) on LB agar. E. coli Mach1-T1R (Invitrogen, Carlsbad, CA) transformed with a pCR2.1-TOPO plasmid was grown in LB medium containing 50 μg/ml kanamycin. A biofilm for spiking experiments and community analysis was harvested from a water reservoir that was not light exposed. The biofilm was resuspended in the original water by pipetting.
Stress conditions.
Culture aliquots (500 μl) of E. coli O157:H7 and Salmonella serovar Typhimurium were heat treated for 30 s to 15 min at 72°C using a standard laboratory heat block and were immediately placed on ice after heat treatment. Alternatively, cells were killed by exposure to isopropanol (final concentration, 70%) for 10 min. Isopropanol was removed by harvesting cells by centrifugation at 5,000 × g for 5 to 7 min prior to resuspension in 500 μl of LB medium. The number of CFU was determined in triplicate by plating 50 μl of serial dilutions on LB agar, followed by overnight incubation at 37°C.
Spiking experiment.
Aliquots (1 ml) of a resuspended drinking water biofilm were spiked with 150 μl of either viable or heat-killed E. coli O157:H7 (optical density at 600 nm, 1). Samples were split in two identical aliquots. One half was subjected to EMA treatment, and the other half remained untreated, followed by isolation of total DNA and quantitative PCR (Q-PCR).
EMA cross-linking.
EMA bromide (phenanthridium, 3-amino-8-azido-5-ethyl-6-phenyl bromide; Molecular Probes Inc., Eugene, Oreg.) was dissolved in water to a stock concentration of 5 mg/ml and stored at −20°C in the dark. EMA was added to cultures to a final concentration of 100 μg/ml, which was reported to be the concentration resulting in the highest signal reduction (18). Following a 5-min incubation in the dark with occasional flipping, samples were light exposed for 1 min using a 650-W halogen light source placed 20 cm from the sample tubes. During exposure, samples were placed on ice to avoid excessive heating. After photoinduced cross-linking, cells were pelleted at 5,000 × g for 5 min prior to DNA isolation.
DNA isolation and quantification.
Genomic DNA was extracted using the Qbiogene (Carlsbad, CA) soil kit according to the manufacturer's instructions. Cell lysis of pure cultures was achieved by bead beating using a FastPrep machine (Qbiogene) for 25 s at a speed setting of 4.5 m/s. For biofilm samples, the bead-beating time and speed were increased to 35 s and 5.5 m/s, respectively. Alternatively, for studying the effect of EMA on the DNA yield of heat- and isopropanol-stressed cultures (see Fig. 1), DNA was extracted using the DNeasy tissue kit (QIAGEN, Valencia, CA) and the PrepMan Ultra sample preparation reagent (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. DNA was quantified using the PicoGreen quantification solution (Molecular Probes Inc.) and a TBS-380 fluorometer (Turner BioSystems Inc., Sunnyvale, CA) using genomic DNA from E. coli O157:H7 as a standard. The quantification results obtained with the fluorometer were compared with band intensities of high-molecular-weight genomic DNA visualized on ethidium bromide-stained 1% agarose gels. Seven to ten percent of the total corresponding eluate volumes was loaded onto the gels.
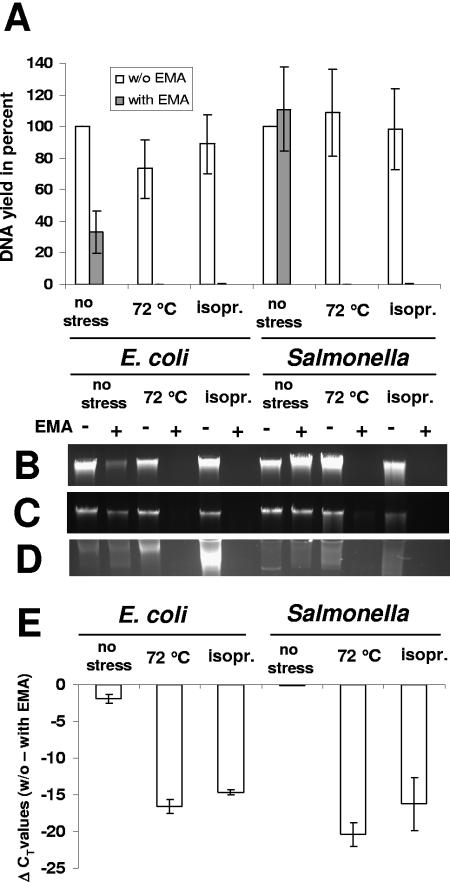
FIG. 1.
Effects of stress and EMA treatment on genomic DNA yield and Q-PCR signal thresholds of E. coli O157:H7 and Salmonella serovar Typhimurium. Cultures were either processed directly without stress exposure, heat treated at 72°C for 5 min, or exposed to 70% isopropanol (isopr.) for 10 min. Error bars, standard deviations from three independent replicates. (A) Genomic DNA yields of unstressed and stress-killed cells without EMA (white bars) and with EMA (gray bars) treatment, expressed as percentages of the DNA yields of the corresponding unstressed and non-EMA-treated cultures. Genomic DNA was extracted using the Qbiogene soil kit. (B to D) Stained agarose gels showing genomic DNA from the same treatments. DNA was extracted using the Qbiogene soil kit (B), the DNeasy tissue kit (C), or the PrepMan Ultra sample preparation reagent (D). (E) Signal reduction as determined by Q-PCR detecting relative differences in stx1 (E. coli O157:H7) and invA (Salmonella) gene copies. CT values derived from EMA-treated cultures were subtracted from the corresponding CT values for untreated cultures.
Quantitative PCR.
For relative PCR quantification of Salmonella, 1 μl of extracted genomic DNA was added to 24 μl of PCR mixture containing Sybr Green PCR master mix (Applied Biosystems, Foster City, CA) and 10 pmol of primers invA2-F (5′-GATTCTGGTACTAATGGTGATGATC-3′) (8) and invA2-R (5′-GCCAGGCTATCGCCAATAAC-3′) (8). The cycling parameters were as follows: 8 min at 95°C (initial polymerase activation and denaturation), followed by 50 cycles of 20 s at 95°C, 30 s at 60°C, and 25 s at 72°C. For melt curve analysis, the temperature was increased in 0.2°C increments from 60 to 94°C.
For relative quantification of E. coli O157:H7, Q-PCR was performed in a total volume of 25 μl containing 1 μl extracted genomic DNA and final concentrations of 1× AmpliTaq GOLD buffer (Applied Biosystems), 5.5 mM MgCl2, 0.3 μM of primer stx1-forward (5′-GACTGCAAAGACGTATGTAGATTCG-3′), 0.3 μM of primer stx1-reverse (5′-ATCTATCCCTCTGACATCAACTGC-3′) (26), 0.15 μM of the stx1 probe (5′-TGAATGTCATTCGCTCTGCAATAGGTACTC-3′) (26), and 2.5 U of AmpliTaq Gold (Applied Biosystems). The stx1-probe had 6-carboxyfluorescein as the 5′ reporter and BHQ1 as the 3′ quencher. The cycling parameters were as follows: 8 min at 95°C, followed by 50 cycles of 20 s at 95°C, 30 s at 55°C, and 25 s at 72°C. The same cycling parameters were applied for the amplification of an internal 244-bp fragment of the pCR2.1 plasmid (Invitrogen, Carlsbad, CA) using 10 pmol each of primers M13-for (5′-GTAAAACGACGGCCAG-3′) and M13-rev (5′-CAGGAAACAGCTATGAC-3′) and Sybr Green PCR master mix (Applied Biosystems, Foster City, CA).
Q-PCR and data analysis were performed with a SmartCycler II (Cepheid, Sunnyvale, CA). Cycle threshold (CT) values were automatically calculated by the SmartCycler software using the second derivative method.
Statistical analysis.
Error bars in diagrams represent standard deviations from three independent replicates.
Community fingerprinting.
Two milliliters of a resuspended biofilm solution was split into two identical aliquots, one of which was treated with EMA. Both aliquots were exposed to light. Approximately 5 ng of extracted genomic DNA served as a template for amplification of an internal fragment of the 16S rRNA coding gene using 10 pg each of primers 1070F (5′-ATGGCTGTCGTCAGCT-3′) (7) and 1392R (5′-ACGGGCGGTGTGTAC-3′) (7); 1392R had a GC clamp at its 5′ end (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCC-3′) (7). The PCR mixture contained 4 mM MgCl2, 4 mM deoxynucleoside triphosphates, 1 U rTth polymerase, and 1× of the buffer supplied with the enzyme (all reagents were from Applied Biosystems) in a total volume of 50 μl. Cycling parameters were as follows: 2 min at 95°C; 30 cycles of 40 s at 95°C, 40 s at 55°C, and 40 s at 72°C; and a 7-min final elongation at 72°C. The 362-bp product was purified using the QIAgen PCR purification kit (QIAGEN, Hilden, Germany). One hundred nanograms of PCR products was analyzed using DGGE. Gels had 8% polyacrylamide and a denaturation gradient of 30% to 70%, where 100% denaturant is defined as 7 M urea and 40% formamide (all reagents were from Sigma-Aldrich, St. Louis, MO). Electrophoresis was carried out at 60 V for 16 h using a DCode system (Bio-Rad, Hercules, CA). Gels were stained with SybrGold (Molecular Probes, Inc.) and documented using a FluorChem 8800 fluorescence imager (Alpha Innotech, Inc., San Leandro, CA).
BacLight live-dead fluorescence microscopy.
Scraped biofilm was resuspended in 1 ml of the original water by pipetting, brief mild vortexing, and a 2-s sonication step. The two-color fluorescence assay-based BacLight bacterial viability kit (Molecular Probes Inc.) was used to stain the organisms for microscopy by adding 1.5 μl each of the dyes SYTO 9 and propidium iodide (both in anhydrous dimethyl sulfoxide) to the suspended biofilm. SYTO 9 stain generally labels all bacteria in a population green, while propidium iodide penetrates only bacteria with compromised membranes and labels them red. After mixing and a 1-h incubation in the dark, a 100-μl sample was filtered through 25-mm polycarbonate filters (Osmonic Inc., Minnetonka, Minn.), washed with 3 ml of filter-sterilized nanopure water, and mounted on slides. Photomicrographs were taken on a Nikon E800 microscope using a 1.4-numerical-aperture 100× oil objective and fluorescein isothiocyanate (480/30 excitation filter, DM505 dichroic mirror, 535/40 emission filter) and tetramethyl rhodamine isocyanate (546/10 excitation filter, DM575 dichroic mirror, 590 emission filter) fluorescence filter sets. The software used for visualization was MetaVue, version 6.1 (Universal Imaging, Downington, Pa.).
RESULTS
Throughout the Results and Discussion, the term “viable” is used. Although there is considerable controversy surrounding the correlation between viable and culturable cells, for E. coli O157:H7 and Salmonella, whose culturability can be tested by plating on standard growth media, we use the term in an operational sense and do not differentiate between “viable” and “culturable.” However, when one is looking at undefined, mixed-population samples from the environment, “viable” cannot be interpreted as “culturable,” because it is well accepted that only a small fraction of bacteria may be cultured. In the case of these data, viability has been related to the relative proportion of live/dead cells as measured using the BacLight live/dead stain.
Effect of stress on DNA yield of EMA-treated cultures.
E. coli O157:H7 and Salmonella cultures were subjected to different stress conditions. Heat exposure at 72°C for 5 min or treatment with 70% isopropanol for 10 min resulted in dramatic losses in DNA yield (below the detection limit of 1 ng/ml) for both species compared to the corresponding non-EMA-treated aliquots (Fig. 1A). In the case of E. coli O157:H7, EMA treatment resulted in a significant decrease in the genomic DNA yield of unstressed cells, but the stress-induced loss was far more pronounced. The loss of DNA correlated with the qualitative observation that during DNA extraction only stressed cultures produced a visible red pellet (caused by the EMA dye) after centrifugation of cell debris. These results, obtained with the Qbiogene soil DNA extraction kit, were confirmed using the DNeasy tissue kit and the PrepMan Ultra sample preparation reagent (used in previous publications [18, 24, 25]). These methods employ different principles for cell lysis and cleanup, excluding the possibility that selective loss of DNA is seen only with a specific DNA extraction procedure. Independently of the extraction method, stress and subsequent EMA treatment resulted in a dramatic loss of genomic DNA as visualized on ethidium bromide-stained agarose gels (Fig. 1B, C, and D). Observations on the loss of DNA correlated with a complete loss of viability from heat and isopropanol treatment as confirmed by plate counts. The stress-induced reduction in DNA yield was reflected in a Q-PCR signal reduction, as determined by subtraction of CT values for EMA-treated cells from those for non-EMA-treated cells (Fig. 1E). For example, the difference in threshold values (ΔCT) approximated 20 cycles in the case of heat-killed Salmonella.
Exposure of pathogens to a stress gradient.
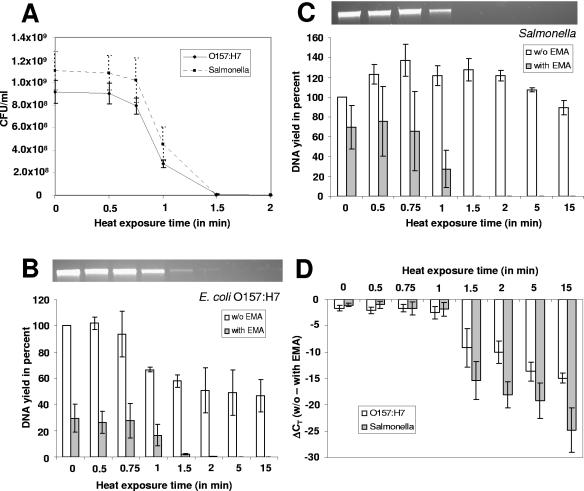
The next step was to study the effect of increasingly long heat exposure on the genomic DNA yield and threshold amplification cycles of E. coli O157:H7 and Salmonella. Increasing stress resulted in an increasing loss in viability (Fig. 2A) as determined by plate counts. Heat exposure of 1.5 to 2 min was the threshold, where no viable colonies could be detected. Genomic DNA extracted from EMA-treated or untreated cultures was normalized to the genomic DNA yield of the corresponding non-EMA-treated and unstressed samples (Fig. 2B and C). Whereas the DNA yields of non-EMA-treated cultures varied about ±60% relative to each other, the yields of EMA-treated aliquots decreased to undetectable levels with prolonged heat exposure. Values obtained by fluorometer quantification of DNA from EMA-treated aliquots correlated nicely with band intensities of high-molecular-weight DNA on stained agarose gels (Fig. 2B and C, top). This was in agreement with qualitative observations that an increasingly long heat exposure resulted in a more intense red color of the pelleted cell debris after centrifugation.
FIG. 2.
Effects of increasingly long heat stress exposures at 72°C on E. coli O157:H7 and Salmonella. Error bars, standard deviations from three independent replicates. (A) Loss of viability with increasing exposure times as determined by plate counts. (B and C) Effects of prolonged heat stress and EMA treatment on the DNA yields of E. coli O157:H7 (B) and Salmonella (C). Yields are expressed as percentages of the yield obtained with an unstressed and non-EMA-treated culture. Genomic DNA from EMA-treated aliquots was also visualized on agarose gels. (D) Signal reduction was determined by Q-PCR detecting relative differences in stx1 (E. coli O157:H7) and invA (Salmonella) gene copies. CT values derived from EMA-treated cultures were subtracted from corresponding CT values from untreated cultures.
Identical volumes of genomic DNA served as the template for Q-PCR for detection of relative differences in stx1 (E. coli O157:H7) and invA (Salmonella) gene copies. The tendencies seen in plate counts and in DNA yields correlated with a gradual increase in the CT values with longer exposure times, meaning that more cycles were needed for amplification. Subtracting the CT values of EMA-treated aliquots from those of the corresponding untreated aliquots showed that longer exposure times resulted in increasingly strong signal reductions for both bacterial species (Fig. 2D).
Effect of EMA on defined ratios of viable and dead cells.
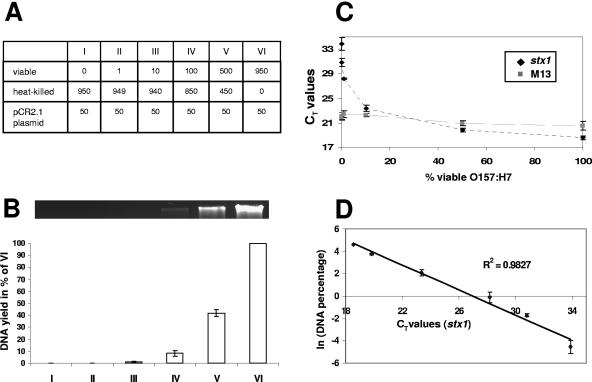
To elucidate the relationship between the proportion of viable cells, the DNA yield, and the real-time PCR signals after EMA treatment, mixtures with defined ratios of viable and heat-killed cells were used. An aliquot of E. coli O157:H7 was subjected to heat treatment at 72°C for 15 min, resulting in a decrease in culturable cell counts to zero. Heat-killed cells were mixed with the untreated original culture in defined ratios, with viable cells representing 0, 0.1, 1, 10, 50, or 100% of the total bacterial cell concentration, respectively (Fig. 3A). Every mixture was supplemented with 5% (vol/vol) E. coli Mach1-T1R (Invitrogen, Carlsbad, CA) transformed with a pCR2.1 plasmid as an amplification control. Increasing proportions of unheated and viable cells led to a substantial increase in the genomic DNA yield (Fig. 3B) and to decreasing CT values in Q-PCR targeting the stx1 gene (Fig. 3C). The highest CT value was observed for sample I, containing only heat-killed E. coli O157:H7 cells. Although the control amplification of an internal sequence of the pCR2.1 plasmid using M13 primers followed a tendency similar to that of the stx1 amplification, the range in CT values was less than 2 cycles compared to a difference of at least 15 cycles for the detection of the stx1 target (Fig. 3C). This observation can be explained by a concentration-dependent coprecipitation of plasmid DNA with cell debris.
FIG. 3.
Effect of EMA treatment on genomic DNA yield and PCR quantification of defined ratios of viable and heat-killed cells. Error bars, standard deviations from three independent replicates. (A) Table showing mixing ratios of viable and heat-killed E. coli O157:H7. Numbers represent volumes in microliters. Every mixture was supplemented with 50 μl of E. coli Mach1-T1R carrying a pCR2.1 plasmid as an amplification control. (B) Genomic DNA yield expressed as a percentage of the concentration obtained from mixture VI. The DNA was also visualized on an agarose gel (top). (C) Real-time PCR was performed using primers specific for the stx1 gene and M13 primers amplifying an internal vector sequence. CT values of the stx1 and M13 (control) amplifications are shown as a function of the normalized total DNA concentration. (D) Correlation between the natural logarithm of the normalized DNA concentrations and the corresponding CT values obtained from stx1 amplification. The R2 value of the linear trendline is given.
The increase in CT values for increasing proportions of heat-killed E. coli O157:H7 seems to be due mainly to a decrease in the DNA template concentrations and not to the presence of EMA-cross-linked DNA as reported previously (24). Indeed, all of the DNA seems to be amplifiable, as reflected in an R2 value of 0.9829 for a plot of the natural logarithm of template concentrations versus CT values (Fig. 3D).
Effect of EMA on defined mixed two-species samples.
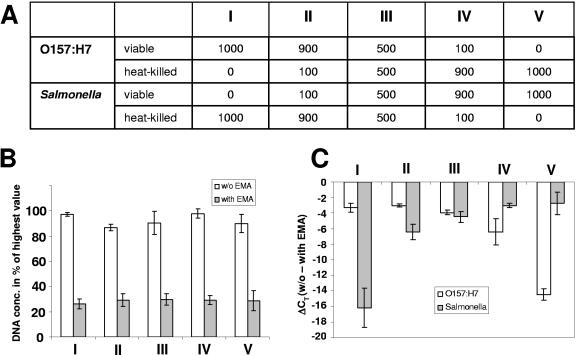
Encouraged by the results obtained from a mixture of viable and heat-killed cells of a single species, we extended the study to a mixture of two bacterial species (E. coli O157:H7 and Salmonella serovar Typhimurium), again with different ratios of viable and heat-killed cells. Each bacterial species made up 50% of the total volume of 2 ml (Fig. 4A). Furthermore, the viable fraction from both species added up to 50%, as did the heat-killed fraction from both species. Quantification of total genomic DNA showed relatively constant levels for non-EMA-treated and EMA-treated samples, although the DNA yields of EMA-treated samples, as expected, were lower, considering the fact that half of the cells were not viable (Fig. 4B). Species-specific PCR amplification of EMA-treated samples resulted in a signal that decreased as the percentage of heat-killed cells increased (Fig. 4C). The total ranges in CT values between viable and heat-killed cells were 12.89 (±0.6) and 15.6 (±3.7) cycles for E. coli O157:H7 and Salmonella, respectively, compared to 1.4 (±0.3) and 2.1 (±0.1) cycles for non-EMA-treated mixtures.
FIG. 4.
Effect of EMA on the amplification of defined two-species mixtures with different ratios of viable and heat-killed cells. Error bars, standard deviations from three independent replicates. (A) Table showing mixing ratios of viable and heat-killed E. coli O157:H7 and Salmonella. Numbers represent volumes in microliters. (B) Genomic DNA yields expressed as percentages of the highest value obtained. (C) Signal reduction as determined by Q-PCR detecting relative differences in stx1 (E. coli O157:H7) and invA (Salmonella) gene copies. CT values derived from EMA-treated cultures were subtracted from the corresponding CT values from untreated cultures.
Distinction between viable and dead pathogens in a spiked biofilm.
We were interested in whether the distinction between viable and dead would also work in a complex mixed-community biofilm background. Aliquots of a resuspended natural drinking-water biofilm were spiked with either viable or heat-killed E. coli O157:H7. The relative differences in amplification efficacies were determined by quantitative PCR followed by subtraction of CT values derived from EMA-treated mixtures from the corresponding CT values from untreated mixtures. EMA treatment led to a signal reduction of 2.8 (±0.2) cycles in the case of unstressed viable cells compared to a signal reduction of 11.8 (±0.2) cycles in the case of heat-killed cells added to the biofilm.
Effect of EMA on community fingerprinting.
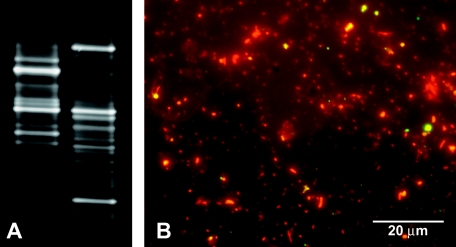
Community fingerprints from a mature biofilm harvested from a water reservoir without light exposure were dramatically different when an EMA-treated aliquot was compared with an untreated aliquot (Fig. 5A). Whereas EMA treatment led to a signal reduction of some bands, the intensities of other bands increased as a result of EMA treatment using DGGE for separation of 16S rRNA gene amplification products. BacLight LIVE/DEAD fluorescence microscopy showed that the majority of the cells stained red or orange with propidium iodide and thus can be considered dead (Fig. 5B). Viable cells staining green with the SYTO 9 dye appeared to be significantly less abundant. Quantification was not attempted due to the presence of abiotic particles and the inability to distinguish between bacteria and other organisms.
FIG. 5.
Effect of EMA treatment on community fingerprints of a mature drinking-water biofilm and qualitative viability analysis of the same biofilm. (A) Community patterns derived from an untreated (left) and an EMA-treated but otherwise identical (right) biofilm by using denaturing gradient gel electrophoresis. (B) BacLight LIVE/DEAD viability staining. Dead cells were stained red or orange with propidium iodide, while live cells were stained green with SYTO 9. Yellow color indicates simultaneous binding of both dyes (most probably to a debris particle).
DISCUSSION
In agreement with previous publications introducing the application of EMA in combination with quantitative PCR to differentiate between viable and dead bacterial cells (18, 24, 25), the efficacy of this DNA-intercalating dye in selecting against DNA from dead cells was confirmed. EMA treatment of mixed bacterial communities might be a promising tool to selectively favor the analysis of the viable fraction.
The method might be an important step to mitigate one of the major drawbacks from which all DNA-based molecular techniques suffer, the inability to differentiate between viable and dead cells. In bacterial diagnostics, this leads to an overestimation of viable cells or even to false-positive results, limiting the suitability of these molecular tools for analysis of microbiological safety in water and food monitoring as well as for clinical diagnostics (for reviews, see references 12 and 23). EMA pretreatment of samples might be a fast and elegant alternative to quantification of RNA, which, due to the rapid degradation of RNA after cell death, is currently the method of choice to avoid overestimation of bacterial numbers. Nevertheless, working with RNA is still technically demanding, due to the risk of RNA degradation and the instability of appropriate standards during storage. Practical problems of extracting detectable levels of intact RNA from small numbers of bacteria and the risk of DNA contamination add to the complexity of RNA-based approaches and are impediments to its utilization in routine diagnostics. Moreover, few studies have investigated the relationship between detection of RNA and viability. Although most prokaryotic mRNA species have half-lives of only a few minutes in living cells (1, 2), Sheridan et al. (27) reported that some mRNA targets were detectable by reverse transcription-PCR 2 to 16 h after heat killing of E. coli and even longer after ethanol treatment. 16S rRNA was detected by reverse transcription-PCR in samples containing dead bacteria, and it did not disappear during a subsequent incubation of 16 h at room temperature. This was confirmed by McKillip et al. (16), who concluded that the presence of 16S rRNA up to 48 h after cell death was not correlated with viability following moderate heat inactivation or UV irradiation of E. coli O157:H7 and Staphylococcus aureus, raising doubts about its potential as a useful indicator of viability.
In contrast to previous studies, we report that EMA-based selection against DNA originating from dead cells seems to occur primarily on the level of genomic DNA extraction. An understanding of the underlying principle might allow the combination of EMA treatment not only with real-time PCR as a downstream analysis tool but also with hybridization-based techniques such as microarray analysis. The selective loss of DNA from dead cells is presumably due to cross-linking of DNA to other cell components. EMA-DNA conjugates are lost during the extraction procedure and are pelleted together with cell debris during the centrifugation step following cell lysis by bead beating. This correlated with qualitative observations that an increasing number of dead cells resulted in a more-intense red color of the pelleted cell debris (caused by the EMA dye) after centrifugation. The observation holds true when different commercially available kits used routinely for extraction of genomic DNA are compared. Comparison with the DNeasy tissue kit and the PrepMan Ultra sample preparation reagent confirmed the results obtained with the Qbiogene kit. Whereas the latter employs bead beating and binding of DNA to a resuspended binding matrix, which is added to the solution, the DNeasy tissue kit relies on cell lysis using proteinase K treatment and selective binding of DNA to a silica gel membrane, followed by washing steps and elution. The PrepMan Ultra sample preparation procedure involves only boiling of samples in the lysis solution provided and removal of debris by simple centrifugation; it does not employ any binding matrix. Theoretically, the chemical properties of EMA could affect the binding efficacy of DNA-EMA conjugates to the binding matrix provided with the Qbiogene extraction kit, but the results did not substantiate this. The molecular composition of EMA conjugates has yet to be elucidated.
Our results do not exclude the possibility that PCR amplification of remaining DNA from dead cells, which was copurified along with DNA from viable cells, is inhibited due to EMA cross-linking. Indeed, we could confirm previous reports that treatment of purified DNA (not protected within a cell) results in strong inhibition of amplification (data not shown). Despite this fact, inhibition of PCR amplification is very likely to play only a minor role, considering the good correlation between the proportion of viable cells, DNA template concentrations, and the signal detection thresholds in Q-PCR shown in this study.
The observation that EMA to some extent also decreases the DNA yield from growing unstressed cultures (mainly from E. coli O157:H7 [Fig. 1A and 2B]) is not likely to be explained by the presence of dead cells in the corresponding log-phase cultures and requires further examination. An intact cell membrane should theoretically be an efficient barrier to EMA due to the charge of the molecule. However, EMA might also penetrate viable cells to a certain extent, though not as efficiently as dead cells. Another speculation would be that the bright light used for inactivation of EMA would result in partial cell lysis and susceptibility to EMA. Special filters allowing the transmission only of that part of the light spectrum relevant for EMA inactivation (around 460 nm [3]) might be useful in this regard. It also remains to be investigated whether the use of other chemicals for live-dead differentiation as an alternative to ethidium monoazide would mitigate this problem. Improvement and further optimization of the method to avoid any loss of DNA from viable cells would allow better quantification of viable organisms. This is especially important in the case of pathogens for which only viable cells are infectious. It is very encouraging, however, that Q-PCR could distinguish between defined communities whose viable cell content showed a difference of only 1% (Fig. 3). Automation of DNA isolation would further improve reproducibility and avoid variations due to manual sample processing. In order to achieve absolute quantification of bacterial pathogens, the use of standards prepared from EMA-treated cells might be considered.
EMA treatment of cells prior to PCR amplification of selected targets may be a valuable method in microbial community analysis, a common procedure in molecular ecology. The study of environmental samples using DNA-based PCR inevitably leads to amplification of genetic material from both viable and dead cells. EMA might allow one to study specifically the viable and thus metabolically active fraction of these mixed communities. Conclusions from fingerprints derived from the viable portion could differ substantially from those originating from the study of total (viable and dead) cells (Fig. 5). It must be mentioned, however, that the community fingerprint shown here should be interpreted with caution, because further studies are needed to answer the questions whether the EMA principle can be applied to a wide spectrum of bacteria and whether the same parameters can be applied to all members of a microbial community. So far EMA pretreatment has been successfully used only with E. coli O157:H7, Salmonella serovar Typhimurium, Campylobacter jejuni, and the gram-positive bacterium Listeria monocytogenes (18, 24, 25).
In summary, EMA treatment of mixed communities might improve the targeting of genomic sequences for determination of the potential pathogenic threat originating from viable bacteria, while avoiding less standardized, more troublesome, and more expensive alternative techniques. The method presented overcomes the currently missing relationship between viability and direct PCR detection of DNA targets without preenrichments. Moreover, it could be an extremely useful method in ecological studies by limiting DNA-based community analysis to the viable fraction.
Acknowledgments
Ray Hozalski is gratefully acknowledged for critical review of the manuscript.
This research has been supported by a grant (DAAD 19-03-1-0198) from the Army Research Office, overseen by Sherry Tove, Chief, Microbiology and Biodegradation, Life Sciences Division.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Army Research Office.
REFERENCES
- 1.Alifano, P., C. B. Bruni, and M. S. Carlomagno. 1994. Control of mRNA processing and decay in prokaryotes. Genetica 94:157-172. [DOI] [PubMed] [Google Scholar]
- 2.Belasco, J. 1993. mRNA degradation in prokaryotic cells: an overview, p. 3-12. In J. Belasco and G. Brawerman (ed.), Control of messenger RNA stability. Academic Press, Inc., San Diego, Calif.
- 3.Bolton, P. H., and D. R. Kearns. 1978. Spectroscopic properties of ethidium monoazide: a fluorescent photoaffinity label for nucleic acids. Nucleic Acids Res. 5:4891-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breeuwer, P., and T. Abee. 2000. Assessment of viability of microorganisms employing fluorescence techniques. Int. J. Food Microbiol. 55:193-200. [DOI] [PubMed] [Google Scholar]
- 5.Coffman, G. L., J. W. Gaubatz, K. L. Yielding, and L. W. Yielding. 1982. Demonstration of specific high affinity binding sites in plasmid DNA by photoaffinity labeling with ethidium analog. J. Biol. Chem. 257:13205-13297. [PubMed] [Google Scholar]
- 6.DeTraglia, M. C., J. S. Brand, and A. M. Tometsko. 1978. Characterization of azidobenzamidines as photoaffinity labeling for trypsin. J. Biol. Chem. 253:1846-1852. [PubMed] [Google Scholar]
- 7.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fey, A., S. Eichler, S. Flavier, R. Christen, M. G. Höfle, and C. A. Guzman. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70:3618-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hixon, S. C., W. E. White, and K. L. Yielding. 1975. Selective covalent binding of an ethidium analog to mitochondrial DNA with production of petite mutants in yeast by photoaffinity labeling. J. Mol. Biol. 92:319-329. [DOI] [PubMed] [Google Scholar]
- 10.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 12.Lemarchand, K., L. Masson, and R. Brousseau. 2004. Molecular biology and DNA microarray technology for microbial quality monitoring of water. Crit. Rev. Microbiol. 30:145-172. [DOI] [PubMed] [Google Scholar]
- 13.López-Amorós, R., S. Castel, J. Comas-Riu, and J. Vives-Rego. 1997. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide and CTC. Cytometry 29:298-305. [DOI] [PubMed] [Google Scholar]
- 14.Masters, C. I., J. A. Shallcross, and B. M. Mackey. 1994. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J. Appl. Bacteriol. 77:73-79. [DOI] [PubMed] [Google Scholar]
- 15.McFeters, G. A., F. P. Yu, B. H. Pyle, and P. S. Stewart. 1995. Physiological assessment of bacteria using fluorochromes. J. Microbiol. Methods 21:1-13. [DOI] [PubMed] [Google Scholar]
- 16.McKillip, J. L., L. A. Jaykus, and M. Drake. 1998. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nebe-von Caron, G., P. Stephens, and R. A. Badley. 1998. Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 84:988-998. [DOI] [PubMed] [Google Scholar]
- 18.Nogva, H. K., S. M. Dromtorp, H. Nissen, and K. Rudi. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease. PCR BioTechniques 810:812-813. [DOI] [PubMed] [Google Scholar]
- 19.O'Brien, M. C., and W. E. Bolton. 1995. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry 19:243-255. [DOI] [PubMed] [Google Scholar]
- 20.Pettipher, G. L., R. Mansell, C. H. McKinnon, and C. Cousins. 1980. Rapid membrane filtration epifluorescent microscopy technique for direct enumeration of bacteria in raw milk. Appl. Environ. Microbiol. 39:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter, J., D. Deere, M. Hardman, C. Edwards, and R. Pickup. 1997. Go with the flow—use of flow cytometry in environmental microbiology. FEMS Microbiol. Ecol. 24:93-101. [Google Scholar]
- 22.Riedy, M. C., K. A. Muirhead, C. P. Jensen, and C. C. Stewart. 1991. Use of a photolabeling technique to identify nonviable cells in fixed homologous or heterologous cell populations. Cytometry 12:133-139. [DOI] [PubMed] [Google Scholar]
- 23.Rompre, A., P. Servais, J. Baudart, M. R. de-Roubin, and P. Laurent. 2002. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J. Microbiol. Methods 49:31-54. [DOI] [PubMed] [Google Scholar]
- 24.Rudi, K., B. Moen, S. M. Drømtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudi, K., K. Naterstad, S. M. Drømtorp, and H. Holo. 2005. Detection of viable and dead Listeria monocytogenes on gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301-306. [DOI] [PubMed] [Google Scholar]
- 26.Sharma, V. K., and E. A. Dean-Nystrom. 2003. Detection of enterohemorrhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and Shiga toxins. Vet. Microbiol. 93:247-260. [DOI] [PubMed] [Google Scholar]
- 27.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vesey, G., N. Ashbolt, E. J. Fricker, D. Deere, K. L. Williams, D. A. Veal, and M. Dorsch. 1998. The use of a ribosomal RNA targeted oligonucleotide probe for fluorescent labeling of viable Cryptosporidium parvum oocysts. J. Appl. Microbiol. 85:429-440. [DOI] [PubMed] [Google Scholar]
- 29.Virta, M., S. Lineri, P. Kankaanpää, M. Karp, K. Peltonen, J. Nuutila, and E.-M. Lilius. 1998. Determination of complement-mediated killing of bacteria by viability staining and bioluminescence. Appl. Environ. Microbiol. 64:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waring, M. J. 1965. Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 13:269-282. [DOI] [PubMed] [Google Scholar]
- 31.Widmer, G., E. A. Orbacz, and S. Tzipori. 1999. β-Tubulin mRNA as a marker of Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 65:1584-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]