Abstract
Comamonas sp. strain CNB-1 grows on 4-chloronitrobenzene (4-CNB) and nitrobenzene as sole carbon and nitrogen sources. In this study, two genetic segments, cnbB-orf2-cnbA and cnbR-orf1-cnbCaCbDEFGHI, located on a newly isolated plasmid, pCNB1 (ca. 89 kb), and involved in 4-CNB/nitrobenzene degradation, were characterized. Seven genes (cnbA, cnbB, cnbCa, cnbCb, cnbD, cnbG, and cnbH) were cloned and functionally expressed in recombinant Escherichia coli, and they were identified as encoding 4-CNB nitroreductase (CnbA), 1-hydroxylaminobenzene mutase (CnbB), 2-aminophenol 1,6-dioxygenase (CnbCab), 2-amino-5-chloromuconic semialdehyde dehydrogenase (CnbD), 2-hydroxy-5-chloromuconic acid (2H5CM) tautomerase, and 2-amino-5-chloromuconic acid (2A5CM) deaminase (CnbH). In particular, the 2A5CM deaminase showed significant identities (31 to 38%) to subunit A of Asp-tRNAAsn/Glu-tRNAGln amidotransferase and not to the previously identified deaminases for nitroaromatic compound degradation. Genetic cloning and expression of cnbH in Escherichia coli revealed that CnbH catalyzed the conversion of 2A5CM into 2H5CM and ammonium. Four other genes (cnbR, cnbE, cnbF, and cnbI) were tentatively identified according to their high sequence identities to other functionally identified genes. It was proposed that CnbH might represent a novel type of deaminase and be involved in a novel partial reductive pathway for chloronitrobenzene or nitrobenzene degradation.
Chlorinated nitroaromatic compounds such as chloronitrobenzenes are massively produced and are widely used as intermediates for chemical syntheses of drugs, herbicides, dyes, etc. The natural formation of chlorinated nitroaromatic compounds is rare, and most of these compounds are from industrial productions and have been introduced into the environment for a relatively short period. Apparently, their occurrence in the environment has selected microorganisms that are able to utilize chlorinated nitroaromatic compounds as carbon and/or nitrogen sources for growth. Examples of such microorganisms are bacterial strain LW1 (15), a coculture of Pseudomonas putida and a Rhodococcus sp. (25), and recently Comamonas sp. strain CNB-1 (38).
Nitroaromatic compounds and chlorinated nitroaromatic compounds are structurally analogs. The microbial degradation of nitroaromatic compounds has been extensively investigated and the removal of the nitro group(s) is carried out via oxidative pathways that initiate with monooxygenases (22, 31, 40) or dioxygenases (8, 16, 20, 19, 32) or a partial reductive pathway that initiates with nitroreductases (7-9, 17, 22, 29, 30). Although structurally related to the nitroaromatic compounds, the chlorinated nitroaromatic compounds are more resistant to microbial degradation due to the simultaneous existence of chlorine and nitro groups, and thus the knowledge of its microbial degradation is very limited.
Previous studies revealed that reductive dehalogenization (35) and partial reduction of nitro groups (15, 39) might be involved in the initial steps during chlorinated nitroaromatic compound degradation. However, these pathways have not been characterized at the genetic and enzymatic levels. This study identified the genes and pathway for 4-chloronitrobenzene degradation by previously isolated Comamonas sp. strain CNB-1.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Comamonas sp. strain CNB-1 (38, 39) was maintained in Luria-Bertani (LB) medium and in MSB (5) containing 2 mM of 4-chloronitrobenzene as the sole carbon and nitrogen source. All Escherichia coli strains were cultured and maintained in LB medium. When necessary, ampicillin at 100 μg/ml was added to the medium.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or primer sequence (restriction enzymes)a | Source or use and reference |
|---|---|---|
| Strains | ||
| Comamonas sp. strain CNB-1 | Isolated from activated sludge, assimilating p-chloronitrobenzene | CGMCC 1028, Wu et al. (38, 39) |
| E. coli BL21(DE3) | Expression host | Stratagene |
| Plasmids | ||
| pET-21a(+) | Expression vector | Novagen |
| pET-28a(+) | Expression vector | Novagen |
| pBG-2 | Plasmid carrying 2-aminophenol 1,6-dioxygenase genes | Wu et al. (39) |
| pCG-13 | Plasmid carrying 2-aminophenol 1,6-dioxygenase genes | Wu et al. (39) |
| pETcnbA | Constructed for expression of 4-chloronitrobenzene nitroreductase | This study |
| pETcnbAB | Constructed for expression of nitroreductase and mutase | This study |
| pETcnbCab | Constructed for expression of 2-amino-5-chlorophenol 1,6-dioxygenase | Wu et al. (39) |
| pETcnbD | Constructed for expression of 2-amino-5-chloromuconic semialdehyde dehydrogenase | This study |
| pETcnbG | Constructed for expression of 2-hydroxy-5-chloromuconate tautomerase | This study |
| pETcnbH | Constructed for expression of 2-amino-5-chloromuconic deaminase | This study |
| Oligonucleotides | ||
| NBPf | GACGTTTCATATGCCGACCAGCCCGTTC (NdeI) | Amplification for cnbA; this study |
| NBPr | TGGGATCCCTATTCGTGGACGAAGGTGG (BamHI) | |
| HabPf | GTCCGAATTCAAGGAGACCCCTTCATGC (EcoRI) | Amplification for cnbB; this study |
| HabPr | GTCAAAGCTTTGCGGGAAGTCTCATGGT (HindIII) | |
| Ps | ATGCAAGGTGAAATCATCG | Amplification for cnbCab; this study |
| Pat | CCGGAATTCTCAGAGTCGGAACTCGATC (EcoRI) | |
| DehPf | GGAAACCCATATGAAGCAATACCGAAATTACATCAACG (NdeI site) | Amplification for cnbD; this study |
| DehPr | GGCCAAGCTTAGACACTGCCTCTTGATCAATTCGG (HindIII site) | |
| cnbGPr | GGAATTCCATATGCCGTTCGCACAGATCTACAT (NdeI site) | Amplification for cnbG; this study |
| cnbGPr | CCCCAAGCTTTCAGCGGCCGAGGTCTTT (HindIII site) | |
| AmdPf | CACGCGCATATGGAACCCCGGCTCAACGCCTACAAG (NdeI site) | Amplification for cnbH; this study |
| AmdPr | GGCGAAGCTTCTATGGCAGGTCGGGCGCGCCCAG (HindIII site) |
The restriction enzyme sites are underlined. The start and stop codons are in bold.
Screening for CNB-negative mutant, plasmid curing, and detection of plasmid.
A mutant, Comamonas sp. strain CNB-2, that could not utilize 4-chloronitrobenzene or nitrobenzene for growth was obtained by curing the plasmid from strain CNB-1 using a modified sodium dodecyl sulfate treatment method of El-Mansi et al. (6). Detection of the megaplasmid in Comamonas sp. strains CNB-1 and CNB-2 was carried out according to Barton et al. (2). Separation of chromosomal and plasmid DNAs was carried out on an agarose gel (1%) under conditions of 6 V/cm and 70 seconds for 22 h using a Bio-Rad pulsed-field gel electrophoresis apparatus (Bio-Rad). Yeast genomic DNA (catalog no. 170-3605, Bio-Rad) was used as DNA molecular weight markers.
DNA extraction and plasmid isolation.
DNAs from Comamonas sp. strain CNB-1 and routine plasmid isolation were carried out following the procedures of Sambrook et al. (27). For large-plasmid isolation, a modified alkaline lysis method was used (28, 36).
DNA sequencing, sequence assembly and analysis.
The 2-aminophenol 1,6-dioxygenase-positive clones pBG-2 and pCG-13, each containing a 35-kb DNA fragment from strain CNB-1, was sequenced with the shotgun method by the Beijing Genome Institute (Huada Corp., Beijing, China). Contigs were assembled using the GCG Wisconsin package.
Cloning and expression of cnb genes in E. coli.
PCR primers (Table 1) were designed according to the DNA sequence obtained in this study, and entire genes were amplified by PCR from the strain CNB-1 genome. Purified PCR products were treated with restriction enzymes and then ligated into the similarly treated pET-21a(+), except for cnbH, which was cloned into pET-28a(+). The resulting plasmids (Table 1) were used to transform E. coli cells for expression of the genes. Expression of the genes in cells of E. coli strains was induced with 1 mM isopropylthiogalactopyranoside (IPTG) when the culture reached an optical density at 600 nm of ca. 0.6.
Preparation of cellular lysates, purification of enzymes, and SDS-PAGE.
Cellular lysates of Comamonas sp. strain CNB-1 or recombinant E. coli actively synthesizing various enzymes of 4-chloronitrobenzene and nitrobenzene degradation were prepared by sonification of cell suspensions in 10 mM phosphate buffer (pH 8). Sonification was conducted (at 200 W, 3 seconds, interval of 5 seconds, for 90 cycles) on ice bath. Cell debris was removed by centrifugation at 12,000 × g for 10 min, and the supernatant was used for purification of various enzymes and for enzymatic activity determination.
The procedures for purification of 2-aminophenol 1,6-dioxygenase from Comamonas sp. strain CNB-1 were previously described (39). Purification of 2-amino-5-chloromuconic semialdehyde dehydrogenase from recombinant E. coli cells was performed with His Bind resin chromatography by following the instructions from the manufacturer (Novagen). The purification efficiency of each step was controlled by running electrophoresis of samples collected from each step, with a 12% polyacrylamide gel containing 0.1% sodium dodecyl sulfate (SDS). To visualize protein bands, the gel was stained with Coomassie brilliant blue. All purified enzymes were stored at −70°C.
Protein concentrations were determined according to Bradford (3).
Enzymatic assays.
4-Chloronitrobenzene nitroreductase activity was determined spectrophotometrically by measuring the decrease of absorption at 340 nm (A340). The reaction mixture contained cellular lysate (4 ng), 4-chloronitrobenzene or nitrobenzene (0.1 mM), NADPH (0.2 mM), and phosphate buffer (10 mM, pH 8) in a final volume of 200 μl. The reaction was started by addition of NADPH.
Hydroxylaminobenzene mutase activities were determined by determination of increase of absorption at 235 nm (A235). The reaction mixture contained the same ingredients as above, except that cellular lysate with 4-chloronitrobenzene nitroreductase was replaced with cellular lysate containing hydroxylaminobenzene mutase. The increase in A235, due to the formation of 2-aminophenol, was used for estimation of the activity of hydroxylaminobenzene mutase. The activity of 1-hydroxylamino-4-chlorobenzene mutase was determined similarly.
The 2-aminophenol 1,6-dioxygenase (39), 2-aminomuconic/2-amino-5-chloromuconic semialdehyde dehydrogenase (13), and 2-aminomuconate deaminase (10, 11) activities were determined according to the methods cited. The 2-amino-5-chloromuconate deaminase activity was determined similarly to that of 2-aminomuconate deaminase (10, 11), except that the wavelength was set at 340 nm (A340).
Determination of molecular weight with SDS-PAGE.
The molecular weights of the recombinant proteins and enzyme subunits were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) with a 15% resolving gel and a 5% stacking gel. Protein molecular weight standards for SDS-PAGE were purchased from the Institute of Biochemistry and Cell Biology (Shanghai, China).
Construction of phylogenetic tree.
Peptide sequences of various deaminases and subunits of Asp-tRNAAsn/Glu-tRNAGln amidotransferases were extracted from NCBI (http://www.ncbi.nlm.nih.gov/). Phylogenetic trees were generated using the neighbor joining method of Saitou and Nei (26) with the AlignX software (Informax, Maryland), and multiple sequence alignment was done using ClustalX (37). The length of each branch pair represents the evolutionary distance between the sequences.
Preparations of 2-amino-5-chloromuconic semialdehyde and 2-amino-5-chloromuconic acid.
2-Amino-5-chloromuconic semialdehyde was prepared by enzymatic cleavage of 2-amino-5-chlorophenol (Sigma). The reaction mixture contained 50 μl of 2-amino-5-chlorophenol (10 mM), 10 ng of partially purified 2-aminophenol 1,6-dioxygenase, and 2.9 ml of phosphate buffer (10 mM, pH 8). After reaction for 5 min, this mixture was used as the substrate without further purification.
The 2-amino-5-chloromuconate was prepared by further enzymatic oxidation of 2-amino-5-chloromuconic semialdehyde (prepared as above), by addition of 4 ng of partially purified 2-amino-5-chloromuconic semialdehyde dehydrogenase to the above reaction mixture. The product, 2-amino-5-chloromuconate, was partially purified according to He and Spain (11).
Preparation, purification, and identification of 2-hydroxy-5-chloromuconic acid with gas chromatography-mass spectroscopy.
The 2-hydroxy-5-chloromuconic acid was prepared by sequential catalysis with partially purified 2-aminophenol 1,6-dioxygenase (0.2 mg/ml), 2-amino-5-chloromuconic semialdehyde dehydrogenase (0.08 mg/ml), and 2-amino-5-chloromuconic acid deaminase (0.13 mg/ml), in phosphate buffer (10 mM, pH 8). After 5 min, the reaction mixture (total 10 ml in volume, initially containing 10 mM 2-amino-5-chlorophenol and 10 mM NAD+) was adjusted to 2.0 with HCl and centrifuged at 12,000 × g for 10 min. The supernatant was extracted with an equal volume of ethyl acetate. After another centrifugation at 12,000 × g for 10 min, the organic phase that contained 2-hydroxyl-5-chloromuconate was pooled and concentrated by evaporation of the organic solvent under a vacuum and then analyzed by UV spectrophotometer and gas chromatography-mass spectroscopy.
2-Hydroxymuconate and 2-hydroxy-5-chloromuconate were identified by liquid chromatography-mass spectrometry (LC-MS) and spectrophotometry. LC-MS analysis was performed on a Finnigan LCQ ion trap mass spectrometer (San Jose, Calif.) equipped with an atmospheric pressure ionization interface. The instrument was operated in a negative electrospray ionization mode. The capillary voltage was fixed at 16 V, and its temperature was maintained at 200°C. The spray voltage was set at 4.25 kV. Liquid chromatography was carried out with an Agilent 1100 system. The sample was separated on a ZORBAX SB-C18 column (particle size, 5 μm; inside diameter, 2.1 by 150 mm; Agilent) and detected by a diode array detector (DAD UV6000). A mobile phase of 40% methanol and 60% water was used with a flow rate of 0.2 ml/min.
Restriction enzymes and chemicals.
All restriction enzymes, Taq and Pfu polymerases for PCR amplification, and T4 DNA ligase were purchased from Promega or TaKaRa. Nitrobenzene, 4-chloronitrobenzene, 2-aminophenol, 2-amino-5-chlorophenol, and other chemicals were purchased from Sigma or Fluka.
Nucleotide sequence accession numbers.
The DNA sequences reported here are available in GenBank under accession numbers AY731710 and DQ207951.
RESULTS
Genetic organization for 4-chloronitrobenzene degradation.
A previous study showed that genes encoding a 2-aminophenol 1,6-dioxygenase and a putative 2-aminohydroxymuconic semialdehyde dehydrogenase were located on a 15-kb fragment of plasmid pBG-2 (39). To identify other genes for the degradation of 4-chloronitrobenzene and nitrobenzene, this plasmid (pBG-2) and a second plasmid, pCG-13 (39), were sequenced with the shotgun method in this study. The results indicated that a ca. 34-kb DNA fragment of pCG-13 covered the 15-kb fragment of pBG-2. This 34-kb DNA fragment contained a genetic cluster (cnbR-orf1-cnbCaCbDEFGHI) that was putatively involved in 4-chloronitrobenzene and nitrobenzene degradation. Two other genes (cnbB-orf2-cnbA) were located on a separate DNA fragment of pBG-2 and were deduced to also be involved in the conversion of 4-chloronitrobenzene and nitrobenzene (Fig. 1). Based on BLAST searches and homology analyses, these genes and their putative functions were tentatively identified and are listed in Table 2.
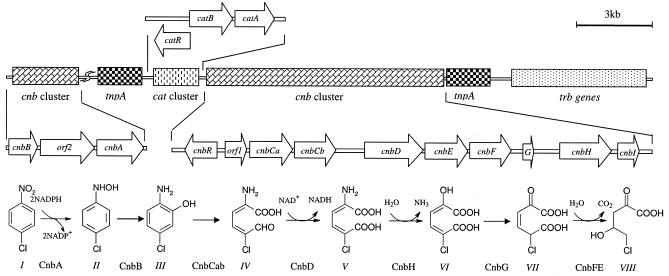
FIG. 1.
Genetic organization of cnb genes involved in 4-chloronitrobenzene (4-CNB) and nitrobenzene degradation and the modified partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. The genes involved in 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1 are organized in two clusters: a larger cluster (cnbR-orf1-cnbCaCbDEFGHI) includes the putative regulator gene (cnbR), the ring cleavage gene (cnbCab), and several genes of the lower pathway; and a smaller cluster (cnbB-orf2-cnbA) includes genes for the upper pathway that converts 4-chloronitrobenzene to 2-amino-5-chlorophenol and nitrobenzene into 2-aminophenol. The decarboxylation and hydration (from compound VII to compound VIII) was catalyzed by CnbE and CnbF, which was confirmed in recombinant E. coli by simultaneous expression of the two genes (data not shown). Upstream of the genetic cluster cnbR-orf1-cnbCaCbDEFGHI is a genetic cluster, catRBA, that putatively encodes the catechol pathway, and the function of catA, encoding catechol 1,2-dioxygenase, was confirmed by expression in E. coli (data not shown). Between the two cnb genetic clusters, there are some putative genes relating to gene transposition and plasmid conjugation. Arrows indicate the direction of transcription. Symbols: I, 4-chloronitrobenzene; II, 1-hydroxylamino-4-chlorobenzene; III, 2-amino-5-chlorophenol; IV, 2-amino-5-chloromuconic semialdehyde; V, 2-amino-5-chloromuconic acid; VI, 2-oxohex-4-ene-5-chloro-1,6-dioate; VII, 2-oxopent-5-chloro-3-enoate; VIII, 5-chloro-4-hydroxy-2-oxovaletate.
TABLE 2.
Annotation of genes involved in 4-chloronitrobenzene and nitrobenzene degradation and some properties of their encoded proteins
| Gene | Position in sequence (bp) | Gene product | Calculated mass, Da (no. of residues) | Homologous protein | Source | % Identity/no. of residues | Accession no. |
|---|---|---|---|---|---|---|---|
| cnbR | 330-845 | Regulatory protein | 18,761 (171) | NbzR | P. putida HS12/pNB1 | 64/114 | AAK26517 |
| MarR | Polaromonas sp. strain JS666 | 38/100 | EAM40420 | ||||
| orf1 | 1131-1550 | Putative ferredoxin | 15,130 (139) | ORF1 | P. pseudoalcaligenes JS45 | 57/125 | AAF03490 |
| NbzJ | P. putida HS12/pNB1 | 56/125 | AAK26518 | ||||
| NbzCa | P. putida HS12/pNB1 | 79/300 | AAK26519 | ||||
| cnbCa | 1623-2561 | 2-Aminophenol-1,6-dioxygenase | 35,037 (312) | AmnB | P. pseudoalcaligenes JS45 | 78/300 | AAB71524 |
| beta subunit | AmnB | Pseudomonas sp. strain AP-3 | 80/295 | BAB03531 | |||
| AmnA | P. pseudoalcaligenes JS45 | 60/271 | AAB71525 | ||||
| cnbCb | 2574-3389 | 2-Aminophenol-1,6-dioxygenase | 29,264 (271) | NbzCb | P. putida HS12/pNB1 | 60/271 | AAK26520 |
| alpha subunit | AmnA | Pseudomonas sp. strain AP-3 | 58/271 | BAB03532 | |||
| AmnC | Pseudomonas sp. strain AP-3 | 75/488 | BAB03533 | ||||
| cnbD | 4581-6053 | 2-Aminomuconic 6-semialdehyde | 54,026 (490) | NbzD | P. putida HS12/pNB1 | 75/487 | AAK26521 |
| dehydrogenase | AmnC | P. pseudoalcaligenes JS45 | 74/320 | AAC33839 | |||
| cnbE | 6197-7036 | 2-Keto-4-pentenoate hydratase | 29,781 (279) | TdnG | P. putida UCC22 | 65/261 | BAB62054 |
| cnbF | 7056-7844 | 4-Oxalocrotonate decarboxylase | 28,077 (262) | CdoK | Comamonas sp. strain JS765 | 69/227 | AAG17138 |
| AphH | Comamonas testosteroni TA441 | 69/227 | BAA88505 | ||||
| cnbG | 8097-8288 | 4-Oxalocrotonate tautomerase | 7,158 (63) | AphI | Comamonas testosteroni TA441 | 76/63 | BAA88507 |
| cnbH | 8728-10011 | 2-Amino-5-chloromuconic acid deaminase | 44,966 (427) | GatA | Agrobacterium tumefaciens C58 | 38/371 | NP_533743 |
| cnbI | 10054-10569 | 2-Oxopent-4-dienoate hydratase | 18,459 (271) | CbzJ | P. putida GJ31 | 98/165 | AAX38586 |
| TdnG | P. putida UCC22 | 37/166 | BAB62054 | ||||
| cnbA | 1869-2552 | 4-Chloronitrobenzene | 25,967 (227) | NbzA | P. putida HS12/pNB1 | 92/227 | AAK26512 |
| nitroreductase | NbzA | P. pseudoalcaligenes JS45 | 88/227 | AAT71308 | |||
| cnbB | 281-733 | Hydroxylaminobenzene mutase | 15,951 (150) | HabB | P. pseudoalcaligenes JS45 | 51/115 | AAB94123 |
| NbzB | P. putida HS12/pNB2 | 50/115 | AAK26516 |
Functional identification of cnbA, cnbB, cnbCa, cnbCb, and cnbD, and sequential conversion of 4-chloronitrobenzene into 2-amino-5-chloromuconic acid.
cnbA, cnbB, cnbCa, cnbCb, and cnbD exhibited high identities to the previously identified nbz or amn or nba genes of Pseudomonas putida strain HS12 (23), Pseudomonas pseudoalcaligenes strain JS45 (4, 29), Pseudomonas sp. strain AP-3 (24, 33, 34), and Pseudomonas sp. strain KU-7 (18), which were involved in degradation of nitrobenzene, aminophenol, or 2-nitrobenzoate (Table 2). Assimilation of chloronitrobenzenes by those strains was not reported, but the high identities of the cnb genes of strain CNB-1 indicated that these cnb genes might function similarly to the nbz, nba, or amn genes of strains HS12, JS45, KU-7, and AP-3.
The cnb genes were individually PCR amplified and cloned into pET21a, the plasmids generated containing each cnb gene (Table 1). E. coli BL21(DE3) harboring the pET derivatives was checked for synthesis of recombinant proteins and assayed for enzymatic activities. The results indicated that the cnbA, cnbB, cnbCab, and cnbD genes encoded chloronitrobenzene nitroreductase, hydroxylaminobenzene mutase, 2-aminophenol 1,6-dioxygenase, and 2-aminomuconic semialdehyde dehydrogenase, respectively (Table 2). Furthermore, when these enzymes were coupled in vitro, they sequentially catalyzed the conversions of 4-chloronitrobenzene to 2-amino-5-chloromuconic acid and nitrobenzene to 2-aminomuconic acid.
Gene cnbH encodes 2-amino-5-chloromuconic acid deaminase and its conversion into 2-hydroxy-5-chloromuconic acid.
The theoretical translational product of gene cnbH shows some identities to the genes encoding subunit A of glutamyl-tRNAGln amidotransferases (Table 2) and no significant identity to the deaminases from Pseudomonas sp. strains AP-3, HS12, and JS45. The entire cnbH was PCR amplified and cloned into pET-28a(+), and the resulting plasmid, pETcnbH, was transformed into E. coli. Recombinant E. coli cells synthesized a new protein with a molecular mass corresponding to the predicted CnbH (45 kDa).
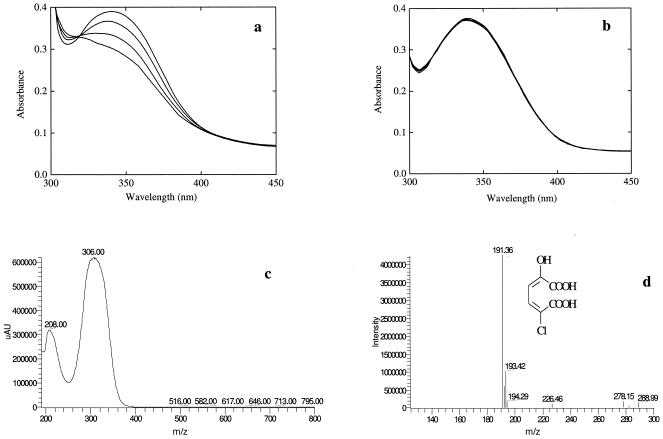
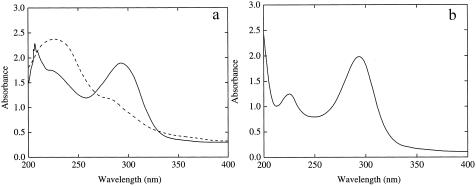
The recombinant CnbH functioned as deaminase (Fig. 2a and b) and catalyzed the conversion of 2-amino-5-chloromuconic acid into a product that had maximal absorption spectrum at 306 nm (Fig. 2c). This product was purified and subjected to LC-MS and spectrophotometric analyses and identified as 2-hydroxy-5-chloromuconic acid (Fig. 2d). Ammonia was nearly stoichiometrically released during the reaction, and 0.18 mM of ammonia was produced from 0.2 mM of 2-amino-5-chloromuconic acid. Similarly, this CnbH catalyzed the formations of 2-hydroxymuconic acid and ammonia from 2-aminomuconic acid. The product of 2-hydroxymuconic acid was isomerized by CnbG into 2-oxalocrotonate, as indicated by the shift of maximal absorption wavelength from 296 to 236 nm (Fig. 3), which was similarly reported by He and Spain (12).
FIG. 2.
Conversion of 2-amino-5-chloromuconic acid (2A5CM) into 2-hydroxy-5-chloromuconic acid (2H5CM), catalyzed by recombinant E. coli/pETcnbH that expressed 2-amino-5-chloromuconic acid deaminase (CnbH) (a) and by E. coli/pET28a as a control (b), and photospectrometry of 2-hydroxy-5-chloromuconic acid (c) and mass spectrometry of 2-hydroxy-5-chloromuconic acid (d). The photospectrum in a and b was recorded at 0 to 90 min after addition of cellular lysate. The specific activities for 2-aminomuconic acid were calculated to be 7 nmol min−1 (mg of protein)−1 for recombinant E. coli expressing CnbH and 120 nmol min−1 (mg of protein)−1 for Comamonas sp. strain CNB-1.
FIG. 3.
Conversion of 2-hydroxymuconic acid into 2-oxalocrotonic acid catalyzed by recombinant E. coli/pETcnbG (a) and by E. coli/pET21a as a control (b). The photospectrum was recorded at time zero and after addition of cellular lysate for 5 seconds.
Detection of plasmid in Comamonas sp. strain CNB-1 and localization of the pathway on the plasmid.
Many degradative pathways are encoded by genes on plasmids, and our previous work revealed that the gene for 2-aminophenol 1,6-dioxygenase was detected at relatively high frequency (three positive clones were obtained out of 300 clones) (39). This high recovery frequency stimulated us to consider that this gene had multicopies and was probably located on a multicopy plasmid. Plasmid curing from strain CNB-1 resulted in a mutant named strain CNB-2 that lost the ability to use 4-chloronitrobenzene and nitrobenzene for growth. Detection of a plasmid with the pulsed-field gel electrophoresis method and extraction of plasmid DNA both indicated that a plasmid of ca. 89 kb (pCNB1) existed in strain CNB-1 but not in CNB-2. The extracted plasmid DNAs were subjected to sequencing, and the results indicated that this pCNB1 was a circular plasmid of ca. 89 kb. The current data from the sequencing indicated that the genes for 2-aminophenol 1,6-dioxygenase (cnbCab), 4-chloronitrobenzene nitroreductase (cnbA), hydroxylaminobenzene mutase (cnbB), and 2-amino-5-chloromuconic acid deaminase (cnbH) were all located on plasmid pCNB1.
DISCUSSION
In this study, we found that both 4-chloronitrobenzene and nitrobenzene were degraded in Comamonas sp. strain CNB-1 via a partial reductive pathway that is similar to but different from that of the nitrobenzene degradative pathways in Pseudomonas sp. strains JS45 and HS12 (partial reductive pathway), and Comamonas sp. strain JS765 (oxidative pathway). The chloronitrobenzene pathway looked more like a combination of the upper pathway (nitroreduction and ring cleavage) of the Pseudomonas strains JS45 and HS12 and the lower pathway (after ring cleavage reactions) of Comamonas sp. strain JS765. Previously, Katsivela et al. (15) proposed a partial reductive pathway of bacterial strain LW1 for chloronitrobenzene degradation based on enzymatic activity assays and on identification of metabolic intermediates.
The novel pathway for chloronitrobenzene degradation in Comamonas sp. strain CNB-1 was identified at the genetic level in this study. The genes involved in the pathway were located on plasmid pCNB1, and two fragments related to chloronitrobenzene degradation were characterized. Among the genes located on the two fragments, seven (cnbA, cnbB, cnbCa, cnbCb, cnbD, cnbG, and cnbH) were functionally identified in recombinant E. coli and involved in chloronitrobenzene degradation. Four other genes (cnbR, cnbE, cnbF, and cnbI) were also tentatively identified as required for chloronitrobenzene degradation, according to the high identities to the genes whose functions are known in other bacteria. The functions of ORF1 and ORF2 were not clear. These nine genes (cnbA, cnbB, cnbCa, cnbCb, cnbD, cnbE, cnbF, cnbG, and cnbH) encoded enzymes that sequentially converted 4-chloronitrobenzene to 5-chloro-4-hydroxy-2-oxovalerate (Fig. 1). The cnbR gene encoded a putative regulator, but how it regulates the cnb genes is not clear at this stage.
The expression of cnbH in E. coli was successful in this study, but CnbH activity was much lower than that of the cellular lysate of Comamonas sp. strain CNB-1. This low activity raises the question of whether this CnbH could support the growth of strain CNB-1 on chloronitrobenzene or nitrobenzene. There might be two explanations for this: the expression of cnbH was not optimized in E. coli, or there was an alternative deaminase in Comamonas sp. strain CNB-1 that allowed this strain to grow on chloronitrobenzene or nitrobenzene. Nevertheless, the cnbH gene was interesting because it encoded a deaminase which is functionally similar to NbzE of strain HS12 (23) and AmnD of strain AP-3 (33) but showed no significant identity to NbzE of strain HS12 or AmnD of strain AP-3. Instead, it showed significant identities to some genes encoding Asp-tRNAAsn/Glu-tRNAGln amidotransferase subunit A (31 to 38%).
Evolutionary analysis indicated that CnbH was more related to Asp-tRNAAsn/Glu-tRNAGln amidotransferase A subunits than to deaminases (Fig. 4). As far as we know, the involvement of such a gene in the biodegradation of xenobiotic compounds has not been reported. We propose that CnbH might represent a novel type of deaminase in the degradation of xenobiotic compounds. The identification of novel genes that are involved in degradation of nitroaromatic compounds of short exposure in this and previous studies (18) should stimulate studies on genetic and metabolic pathway evolution. Currently, the details of the evolutionary relationship between this gene and nbzE/amnD and amidotransferase genes are under investigation.
FIG. 4.
Evolutionary dendrogram of deaminases involved in nitrobenzene and chloronitrobenzene degradation and homologs selected from results of a BLASTP search of GenBank, performed according to Altschul et al. (1). The phylogenetic tree was generated using the neighbor joining method of Saitou and Nei (26) with the AlignX software (Informax, Maryland), and multiple sequence alignment was done using ClustalX (37). The length of each branch pair represents the distance between the sequences.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30230010) and the Chinese Academy of Sciences (KSCX2-SW-113).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barton, B. M., P. H. Gordon, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Davis, J. K., G. C. Paoli, Z. He, L. J. Nadeau, C. C. Somerville, and J. C. Spain. 2000. Sequence analysis and initial characterization of two isozymes of hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenes JS45. Appl. Environ. Microbiol. 66:2965-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 6.El-Mansi, M., K. J. Anderson, C. A. Inche, L. K. Knowles, and D. J. Platt. 2000. Isolation and curing of the Klebsiella pneumoniae large indigenous plasmid using sodium dodecyl sulphate. Res. Microbiol. 151:201-208. [DOI] [PubMed] [Google Scholar]
- 7.Groenewegen, P. E., P. Breeuwer, J. M. van Helvoort, A. A. Langenhoff, F. P. de Vries, and J. A. de Bont. 1992. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J. Gen. Microbiol. 138:1599-1605. [DOI] [PubMed] [Google Scholar]
- 8.Haigler, B. E., W. H. Wallace, and J. C. Spain. 1994. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol. 60:3466-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa, Y., T. Muraki, T. Tokuyama, H. Iwaki, M. Tatsuno, and P. C. K. Lau. 2000. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol. Lett. 190:185-190. [DOI] [PubMed] [Google Scholar]
- 10.He, Z., and J. C. Spain. 1997. Studies of the catabolic pathways of degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45: removal of the amino group from 2-aminomuconic semialdehyde. Appl. Environ. Microbiol. 63:4839-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Z., and J. C. Spain. 1998. A novel 2-aminomuconate deaminase in the nitrobenzene degradation pathway of Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 180:2502-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, Z., and J. C. Spain. 1999. Comparison of the downstream pathways for degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45 (2-aminophenol pathway) and by Comamonas JS765 (catechol pathway). Arch. Microbiol. 171:309-316. [DOI] [PubMed] [Google Scholar]
- 13.He, Z., J. K. Davis, and J. C. Spain. 1998. Purification, characterization, and sequence analysis of 2-aminomuconic 6-semialdehyde dehydrogenase from Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 180:4591-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, Z., L. J. Nadeau, and J. C. Spain. 2000. Characterization of hydroxylaminobenzene mutase from pNBZ139 cloned from Pseudomonas pseudoalcaligenes JS45. A highly associated SDS-stable enzyme catalyzing an intramolecular transfer of hydroxy groups. Eur. J. Biochem. 267:1110-1116. [DOI] [PubMed] [Google Scholar]
- 15.Katsivela, E., V. Wray, D. H. Pieper, and R. M. Wittich. 1999. Initial reactions in the biodegradation of 1-chloro-4-nitrobenzene by a newly isolated bacterium, strain LW1. Appl. Environ. Microbiol. 65:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lessner, D. J., R. E. Parales, S. Narayan, and D. T. Gibson. 2003. Expression of the nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds. J. Bacteriol. 185:3895-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meulenberg, R., M. Pepi, and J. A. de Bont. 1996. Degradation of 3-nitrophenol by Pseudomonas putida B2 occurs via 1,2,4-benzenetriol. Biodegradation 7:303-311. [DOI] [PubMed] [Google Scholar]
- 18.Muraki, T., M. Taki, Y. Hasegawa, H. Iwaki, and P. C. K. Lau. 2003. Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 69:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadeau, L. J., Z. He, and J. C. Spain. 2003. Bacterial conversion of hydroxylamino aromatic compounds by both lyase and mutase enzymes involves intramolecular transfer of hydroxyl groups. Appl. Environ. Microbiol. 69:2786-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino, S. F., and J. C. Spain. 1995. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl. Environ. Microbiol. 61:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino, S. F., G. C. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishino, S. F., and J. C. Spain. 1993. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, H.-S., and H.-S. Kim. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, H. S., and H. S. Kim. 2001. Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process. J. Bacteriol. 183:5074-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, H. S., S. J. Lim, Y. K. Chang, A. G. Linvingston, and H. S. Kim. 1999. Degradation of chloronitrobenzenes by a coculture of Pseudomonas putida and a Rhodococcus sp. Appl. Environ. Microbiol. 65:1083-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sinnett, D., C. Richer, and A. Baccichet. 1998. Isolation of stable bacterial artificial chromosome DNA using a modified alkaline lysis method. BioTechniques 24:752-754. [DOI] [PubMed] [Google Scholar]
- 29.Somerville, C. C., S. F. Nishino, and J. C. Spain. 1995. Purification and characterization of nitrobenzene nitroreductase from Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 177:3837-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spain, J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523-555. [DOI] [PubMed] [Google Scholar]
- 31.Spain, J. C., and D. T. Gibson. 1991. Pathway for biodegradation of para-nitrophenol in a Moraxella sp. Appl. Environ. Microbiol. 57:812-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spanggord, R. J., J. C. Spain, S. F. Nishino, and K. E. Mortelmans. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takenaka, S., S. Murakami, Y.-J. Kim, and K. Aoki. 2000. Complete nucleotide sequence and functional analysis of the genes for 2-aminophenol metabolism from Pseudomonas sp. AP-3. Arch. Microbiol. 174:265-272. [DOI] [PubMed] [Google Scholar]
- 34.Takenaka, S., S. Murakami, R. Shinke, and K. Aoki. 1998. Metabolism of 2-aminophenol by Pseudomonas sp. AP-3: modified meta-cleavage pathway. Arch. Microbiol. 170:132-137. [DOI] [PubMed] [Google Scholar]
- 35.Thiele, J., R. Mueller, and F. Lingens. 1988. Enzymatic dehalogenation of chlorinated nitroaromatic compounds. Appl. Environ. Microbiol. 54:1199-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, N. R., S. Koshy, M. Simsek, and A. K. Abraham. 1988. A precaution when preparing very large plasmids by alkaline lysis procedure. Appl. Biochem. Biotechnol. 10:402-407. [PubMed] [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J.-F., X.-H. Shen, Y.-G. Zhou, and S.-J. Liu. 2004. Characterization of p-chloronitrobenzene-degrading Comamonas CNB-1 and its degradation of p-chloronitrobenzene. Acta Microbiol. Sin. 44:8-12. [Google Scholar]
- 39.Wu, J.-F., C.-W. Sun, C.-Y. Jiang, Z.-P. Liu, and S.-J. Liu. 2005. A novel 2-aminophenol 1,6-dioxygenase involved in the degradation of p-chloronitrobenzene by Comamonas sp. strain CNB-1: purification, properties, genetic cloning and expression in Escherichia coli. Arch. Microbiol. 183:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Zeyer, J., and P. C. Kearney. 1984. Degradation of ortho-nitrophenol and meta-nitrophenol by a Pseudomonas putida. J. Agric. Food Chem. 32:238-242. [Google Scholar]