Abstract
The species diversity, phylogenetic affiliations, and physiological activity rates of carbon monoxide-oxidizing microorganisms were investigated, using new isolates from surface waters collected from the coast of New England and type strains from established collections. A direct isolation method allowed the simultaneous recovery of organisms with different growth rates and nutritional requirements and the identification of marine microorganisms that oxidize CO at an environmentally relevant concentration (42 nM CO). Isolates that oxidized CO at environmentally relevant rates (>4.5 × 10−11 nmol CO oxidized cell−1 h−1) were taxonomically diverse, with representatives in the alpha and gamma subclasses of the Proteobacteria and the phylum Bacteroidetes, and represent a hitherto unreported metabolic function for several diverse microbial types. Isolates and type strains having the greatest specific rates of CO metabolism (1.1 × 10−10 to 2.3 × 10−10 nmol CO oxidized cell−1 h−1) belonged to the Roseobacter-associated clade (RAC) of the alpha subclass of the Proteobacteria. By using triple-labeled slide preparations, differential counts of active CO-oxidizing RAC cells, total RAC cells, and total bacterial cell counts in environmental samples were obtained. RAC organisms were a major component of total cell numbers (36%). Based on the density of active CO-oxidizing RAC cells in natural samples and RAC-specific metabolic activities determined for pure cultures, active CO-oxidizing RAC cells may contribute up to 15% of the total CO oxidation occurring in coastal waters.
Sunlight-initiated photodegradation of colored dissolved organic matter is primarily responsible for producing carbon monoxide in sunlit waters (27). The surface waters of the world's oceans are saturated with CO with respect to the atmosphere and are therefore a source of atmospheric CO (8, 32). It is likely that microorganisms within the water column consume most autochthonous CO (7), and several biogeochemical studies have estimated the diel, annual, and global rates of microbial CO oxidation as an oceanic CO sink (7, 8, 17, 18, 38, 39). CO production is typically greater near coastal waters than at open-ocean sites, due to higher colored dissolved organic matter content (19). The mid-day CO concentration of 12 nM (36, 37) at our coastal sampling location in Vineyard Sound, Mass., for example, is up to fivefold higher than the maximum measured in the Sargasso Sea (18, 39). Microbial CO oxidation rate coefficients for coastal waters (0.01 to 0.11 h−1) can be an order of magnitude greater than those measured in oligotrophic environments (0.01 to 0.02 h−1) (36, 37), suggesting the presence of an active CO-oxidizing microbial community near shore.
The question of which microorganisms are important contributors to CO bio-oxidation observed in coastal or oceanic surface waters has not been satisfactorily resolved. The identities of the microbes responsible for CO bio-oxidation in marine environments remain unknown, and there exists only circumstantial evidence for CO-metabolizing marine organisms based on the ancillary ability of certain bacteria to metabolize CO. The apparent half-saturation constant (Kapp) values for CO of carboxydotrophic organisms obtained in previous isolation programs from terrestrial enrichments containing >500 nM CO (6, 25) have been 1 to 2 orders of magnitude higher than the average Kapp values for CO consumption in soil or freshwater (5 to 50 nM) (6, 8, 20) or in marine environments (2.0 to 5.4 nM) (36, 37). The high Kapp values for CO reported for most terrestrial carboxydotrophic strains are likely a bias created by the high [CO] used in the enrichments and for their isolation. This highlights the importance of cultivating CO-oxidizing bacteria at CO concentrations close to environmental levels to obtain microbes that are most active in oxidizing CO in the natural setting.
Providing an atmosphere enriched with CO and removing dissolved organic substances specify the conditions for the growth of facultative lithotrophs that are able to utilize CO oxidation as a source for energy, while selecting against a vast majority of CO-sensitive organisms. One objective of this study was to provide growth conditions for the cultivation of organisms capable of CO metabolism under slightly elevated environmental levels of CO [50 ppmv CO:air = ∼42 nM CO (aq)] and in an oligotrophic mineral medium. Macroautoradiographic screening of colonies grown in the above atmosphere containing tracer levels of 14CO allowed the detection and isolation of microorganisms incorporating labeled carbon from CO and initial estimates of their abundance. The identification of some marine microorganisms that oxidize CO at environmentally relevant concentrations and their specific CO oxidation activities are provided in this study.
A second objective was to determine the proportion of CO-metabolizing cells on a group (genus)-specific level by combining microautoradiography with in situ hybridization (26), using oligonucleotide probes that target the phylogenetic group represented most by the inferred genetic identities of isolated CO-oxidizing strains. The substrate-tracking microautoradiography-fluorescent in situ hybridization (STAR-FISH) assay is a triple-labeling technique that allows simultaneous microscopic visualization of (i) DNA-stained cells (stained with 4′,6′-diamidino-2-phenylindole [DAPI]), (ii) cells that have incorporated labeled carbon from 14CO and have formed silver grains in microradiographic emulsion, and (iii) cells which have hybridized with a fluorescently labeled oligonucleotide probe (23) specific for the Roseobacter-associated clade (RAC) (3). We determined the proportion of RAC cells in a natural seawater assemblage and the distribution of substrate uptake within the target group. In combination with activity measurements of CO oxidation by RAC seawater isolates and type cultures, we determined the proportion of total environmental CO oxidation that can be attributed to RAC microorganisms.
Organisms of the Roseobacter-associated clade are emerging as numerically dominant and metabolically versatile in various marine habitats. It is now apparent that some otherwise well-characterized microorganisms within the RAC have a previously unrecognized capability and play an important role in the cycling of CO in the marine environment (28). This seemingly ubiquitous group of microorganisms may significantly contribute to the bio-oxidation of autochthonous CO in coastal marine environments, and their potential CO metabolic activity has previously been overlooked in other characterization studies. The ability of some species within the RAC to oxidize CO has only recently been documented (28), and we confirmed this phenotype of RAC directly in environmental samples by STAR-FISH. The proportion of total in situ CO oxidation for which RAC organisms are accountable has hitherto been unreported.
MATERIALS AND METHODS
Sampling.
The primary coastal sampling site for this study was the WHOI Shore Lab 1 km east of Nobska Light, Woods Hole, on a southeast-facing beach on Vineyard Sound, Cape Cod, Massachusetts. This location was chosen for its distance away from coastal pond and freshwater discharge (1 km), proximity to the laboratory, and ease of sampling. Syringe or bottle samples were collected 0.5 m below the surface to avoid atmospheric CO contamination. The water depth at the sampling site was approximately 1 m.
Isolation program.
Using a modification of the method of Dunbar et al. (10), the microbial assemblage in seawater was deposited by filtration onto autoclaved 47-mm-diameter, 0.22-μm polycarbonate membrane filters previously washed with methanol and distilled water to remove potential organic contaminants. The filters were placed atop a combusted glass-fiber filter saturated with liquid oligotrophic growth medium that consisted of a six-salt base (0.52 M NaCl, 14.8 mM MgCl2 · 6H2O, 24 mM MgSO4 · 7H2O, 6.8 mM KCl, 2.4 mM NaHCO3, 2.0 mM CaCl2 · 2H2O, 0.2 mM K2HPO4) and 1 ml liter−1 Pfennig's SL-8 solution (29). A trace element solution which included molybdenum (0.15 μM Na2MoO4 · 2H2O [final concentration]) and selenium (0.12 μM Na2SeO [final concentration]) was provided in the medium for the formation of fully active carbon monoxide dehydrogenase (25). In the event that there existed a group of organisms in natural coastal seawater that cooxidized CO while obtaining their energy requirements from unknown organic substrates contained in seawater, a set of membrane incubations were run, using natural filtered (0.22 μm) coastal seawater from the sampling site as the medium.
Membranes containing filtered samples were incubated in a 14CO-containing atmosphere for sufficient time (>2 weeks) to allow for the induction of enzymes necessary for CO metabolism and visible colony formation. The atmosphere was maintained by injecting the 14CO and air mixture into an evacuated desiccator containing the samples. A slight negative pressure maintained within the chamber provided a tight seal. 14C-labeled CO was generated by dehydration of 14C-labeled formate, as described previously (37). 14C activity was measured via liquid scintillation spectroscopy in the laboratory (Scintiverse II scintillation fluid).
Macroautoradiographic screening.
After visible colonies had developed on the filters (master filters), they were duplicated by placing them colony side up on a sterile square of Whatman no. 1 filter paper. A sterile, moistened 0.22-μm Nuclepore filter was placed atop each master filter, covered with sterile Whatman filter paper, and evenly compressed with a surface-sterilized polycarbonate disk. Reference notches were made in the master and duplicate filters with a sterile razor blade to record their relative orientations. Filter pairs were separated, and each was placed colony side up on fresh mineral medium and regrown for several days in a 14CO-containing atmosphere of the same composition used to support original growth. Following the second growth period, the duplicate filters were removed from the medium and allowed to air dry. Using a sterile toothpick, the duplicate filters were spotted with NaH14CO3 tracer at the locations of the registration notches. The dry duplicate filters were attached colony side up to 8-in. by 10-in. sheets of absorbent Bench-Kote paper with small pieces of transparent tape at the edges of the filter. In a photographic darkroom, an autoradiography cassette was prepared by overlaying the filter-containing 8-in. by 10-in. sheets with a Kodak BioMax LE (low energy) intensifying screen, followed by Kodak XAR paper placed on top of the intensifying screen. The film was exposed for 5 to 7 days at −80°C and developed under darkroom conditions following standard procedures for X-ray film development (3 min with developer, 30-s water rinse, 3 min with acid fixer, and 20-min water wash). The 14C-labeled orientation notches visible on the autoradiograph were used to orient corresponding master filters. When viewed on a light table, the exposed areas on the radiographic film revealed the locations of 14C-positive colonies on the master filters. Duplicate filters were wrapped with cellophane and archived.
Colonies that were positive for 14C uptake and exposed on the X-ray film were selected from the master filters and streaked onto the solid mineral medium originally used for isolation, i.e., six-salt base plus trace elements (described above), amended with either nitrate (3.06 mM) or ammonium (3.04 mM), or onto 2216 marine agar. The mineral medium plates yielded no further detectable growth; only the marine agar plates supported visible colonies. Isolates selected from the marine agar plates then underwent a second round of 14CO incubation and autoradiography, as described above, to confirm CO-borne carbon assimilation conferring the original autoradiographic signal. Fifty microliters of turbid liquid culture (2216 marine broth) was diluted in 3 ml aged, filtered (0.22 μm) Sargasso seawater for filter deposition onto 25-mm-diameter, 0.22-μm Nuclepore filters. Each filter was placed on a GFF pad saturated with either 2216 marine broth or mineral medium amended with nitrate or ammonium. Membranes were incubated in a 14CO-containing atmosphere (50 ppm CO) for 1 week and used for exposure on XAR film as previously described.
Specific CO oxidation rates of environmental isolates and type cultures.
The type cultures assayed for CO oxidation activity were Paracoccus versatus DSMZ-582, Ruegeria atlantica DSMZ-5823, Ruegeria gelatinovorans DSMZ-5887, Paracoccus denitrificans DSMZ-7001, Roseobacter litoralis DSMZ-6996, Paracoccus alcaliphilus DSMZ-8512, Paracoccus aminophilus DSMZ-8538, Ruegeria algicola DSMZ-10253, Paracoccus pantotrophus DSMZ-11072, Silicibacter lacuscaerulensis DSMZ-11314, Sulfitobacter brevis DSMZ-11443, Roseovarius tolerans DSMZ-11457, Sulfitobacter mediterraneus DSMZ-12244, Roseobacter gallaeciensis DSMZ-12440, Pseudomonas aeruginosa ATCC 10145, and an unidentified organism (LFR), ATCC 51258 (W. Dacey, personal communication). Hydrothermal vent strains NF18, TB66, AIII3, DI4, and DIII4 were selected from the Woods Hole Oceanographic Institution collection (34). Silicibacter pomeroyi strain DSS-3, Silicibacter lacuscaerulensis, Roseovarius nubinhibins ISM, and Sulfitobacter sp. strain EE36 were supplied by Mary A. Moran, University of Georgia.
To determine cell-specific rates of CO oxidation, we utilized the 14CO oxidation method. Pure isolates were grown for 1 to 2 days in 15 ml 2216 marine broth to a light turbidity (1 × 107 cells ml−1). Liquid cultures were centrifuged for 10 min at 10,000 rpm, and the supernatants were decanted. Each pellet was resuspended in 15 ml filtered (0.22 μm) Sargasso seawater as a rinse and then centrifuged for 10 min. The rinse was repeated twice, and the pellet was resuspended in 150 ml of aged, filtered (0.22 μm) Sargasso seawater to a final density of 1 × 106 cells ml−1. Unlabeled CO was injected into the headspace of each sealed bottle in an amount sufficient to maintain a dissolved [CO] of approximately 40 nM to induce CO metabolic enzymes. The cell density of this suspension was determined by direct microscopy (16) after 2 days of incubation with CO and immediately prior to the 14CO oxidation assay. One-milliliter aliquots of 14CO stock (∼45 mCi mmol−1) were injected into eight septum-sealed vials, each containing 25 ml of cell suspension, resulting in a dissolved [CO] of 1.5 nM. Paired subsamples were sacrificed at 2-h intervals over an 8-h incubation period. 14C activity was assessed in the sacrificed subsamples, where the 14CO2 product was released, trapped, and assayed by previously described methods (15). Specific CO oxidation rates (Rsp) were calculated on a per-cell basis for each isolate (nmol CO oxidized cell−1 h−1) by use of the following equation: Rsp = n25 × (25 × N × t)−1, where n25 is the molar amount of CO oxidized in each 25-ml sample (nmol), N is the cell density (cells ml−1), and t is the duration of incubation (h).
Molecular analysis by PCR amplification of 16S rRNA gene.
CO-metabolizing microorganisms were isolated, and the 16S rRNA gene was amplified by PCR using the universal bacterial primers 8f and 1492r (22). Isolated strains provided amplifiable templates after picking of solitary colonies with a sterile toothpick and incubation for 2 min at 95°C in 5 μl Lyse-N-Go PCR reagent (Pierce) prior to the addition of primers, nucleotides, and Taq polymerase. In all amplification runs, a negative control containing no template and a positive control using 0.5 μl of a 1 × 107 cell ml−1 Escherichia coli liquid culture were run to assess reagent quality and freedom from contamination. PCR products were cleaned using a QIAquick PCR purification kit (QIAGEN). Sequencing was performed with an ABI PRISM 3700 DNA analyzer/sequencer located at the Josephine Bay Paul Center at the Marine Biological Laboratory, Woods Hole, Mass. Sequences from 30 isolates were edited and assembled using the program Sequencher 4.0.5 (Gene Codes Corporation, Ann Arbor, MI). Sequencing resulted in double-stranded sequence fragments that were checked against GenBank using the BLAST search program (1) for identification.
The ClustalW software package (35) was used for sequence alignment and construction of a neighbor-joining tree with 1,000 bootstrap replicates. The aligned sequences of the experimental isolates and closely related organisms were imported into PAUP, version 4.0b10 (Sinauer Associates, Sunderland, Mass.), for further phylogenetic analyses. Trees constructed with other reconstruction algorithms (parsimony and maximum likelihood) resulted in the same overall topology, with few minor rearrangements within the Roseobacter clade. The phylogenetic trees were viewed with the TREEVIEW (Win32) program, version 1.6.5.
STAR-FISH enumeration. (i) Direct counting.
Seawater samples (1.8 ml) were stained for 5 min by the addition of DAPI (final concentration, 1.0 μg/ml) within 1 hour of sample collection. The cells were gently vacuum filtered (5 mm Hg) onto a Sudan Black-stained 0.22-μm Nuclepore membrane and enumerated by epifluorescence microscopy (16).
(ii) FISH.
Liquid suspensions of the type species Paracoccus denitrificans (DSMZ-6610) and the type strain of Roseobacter litoralis (DSMZ-6996) grown in 2216 marine broth for 24 h at 20°C were used to determine optimal stringency conditions for FISH assays. The cultures were centrifuged at 9,000 rpm for 5 min, and the pellets were resuspended in filtered (0.22 μm) coastal seawater to rinse. The suspension was again centrifuged at 9,000 rpm for 5 min, and the pellets were resuspended in 250 μl (pH 7.2) or 750 μl 4% paraformaldehyde solution. FISH was performed under various hybridization conditions to find the optimal formamide concentration for discrimination between Roseobacter target cells and non-RAC Paracoccus cells having a single and consistent mismatch (indicated in italics below) to the probe sequence. The Roseobacter group-specific oligonucleotide probe used for hybridization was ROSEO536R (3). The probe sequence, 5′-CAACGCTAACCCCCTCCG-3′, was constructed with a 5′ Cy3 label and purified by high-performance liquid chromatography by Thermo Hybaid (formerly Interactiva, Ulm, Germany). A high formamide concentration (40%) in the hybridization buffer allowed discrimination between Roseobacter target cells and Paracoccus cells having the single mismatch to the labeled probe. Use of the competitor probe ROSEOC536R (5′-CAACGCTAGCCCCCTCCG-3′) (3) further suppressed false-positive signals. The ROSEO536R probe specificity was checked against the RDP II database (http://rdp.cme.msu.edu/), using the “Probe Match” function, and was found to be highly specific for RAC. Type discrimination was confirmed via FISH with a mixed binary culture of Roseobacter litoralis and Paracoccus denitrificans and the ROSEO536R/ROSEOC536R probe combination. The specificity of the competitor probe ROSEOC536R was checked against the RDP II database, and the probe was inclusive of Paracoccus and several other alphaproteobacterial sequences, including those from Sphingomonas, Rhodobacter, Rhodovulum, and Caulobacter. Specific counts of Roseobacter using the ROSEO536R/ROSEOC536R combination are therefore well supported.
Filter mounts of the environmental microbial community were prepared by mixing of 20 μl of prefiltered (10 μm) coastal seawater with 1 ml 1× phosphate-buffered saline and gentle vacuum (−5 lb/in2) filtration onto 0.22-μm Nuclepore filters. The filters were placed temporarily on a Whatman no. 4 filter paper pad saturated with 4% paraformaldehyde solution in filtered seawater and cut in half with a sterile razor blade. Wet filter halves were mounted sample side down onto a layer of 0.1% gelatin that was deposited onto the surfaces of ethanol-cleaned slides and allowed to completely air dry. The filters were gently peeled from the gelatin layer, leaving the filtered cells partially embedded in the gelatin layer.
Fluorescent in situ hybridization was performed in accordance with a previously published protocol (30), with stringency conditions determined empirically (40% formamide concentration at 46°C). Hybridizations were performed by the application of 20 μl hybridization buffer (18 μl hybridization buffer, 1 μl ROSEO536R, and 1 μl ROSEOC536R [final probe concentration, 10 ng μl−1]) directly onto the embedded samples, which were then covered with coverslips. Hybridized slides were air dried in the dark, treated with 20 μl of a 1.0-μg μl−1 DAPI solution for 1 min in the dark, given a final rinse by soaking in double-distilled H2O for 5 min in a staining jar, and allowed to air dry again. Dry samples were mounted in Citifluor and covered with glass coverslips.
Autoradiographic procedure.
For STAR-FISH performed with environmental samples, 100 ml of filtered (10 μm) sample water was sealed within a 120-ml glass serum vial and injected with 14CO to result in a dissolved [CO] of 42 nM. Subsamples were removed at 0 h, 2 h, 4 h, and 6 h and were fixed with formaldehyde to a final concentration of 4%. These subsamples were filtered onto 0.22-μm filters and transferred to emulsion-coated glass microscope slides to undergo STAR-FISH. The uptake of labeled substrate by microorganisms was determined by a microautoradiographic technique based primarily on the “MARGE-E” procedure developed by Tabor and Neihof (33) and the general procedures for autoradiography with liquid emulsions described by various authors (2). LM-1 hypercoat emulsion was the photographic medium used in our STAR-FISH assays (Amersham Pharmacia Biotech). An ethanol-cleaned 3-in. by 1-in. glass microslide was dipped in 0.1% gelatin at 46°C to create a gelatin sublayer. After the gelatin layer was dry, the coated slide was immersed for 15 s in diluted LM-1 (1 part emulsion to 1 part 0.1% gelatin solution improved cell adhesion compared to layering with emulsion only) maintained at 46°C under darkroom conditions (Kodak safelight filter GBX-2 placed 1 m away from the workspace). Later, a NightVision head-mounted infrared illumination system was used in lieu of the safelight when manipulating unexposed emulsion-coated slides so as to further reduce the autoradiographic background. The slide was drained horizontally for 20 s to produce a thin emulsion film. The emulsion was allowed to dry in the dark for 5 min at room temperature. Wet sample filters were applied with the sample side placed on the emulsion film carefully to exclude bubbles and to avoid lateral movement of the filter against the semisolid emulsion. The slides were placed in a light-tight slide box containing a packet of silicon desiccant and allowed to set for another 3 h, following the manufacturer's recommendations. After the emulsion was completely dry, the wooden light-tight slide box was wrapped in aluminum foil and placed at 4°C for radiographic exposure.
After the emulsion slides had been exposed for 2 days, they were developed using a standard photographic procedure. In the dark, slides were immersed in a staining jar containing Kodak GBX developer at ambient lab temperature for 5 min, the reactions were stopped with 0.5% acetic acid in deionized water as a stop solution for 30 s, and the slides were fixed for 10 min in Kodak GBX fixer solution. The developer and fixer solutions were diluted per the manufacturer's directions. The slides were rinsed for 20 min in gently flowing tap water in a destaining tray. After being air dried, the filter was gently peeled away from the emulsion film. Cells were dehydrated in an ethanol series (50%, 80%, and 98% aqueous ethanol [vol/vol]; 3 min each) (30). Developed emulsion slides were subjected to FISH and DAPI stained as previously described.
Microscopy.
Microscopic counts were made using a Zeiss Axioplan 2 imaging microscope with a Zeiss αPlan-FLUAR 100× oil objective. DAPI counts were made with a UV lamp and a filter set (Choma Technology Corp.) comprised of exciter D360/40 and emitter GG420. Cy3 counts were made with an Endow GFP long-pass emission filter set (Choma Technology Corp.) comprised of exciter HQ470/40 and emitter HQ500lp. Autoradiographic counts were made with transmitted incandescent light simultaneously with epifluorescent fields. For each microscopic field, Cy3 signals, DAPI signals, and Cy3/emulsion exposures were counted and recorded. Ten random microscopic fields were counted on each slide for statistical purposes.
Calculations.
The in situ cell density for each probe-targeted group was estimated by calculating the ratio of Cy3 signals to DAPI signals per microscopic field. This ratio was multiplied by the DAPI direct count of the sample, resulting in a group-specific cell density for RAC cells in the natural sample. The ratio of Cy3- and 14C-positive cells to DAPI-positive cells on the STAR-FISH slides multiplied by the cell density, determined by separate DAPI direct counts, yielded the relative density of active CO-metabolizing cells belonging to the Roseobacter group within the incubated sample. These activity- and group-specific cell densities (cells ml−1) were multiplied by the specific CO oxidation activity term (nmol CO oxidized cell−1 h−1), incubation time (h), and sample volume (25 ml) to calculate the relative proportion of total CO oxidation that can be attributed to RAC organisms.
Nucleotide sequence accession numbers.
16S rRNA gene sequences for all CO-metabolizing strains resulting from this study were submitted to GenBank under accession numbers AY349458 to AY349466.
RESULTS
Isolation, identity, and phylogeny of CO-oxidizing marine microbes.
Positive signals on XAR film were counted after 2 weeks of incubation in a 14CO-containing atmosphere at environmentally relevant levels of CO (50 ppmv) to yield the CFU density (CFU ml−1) that had incorporated 14C from CO. In autoradiographic plate counts, the CO-active CFU density was 395 ± 55 CFU ml−1 for Vineyard Sound water (mean ± standard error of the mean [SEM]; n = 6). Direct counts of Vineyard Sound water showed a total cell density of 2.71 × 106 ± 5.5 × 104 cells ml−1 (mean ± SEM; n = 10). As expected, only a small fraction of cells present in the sample (<0.01% of the total) grew to visually distinct colonies under the experimental growth conditions, e.g., low organic carbon and elevated CO. Thirty colonies conferring distinct autoradiographic signals on XAR film were selected for purification and further CO oxidation analyses.
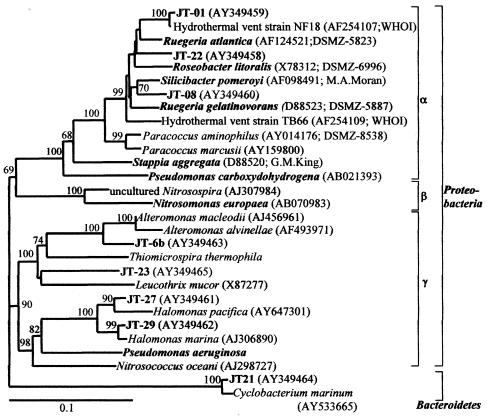
16S rRNA gene sequences were determined for 30 isolated strains, and putative identifications based on sequence and BLAST search returns were compiled. While all isolates were tolerant to elevated concentrations of CO (40 to 50 nM) and were phylogenetically diverse, none of the 21 previously described “carboxydotrophic” organisms or ammonium monooxygenase-containing ammonia oxidizers known to also oxidize CO in pure cultures are represented among our isolates. Furthermore, isolates initially recovered on oligotrophic medium did not survive multiple transfers to oligotrophic medium; they were revived and maintained on 2216 marine agar. Phylogenetic analysis based on the 16S rRNA gene sequences of positive CO-oxidizing isolates placed the most active strains (JT-01, JT-08, and JT-22) in the Roseobacter-associated clade of the alpha subclass of the Proteobacteria (Fig. 1). We obtained and tested type strains belonging to the RAC for CO oxidation activity, using the 14CO oxidation assay, and found three other RAC organisms that oxidized CO at environmentally relevant levels, namely, Roseobacter litoralis, Ruegeria atlantica, and Ruegeria gelatinovora, and we also confirmed the phenotype in Silicibacter pomeroyi (28). Other strains isolated in this study are representatives of the gamma subclass of Proteobacteria (JT-6b, JT-11, JT-23, JT-27, and JT-29) and the phylum Bacteroidetes (JT-21) (Fig. 1).
FIG. 1.
Phylogenetic tree based on the neighbor-joining method for partial 16S rRNA gene sequences showing the relationship of CO-oxidizing isolates (with a “JT-” prefix) to select cultured and environmental sequences. Labels depicted in bold are CO-oxidizing isolates and type cultures or organisms that have been previously implicated in environmental CO oxidation. Bootstrap values above 50% are shown at the nodes. Sequence accession numbers and culture sources or references follow parenthetically. Bar, 10% estimated change per nucleotide.
CO oxidation activities of isolated strains.
The specific CO oxidation activity was determined for each isolate in mineral medium amended with either nitrate or ammonium. The mean specific CO oxidation activity of all CO-oxidizing isolates was 1.17 × 10−10 ± 2.19 × 10−11 nmol CO oxidized cell−1 h−1 (mean ± SEM; n = 10), and values ranged between 3.4 × 10−11 and 2.3 × 10−10 nmol CO oxidized cell−1 h−1. The mean cell-specific activity of the seven CO-oxidizing RAC strains assayed (JT-01, JT-08, JT-22, Roseobacter litoralis, Ruegeria gelatinovora, Silicibacter pomeroyi, and Ruegeria atlantica) was 1.2 × 10−10 ± 2.2 × 10−11 nmol CO cell−1 h−1 (mean ± SEM; n = 10). The mean CO oxidation activity of the two most active isolates, JT-01 and JT-08, was 1.9 × 10−10 ± 2.3 × 10−11 nmol CO oxidized cell−1 h−1 (n = 4) (Table 1).
TABLE 1.
Specific CO oxidation rates of CO-oxidizing microorganisms isolated during this study, RAC type cultures obtained from various sources, and some known carboxydotrophic organisms
| Organism or isolate no. | Nitrogen source | CO oxidation rate (nmol CO cell−1 h−1) (mean ± SEM)a | Source and/or reference(s) |
|---|---|---|---|
| JT-01 | -NO3− | 2.2 × 10−10 ± 3.1 × 10−12 | This study |
| -NH4+ | 2.3 × 10−10 ± 1.2 × 10−11 | This study | |
| JT-6b | -NO3− | 4.3 × 10−11 ± 1.2 × 10−13 | This study |
| -NH4+ | 4.6 × 10−11 ± 3.2 × 10−12 | This study | |
| JT-08 | -NO3− | 2.0 × 10−10 ± 1.1 × 10−11 | This study |
| -NH4+ | 1.1 × 10−10 ± 4.4 × 10−12 | This study | |
| JT-22 | -NO3− | 1.1 × 10−10 ± 3.1 × 10−12 | This study |
| -NH4+ | 7.4 × 10−11 ± 3.9 × 10−12 | This study | |
| JT-23 | -NO3− | 3.0 × 10−11 ± 2.3 × 10−13 | This study |
| -NH4+ | 4.3 × 10−11 ± 5.9 × 10−13 | This study | |
| JT-27 | -NO3− | 5.9 × 10−11 ± 1.4 × 10−11 | This study |
| -NH4+ | 6.6 × 10−11 ± 1.7 × 10−11 | This study | |
| JT-29 | -NO3− | 5.5 × 10−11 ± 4.7 × 10−12 | This study |
| -NH4+ | 5.7 × 10−11 ± 4.2 × 10−12 | This study | |
| Pseudomonas aeruginosa | -NO3− | 2.0 × 10−11 ± 1.3 × 10−12 | This study |
| -NH4+ | 1.5 × 10−11 ± 1.4 × 10−12 | This study | |
| Ruegeria atlantica | -NO3− | 3.4 × 10−11 ± 5.1 × 10−14 | DSMZ-5823; this study |
| Roseobacter litoralis | -NO3− | 6.5 × 10−11 ± 3.1 × 10−13 | DSMZ-6996; this study |
| Ruegeria gelatinovorans | -NO3− | 5.4 × 10−11 ± 3.3 × 10−13 | DSMZ-5887; this study |
| Silicibacter pomeroyi | -NO3− | 7.2 × 10−11 ± 1.3 × 10−14 | M. A. Moran; this study |
| Pseudomonas carboxydoflava | -NO3− | 1.8 × 10−5 | 9 |
| Comamonas compransoris | -NO3− | 1.6 × 10−5 | 9 |
| Pseudomonas carboxydohydrogena | -NO3− | 7.9 × 10−10 | 9 |
| Vineyard Sound total assemblage | Seawater | 4.5 × 10−11-6.0 × 10−11 | This study |
CO oxidation rates were calculated on a per-cell basis for each isolate (nmol CO oxidized cell−1 h−1) by the equation n25 × (25 × N × t)−1, where n25 is the molar amount of CO oxidized in a 25-ml sample (nmol), N is the cell density (cells ml−1), and t is the duration of 14CO incubation (h).
Substrate-tracking microautoradiography.
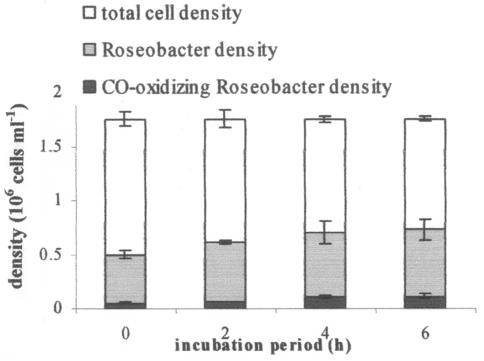
In sample water collected from Vineyard Sound in April 2003 for the STAR-FISH assay, direct DAPI counts exhibited a total cell density of 1.75 × 106 ± 3.96 × 104 cells ml−1 (mean ± SEM; n = 10 fields). In a time series of 14CO incubation, subsamples were fixed, mounted, exposed to a radiographic emulsion, hybridized with a Cy3-labeled RAC-specific probe, and stained with DAPI. Microscopic counts of cells under different regimens of illumination per field were recorded. With RAC-specific CO oxidation activities determined by physiological measurements of seawater isolates and type strains, in combination with CO-active RAC cell densities, the relative proportion of environmental CO oxidation that is performed by RAC organisms was calculated. The RAC comprised 36% ± 2.4% (mean ± SEM) of the total microbial assemblage, and 12.8% ± 1.0% of Cy3-labeled RAC cells formed silver grains in autoradiographic emulsion in STAR-FISH (Fig. 2).
FIG. 2.
Differential cell counts in STAR-FISH preparations. Epifluorescent signals were counted under different lighting regimens to illuminate all cells (DAPI stained) (total lengths of bars), all RAC cells (Cy3 labeled) (gray portions of bars), and CO-oxidizing RAC cells (Cy3 plus 14C labeled) (black portions of bars). Cell retention during the triple-staining process was ∼70%, and counts were scaled up to reflect incubation densities. Error bars represent SEM (n = 10 fields per illumination regimen).
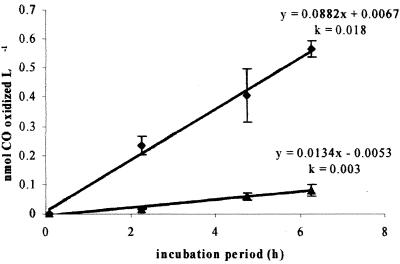
The gross rate of CO oxidation in sample water, determined by a direct 14CO oxidation assay, was 0.0882 nmol liter−1 h−1 (kco = 0.018 h−1). When the relative proportion of label-containing RAC cells was factored in with the mean specific RAC activity, the rate of CO oxidation attributable to RAC cells was 0.0134 nmol liter−1 h−1 (kco = 0.003 h−1). By this calculation, then, 15% of total environmental CO oxidation occurring at this coastal location can be attributed to RAC organisms (Fig. 3).
FIG. 3.
Total CO oxidation occurring in seawater sample (⧫) and activities of CO-oxidizing RAC isolates (▴). The linear equations of time series of CO oxidation and the rate coefficients (k) for CO oxidation normalized to 1 nM CO (aq) are shown. CO-oxidizing RAC microorganisms may contribute 15% of total environmental CO oxidation, as determined by 14CO oxidation in bulk water samples.
DISCUSSION
In this effort to identify microbial species in the marine environment that oxidize CO at environmentally relevant concentrations, we found an unexpected diversity of active CO metabolizers spanning multiple phyla. Although the microbial strains isolated from coastal seawater that oxidize CO at environmental concentrations are phylogenetically diverse, the most active strains are closely related to the Roseobacter-associated clade of alphaproteobacteria. This ability has also been detected in isolates closely related to Halomonas (a gammaproteobacterium) and Cyclobacterium (phylum Bacteroidetes) and is a newly discovered feature within these groups. The CO-oxidizing isolates and type cultures in this study are phylogenetically unrelated to functionally characterized carboxydotrophic organisms (9, 40) and, with the exception of Silicibacter pomeroyi, microorganisms that have been previously reported to oxidize CO or implicated in environmental CO oxidation, including certain ammonium- and methane-oxidizing bacteria (5, 20).
The most active CO oxidizers in northeastern coastal waters belong to the Roseobacter-associated clade, or “Roseobacter group,” as recently used by González et al. (13). The RAC encompasses a physiologically and geographically diverse group of bacteria within the alpha subclass of the Proteobacteria (3) and, together with Paracoccus and other genera within the Rhodobacteraceae, comprises the “marine alpha group.” Organisms belonging to the RAC, including the genera Rhodovulum, Roseivivax, Roseobacter, Roseovarius, Rubrimonas, Ruegeria, Sagittula, Stayela, Stappia, Silicibacter, and Sulfitobacter, are often numerically dominant in the bacterioplankton in coastal waters. The RAC has been reported to contribute up to 28% of the total 16S rRNA gene sequences in seawater samples from the coast of the southeastern United States (12), as being a dominant prokaryotic component during a North Sea coccolithophore bloom, contributing 24% of the bacterioplankton numbers (14, 41), and to contribute >30% of the total small-subunit rRNA genes in the surface waters of the Monterey Bay upwelling plume (31).
Recent sequencing of the Silicibacter pomeroyi genome has led to the identification of all the genes necessary for CO oxidation (28), lending support to observations of CO oxidation by S. pomeroyi and other RAC strains. Given the high rates of CO oxidation by some RAC isolates and their observed density in the study location, RAC organisms are important contributors to total CO oxidation in coastal waters. Many members of the RAC acquired from various sources tested negatively for CO oxidation activity; CO oxidation is therefore not a characteristic phenotype of the RAC under these defined growth conditions. However, the most active of the organisms isolated in this study are representatives of genera within the RAC.
To compare our direct isolation approach with other studies and to confirm that our CO-metabolizing isolates provide a plausible contribution to marine CO oxidation, we researched primary literature sources to find activities of known CO-oxidizing strains. For an initial estimate of the proportion of total CO oxidation that can be attributed to various CO-oxidizing microorganisms, we used literature values for uptake efficiency (4%) (40), cellular carbon content (20 fg C cell−1) (4), and a protein-to-dry weight conversion factor (1.4 mg protein:3.1 mg dry weight) (9). The amount of CO oxidized in a 25-ml liquid culture of characterized strains at a cell density of 1 × 106 cells ml−1 can be compared with the CO oxidation rates we observed in natural seawater samples (Table 1). Cell-specific CO oxidation activities in bulk water samples at our experimental site ranged from 4.5 × 10−11 to 6.0 × 10−11 nmol cell−1 h−1 (total CO oxidation rate/total cell density by direct microscopy) (36, 37). This value is of limited importance since it assumes that all cells present are oxidizing CO; however, it provides a first approximation of environmentally relevant activity and a minimum activity that we should observe in CO-oxidizing strains. The CO oxidation rate of one well-characterized carboxydotrophic organism, Pseudomonas carboxydohydrogena, is calculated to be 7.9 × 10−11 nmol cell−1 h−1 (9) and is only slightly greater than the cell-specific CO oxidation activity observed at our sampling site. Our calculation of RAC-specific CO oxidation activity inherently assumes that the CO oxidation activities of pure isolates growing under a slightly elevated [CO] are representative of uncultured organisms growing in mixed communities at a lower [CO].
Our initial macroautoradiographs exhibited approximately 400 CFU ml−1 actively metabolizing CO in the environmental samples; this is a conservative estimate given our current inability to cultivate the vast majority of environmental microorganisms. Using this value as the environmental density of CO-oxidizing cells, the cell-specific CO oxidation is increased to 3.07 × 10−7 to 4.06 × 10−7 nmol cell−1 h−1. The specific CO oxidation activity for the most active carboxydotrophic organism characterized to date, Pseudomonas carboxydoflava, is 1.8 × 10−5 nmol cell−1 h−1 (9) and represents an upper bound for cell-specific CO oxidation activities, although we have not observed activity of this magnitude in any of the isolates or type cultures measured in this study. The specific CO oxidation activities that we observed are well within these minimum and maximum values. It is probable that there exist other microorganisms with high rates of CO oxidation that were not detected in our screening and isolation program; these putative contributors to CO oxidation in seawater remain to be discovered.
Other seawater isolates that oxidize CO are not closely related to the RAC but are representatives of the gamma subclass of the Proteobacteria (Halomonas spp.) and across phyla in the Bacteroidetes (Cyclobacterium sp.). The relative proportion of environmental CO oxidation performed by these groups has yet to be resolved for natural samples via the STAR-FISH assay. By developing new or using extant general oligonucleotide probes for these phylogenetic groups, it is a viable and practical endeavor to enumerate CO-active members of these groups in natural waters and to determine their relative contributions to the total CO metabolism occurring there. With the use of only RAC-specific probes in STAR-FISH, the efficacy of this assay was demonstrated for quantifying a numerically and metabolically important phylogenetic group contributing to a natural chemical transformation that occurs in marine environments. A more complete picture of the CO-oxidizing marine community may be obtained when more CO-oxidizing taxa have been quantified, using additional probe sets in STAR-FISH, and included in the CO oxidation budget.
The active enzyme in the aerobic oxidation of CO by many described carboxydotrophic microorganisms is carbon monoxide dehydrogenase (CODH), which catalyzes the oxidation of CO to CO2 (24) and is a product of the coxSML genes. The similarity among the CODHs from otherwise phenotypically diverse carboxydobacteria is striking (21) and suggests a common evolutionary origin for the CODH genes and a relatively recent dispersal of the trait, perhaps by plasmid-mediated genetic exchange. Primer sequences specific for coxL genes, required for active CODH and the ability to oxidize CO, may be used to amplify and confirm the presence of these genes in marine isolates known to oxidize environmental CO (11). If the CO oxidation phenotype is always coincident with the presence of the coxL gene and product, then the biochemical mechanism of CO oxidation in marine environments will be better understood. The phylogenetic distribution of the phenotype may also be explained by molecular analysis of coxL sequences from CO-oxidizing isolates or cloned from environmental DNA extracts. Furthermore, molecular probes specific for conserved coxL genes used in STAR-FISH may potentially enable a direct quantification of diverse and metabolically active CO-oxidizing cells in marine environments.
Acknowledgments
We gratefully acknowledge M. A. Moran for providing live cultures of S. pomeroyi DSS-3, S. lacuscaerulensis, ISM, and EE-36 and Erich Horgan for collecting Sargasso seawater for use in our media. We thank Gary King, Andreas Teske, and John Waterbury for helpful discussions and advice. We also appreciate the comments and critiques of two diligent reviewers who contributed to the manuscript.
Funding was provided by National Science Foundation grant OCE-0136876, Coastal Ocean Institute and Rinehart Coastal Research Center grant BI10918, and the Woods Hole Oceanographic Institution Academic Programs Office.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andreasen, K., and P. H. Nielsen. 1997. Application of microautoradiography to the study of substrate uptake by filamentous microorganisms in activated sludge. Appl. Environ. Microbiol. 63:3662-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkmeyer, R., M. Rappe, S. Gallacher, and L. Medlin. 2000. Development of clade (Roseobacter and Alteromonas) and taxon-specific oligonucleotide probes to study interactions between toxic dinoflagellates and their associated bacteria. Eur. J. Phycol. 35:315-329. [Google Scholar]
- 4.Cho, B. C., and F. Azam. 1990. Biogeochemical significance of bacterial biomass in the ocean's euphotic zone. MEPS 63:253-259. [Google Scholar]
- 5.Conrad, R., and W. Seiler. 1980. Photooxidative production and microbial consumption of carbon monoxide in seawater. FEMS Microbiol. Lett. 9:61-64. [Google Scholar]
- 6.Conrad, R., O. Meyer, and W. Seiler. 1981. Role of carboxydobacteria in consumption of atmospheric carbon monoxide by soil. Appl. Environ. Microbiol. 42:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad, R., W. Seiler, G. Bunse, and H. Giehl. 1982. Carbon monoxide in seawater (Atlantic Ocean). J. Geophys. Res. 87:8839-8852. [Google Scholar]
- 8.Conrad, R., and W. Seiler. 1982. Utilization of traces of carbon monoxide by aerobic oligotrophic microorganisms in ocean, lake, and soil. Arch. Microbiol. 132:41-46. [Google Scholar]
- 9.Cypionka, H., O. Meyer, and H. G. Schlegel. 1980. Physiological characteristics of various species of strains of carboxydobacteria. Arch. Microbiol. 127:301-307. [Google Scholar]
- 10.Dunbar, J., D. C. L. Wong, M. J. Yarus, and L. J. Forney. 1996. Autoradiographic method for isolation of diverse microbial species with unique catabolic traits. Appl. Environ. Microbiol. 62:4180-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunfield, K., and G. M. King. 2004. Molecular analysis of carbon monoxide-oxidizing bacteria associated with recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 70:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosink, J. J., R. P. Herwig, and J. T. Stayley. 1997. Octadecabacter arcticus gen. nov., sp. nov. and O. antarcticus, sp. nov., nonpigmented, psychrophilic gas vacuolated bacteria from polar sea ice and water. Syst. Appl. Microbiol. 20:356-365. [Google Scholar]
- 15.Griffiths, R. P., B. A. Caldwell, J. D. Cline, W. A. Broich, and R. Y. Morita. 1982. Field observations of methane concentrations and oxidation rates in the southeastern Bering Sea. Appl. Environ. Microbiol. 44:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by epifluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. E., and T. S. Bates. 1996. Sources and sinks of carbon monoxide in the mixed layer of the tropical South Pacific Ocean. Global Biogeochem. Cycles 10:347-359. [Google Scholar]
- 18.Jones, R. D. 1991. Carbon monoxide and methane distribution and consumption in the photic zone of the Sargasso Sea. Deep-Sea Res. II 38:625-635. [Google Scholar]
- 19.Jones, R. D., and J. A. Amador. 1993. Methane and carbon monoxide production, oxidation, and turnover times in the Caribbean Sea as influenced by the Orinoco River. J. Geophys. Res. 98:2353-2359. [Google Scholar]
- 20.Jones, R. D., and R. Y. Morita. 1983. Carbon monoxide oxidation by chemolithotrophic ammonium oxidizers. Can. J. Microbiol. 29:1545-1551. [Google Scholar]
- 21.Kim, Y., S. Kirkconnell, and G. Hegeman. 1982. Immunological relationships among carbon monoxide dehydrogenases of carboxydobacteria. FEMS Microbiol. Lett. 13:219-223. [Google Scholar]
- 22.Lane, D., B. Pace, G. Olsen, D. Stahl, M. Sogin, and N. Pace. 1985. Rapid determination of 16S ribosomal sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, N., P. H. Nielsen, K. H. Andreason, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, O. 1982. Chemical and spectral properties of carbon monoxide: methylene blue oxidoreductase. The molybdenum-containing iron-sulfur flavoprotein from Pseudomonas carboxydovorans. J. Biol. Chem. 257:1333-1341. [PubMed] [Google Scholar]
- 25.Meyer, O., and H. G. Schlegel. 1983. Biology of aerobic carbon monoxide oxidizing bacteria. Annu. Rev. Microbiol. 37:277-310. [DOI] [PubMed] [Google Scholar]
- 26.Meyer-Reil, L.-A. 1978. Autoradiography and epifluorescence microscopy combined for the determination of number and spectrum of actively metabolizing bacteria in natural waters. Appl. Environ. Microbiol. 36:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mopper, K., X. Zhou, R. J. Kieber, D. J. Kieber, R. J. Sikorski, and R. D. Jones. 1991. Photochemical degradation of dissolved organic carbon and its impact on the oceanic carbon cycle. Nature 353:60-62. [Google Scholar]
- 28.Moran, M. A., A. Buchan, J. M. González, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequencing of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 29.Pfennig, N., and H. G. Trüper. 1981. Isolation of members of the families Chromatiaceae and Chlorobiaceae, p. 279-289. In M. P. Starr (ed.), The prokaryotes, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 30.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 31.Suzuki, M., C. Preston, F. Chavez, and E. DeLong. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat. Microb. Ecol. 24:117-127. [Google Scholar]
- 32.Swinnerton, J. W., V. J. Linnenbom, and R. A. Lamontagne. 1970. The ocean: a natural source of carbon monoxide. Science 167:984-986. [DOI] [PubMed] [Google Scholar]
- 33.Tabor, P., and R. A. Neihof. 1982. Improved microautoradiographic method to determine individual microorganisms active in substrate uptake in natural waters. Appl. Environ. Microbiol. 44:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2002. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolli, J. D. 2003. Identity and dynamics of the microbial community responsible for carbon monoxide oxidation in marine environments. Ph.D. thesis. Woods Hole Oceanographic Institution/Massachusetts Institute of Technology Joint Program, Woods Hole, Mass.
- 37.Tolli, J. D., and C. D. Taylor. 2005. Biological CO-oxidation in the Sargasso Sea and in Vineyard Sound, Massachusetts. Limnol. Oceanogr. 50:1205-1212. [Google Scholar]
- 38.Xie, H., O. C. Zafiriou, T. P. Umile, and D. J. Kieber. 2005. Biological consumption of carbon monoxide in Delaware Bay, NW Atlantic, and Beaufort Sea. Mar. Ecol. Prog. Ser. 290:1-14. [Google Scholar]
- 39.Zafiriou, O. C., S. S. Andrews, and W. Wang. 2003. Concordant estimates of oceanic carbon monoxide source and sink processes in the Pacific yield a balanced global “blue-water” CO budget. Global Biogeochem. Cycles 17:1015. [Google Scholar]
- 40.Zavarzin, G. A., and A. N. Nozhevnikova. 1977. Aerobic carboxydobacteria. Microb. Ecol. 3:305-326. [DOI] [PubMed] [Google Scholar]
- 41.Zubkov, M., B. Fuchs, S. Archer, R. Kiene, R. Amann, and P. Burkill. 2002. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]