Abstract
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:NM (nonmotile) is a unique clone that causes outbreaks of hemorrhagic colitis and hemolytic-uremic syndrome. In well-defined clusters of cases, we have observed significant variability in pulsed-field gel electrophoresis (PFGE) patterns which could indicate coinfection by different strains. An analysis of randomly selected progeny colonies of an outbreak strain after subcultivation demonstrated that they displayed either the cognate PFGE outbreak pattern or one of four additional patterns and were <89% similar. These profound alterations were associated with changes in the genomic position of one of two Shiga toxin 2-encoding genes (stx2) in the outbreak strain or with the loss of this gene. The two stx2 alleles in the outbreak strain were identical but were flanked with phage-related sequences with only 77% sequence identity. Neither of these phages produced plaques, but one lysogenized E. coli K-12 and integrated in yecE in the lysogens and the wild-type strain. The presence of two stx2 genes which correlated with increased production of Stx2 in vitro but not with the clinical outcome of infection was also found in 14 (21%) of 67 SF EHEC O157:NM isolates from sporadic cases of human disease. The variability of PFGE patterns for the progeny of a single colony must be considered when interpreting PFGE patterns in SF EHEC O157-associated outbreaks.
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:NM (nonmotile) strains have emerged as causes of hemolytic-uremic syndrome (HUS) and diarrhea in Europe (1, 4, 14, 17, 27, 28, 43) and Australia (3). Multilocus enzyme electrophoresis (11), sequence typing (42), and identification of additional inserted loci (15, 16, 22, 23, 24) indicated that SF EHEC O157:NM strains are closely related to but clearly distinct from EHEC O157:H7. In contrast to E. coli O157:H7, SF EHEC O157:NM strains ferment sorbitol overnight, produce β-d-glucuronidase, possess genes encoding Shiga toxin 2 (Stx2) but not those encoding Stx1 or Stx2c, and do not contain the tellurite resistance- and adherence-conferring island (6, 27, 49). Also in contrast to EHEC O157:H7, SF EHEC O157:NM strains do possess a mosaic genomic island composed of segments of the Shigella resistance locus and the E. coli O157:H7 strain EDL933 genome (24) and a plasmid-carried sfp cluster encoding Sfp fimbriae (15).
Stxs presumably account for the most severe consequences of human EHEC infection (5). The structural genes for Stx1, Stx2, and Stx2c in EHEC O157:H7 are carried in the genomes of temperate lambdoid bacteriophages (26, 32, 33, 36, 40, 45, 47, 54) that are quite variable in structure (33, 36, 40, 47, 54). stx genes are typically located in the late-phase phage region, between the Q-like gene, which encodes a transcription antiterminator that enables stx transcription after induction with mitomycin C, and the S holin gene, which is responsible for the release of Stx from the host cell (26, 32, 33, 40, 52, 54). Chromosomal integration sites for stx2-converting phages in EHEC O157:H7 include wrbA (32, 40, 45), sbcB (37), and yecE (9), whereas stx1-converting phages integrate in yehV (39, 54). stx2 genes in SF EHEC O157:NM strains have also been reported to be phage borne (25), but the toxin-converting phages and their chromosomal integration sites have not been characterized.
Pulsed-field gel electrophoresis (PFGE) has been successfully used to identify outbreaks caused by EHEC O157:H7 (2, 13, 18, 31, 48). This technology has also been applied to SF EHEC O157:NM clusters. In several outbreaks, the SF EHEC O157:NM isolates displayed identical outbreak-specific PFGE patterns (1, 31), whereas in two epidemiologically well-documented clusters, there was substantial variability in PFGE patterns (4, 43). In this study, we investigated possible reasons for the variability of PFGE patterns among SF EHEC O157:NM isolates and demonstrate potential consequences of this phenomenon for outbreak investigations.
MATERIALS AND METHODS
Bacterial strains.
Three SF EHEC O157:NM strains were isolated from a 15-month-old boy with HUS (strain 258/98), his 6-year-old brother with diarrhea (strain 269/98), and a cow that was epidemiologically associated with the human infections (strain 550/98) (4). Sixty-seven SF EHEC O157:NM isolates from sporadic human infections (Table 1) were randomly selected from 148 SF EHEC O157:NM strains recovered between 1996 and 2004 at the Institute for Microbiology and Hygiene, University of Würzburg, Würzburg, the Institute for Hygiene, University of Münster, Münster, and the Robert Koch Institute, Wernigerode, Germany, during routine diagnostic efforts and epidemiological investigations. After isolation, the strains were characterized for virulence genes and phenotypes (4, 14, 15, 46, 55) and then frozen at −70°C. All strains fermented sorbitol after overnight culture on sorbitol MacConkey agar and possessed stx2 as their sole stx gene, the eae gene encoding intimin γ, and sfpA, which uniquely marks the SF EHEC O157:NM clonal lineage (15). Each strain displayed a fliC restriction fragment length polymorphism pattern identical to that of E. coli O157:H7 strain EDL933 (data not shown), demonstrating that although they were nonmotile, all strains belonged to the H7 clone complex.
TABLE 1.
SF EHEC O157:NM strains investigated in this study
| No. of strainsa | Disease associationb (no. of strains)
|
No. of stx2 copiesc | Location of stx2 (XbaI fragment [kb])c | ||
|---|---|---|---|---|---|
| HUS | BD | WD | |||
| 47 | 39 | 3 | 5 | 1 | 320 |
| 1 | 1 | 0 | 0 | 1 | 300 |
| 1 | 1 | 0 | 0 | 1 | 210 |
| 4 | 3 | 1 | 0 | 1 | 150 |
| 10d | 6 | 1 | 2 | 2 | 490,320 |
| 1 | 1 | 0 | 0 | 2 | 490,210 |
| 1 | 1 | 0 | 0 | 2 | 490,170 |
| 3e | 2 | 0 | 1 | 2 | 450,320 |
| 1 | 1 | 0 | 0 | 2 | 370,320 |
| 1 | 0 | 0 | 1 | 2 | 310,300 |
Three strains were from a cluster of SF EHEC O157:NM infections (4) and 67 were from sporadic infections.
HUS, hemolytic-uremic syndrome; BD, bloody diarrhea; WD, watery diarrhea without visible blood.
As determined by hybridization of XbaI-digested, PFGE-separated genomic DNAs with the stxA2 probe.
One strain was from a cow epidemiologically associated with infections of two siblings (4).
Two strains were from siblings epidemiologically associated with a cow (4).
Stx assay.
Stx production was quantified by the Vero cell cytotoxicity assay (29), without and after induction with mitomycin C (0.5 μg/ml; Sigma-Aldrich, Deisenhofen, Germany) (44). Strains harboring one or two stx2 genes were tested in the same experiment, allowing a direct comparison of their cytotoxicities. For each strain, a mean cytotoxicity titer was calculated from three repeated tests.
PCR techniques.
PCRs were performed in an iCycler instrument (version 1.259; Bio-Rad, München, Germany) or a Biometra TGradient 96 cycler (Biometra GmbH, Göttingen, Germany) as described previously (46), using PCR reagents from PEQLAB Biotechnologie (Erlangen, Germany). The PCR primers, target sequences, conditions, and positive control strains used for this study are listed in Table 2. DNA regions of ≥3.0 kb were amplified using an AccuPrime Taq DNA polymerase high-fidelity kit (Invitrogen, Karlsruhe, Germany) and genomic DNA (500 ng) according to the manufacturer's instructions.
TABLE 2.
PCR primers and conditions used in this study
| Primer | Sequence (5′-3′) | Target | PCR conditiona
|
PCR product size (bp) | Reference | Positive control | ||
|---|---|---|---|---|---|---|---|---|
| Denaturing | Annealing | Extension | ||||||
| LP43 | ATCCTATTCCCGGGAGTTTACG | stxA2 | 94°C, 30 s | 57°C, 60 s | 72°C, 60 s | 584 | 14 | EDL933 |
| LP44 | GCGTCATCGTATACACAGGAGC | |||||||
| GK3 | ATGAAGAAGATGTTTATG | stxB2 | 94°C, 30 s | 52°C, 60 s | 72°C, 60 s | 260 | 14 | EDL933 |
| GK4 | TCAGTCATTATTAAACTG | |||||||
| SCab | CGGGTCTGGTGCTGATTACT | Whole stx2 | 94°C, 30 s | 55°C, 60 s | 72°C, 90 s | 1,462 | This study | E32511 |
| SCbb | ATCTCGCTCCAGGTTGTCGT | |||||||
| Stx2B-fwdc | ATGAAGAAGATGTTTATGGCG | stxB2-S | 94°C, 30 s | 57°C, 60 s | 69°C, 180 sd | 3,907 or 2,813e | This study | EDL933 |
| S-revc | AAACAGCAGACTCCCCAGCAC | |||||||
| A | AAGTGGCGTTGCTTTGTGAT | Intact yehV | 94°C, 30 s | 47°C, 60 s | 72°C, 90 s | 340 | 45 | C600 |
| B | AACAGATGTGTGGTGAGTGTCTG | |||||||
| wrbA1 | ATGGCTAAAGTTCTGGTG | Intact wrbA | 94°C, 30 s | 47°C, 60 s | 72°C, 60 s | 600 | 50 | C600 |
| wrbA2 | CTCCTGTTGAAGATTAGC | |||||||
| EC10 | GCCAGCGCCGAGCAGCACAATA | Intact yecE | 94°C, 30 s | 63°C, 60 s | 72°C, 60 s | 400 | 9 | C600 |
| EC11 | GGCAGGCAGTTGCAGCCAGTAT | |||||||
| yecD-fwd | CGAAGACGCCTGTAGTGCC | Intact yecE | 94°C, 30 s | 47°C, 60 s | 72°C, 90 s | 1,371 | 50 | C600 |
| yecN-rev | CGCAGGGAGAAAACCAACTC | |||||||
| sbcB1 | CATGATCTGTTGCCACTCG | Intact sbcB | 94°C, 30 s | 47°C, 60 s | 72°C, 90 s | 1,800 | 50 | C600 |
| sbcB2 | AGGTCTGTCCGTTTCCACTC | |||||||
| Int-297fwdf | GGAGGTTTTATGAGTAACGCATCATACCC | intΦ297 | 94°C, 30 s | 58°C, 60 s | 72°C, 90 s | 1,297 | This study | EH297g |
| Int-297revf | TTCAGAGTCTGGCTCTATGGGGCATGTA | |||||||
| Int-258320h | CATAGCAAACCAAATGGGCCA | intΦ258320-yecE | 94°C, 30 s | 57°C, 60 s | 72°C, 60 s | 425 | This study | 258/98 |
| EC11h | GGCAGGCAGTTGCAGCCAGTAT | |||||||
All PCRs included 30 cycles under the indicated conditions, followed by a final extension of 5 min at 72°C, unless otherwise stated.
Primers SCa and SCb were derived from stx2c-flanking regions of E. coli O157:NM strain E32511 (GenBank accession numbers M59432 [positions 153 to 172] and AJ251452 [positions 448 to 429], respectively).
Primer Stx2B-fwd was derived from the stxB2 gene of phage Φ933W (positions 1 to 21), and primer S-rev was derived from the S gene of Φ933W (positions 141 to 121) (GenBank accession number AF125520).
This amplification step was carried out for 10 cycles; in the following 20 cycles, the elongation time was 3 min plus a 20-s elongation step for each cycle.
The length of this PCR product from strain EDL933 was 3,538 bp.
Primers Int-297fwd and Int-297rev were derived from the sequence of Φ297 (GenBank accession number AJ431361) (positions 585 to 613 and 1881 to 1854, respectively).
Strain EH297 (9) was kindly provided by Henri De Greve (Vrije Universiteit Brussel, Brussels, Belgium); the other control strains were from our collection.
PFGE and Southern hybridization.
PFGE was performed according to the E. coli O157:H7 PulseNet protocol (19), using XbaI-digested DNAs of Salmonella enterica serovar Braenderup strain H9812 (19) and E. coli O157:H7 strain G5244 (Centers for Disease Control and Prevention [CDC], Atlanta, Ga.) as reference standards. Restriction fragment patterns of genomic DNAs were analyzed with BioNumerics, version 4.0 (Applied Maths BVBA, Belgium). XbaI-digested, PFGE-separated genomic DNAs were vacuum blotted on a nylon membrane (no. 1417240; Roche Molecular Biochemicals, Mannheim, Germany) and hybridized with a digoxigenin-11-dUTP-labeled (DIG High Prime kit; Roche Molecular Biochemicals) stxA2 probe (see below); hybridization was detected using a DIG Luminescent detection kit (Roche Molecular Biochemicals).
Detection of stx2 in phage DNA.
Phage DNA isolated from strain 258/98 after induction with mitomycin C by polyethylene glycol concentration and phenol-chloroform extraction (41) was digested with HpaI (New England Biolabs, Frankfurt, Germany), separated in 0.6% agarose, transfered to a nylon membrane (Zeta-probe GT; Bio-Rad), and hybridized with digoxigenin-labeled stxA2 and stxB2 probes using a DIG DNA labeling and detection kit (Roche Molecular Biochemicals). The stxA2 and stxB2 probes were derived from E. coli EDL933 by PCRs with primer pairs LP43-LP44 and GK3-GK4 (Table 2), respectively.
Induction of stx2-converting phages and transduction experiments.
Bacteriophages were induced with mitomycin C (0.5 μg/ml) (44), and sterile filtrates of the induced cultures were subjected to a plaque assay, using E. coli C600, DH5α, and HB101 as indicators (44). The resulting plaques were hybridized with the stxA2 probe (44). In transduction experiments, 100 μl of mitomycin C phage lysate (103 PFU) was mixed with 100 μl of the log-phase (106 to 107 CFU) E. coli C600 or DH5α recipient strain and 125 μl of 0.1 M CaCl2 solution and incubated (2 h, 37°C) without shaking. The mixtures were then transferred to 4 ml of Luria-Bertani (LB) broth and incubated (24 h, 37°C) with shaking at 180 rpm. Tenfold dilutions of the cultures were plated on LB agar, and overnight growths harvested into 1 ml of saline were screened for stx2 by PCR with primers LP43 and LP44 (Table 2). To identify lysogens in PCR-positive cultures, the cultures were restreaked on LB agar, and plates containing 300 to 400 well-separated colonies were subjected to colony blot hybridization with the stxA2 probe (14). stx2-positive colonies were subcultured three times on LB agar and tested by PCR for stx2 after the third passage to identify stable lysogens.
Nucleotide sequencing.
Nucleotide sequencing was performed with an automated ABI Prism 3100 Avant genetic analyzer (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany) and an ABI Prism BigDye Terminator ready reaction cycle sequencing kit (Perkin-Elmer Applied Biosystems). For sequence analysis of two stx2 genes located on different PFGE XbaI fragments in the same strain, the XbaI fragments which hybridized with the stxA2 probe were excised from the gel, and DNA from each of these fragments was purified using a Prep-A-Gene DNA purification kit (Bio-Rad) and used as a template for PCR with primers SCa and SCb (Table 2), which amplify the whole stx2 gene. The amplification products were purified (QIAquick PCR purification kit; QIAGEN, Hilden, Germany) and directly sequenced. Direct sequencing was also applied to purified amplicons of the phage integrase gene (int) and the int-yecE connection. PCR amplicons that spanned the region between the stxB2 and S genes were ligated into the pCR 2.1 vector (original TA cloning kit; Invitrogen), transformed into E. coli InvVαF′ competent cells as recommended by the manufacturer, and sequenced with universal and reverse primers for pUC/M13 vectors and with customized primers. Sequences were analyzed with the DNASIS program, and homology searches were performed using the EMBL-GenBank database (http://www.ncbi.nlm.nih.gov/BLAST).
Statistical analysis.
Comparisons of the means of Stx titers of strains containing a single stx2 gene or two stx2 genes were performed using the F test for independent variables and the t test for equal variances (35). Differences in means of the Stx titers without and after mitomycin C induction were assessed with the paired t test for dependent variables (35). Differences between groups were assessed using the Yate's corrected χ2 test and one-tailed Fisher's exact test for small numbers (35). OpenStat3 (William G. Miller; http://www.statpages.org/miller/openstat/OS3.html) was used to perform calculations. P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
Nucleotide sequences from this study have been deposited in EMBL-GenBank under the accession numbers listed in Table 3.
TABLE 3.
Characteristics and accession numbers of sequences determined in this study
| Strain | Sequence description (size of XbaI fragment carrying gene [kb])a | Sequence length (bp) | Accession no. |
|---|---|---|---|
| 258/98 | stx2 (450) | 1,422 | AF525040 |
| stx2 (320) | 1,422 | AF524944 | |
| 269/98 | stx2 (450) | 1,422 | AF521641 |
| stx2 (320) | 1,422 | AF525039 | |
| 550/98 | stx2 (490) | 1,422 | AF525041 |
| stx2 (320) | 1,422 | AF524945 | |
| T2 lysogen | stx2 (260) | 1,278 | DQ231586 |
| T3 lysogen | stx2 (260) | 1,278 | DQ231587 |
| T4 lysogen | stx2 (260) | 1,278 | DQ231588 |
| 164/01 | stx2 (490) | 1,422 | DQ231582 |
| stx2 (320) | 1,422 | DQ231583 | |
| 1/03 | stx2 (490) | 1,422 | DQ231584 |
| stx2 (170) | 1,422 | DQ231585 | |
| 2584/99 | stx2 (210) | 1,404 | DQ231589 |
| stx2 (490) | 1,404 | DQ231590 | |
| 258/98 | stxB2 flanks (320): stxB2, orf107, orf631, orf59, orf90, partial S gene | 3,907 | DQ231591 |
| stxB2 flanks (450): stxB2, orf107, orf344, partial S gene | 2,813 | DQ231595 | |
| T2 lysogen | stxB2 flanks: stxB2, orf107, orf631, orf59, orf90, partial S gene | 3,907 | DQ231592 |
| T3 lysogen | stxB2 flanks: stxB2, orf107, orf631, orf59, orf90, partial S gene | 3,907 | DQ231593 |
| T4 lysogen | stxB2 flanks: stxB2, orf107, orf631, orf59, orf90, partial S gene | 3,907 | DQ231594 |
| 258/98 | Φ258320int gene and int-yecE linkage | 1,562 | DQ231596 |
| T2 lysogen | Φ258320int gene and int-yecE linkage | 1,562 | DQ231597 |
None of the stx2 sequences contains an XbaI restriction site.
RESULTS
SF EHEC O157:NM strains isolated during a family outbreak contain two stx2 genes in different genomic positions.
To investigate PFGE differences observed during a family SF EHEC O157:NM outbreak (4), two human strains and one bovine strain were subjected to PFGE (Fig. 1A) and Southern hybridization with a stxA2 probe (Fig. 1B). For each strain, the probe hybridized to two XbaI fragments, which were 450 kb and 320 kb in length for the two human isolates (Fig. 1B, lanes 1 and 2) and 490 kb and 320 kb in length for the cattle isolate (Fig. 1B, lane 3), which lacked a 450-kb fragment in its PFGE profile (Fig. 1A, lane 3).
FIG. 1.
PFGE of XbaI-digested genomic DNAs (A) and Southern blot hybridization with an stxA2 probe (B) of SF EHEC O157:NM strains isolated during a family outbreak from a patient with HUS (strain 258/98; lanes 1), his brother with diarrhea (strain 269/98; lanes 2), and an epidemiologically associated cow (strain 550/98; lanes 3). Lanes S, molecular size standard (E. coli O157:H7 strain G5244; CDC). The sizes of XbaI fragments that hybridized with the stxA2 probe are indicated in panel B. Designations of PFGE and stxA2 hybridization patterns are given below the figure.
stx2 sequences.
The nucleotide sequences of the six stx2 genes in these three isolates (GenBank accession numbers are given in Table 3) were identical. Their A and B subunits differed from the corresponding subunits of the classical stx2 gene from bacteriophage Φ933W (GenBank accession number AF125520) (40) by seven and one nucleotides, respectively, and encoded an Stx2 protein which differed from Stx2 from Φ933W by one amino acid residue in each of the subunits.
Variability in stx2 genomic position and stx2 loss in strain 258/98 in vitro.
To determine if the variability in the genomic position of stx2 in the larger XbaI fragment observed in the outbreak isolates (Fig. 1B, lanes 1 to 3) occurs in vitro, a single colony of strain 258/98 was streaked on sorbitol MacConkey agar, and 10 randomly selected progeny colonies were analyzed by PFGE and stxA2 hybridization. The PFGE and stxA2 hybridization patterns of the progenies are shown in Fig. 2. stx2 in the 320-kb XbaI fragment from the original isolate, 258/98 (Fig. 1B, lane 1), was stable in its genomic position in the progenies (Fig. 2B, lanes 1 to 5). In contrast, the stx2 gene in the 450-kb XbaI fragment from strain 258/98 (Fig. 1B, lane 1) was found in four different positions in the progenies, including the original position in the 450-kb fragment (Fig. 2B, lane 1), the position in the 490-kb fragment (Fig. 2B, lane 2) seen for the cattle isolate (Fig. 1B, lane 3), and two other positions, in the 650-kb (Fig. 2B, lane 3) and 390-kb (Fig. 2B, lane 4) fragments. Moreover, one additional progeny lost this stx2 copy, as demonstrated by a single hybridization signal for the 320-kb fragment (Fig. 2B, lane 5).
FIG. 2.
PFGE of XbaI-digested genomic DNAs (A) and stxA2-specific Southern blot hybridization (B) of representative progenies of strain 258/98. Lanes S, molecular size standard (E. coli O157:H7 strain G5244; CDC). In lanes 1 to 5, the following progeny colonies (genomic positions of stx2 on XbaI fragments [sizes of fragments are given in kilobases] are given in parentheses) are shown: lanes 1, progeny 3 (450 and 320); lanes 2, progeny 7 (490 and 320); lanes 3, progeny 1 (650 and 320); lanes 4, progeny 9 (390 and 320); and lanes 5, progeny 2 (320). The XbaI fragments which hybridized with the stxA2 probe are indicated in panel A, and their sizes are given in panel B. Designations of PFGE and stxA2 hybridization patterns and the numbers of progenies that displayed the respective patterns are indicated below the figure.
Impact of change of stx2 genomic position and stx2 loss on PFGE patterns.
The progeny of strain 258/98 which contained stx2 copies in the 450-kb and 320-kb XbaI fragments (Fig. 2B, lane 1) had PFGE pattern (Fig. 2A, lane 1) identical to that of the original 258/98 isolate (Fig. 1A, lane 1), while progenies with stx2 copies in the 490-kb and 320-kb fragments (Fig. 2B, lane 2) had PFGE patterns (Fig. 2A, lane 2) identical to that of the cattle isolate 550/98 (Fig. 1A, lane 3). Moreover, three additional PFGE patterns were observed among the 258/98 progenies (Fig. 2A, lanes 3 to 5), each of which was associated with a particular change in the position of the stx2 copy that was originally in the 450-kb XbaI fragment (Fig. 2B, lanes 3 and 4) or with the loss of this stx2 gene (Fig. 2B, lane 5). The PFGE patterns of the progenies that contained an stx2 gene in the 490-, 650-, or 390-kb XbaI fragment or had lost this gene differed from that of the progeny containing this stx2 copy in the 450-kb XbaI fragment by four or five bands (Fig. 3A); the similarity between PFGE patterns of these progenies was <89% (Fig. 3B). The change in genomic position of one of the two stx2 copies or the loss of this stx2 gene from strain 258/98 was thus associated with a profoundly altered PFGE pattern.
FIG. 3.
Cluster analysis of PFGE patterns of representative progenies of strain 258/98 which harbored stx2 on different XbaI genomic fragments or had lost one stx2 copy. (A) PFGE gel; (B) dendrogram derived from the PFGE data with BioNumerics software. The analysis of the bands generated was performed using the Dice coefficient and the unweighted-pair group method using average linkages. Lanes S, molecular size standard (S. enterica serovar Braenderup strain H9812; CDC). The progeny colonies investigated (P) and their PFGE pattern designations (A to E) are indicated. The bands which hybridized with the stxA2 probe are shown on the PFGE gel.
Investigation of stx2-converting phages in strain 258/98.
To investigate if the two stx2 genes in strain 258/98 are carried by bacteriophages, phage lysate from bacteria induced with mitomycin C underwent plaque assay with E. coli strains HB101, DH5α, and C600, and the resulting plaques were hybridized with the stxA2 probe. Between 100 and 1,000 plaques were produced on the lawns of the indicators used (phage titers between 1 × 103 and 1 × 104 PFU/ml), but none hybridized with the stxA2 probe, demonstrating that none contained an stx2-converting bacteriophage. In contrast, most of the plaques produced by a phage lysate of the control strain E. coli O157:H7 EDL933 hybridized with the probe (data not shown).
Transduction of stx2.
The transduction of E. coli strains C600 and DH5α with phage lysate from strain 258/98 gave rise to three stable lysogens (designated T2, T3, and T4). Each of these lysogens contained a single stx2 gene in the 260-kb XbaI fragment (Fig. 4B, lanes 11 to 13), and this gene was identical to stx2 from the parental strain 258/98 (GenBank accession numbers are given in Table 3). All lysogens produced Stx2 (titers, 1:32 to 1:128). Thus, at least one stx2-converting phage from strain 258/98 can lysogenize E. coli C600 and E. coli DH5α, although it does not lyse these strains.
FIG. 4.
PFGE of XbaI-digested genomic DNAs (A) and stxA2-specific Southern blot hybridization (B) of representative SF EHEC O157:NM strains which contain one or two stx2 genes in different genomic positions and of lysogens of strain 258/98. Lanes S, molecular size standard (E. coli O157:H7 strain G5244; CDC). The following strains (stx2 positions on XbaI fragments [sizes of fragments are given in kilobases] are given in parentheses) are depicted in each panel: lanes 1, 164/01 (490 and 320); lanes 2, 25/99 (450 and 320); lanes 3, 2584/99 (490 and 210); lanes 4, 1/03 (490 and 170); lanes 5, 491/03 (370 and 320); lanes 6, 8082/02 (310 and 300); lanes 7, 493/89 (320); lanes 8, 221/95 (300); lanes 9, 703/88 (210); lanes 10, 3072/96 (150); lanes 11, lysogen T2 (260); lanes 12, lysogen T3 (260); and lanes 13, lysogen T4 (260). The sizes of the XbaI fragments that hybridized with the stxA2 probe are indicated in panel B.
Analysis of phage DNA from strain 258/98.
To investigate whether both stx2 copies in strain 258/98 are phage borne, phage DNA was isolated after induction with mitomycin C, digested with HpaI (which does not cut within stx2), separated electrophoretically, and hybridized with stxA2 and stxB2 probes. A 6- and a 7-kb fragment hybridized with each probe (data not shown), suggesting that each of the two stx2 genes in strain 258/98 belongs to an inducible prophage.
Analysis of stx2-flanking regions in strain 258/98.
To characterize sequences flanking the two stx2 copies in strain 258/98, DNA isolated from each of the 320-kb and the 450-kb XbaI fragments was subjected to PCR with primers Stx2B-fwd and S-rev (Table 2), which span the DNA between the stxB2 subunit gene and the S gene in the late-phase region of stx-converting bacteriophages. Amplicons of 3,907 bp and 2,813 bp were obtained from the 320-kb and 450-kb fragments, respectively. Each amplicon starts with stxB2 and ends with the S gene, but their intergenic sequences differ (Fig. 5). Open reading frames (ORFs) 107 (324 bp), 631 (1,896 bp), 59 (180 bp), and 90 (273 bp) were identified between stxB2 and the S gene in the 3,907-bp amplicon (GenBank accession number DQ231591) (Fig. 5). These ORFs showed high nucleotide sequence identities to the four respective ORFs between stxB2 and the S gene in an stx2-carrying phage from EHEC O145:NM strain 3985/96 (GenBank accession number AJ251520) (51) (Fig. 5). ORF 631 was also 93% identical to ORF L0105, the major ORF located in this region in Φ933W (GenBank accession no. AF125520) (40). In contrast, the 2,813-bp amplicon (GenBank accession number DQ231595) contained only two ORFs between the stxB2 and S genes (Fig. 5), one of which was 100% identical to ORF 107 from the 3,907-bp fragment while the other, ORF 344 (1,035 bp), was 97% identical to ORF 616, located downstream of stxB1 in an stx1-carrying phage from EHEC O103:H2 strain 1858/96 (GenBank accession number AJ251754) (51) (Fig. 5). The 3,907-bp and 2,813-bp amplicons from strain 258/98 were 100% identical in their stxB2 genes and ORF 107 and 92% identical in their S genes and the initial 710-bp stretches of ORFs 631 and 344. However, they showed no significant homology in the remaining sequences. Their overall nucleotide sequence identity is 77% (Fig. 5). Thus, the two stx2 copies in strain 258/98 are carried in the genomes of two different phages.
FIG. 5.
Structures of stxB2-flanking regions of two stx2 genes located on the 320-kb and 450-kb XbaI fragments of strain 258/98. Analyzed sequences and their lengths are given on the left side. Arrows indicate the lengths and directions of ORFs. Significant nucleotide sequence identities between the two 258/98 stxB2-flanking regions and their identities with the most closely related sequences in GenBank are shown in light shaded boxes. The overall nucleotide sequence identity between the two stxB2-flanking regions of strain 258/98 is shown in a hatched box. Bar, 1 kb.
Analysis of stx2-flanking regions in lysogens.
Amplicons of 3,907 bp were obtained, using primers Stx2B-fwd and S-rev, from the T2, T3, and T4 lysogens; they were 100% identical to each other and to the 3,907-bp amplicon from the 320-kb XbaI fragment of strain 258/98 (GenBank accession numbers are given in Table 3). This suggests that the bacteriophage carrying stx2 on the 320-kb XbaI fragment of strain 258/98 (described hereafter as Φ258320) has been transduced into each of these lysogens. None of the lysogens contained the phage carrying stx2 on the 450-kb XbaI fragment of strain 258/98 (Φ258450).
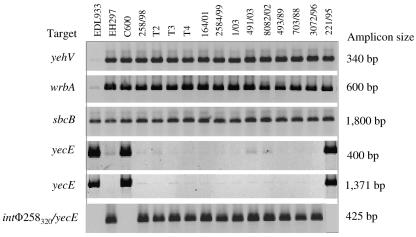
Phage integration sites.
To identify integration sites for Φ258320 and Φ258450, strain 258/98 was screened for the occupation of presently known phage integration sites in EHEC O157 by PCR (Table 2). Amplicons of expected sizes were obtained in each of the PCRs targeting yehV, wrbA, and sbcB (Fig. 6, lanes 4), indicating that these genes are intact and therefore not occupied by a phage. In contrast, the absence of specific amplicons in two different PCRs targeting yecE (Fig. 6, lanes 4) indicated that this gene is occupied in strain 258/98. The same results for each of the T2, T3, and T4 lysogens harboring Φ258320 (Fig. 6, lanes 5 to 7) demonstrated that yecE is also occupied in each of them. To determine whether Φ258320 occupies yecE, the phage int gene in lysogen T2 was amplified by PCR with primers Int-297fwd and Int-297rev (Table 2), derived from the int flanks of the stx2-converting phage Φ297, which integrates in yecE in E. coli O157:H7 (9). Sequence analysis demonstrated that the resulting amplicon contained the complete int gene (1,287 bp; GenBank accession number DQ231597), which has 98% nucleotide sequence identity to int of Φ297 (GenBank accession number AJ431361) and encodes a protein with 99% identity to the Int protein from this phage. The linkage between int of Φ258320 and yecE in lysogen T2 was determined by PCR with primers Int-258320 and EC11 (Table 2), derived from the int gene of Φ258320 and yecE of E. coli K-12, respectively. This amplicon (GenBank accession number DQ231597) contained 133 C-terminal nucleotides of the Φ258320 int gene, an attR core bacteriophage attachment site (CCGTCACATTAACTGCGTG) identical to that of Φ297 (GenBank accession number AJ431361), and 217 nucleotides (positions 97 to 313) of E. coli K-12 yecE (GenBank accession number AE000280). Thus, Φ258320 integrates in yecE in lysogen T2.
FIG. 6.
PCR analyses of phage integration sites in SF EHEC O157:NM. The strains tested, loci examined, and lengths of resulting amplicons are listed across the top and to the left and right of the rows of amplicons, respectively. Primers for the respective PCRs are listed in Table 2. Strains EDL933 (with stx1- and stx2-converting phages integrated in yehV and wrbA, respectively) (39, 40), EH297 (with a stx2-converting phage integrated in yecE) (9), and E. coli K-12 C600 (all of the genes investigated as putative phage integration sites are intact) (7) were used as controls.
To confirm that Φ258320 is also integrated in yecE in strain 258/98, the above PCR and sequence analyses were applied to the 320-kb XbaI fragment of this strain. Amplicons spanning the int gene and the int-yecE region (GenBank accession number DQ231596) were 100% and 99% (two nucleotide differences in yecE), respectively, identical to those from lysogen T2, demonstrating that Φ258320 is integrated in yecE in strain 258/98. No amplification products were elicited from DNA isolated from the 450-kb XbaI fragment of strain 258/98 in PCRs targeting the int gene and the int-yecE linkage. The integration site for Φ258450 carrying stx2 at this position could not be identified.
stx2 genes in SF EHEC O157:NM isolates from sporadic infections.
Sixty-seven SF EHEC O157:NM strains from sporadic infections (Table 1) were investigated by PFGE and stxA2 hybridization for the number and genomic positions of stx2 genes. In 53 (79%) of these strains, a single XbaI fragment of between 150 kb and 320 kb hybridized with the probe (Table 1; Fig. 4B). In the remaining 14 (21%) strains, two XbaI fragments hybridized (Table 1). These fragments were 490 kb and 320 kb long in most of the strains (Table 1), but six additional positions of stx2 were also observed (Table 1; Fig. 4B). Nucleotide sequence analysis demonstrated that the two stx2 genes located on the 490-kb and 320-kb XbaI fragments of strain 164/01 (Fig. 4B, lane 1) and on the 490-kb and 170-kb XbaI fragments of strain 1/03 (Fig. 4B, lane 4) were identical to each other and to stx2 from strain 258/98. The two stx2 genes located on the 490-kb and 210-kb XbaI fragments of strain 2584/99 (Fig. 4B, lane 3) were also identical to each other, but they differed from stx2 from strain 258/98 by six and one nucleotides in their A and B subunits, respectively (GenBank accession numbers are given in Table 3). In eight of nine SF EHEC O157 strains which harbored one or two stx2 genes in different genomic positions (Fig. 4B, lanes 1 and 3 to 10), stx2-carrying phages integrated into yecE (Fig. 6, lanes 8 to 15). PFGE analysis of 10 randomly selected progeny colonies from each of 10 strains after subculture demonstrated that none of 6 strains with a single stx2 gene changed its PFGE pattern. In contrast, for two of four strains with two stx2 genes, changes in PFGE patterns similar to those observed for strain 258/98 occurred (data not shown).
Stx2 production by SF EHEC O157:NM strains harboring one or two stx2 genes.
Stx2 titers in culture supernatants of the 14 strains harboring two stx2 genes were compared with those of the 53 strains containing a single stx2 gene, without and after induction with mitomycin C (Table 4). In noninduced cultures, the Stx2 titers of strains harboring two stx2 genes were significantly higher than those containing a single stx2 gene. Induction with mitomycin C dramatically increased Stx2 production both in strains harboring two stx2 genes and in strains harboring a single stx2 gene, but this increase was significantly higher for strains with one stx2 gene than for those with two stx2 genes (Table 4). Consequently, in contrast to the case with noninduced cultures, there was no significant difference between Stx2 titers in strains with two or one stx2 genes after induction with mitomycin C (Table 4). Among the patients from whom these strains were recovered (Table 1), the development of HUS was significantly associated with the presence of a single stx2 copy in the infecting strain (χ2 = 10.63; P = 0.00235; 95% confidence interval, 1.70 to 32.24).
TABLE 4.
Comparison of Stx2 titers of SF EHEC O157:NM strains harboring one or two stx2 genes using the Vero cell cytotoxicity assay
| Mitomycin C induction | Stx2 titersa of strains with the indicated number of stx2 genes
|
P value | t | df | |||
|---|---|---|---|---|---|---|---|
| 1 stx2 gene (n = 53)
|
2 stx2 genes (n = 14)
|
||||||
| Range | Median | Range | Median | ||||
| Noninduced | 192-2,133b | 714 | 853-6,826c | 2,112 | 0.0032d | 315.25 | 4 |
| Induced | 163,840-983,040b | 450,560 | 491,520-1,310,720c | 819,200 | 0.3849e | 1.25 | 8 |
| Fold increase in titer after induction | 154-3,413 | 593 | 72-886 | 373 | 0.0034f | 3.64 | 13 |
The Stx2 titer of each strain used for calculations was a mean value from three repeated tests.
Stx2 titers in strains with one stx2 gene. Noninduced cultures were compared to cultures induced with mitomycin C (in paired t test for dependent variables, t = 933.88; P < 0.00001; df = 21).
Stx2 titers in strains with two stx2 genes. Noninduced cultures were compared to cultures induced with mitomycin C (in paired t test for dependent variables, t = 348.17; P < 0.00001; df = 27).
Stx2 titers in noninduced cultures. Strains with one stx2 gene were compared to strains with two stx2 genes (F test for independent variables).
Stx2 titers in mitomycin C-induced cultures. Strains with one stx2 gene were compared to strains with two stx2 genes (F test for independent variables).
Increase in Stx2 titers after mitomycin C induction. Strains with one stx2 gene were compared to strains with two stx2 genes (t test for equal variances).
DISCUSSION
All sequenced genomes contain duplicated genes. We demonstrate here the frequent duplication of the gene encoding Stx2, the cardinal virulence factor of EHEC (14). This duplication does not increase the virulence of strains, as measured by their ability to cause HUS, but has consequences for the integrity of the genome. Progenies of a single colony of the outbreak strain displayed significantly altered PFGE patterns, suggesting the occurrence of genomic rearrangement. These profound alterations were associated with changes in genomic position of one of two stx2 copies or with the loss of this gene. The observation of similar PFGE alterations in the progenies of two of four additional strains with two stx2 genes, but not in the progenies of six strains with a single stx2 gene, further supports the suggestion that stx2 duplication is associated with decreased genomic stability in SF EHEC O157:NM.
Our findings have implications for epidemiological investigations based on PFGE subtyping. PFGE performed according to standardized protocols is presently widely used internationally for subtyping pathogenic bacteria, including E. coli O157:H7 (2, 13, 18, 48). We showed that the variations in PFGE patterns observed among the outbreak SF EHEC O157:NM isolates upon the first PFGE analysis (4) could be reproduced by subculture of strain 258/98 in vitro. This strongly indicates that all of these isolates were indeed the same strain, which probably changed its DNA fingerprints either in vivo, during infection, or in vitro (i.e., after isolation) before PFGE was performed, simulating infection with different strains. Our findings thus provide an explanation for some PFGE differences which might be observed during SF EHEC O157:NM outbreaks. They also demonstrate that it may be useful to perform PFGE on several colonies from stool cultures of a few patients known to be part of a cluster to identify other profiles involved.
PFGE differences are attributable to discrete insertions and deletions that contain XbaI sites rather than to single-nucleotide polymorphisms in XbaI sites themselves (30). The insertions and deletions containing XbaI sites in the genomes of E. coli O157:H7 strains were found within stx2-carrying Φ933W, within O islands that contain cryptic prophage genes, and within virulence plasmid pO157 (30). Thus, differences in PFGE profiles can arise from acquired DNA (21, 30) or from lost phages and/or plasmids (30, 34).
In contrast to E. coli O157:H7, where stx loss results in minor PFGE variations (one- to two-band difference), mostly limited to bands containing stx genes (12, 34), stx2 loss in SF EHEC O157:NM was associated with a five-band difference, including alterations in fragments that do and do not contain stx2. The rapidity with which these rearrangements occurred contrasts with PFGE pattern alterations for E. coli O157:H7, which are detectable after multiple subcultures (20) or prolonged storage (20, 34). In accordance with reports that stx2 loss in E. coli O157:H7 results from the loss of stx2-converting bacteriophages (33, 34), we demonstrated that stx2 on the 450-kb XbaI fragment in strain 258/98, which was lost after subculture, is also carried within a phage, suggesting a similar scenario for its loss. In contrast, alterations in PFGE patterns associated with changes in the genomic position of an stx gene, similar to those observed for the strain 258/98 progenies, have not been described, to our knowledge. This phenomenon might be attributed to the potential absence of a stable integration site for the phage harboring this stx2 copy in strain 258/98, resulting in only transitory phage integrations in different genomic sites.
Gene duplication is believed to play an important role in evolution because one copy maintains its original function in response to selective constraints, thereby freeing the other to generate possibly advantageous mutations and new functions (53). These processes are believed to facilitate the formation of antigenically variant families of proteins and of proteins with novel functions, thus enabling organisms to evade host immune responses and adapt to different microenvironments. Different genes have different duplicabilities. Papp et al. (38) proposed the dosage balance hypothesis, which postulates that genes coding for subunits of protein complexes tend to have lower duplicabilities than do genes coding for monomers, because duplication of a single subunit may cause a dosage imbalance among the subunits of the protein complex. Yang et al. (53) hypothesized that dosage sensitivity increases, while gene duplicability decreases, with the number of subunits in a protein. We demonstrated that each of the A and B subunits of two different stx2 alleles occurs as two identical copies in SF EHEC O157:NM. Two stx2 genes were present in 21% of SF EHEC O157:NM clinical isolates, and these two genes were identical in each of five sequenced strains, suggesting frequent stx2 duplicability in these pathogens. The frequency of stx duplication among other EHEC strains, including E. coli O157:H7, is unknown because detection methodologies such as PCR or colony hybridization will not discern duplicated copies. Indeed, the duplicated stx2 genes in SF EHEC O157 strains were not detected by either our PCR protocols or hybridization of chromosomal DNA after conventional (non-PFGE) gel electrophoresis (4). Two stx2 genes which were both identical to the classical stx2 gene from Φ933W were recently identified by sequencing of an E. coli O157:H7 outbreak strain in Spain (33).
We hypothesize that a possible mechanism for the stx2 duplication in SF EHEC O157:NM involves the acquisition of two different stx2-converting bacteriophages which, coincidentally, contained identical copies of the toxin gene. Such a scenario is supported by a report that the two stx2 copies in E. coli O157:H7 are also borne by different phages (33), and it parallels the scenario proposed for the acquisition of different stx genes, stx1 and stx2, by EHEC O157:H7 (11, 45). In E. coli O157:H7 strains EDL933 and Sakai, Stx1-encoding phages integrate in yehV (39, 54) and Stx2-encoding phages integrate in wrbA (32, 40), which, as we showed here, are intact in SF EHEC O157:NM. One of the two stx2-converting phages in strain 258/98 integrates in the yecE gene, which was previously shown to be the integration site for the stx2-converting bacteriophage Φ297 in E. coli O157:H7 (9) and for the stx2e-converting bacteriophage ΦP27 in a non-O157 EHEC strain (41). This is the first report of the integration site for a stx2-converting bacteriophage in SF EHEC O157:NM. Moreover, yecE was a phage integration site in eight of nine other SF EHEC O157:NM strains which contained one or two stx2 copies at different genomic positions. These data indicate that yecE may be a common integration site for stx2-converting bacteriophages in SF EHEC O157:NM strains.
The level of Stx production might be critical for EHEC pathogenicity. In two European studies (10, 33), Stx quantities produced by infecting EHEC O157 and non-O157 strains correlated with the clinical outcomes of infection, although this relationship was not observed in a North American study (8). We did not observe an association between the increased Stx2 production in vitro by SF EHEC O157:NM strains which possess two stx2 genes and their ability to cause HUS. On the contrary, HUS development in patients in our study correlated with infection by strains harboring a single stx2 copy and producing less Stx2 in vitro. One explanation could be that in the latter strains, mitomycin C induced Stx2 production significantly more than in strains harboring two stx2 genes. Thus, because the human body contains other prophage-inducing agents, such as H2O2 released by neutrophils (52), strains with a single stx2 gene might produce large amounts of Stx2 in vivo, thereby promoting their ability to cause HUS. These observations suggest that both virulence factors of the infecting strain and environmental factors in the diverse milieus in which EHEC strains are found must be considered when the outcome of E. coli O157 infection is examined.
Acknowledgments
This study was supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF) Project Network of Competence Pathogenomics Alliance program “Functional genomic research on enterohemorrhagic Escherichia coli” (number 119523).
We thank Phillip I. Tarr (Washington University School of Medicine, St. Louis, Mo.) and Mohamed A. Karmali (Laboratory for Food-Borne Zoonoses, Public Health Agency of Canada, Guelph, Ontario, Canada) for fruitful and extensive discussions of the manuscript and Henri De Greve (Vrije Universiteit Brussel, Brussels, Belgium) for providing us with strain EH297. The skillful technical assistance of Dagmar Mense and Nadine Brandt (Münster) and of Gerlinde Bartel, Evelyn Skiebe, and Maria Stöckel (Wernigerode) is highly appreciated.
REFERENCES
- 1.Ammon, A., L. R. Peterson, and H. Karch. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of E. coli O157:H−. J. Infect. Dis. 179:1274-1277. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, and J. G. Wells. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettelheim, K. A., M. Whipp, S. P. Djordjevic, and V. Ramachandran. 2002. First isolation outside Europe of sorbitol-fermenting verocytotoxigenic Escherichia coli (VTEC) belonging to O group O157. J. Med. Microbiol. 51:713-714. [DOI] [PubMed] [Google Scholar]
- 4.Bielaszewska, M., H. Schmidt, A. Liesegang, R. Prager, W. Rabsch, H. Tschäpe, A. Cizek, J. Janda, K. Blahova, and H. Karch. 2000. Cattle can be a reservoir of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J. Clin. Microbiol. 38:3470-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., and H. Karch. 2005. Consequences of enterohaemorrhagic Escherichia coli infection for the vascular endothelium. Thromb. Haemost. 94:312-318. [DOI] [PubMed] [Google Scholar]
- 6.Bielaszewska, M., P. I. Tarr, H. Karch, W. Zhang, and W. Mathys. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, V. J. Collado, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Cornick, N. A., S. Jelacic, M. A. Ciol, and P. I. Tarr. 2002. Escherichia coli O157:H7 infections: discordance between filterable fecal Shiga toxin and disease outcome. J. Infect. Dis. 186:57-63. [DOI] [PubMed] [Google Scholar]
- 9.De Greve, H., C. Qizhi, F. Deboeck, and J. P. Hernalsteens. 2002. The Shiga-toxin VT2-encoding bacteriophage Φ297 integrates at a distinct position in the Escherichia coli genome. Biochim. Biophys. Acta 1579:196-202. [DOI] [PubMed] [Google Scholar]
- 10.Eklund, M., K. Leino, and A. Siitonen. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750-1753. [DOI] [PubMed] [Google Scholar]
- 12.Feng, P., M. Dey, A. Abe, and T. Takeda. 2001. Isogenic strain of Escherichia coli O157:H7 that has lost both Shiga toxin 1 and 2 genes. Clin. Diagn. Lab. Immunol. 8:711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson, D. D., J. Scheftel, A. Cronquist, K. Smith, A. Woo-Ming, E. Anderson, J. Knutsen, A. K. De, and K. Gershman. 2005. Temporally distinct Escherichia coli O157 outbreaks associated with alfalfa sprouts linked to a common seed source—Colorado and Minnesota, 2003. Epidemiol. Infect. 133:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich, A. W., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich, A. W., K. V. Nierhoff, M. Bielaszewska, A. Mellmann, and H. Karch. 2004. Phylogeny, clinical associations, and diagnostic utility of the pilin subunit gene (sfpA) of sorbitol-fermenting, enterohemorrhagic Escherichia coli O157:H−. J. Clin. Microbiol. 42:4697-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich, A. W., R. Köck, M. Bielaszewska, W. Zhang, H. Karch, and W. Mathys. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J. Clin. Microbiol. 43:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber, A., H. Karch, F. Allerberger, H. M. Verweyen, and L. B. Zimmerhackl. 2002. Clinical course and the role of Shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997-2000, in Germany and Austria: a prospective study. J. Infect. Dis. 186:493-500. [DOI] [PubMed] [Google Scholar]
- 18.Gerner-Smidt, P., J. Kincaid, K. Kubota, K. Hise, S. B. Hunter, M. A. Fair, D. Norton, A. Woo-Ming, T. Kurzynski, M. J. Sotir, M. Head, K. Holt, and B. Swaminathan. 2005. Molecular surveillance of Shiga toxigenic Escherichia coli O157 by PulseNet USA. J. Food Prot. 68:1926-1931. [DOI] [PubMed] [Google Scholar]
- 19.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iguchi, A., R. Osawa, J. Kawano, A. Shimizu, J. Terajima, and H. Watanabe. 2002. Effects of repeated subculturing and prolonged storage at room temperature of enterohemorrhagic Escherichia coli O157:H7 on pulsed-field gel electrophoresis profiles. J. Clin. Microbiol. 40:3079-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iguchi, A., R. Osawa, J. Kawano, A. Shimizu, J. Terajima, and H. Watanabe. 2003. Effects of lysogeny of Shiga toxin 2-encoding bacteriophages on pulsed-field gel electrophoresis fragment pattern of Escherichia coli K-12. Curr. Microbiol. 46:224-227. [DOI] [PubMed] [Google Scholar]
- 22.Janka, A., M. Bielaszewska, U. Dobrindt, and H. Karch. 2002. Identification and distribution of the enterohemorrhagic Escherichia coli factor for adherence (efa1) gene in sorbitol-fermenting Escherichia coli O157:H−. Int. J. Med. Microbiol. 292:207-214. [DOI] [PubMed] [Google Scholar]
- 23.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. The cytolethal distending toxin (cdt) gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janka, A., G. Becker, A.-K. Sonntag, M. Bielaszewska, U. Dobrindt, and H. Karch. 2005. Presence and characterization of a mosaic genomic island which distinguishes sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H− from E. coli O157:H7. Appl. Environ. Microbiol. 71:4875-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karch, H., H. Böhm, H. Schmidt, F. Gunzer, S. Aleksic, and J. Heesemann. 1993. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J. Clin. Microbiol. 31:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karch, H., H. Schmidt, C. Janetzki-Mittmann, J. Scheef, and M. Kroger. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol. Gen. Genet. 262:600-607. [DOI] [PubMed] [Google Scholar]
- 27.Karch, H., and M. Bielaszewska. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 39:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405-418. [DOI] [PubMed] [Google Scholar]
- 29.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 30.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liesegang, A., U. Sachse, R. Prager, H. Claus, H. Steinrück, S. Aleksic, W. Rabsch, W. Voigt, A. Fruth, H. Karch, J. Bockemühl, and H. Tschäpe. 2000. Clonal diversity of Shiga toxin-producing Escherichia coli O157:H7/H− in Germany—a ten-year study. Int. J. Med. Microbiol. 290:269-278. [DOI] [PubMed] [Google Scholar]
- 32.Makino, K., K. Yokoyama, Y. Kubota, C. H. Yutsudo, S. Kimura, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, T. Iida, K. Yamamoto, M. Onishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 33.Muniesa, M., M. De Simon, G. Prats, D. Ferrere, H. Panella, and J. Jofre. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murase, T., S. Yamai, and H. Watanabe. 1999. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr. Microbiol. 38:48-50. [DOI] [PubMed] [Google Scholar]
- 35.Norman, G. R. 2003. Statistics, p. 41-52. In G. R. Norman and D. L. Streiner (ed.), Statistics, 3rd ed. BC Decker Inc., Hamilton, Canada.
- 36.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papp, B., C. Pal, and L. D. Hurst. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424:194-197. [DOI] [PubMed] [Google Scholar]
- 39.Perna, N. T., G. Plunket III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 40.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage ΦP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 43.Robert Koch Institute. 2003. Ein HUS-Ausbruch durch sorbitol-fermentierende EHEC des Serovars O157:H−: Untersuchungsergebnisse und Lehren für die Surveillance. Epidemiologisches Bull. 22:171-175. [Google Scholar]
- 44.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage Φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonntag, A.-K., R. Prager, M. Bielaszewska, W. Zhang, A. Fruth, H. Tschape, and H. Karch. 2004. Phenotypic and genotypic analyses of enterohemorrhagic Escherichia coli O145 strains from patients in Germany. J. Clin. Microbiol. 42:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauch, E., C. Schaudinn, and L. Beutin. 2004. First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157. Infect. Immun. 72:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and the CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarr, P. I., S. S. Bilge, J. C. Vary, S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toth, I., H. Schmidt, M. Dow, A. Malik, E. Oswald, and B. Nagy. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, J., R. Lusk, and W. H. Li. 2003. Organismal complexity, protein complexity, and gene duplicability. Proc. Natl. Acad. Sci. USA 100:15661-15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokoyama, K., K. Makino, Y. Kubota, M. Watanabe, S. Kimura, C. H. Yutsudo, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, M. Yoh, T. Iida, M. Ohnishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 2000. Complete nucleotide sequence of the prophage VT1-Sakai carrying the Shiga toxin 1 genes of the enterohemorrhagic Escherichia coli O157:H7 strain derived from the Sakai outbreak. Gene 258:127-139. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, W.-L., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]