Abstract
Evidence suggests that the Pseudomonas aeruginosa PAO1 gnyRDBHAL cluster, which is involved in acyclic isoprenoid degradation (A. L. Díaz-Pérez, N. A. Zavala-Hernández, C. Cervantes, and J. Campos-García, Appl. Environ. Microbiol. 70:5102-5110, 2004), corresponds to the liuRABCDE cluster (B. Hoschle, V. Gnau, and D. Jendrossek, Microbiology 151:3649-3656, 2005). A liu (leucine and isovalerate utilization) homolog cluster was found in the PAO1 genome and is related to the catabolism of acyclic monoterpenes of the citronellol family (AMTC); it was named the atu cluster (acyclic terpene utilization), consisting of the atuCDEF genes and lacking the hydroxymethyl-glutaryl-coenzyme A (CoA) lyase (HMG-CoA lyase) homolog. Mutagenesis of the atu and liu clusters showed that both are involved in AMTC and leucine catabolism by encoding the enzymes related to the geranyl-CoA and the 3-methylcrotonyl-CoA pathways, respectively. Intermediary metabolites of the acyclic monoterpene pathway, citronellic and geranic acids, were accumulated, and leucine degradation rates were affected in both atuF and liuD mutants. The alpha subunit of geranyl-CoA carboxylase and the alpha subunit of 3-methylcrotonyl-CoA carboxylase (α-MCCase), encoded by the atuF and liuD genes, respectively, were both induced by citronellol, whereas only the α-MCCase subunit was induced by leucine. Both citronellol and leucine also induced a LacZ transcriptional fusion at the liuB gene. The liuE gene encodes a probable hydroxy-acyl-CoA lyase (probably HMG-CoA lyase), an enzyme with bifunctional activity that is essential for both AMTC and leucine degradation. P. aeruginosa PAO1 products encoded by the liuABCD cluster showed a higher sequence similarity (77.2 to 79.5%) with the probable products of liu clusters from several Pseudomonas species than with the atuCDEF cluster from PAO1 (41.5%). Phylogenetic studies suggest that the atu cluster from P. aeruginosa could be the result of horizontal transfer from Alphaproteobacteria. Our results suggest that the atu and liu clusters are bifunctional operons involved in both the AMTC and leucine catabolic pathways.
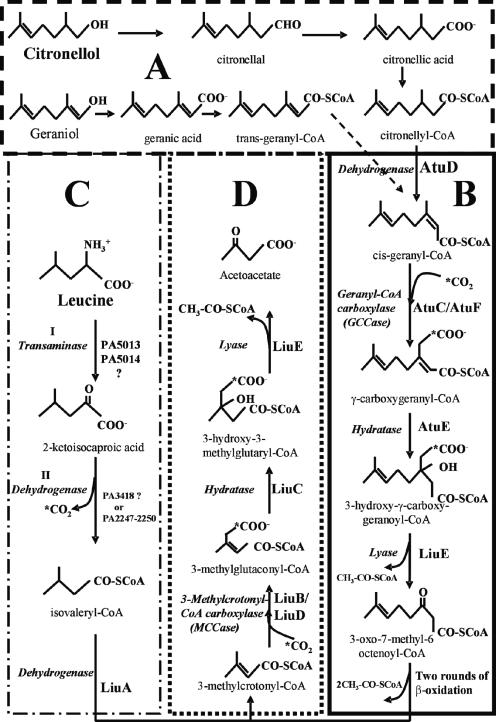
The 3-methyl branched alkanes (3-MBA) have been described as environmentally persistent compounds, although several species of microorganisms can use 3-MBA as the sole carbon source (28). Acyclic monoterpenes of the citronellol family (AMTC) are 3-MBA compounds that can be used by the Pseudomonadaceae (Pseudomonas citronellolis, Pseudomonas aeruginosa, and Pseudomonas mendocina) (2, 3). The pathways for the degradation of AMTC in P. citronellolis (3) and plants (9) have been reported, but only for P. aeruginosa have the genes involved been identified (5, 12). The proposed general route for citronellol degradation in P. aeruginosa first involves the oxidation of the alcohol to citronellic acid (called the upper pathway). In the lower pathway, citronellic acid is transformed to acetyl coenzyme A (acetyl-CoA) and 3-oxo-7-methyl-octenoate, a suitable substrate for the β-oxidation route. These transformations are suggested to be carried out by the enzymes citronellyl-CoA dehydrogenase, geranyl-CoA carboxylase (GCCase), γ-carboxygeranyl-CoA hydratase, and 3-hydroxy-γ-carboxygeranyl-CoA lyase (5, 12). After two rounds of β-oxidation, 3-methylcrotonyl-CoA (MC-CoA) is generated (Fig. 1). This metabolite may be transformed to acetyl-CoA and acetoacetate by the 3-methylcrotonyl-CoA carboxylase (MCCase), which is involved in leucine and AMTC catabolic pathways, as has been shown to occur in bacteria (5, 6, 12), fungi (26), and plants (1, 9, 20). Two pathways have been proposed for leucine catabolic breakdown in microbes; the most common involves the MC-CoA pathway to produce acetyl-CoA and acetoacetate (22). In P. aeruginosa, Pseudomonas putida, and Pseudomonas fluorescens, leucine catabolism occurs by the MC-CoA pathway (4, 12, 17, 18, 19). Leucine transaminase first converts leucine to 2-ketoisocaproic acid, and 2-ketoisocaproic dehydrogenase renders isovaleryl-CoA. This metabolite is converted to 3-methylcrotonyl-CoA by isovaleryl-CoA dehydrogenase; 3-methylcrotonyl-CoA carboxylase then produces 3-methylglutaconyl-CoA, which is converted to 3- hydroxy-3-methylglutaryl-CoA by 3-methylglutaconyl-CoA hydratase; and finally acetyl-CoA and acetoacetate are produced by 3-hydroxy-3-methylglutaryl-CoA lyase (12, 17, 18, 19, 26) (Fig. 1).
FIG. 1.
Catabolic pathways and proposed functions of the atu and liu cluster products from P. aeruginosa PAO1. (A and B) Upper and lower pathways of acyclic monoterpene catabolism; (C and D) Upper and lower pathways of leucine catabolism. AtuD, citronellyl-CoA dehydrogenase; AtuC/AtuF, geranyl-CoA carboxylase; AtuE, γ-carboxygeranyl-CoA hydratase; LiuE, 3-hydroxy-γ-carboxygeranyl-CoA and 3-hydroxy-3-methylglutaryl-CoA lyase; I, leucine transaminase; II, 2-ketoisocaproic dehydrogenase; LiuA, isovaleryl-CoA dehydrogenase; LiuB/LiuD, 3-methylcrotonyl- CoA carboxylase; LiuC, 3-methylglutaconyl-CoA hydratase. (Adapted from references 5, 9, 12, and 26.)
We previously described the P. aeruginosa gnyRDBHAL gene cluster, which encodes the enzymes for the AMTC degradation lower pathway (5); in a recent paper this was renamed the liuRABCDE cluster (12). Following our study (5), a highly conserved homologous cluster corresponding to the atu cluster was identified (12). In this study, we characterized the atu cluster, and we propose that the atu and liu clusters encode the enzymes involved in both AMTC and leucine utilization; the evolutionary relation of these gene clusters is also analyzed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this work are shown in Table 1. The strains were grown at 30°C in Luria broth or in M9 minimal medium (27). Solid media were prepared by adding 1.5% agar. Strains were tested for their ability to grow on M9 agar plates supplemented with citronellol (Merck) as the sole carbon and energy source (added as vapor) after an incubation for 48 h at 30°C. The growth of strains on branched-chain amino acids was tested as described by Martin et al. (18), using 0.3% (wt/vol) l-leucine or l-isoleucine supplemented with l-valine and l-isoleucine at 0.005% each (obtained from Sigma and Merck Co.). Antibiotic concentrations used for P. aeruginosa strains were as follows: streptomycin, 200 μg ml−1; carbenicillin, 100 μg ml−1; gentamicin (Gm), 100 μg ml−1; and tetracycline (Tc), 100 μg ml−1. Antibiotic concentrations used for E. coli strains were as follows: ampicillin, 100 μg ml−1; Gm, 20 μg ml−1; and Tc, 15 μg ml−1.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa PAO1SM | Spontaneous streptomycin-resistant mutant derived from strain PAO1 | 33 |
| P. fluorescens L1 | Environmental strain | Donated by E. Valencia |
| P. putida | Environmental strain | Donated by S. Aguilera |
| PAO atuC | PAO1::ISlacZ/hah (ID 10287), transposon insertion in ORF PA2888 | 13 |
| PAO atuD | PAO1::ISlacZ/hah (ID 15357), transposon insertion in ORF PA2889 | 13 |
| PAO atuE | PAO1::ISlacZ/hah (ID 8388), transposon insertion in ORF PA2890 | 13 |
| PAO atuF | PAO1::ISlacZ/hah (ID 10788), transposon insertion in ORF PA2891 | 13 |
| PAO2893 | PAO1::ISlacZ/hah (ID 8377), transposon insertion in ORF PA2893 | 13 |
| PAO liuD | PAO1::ISlacZ/hah (ID 11254), transposon insertion in ORF PA2012 | 13 |
| PAM1529 (atuF) | PAO1SM mutant obtained by Himar1::Gmr transposition in ORF PA2891, unable to grow on citronellol | This work |
| PAE80 (liuD) | PAO1SM mutant obtained by Himar1::Gmr transposition in ORF PA2012, unable to grow on citronellol | 5 |
| PAO atuF liuD | Double mutant strain derived from PAO atuF, mutated in liuD gene | This work |
| PAO atuF liuB | Double mutant strain derived from PAO atuF, mutated in liuB gene | This work |
| PAO atuF liuE | Double mutant strain derived from PAO atuF, mutated in liuE gene | This work |
| PAO atuC liuE | Double mutant strain derived from PAO atuC, mutated in liuE gene | This work |
| PAM liuA | PAO1SM liuA::Gmr mutant obtained by recombination in ORF PA2015 | 5 |
| PAM liuB | PAO1SM liuB::Gmr mutant obtained by recombination in ORF PA2014 | 5 |
| PAM liuD | PAO1SM liuD::Gmr mutant obtained by recombination in ORF PA2012 | 5 |
| PAM liuE | PAO1SM liuE::Gmr mutant obtained by recombination in ORF PA2011 | 5 |
| Plasmids | ||
| pUCP20 | pUC19-derived E. coli-Pseudomonas shuttle vector; Apr Cbr | 32 |
| pMO013850 | pLA2917 cosmid plus 25 kb of PAO1 genome, including the liu cluster | Pseudomonas Genetic Stock Center |
| pMO011609 | pLA2917 cosmid plus 25 kb of PAO1 genome, including the atu cluster | Pseudomonas Genetic Stock Center |
| pAL-22 | pUCP20 with 7.42-kb HindIII-EcoRV DNA fragment containing liuABCDE genes from pMO013850 cosmid | 5 |
| pAL-23 | pUCP20 with 7.84-kb KpnI-XbaI DNA fragment containing atuCDEF genes from pMO011609 cosmid | This work |
| pLP170 | lacZ transcriptional fusion vector able to replicate in P. aeruginosa | 23 |
| pANP2 | P2 liuB::lacZ fusion derived from pLP170 | 5 |
Genetic tests.
Construction of double mutants (Table 1) was done using triparental conjugation among the P. aeruginosa PAO1 mutant strain PAO atuF (open reading frame [ORF] PA2891) (13), the pRK2013 helper plasmid (8), and one of the pKAAA, pKAAB, and pKAAL plasmids, which contain the disrupted liuD (ORF PA2012), liuB (ORF PA2014), and liuE (ORF PA2011) genes, respectively, as described by Díaz-Pérez et al. (5). Transconjugants were selected on plates with Tc and Gm, and their growth ability on M9 medium with the appropriate carbon source was evaluated. For genetic complementation, the complete wild-type operons were obtained from a cosmid genomic library of P. aeruginosa PAO1 (Pseudomonas Genetic Stock Center of the Pseudomonas Genome Project [PGP]). Cosmids pMO013850 (containing the liu cluster) and pMO011609 (containing the atu cluster) carried a 25-kb PAO1 chromosomal DNA fragment. Fragments of 7.4 and 7.8 kb containing the liu and atu clusters were subcloned into the pUCP20 vector, rendering the pAL-22 and pAL-23 plasmids, respectively. Genetic complementation was assayed either by conjugation or by heat shock transformation of mutant strains (5), using the wild-type operons from the plasmids mentioned above (Table 1). Recombinants were selected on plates with the appropriate antibiotics and subjected to growth tests.
Quantification of β-galactosidase.
The β-galactosidase activities of P. aeruginosa strains harboring a LacZ transcriptional fusion were quantified in cultures grown at 30°C to the exponential phase (optical density at 600 nm of 0.6) in M9 medium supplemented with glucose (0.2%, wt/vol); cells were harvested by centrifugation, washed twice with M9 salts solution, and suspended in M9 salts. The inducer compound at 0.005% or 0.2% (wt/vol) was added to the cell suspensions and incubated at 30°C, aliquots were taken at intervals, and β-galactosidase activity was measured as described previously (27).
Detection of biotin-containing proteins.
Samples of 100 μg of total protein of cell-free extracts from cultures of P. aeruginosa strains were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) and electrophoretically transferred to nitrocellulose membranes (Amersham Biosciences). The membranes were blotted using avidin-horseradish peroxidase (HRP) (Bio-Rad) as indicated by the provider. HRP color development was done using 4-chloro-1-naphtol (Sigma) and H2O2. Biotinylated sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein standards (Bio-Rad) were used as molecular markers.
Determination of carboxylase activity.
Cultures of the P. aeruginosa PAO1 strain were grown in 50 ml of M9 with 0.2% of the appropriate carbon source for 48 h to 30°C with shaking. Cells were harvested by centrifugation and washed with 50 ml 100 mM K2HPO4, pH 8.0. Pellets were suspended in 5 ml of the same buffer, disrupted by sonication, and centrifuged for 10 min at 15,000 × g and 4°C to eliminate unlysed cells. Protein content was determined as described previously (27). Acetyl-CoA carboxylase (ACCase), propionyl-CoA carboxylase (PCCase), and MCCase activities in the crude extracts were measured by the incorporation of 14CO2 into acid nonvolatile material as previously described (10, 25). The reaction mixture contained 100 mM K2HPO4 (pH 8.0), 300 μg bovine serum albumin, 10 mM ATP, 10 mM MgCl2, 50 mM NaH14CO3 (specific activity, 1.96 GBq/mmol; 53 mCi/mmol; Amersham), cell extract (300 μg of protein), and 0.5 mM of the appropriate substrate (acetyl-CoA, propionyl-CoA, or 3-methylcrotonyl-CoA, from Sigma Co.) in a total reaction volume of 100 μl. The reaction was started by the addition of NaH14CO3, the mixture was incubated at 37°C for 10 min, and the reaction was stopped by adding 200 μl of 6 M HCl. The contents of the tubes were then evaporated to dryness at 90°C, and the residue was suspended in 100 μl of distilled water. Radioactivity was quantified using a liquid scintillation counter (Beckman LS Analyzer LS6000IC). Nonspecific CO2 fixation by crude extracts was assayed in the absence of substrate.
Metabolite analyses and degradation rates.
Strains were grown in 100 ml of M9 medium with 0.2% (wt/vol) succinic acid and 0.05% (wt/vol) casein peptone at 30°C with shaking for 18 h. Cells were harvested by centrifugation and washed twice with M9 salts solution. The pellet was suspended in the original volume with M9 salts, and citronellol was added to a final concentration of 0.1% (vol/vol). The suspension was incubated at 30°C with shaking, and aliquots of 25 ml were withdrawn at intervals and centrifuged at 10,000 × g for 10 min. Supernatants were saturated with NaCl (7.5 g) and extracted three times with 10 ml of ethyl acetate. Samples were collected, dried with anhydrous Na2SO4, concentrated by evaporation in a fume hood at 50°C, and suspended in 500 μl methanol. Samples of 1 μl were analyzed by gas chromatography (GC) (Perkin-Elmer Auto System gas chromatograph), using an Agilent/J&W HP-FFAP GC column (length, 30 m; inner diameter, 0.25 mm; film, 0.25 μm). The analyses were carried out with a 200°C injection temperature and a 250°C detection temperature, with a method start to 60°C for 2 min, an increase to 135°C (25°C/min), and then an increase to 210°C (5°C/min). Commercial citronellol, citral, citronellic acid, and geranic acid mix isomers were used in the GC as standard identification compounds.
Leucine and citronellol degradation rates.
Strains were grown in 20 ml of M9 medium with 0.2% (wt/vol) citric acid plus 0.2% (wt/vol) leucine and 0.075% (vol/vol) citronellol at 30°C with shaking for 18 h. Cells were harvested by centrifugation and washed twice with M9 salts solution. The pellet was suspended in the original volume with M9 salts with leucine plus citronellol (0.2% each). The bacterial suspension was incubated at 30°C with shaking, and aliquots of 1 ml were withdrawn at intervals and centrifuged at 10,000 × g for 10 min. Leucine and citronellol concentrations in supernatants were determined by high-pressure liquid chromatography. Samples of 50 μl were analyzed using a C18 reverse-phase analytical column (Alltech), carried out with isocratic runs using deionized water for 20 min for leucine and with a water-acetonitrile mix as the mobile phase for 5 min (60:40), 10 min (20:80), and 10 min (0:100) for citronellol quantification runs. Detection was done at a UV wavelength of 210 nm, using a Perkin-Elmer high-pressure liquid chromatograph.
Sequence analysis.
The nucleotide and amino acid sequences analyzed are designated ORFs PA2011 to PA2016 and ORFs PA2888 to PA2891 in PGP (http://www.pseudomonas.com) (30) and were renamed the liu and atu clusters, respectively (12).
Phylogenetic analysis.
Protein sequence data were obtained from the ortholog clusters of the SSBD KEGG (Kyoto Encyclopedia of Genes and Genomes) and MBGD (Microbial Genome Database for Comparative Analysis) databases by using LiuD (PA2012) and AtuF (PA2891) as queries. Redundant protein sequences were removed, resulting in 201 protein sequences. Progressive multiple protein sequence alignment was calculated with the ClustalW version 1.83 software (11), and the alignment was refined with the MUSCLE version 3.51 program (24) and was later corrected according to the results of gapped BlastP (21). Phylogenetic and molecular evolutionary analyses were conducted with MEGA version 2.1 (16) using distance-based methods, i.e., unweighted pair group method with arithmetic mean (UPGMA), neighbor joining (NJ), and minimum evolution (ME), with the Poisson correction distance method and with gaps treated by pairwise deletion. Confidence limits of branch points were estimated by 1,000 bootstrap replications.
RESULTS AND DISCUSSION
Identification of the atu cluster.
In a previous work, we characterized the P. aeruginosa gnyRDBHAL gene cluster, which is involved in AMTC catabolism; it was renamed the liuRABCDE cluster because of its involvement in leucine and isovalerate utilization (12). From a library of about 1 × 106 P. aeruginosa Gmr mutants generated by transposition (5), about 5,000 mutants were screened for their ability to grow on AMTC as the sole carbon source. Genomic regions interrupted by the transposon in the mutants were cloned in the pBluescript SK vector, and the flanking DNA fragments were sequenced (data not shown). Sequence analysis from strain PAM1529 showed that transposon insertion occurred in the PA2891 ORF from the PGP database. By further analysis of the regions flanking ORF PA2891 in the PAO1 genome, a putative atu cluster containing ORFs PA2888, PA2889, PA2890, and PA2891 was located, which corresponded with the atu cluster named for its involvement in AMTC utilization (12).
Sequence analysis of the atu cluster.
Nucleotide sequence analysis of the atu cluster showed that it could be constituted by eight probable structural genes, the ORFs PA2886 to PA2893 (Fig. 2). ORFs PA2888 (AtuC) and PA2891 (AtuF) showed similarity with the β and α subunits of the acyl-CoA carboxylases family (ACCase, PCCase, and MCCase) and 63% and 64% of similarity with the LiuB (PA2014) and LiuD (PA2012) proteins from P. aeruginosa, respectively. AtuF showed the ATP-binding site (GxGxxG), the sequence implicated in CO2 fixation (RDCS), the catalytic site of the biotin-dependent carboxylase family (EMNTR), and the biotin-carboxyl carrier domain (AMKM) (5, 14, 15, 29). AtuC showed the acyl- CoA-binding and the carboxybiotin-binding domains (5, 14, 29). These data suggest that the atuC and atuF genes could encode the subunits of a novel acyl-CoA carboxylase, as also proposed by Hoschle et al. (12).
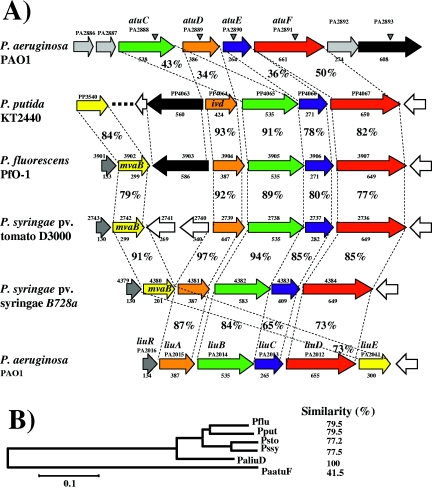
FIG. 2.
Sequence analysis of products encoded in the atu and liu gene clusters. (A) Genetic arrangement of the atu/liu homolog clusters from Pseudomonas species. The species and strain names are shown at the left. ORF arrangement and transcription direction are shown by arrows. Locations of transposon insertions in the P. aeruginosa mutants are indicated with shaded arrowheads. Numbers above arrows show the ORF number assigned in the respective genome sequencing project. Identified genes are also shown. The amino acid number of the ORF is indicated below the arrows. Corresponding ortholog proteins are indicated by the same color. Orange, LiuA; green, LiuB; blue, LiuC; red, LiuD; yellow, lyase (LiuE); black, PA2893.The percentages of amino acid identity between upper and lower ORFs are indicated. (B) Phylogenetic tree of the Atu/Liu homolog proteins of Pseudomonas species. The amino acid sequences of LiuABCD and AtuDCEF were put together as one sequence and aligned with the homologous sequences from the strains indicated. The alignment was done using the Clustal W software, and the tree was done by the neighbor joining in the Mega2 software with a bootstrap of 1,000 replicates. The numbers indicate the percent similarities between the amino acids sequences aligned. Pflu, P. fluorescens PfO-1; Pput, P. putida KT2440; Psto, P. syringae pv. tomato D3000; Pssy, P. syringae pv. syringae B728a; PaatuF, P. aeruginosa PAO1 atu cluster; PaliuD, P. aeruginosa PAO1 liu cluster. The evolutionary distance scale bar is shown at the bottom.
ORF PA2889 (AtuD) showed high amino acid sequence similarity with acyl-CoA dehydrogenases and 34% of identity with the LiuA protein of P. aeruginosa. ORF PA2890 (AtuE) showed similarity with enoyl-CoA hydratase/isomerase and 38% identity with the LiuC protein of the PAO1 strain. ORFs PA2886, PA2887, and PA2892 showed similarity with predicted hypothetical proteins and short-chain dehydrogenase family proteins, respectively, whereas ORF PA2893 (AtuH) showed similarity with very-long-chain acyl-CoA synthetases, suggesting that these three ORFs could be related to the atu cluster, although mutagenesis results (see below) do not indicate that this is so. These data suggest that the atuCDEF gene may code for products homologous to those of the liuABCD genes (Fig. 2).
atu/liu homologous clusters among Pseudomonas species.
The amino acid sequences encoded by the most conserved gene homologs (AtuC, AtuD, AtuE, and AtuF) corresponding to several Pseudomonas species were analyzed by multiple alignments. Probable homologs were found in genomes from P. putida KT2440, P. fluorescens PfO-1, Pseudomonas syringae pv. tomato D3000, and P. syringae pv. syringae B728a (Fig. 2A). Interestingly, only one homolog cluster was found in these genomes, making evident the catabolic versatility of P. aeruginosa, where two homolog clusters were found.
The resultant alignments and phylogenetic tree showed that the liu cluster products from P. aeruginosa had more similarity with the products of the liu homolog clusters from the other Pseudomonas species (77.2 to 79.5%) than with the products of the atu cluster from P. aeruginosa (41.5%) (Fig. 2B). This tendency was also observed when the ORFs were analyzed separately, where amino acid identities ranged from 65% to 94% with the liu cluster versus 34% to 50% with the atu cluster (Fig. 2A). These analyses showed that the homologous proteins encoded by the liu genes are conserved in both amino acid sequence and genetic arrangement in the Pseudomonas species studied.
Interestingly, a liuE homolog gene was not found in the atu cluster or elsewhere in the PAO1 genome (Fig. 2A), suggesting that LiuE may have a dual function, being used in both the AMTC and the leucine catabolic pathways. This is the first study to the genetic level in which a hydroxymethyl-glutaryl-CoA lyase (HMG-CoA lyase) homolog (LiuE) has been found to be involved in both AMTC and leucine catabolism in bacteria (see below).
Growth analysis of atu and liu mutants.
The P. aeruginosa PAO1 wild-type strain was able to grow on leucine or isoleucine as well as on citronellol as the sole carbon and energy source, while P. fluorescens and P. putida strains were unable to grow on citronellol (Table 2). The atu mutants with an outward-facing promoter, which reduces polar effects on downstream gene expression (PAO atuC, PAO atuD, PAO atuE, and PAO atuF), were unable to grow on citronellol but were able to grow on leucine, except for the PAO atuC and PAO atuF mutants, whose growth was impaired. Growth on leucine, isoleucine, and citronellol was not affected in the PAO2893 mutant, which contains a transposon insertion in ORF PA2893 (Table 2), suggesting that this ORF is not involved in leucine or AMTC catabolic pathways.
TABLE 2.
Growth phenotypes of Pseudomonas strains
| Strain | Growth ina:
|
Doubling time (h)e | ||
|---|---|---|---|---|
| Citronellol | l-Leucine | l-Isoleucine | ||
| P. aeruginosa PAO1SM | +++ | +++ | +++ | 8.2 |
| P. fluorescens L1 | − | +++ | +++ | NDf |
| P. putida | − | +++ | +++ | ND |
| PAO2893b | +++ | +++ | +++ | 8.0 |
| PAO atuDb | − | +++ | +++ | 7.5 |
| PAO atuEb | − | +++ | +++ | 11.2 |
| PAO atuCb | − | ++ | +++ | 13.1 |
| PAO atuFb | − | + | +++ | 13.5 |
| PAO atuF(pAL-22)b | − | ++ | +++ | 6.5 |
| PAO atuF(pAL-23)b | +++ | +++ | +++ | 7.0 |
| PAM1529 (atuF mutant)c | − | +++ | +++ | 13.1 |
| PAM1529(pAL-22)c | − | +++ | +++ | 6.5 |
| PAM1529(pAL-23)c | +++ | +++ | +++ | 6.5 |
| PAE80 (liuD mutant)c | − | − | +++ | ND |
| PAE80(pAL-22)c | +++ | +++ | +++ | 9.0 |
| PAE80(pAL-23)c | − | − | +++ | ND |
| PAM liuDd | − | − | +++ | ND |
| PAM liuD(pAL-22)d | +++ | +++ | +++ | 9.7 |
| PAM liuD(pAL-23)d | − | − | +++ | ND |
| PAO liuDb | − | +++ | +++ | ND |
| PAM liuAd | − | − | +++ | ND |
| PAM liuBd | − | − | +++ | ND |
| PAM liuEd | − | − | +++ | ND |
| PAO atuF liuD | − | − | +++ | ND |
| PAO atuF liuB | − | − | +++ | ND |
| PAO atuF liuE | − | − | + | ND |
| PAO atuC liuE | − | − | + | ND |
The strains were inoculated on M9 plates with the compounds indicated as sole carbon sources and incubated for 48 h at 30°C as described in Materials and Methods. +++, good growth; ++, medium growth; +, low growth; −, no growth.
Mutant obtained by Jacobs et al. (13) (no polar effects over downstream gene expression).
Mutant described in this work (polar effects over downstream gene expression).
Mutant obtained by homologous recombination (polar effects over downstream gene expression).
Doubling times of cultures grown in M9 minimal liquid medium with leucine (0.2%, wt/vol) as the sole carbon source, incubated at 30°C with shaking at 150 rpm. The growth was measured by optical density at 600 nm and calculated in the exponential growth phase.
ND, not determined.
Mutants obtained with the mutagenesis system that provokes polar effects over downstream gene expression by transposition or homologous recombination, i.e., PAE80 (affected in the liuD gene), PAM liuA, PAM liuB, PAM liuD, and PAM liuE, were unable to grow on both citronellol and leucine, whereas PAM1529 (affected in the atuF gene) was unable to grow on citronellol. When the PAM1529, PAE80, and PAM liuD mutants were transformed with plasmid pAL-22 (contains the liu cluster) or pAL-23 (contains the atu cluster), only pAL-23 complemented the growth on citronellol in the PAM1529 mutant, while in the PAE80 and PAM liuD mutants, only pAL-22 restored the ability to grow on both leucine and citronellol (Table 2). In contrast, for the PAO atuF mutant, unable to grow on citronellol and impaired in leucine growth, leucine growth was improved with both plasmids, but only with pAL-23 was growth on citronellol recovered (Table 2). These results suggest that both the atu and liu clusters are related to the ability to grow on both citronellol and leucine and also suggest that their respective acyl-CoA carboxylases encoded in the atu and liu clusters may be shared in both AMTC and leucine catabolism. In addition, all single mutant strains tested were able to grow on isoleucine (Table 2), suggesting that the phenotypes shown in the mutants were exclusively associated with leucine or citronellol metabolism.
The phenotypes of double mutants affected in the atu and liu cluster genes (atuF liuD, atuF liuB, atuF liuE, and atuC liuE mutants) showed a clear deficiency in growth on both AMTC and leucine (Table 2). The impaired growth on isoleucine in the last two strains is still unclear. These results show that the atu and liu clusters are absolutely necessary for AMTC catabolism and that mutations in some of their genes affect leucine utilization.
Growth rates of the atu and liu mutants.
The PAO1 strain grown on leucine as the sole carbon and energy source had a doubling time of 8.2 h, whereas the PAO atuE, PAO atuC, and PAO atuF mutants showed impaired growth in leucine, with doubling times of 11.2, 13.1, and 13.5 h, respectively (Table 2). When the PAO atuF mutant strain was transformed with the pAL-22 (containing the liu cluster) or pAL-23 (containing the atu cluster) plasmid, the growth rate in leucine was even better in the transformed mutants than in the wild-type strain, showing doubling times of 6.5 h and 7.0 h, respectively (Table 2), probably due to protein overexpression. For the PAE80 strain (liuD gene mutant), leucine growth was recovered with the pAL-22 plasmid, in the same proportion as with the PAO1 strain, but not with pAL-23 (Table 2). As expected, the atuF liuD, atuF liuB, atuF liuE, and atuC liuE double mutants and the PAM liuE mutant did not grow on leucine (Table 2). The leucine complementation results for both the atuF and liuD mutants with the atu or liu clusters (contained in the pAL-23 or pAL-22 plasmid, respectively) suggest that the leucine catabolic pathway could share gene products. The PAM1529 (atuF mutant) and PAE80 (liuD mutant) strains were unable to grow on citronellol, but they recovered the growth on citronellol in similar proportion as the PAO1 strain when were transformed with the pAL-23 and pAL-22 plasmids, respectively (Table 2).
In addition, the PAM liuE mutant was complemented in its growth on both leucine and citronellol by the pMO013850 cosmid (containing the liu cluster), but not by the pMO011609 cosmid (containing the atu cluster) (data not shown), confirming that LiuE (an HMG-CoA lyase homolog) is located in the liu cluster in P. aeruginosa and that it is absolutely necessary for the degradation of AMTC and leucine.
When cosmids pMO011609 and pMO013850 were transferred to P. putida and P. fluorescens strains (both wild-type strains are able to grow on leucine [Table 2]), neither strain was able to grow on citronellol (data not shown), suggesting that additional genes, not contained in their genomes or in the liu and atu clusters, are required for AMTC degradation.
Metabolite accumulation.
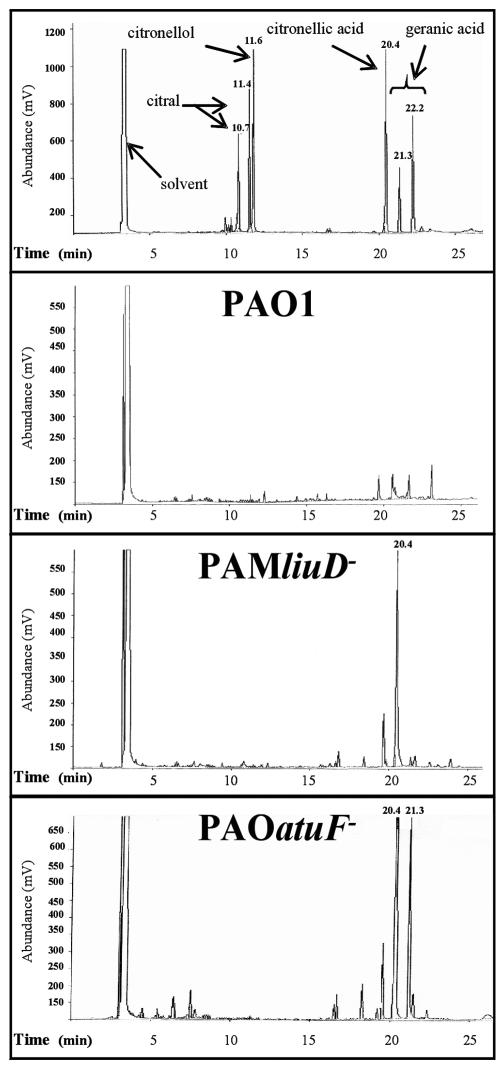
P. aeruginosa liuD and liuB mutants have been shown to accumulate citronellic and geranic acids when incubated with citronellol (5). The liuD mutant grown in the presence of citronellol accumulated principally citronellic acid, while the atuF mutant accumulated both citronellic and geranic acids, compared to the wild-type strain (Fig. 3). Accumulation of citronellic and geranic acids in liuD and atuF mutants suggests that the activities of both GCCase and MCCase are indispensable for AMTC degradation (Fig. 1).
FIG. 3.
Metabolite accumulation in cultures of P. aeruginosa grown in citronellol. Cultures were grown in M9 medium with succinic acid as the carbon source for 18 h, and citronellol was then added. The metabolites were extracted from the supernatants and analyzed at 48 h after the addition of citronellol by gas chromatography as described in Materials and Methods. Commercial citral (10.7 and 11.4 min), citronellol (11.6 min), citronellic acid (20.4 min), and geranic acid mix isomers (21.3 and 22.2 min) were used as standard compounds. Peaks with retention times of 20.4 and 21.3 min correspond to citronellic acid and geranic acid, respectively.
Leucine and citronellol assimilation rates.
Assimilation kinetics of the P. aeruginosa wild-type PAO1 strain showed utilization of leucine and citronellol in liquid cultures with these compounds as carbon sources of 48% and 62%, respectively, after 24 h of incubation (data not shown). Utilization of leucine and citronellol by the PAO liuD mutant was 3% and 40%, and that by the PAO atuF mutant was 17% and 4%, respectively (data not shown). The assimilation of leucine by the PAO liuD mutant was slower than that of the PAO atuF mutant. In contrast, the ability to assimilate citronellol was more severely affected in the PAO atuF mutant than in the PAO liuD mutant. These results further confirm that the liu cluster is principally involved in leucine assimilation and the atu cluster in citronellol catabolism and also suggest that both clusters are involved in leucine and AMTC catabolic pathways and that the liu and atu clusters probably share gene products. These results are in disagreement with those described by Hoschle et al. (12), who concluded that the GCCase is needed only for utilization of AMTC and that the MCCase cannot replace GCCase (12).
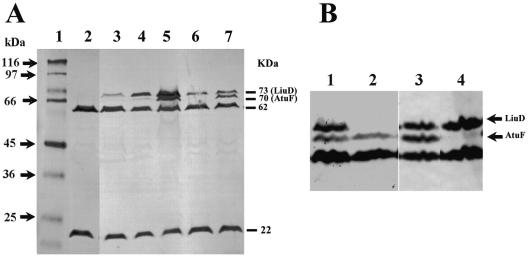
Expression of liuD and atuF genes.
Western blotting analysis from cultures of PAO1 grown in minimal medium with citronellol as the sole carbon source usually shows four major biotinylated proteins with molecular masses of 73, 70, 62, and 22 kDa (5) (Fig. 4a). The 73- and 70-kDa proteins have been proposed to correspond to α-GCCase and α-MCCase subunits, respectively (5). However, Hoschle et al. (12) recently reported that P. aeruginosa PAO1 showed three biotinylated proteins (74, 71, and 63 kDa) when grown on citronellate, suggesting by trypsin fingerprint analysis that these corresponded to the ORFs PA2012 (71.28 kDa), PA2891 (71.74 kDa), and PA5435 (66.09 kDa), respectively. The 74- and 71-kDa proteins were named LiuD and AtuF, respectively (12). When the wild-type PAO1 strain was grown on glucose, the LiuD (73 kDa) and AtuF (70 kDa) proteins were not observed (Fig. 4A, lane 2); with isoleucine as the sole carbon and energy source, a slight expression of LiuD was observed (Fig. 4A, lane 3). With leucine, LiuD, but not AtuF, was present (Fig. 4A, lane 4), suggesting that LiuD is related to leucine and isoleucine metabolism. When citronellol was used for growth, both the LiuD and AtuF proteins were overexpressed (Fig. 4A, lane 5). These results show that citronellol, leucine, and isoleucine, in decreasing order, were able to induce the expression of the LiuD protein, whereas only citronellol was able to induce the expression of both the LiuD and AtuF proteins, confirming further that both liu and atu cluster genes are required for AMTC catabolism. In addition, in extracts obtained from cultures of the PAO1 strain grown on glucose plus either leucine or citronellol, the expression levels of the LiuD and AtuF proteins were somewhat decreased (Fig. 4A, lanes 6 and 7), sug- gesting that glucose exerts a moderate catabolite repression effect on the expression of both liuD and atuF genes.
FIG. 4.
Western blot analysis of biotinylated proteins from P. aeruginosa PAO1 derivatives. (A) Lanes: 1, molecular masses of protein standards; 2 to 7, cell extracts from the PAO1 strain grown on glucose (lane 2), isoleucine (lane 3), leucine (lane 4), citronellol (lane 5), glucose plus leucine (lane 6), and glucose plus citronellol (lane 7). Proteins of 73, 70, 62, and 22 kDa identified with avidin-HRP conjugate, corresponding to biotinylated subunits from alpha subunits of geranyl-CoA carboxylase (AtuF), 3-methylcrotonyl-CoA carboxylase (LiuD), a putative acyl-CoA carboxylase, and acetyl-CoA carboxylase, respectively, are indicated. (B) Biotinylated proteins from cultures grown in glucose plus citronellol. Lanes: 1 and 3, PAO1 wild-type strain; 2, PAM liuD mutant; 4, PAO atuF mutant. The positions of the LiuD and AtuF proteins are indicated.
In extracts from the PAM liuD and PAO atuF mutants grown on citronellol plus glucose, only the 70- and 73-kDa proteins were detected, respectively (Fig. 4B). This confirms that the LiuD and AtuF proteins are encoded by the liuD and atuF genes, respectively, as was also found by trypsin fingerprint protein analysis (12). These results suggest that the LiuD protein corresponds to the alpha subunit of MCCase (α-MCCase) and that the AtuF protein corresponds to α-GCCase.
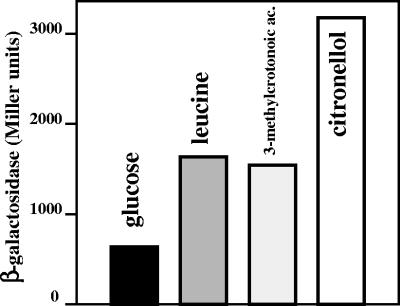
The expression of the liu cluster in PAO1 was also tested using a liuB::lacZ transcriptional fusion. The reporter gene was induced threefold by either leucine or 3-methylcrotonoic acid (a compound related to the leucine catabolic pathway), whereas citronellol induced it about sixfold, compared to the glucose-grown cultures (Fig. 5). This induction was also reflected at the translational level, since the LiuD protein showed a higher level in cultures grown on citronellol than in those grown on leucine (Fig. 4A). These results show that citronellol is a better inducer of the liu cluster than leucine.
FIG. 5.
Induction of the liuB::lacZ transcriptional fusion. Cultures of the PAO1 strain with plasmid pANP2 (P2liuB::lacZ) were grown in M9 medium with glucose. β-Galactosidase activity was measured after the addition of the inducer compounds glucose (0.2%), leucine (0.005%), 3-methylcrotonoic acid (0.005%), and citronellol (0.005%). Data given correspond to Miller units after 3 h of induction with the compound indicated and are the averages of two independent experiments done in duplicate; the standard deviation was less than 5% of the given value.
Carboxylase activity.
In cell extracts from P. aeruginosa PAO1 obtained from cultures grown on glucose or on glucose plus citronellol or leucine, the MCCase enzymatic activity was low (Table 3). These results are in agreement with our previous finding that glucose exerts catabolite repression on liuD expression. The activity of MCCase increased 6-fold in cultures of the PAO1 strain grown on leucine compared to extracts from cultures grown on glucose; however, cultures grown on citronellol showed a 16-fold increase in MCCase activity (Table 3). Growth on citronellol had no significant differences from growth on glucose when the ACCase and PCCase enzyme activities were measured, while growth on glucose plus leucine showed only slight differences (Table 3). These results support our data that the MCCase, but not the ACCase and PCCase activities, are induced by citronellol (best inducer) as well as leucine. The differences in enzymatic activity of MCCase between the extracts from cultures grown on citronellol and those grown on leucine could be due to an increase of the expression level of the MCCase (as it was shown in the Western blot analysis) or to unspecific activity of GCCase by geranyl-CoA or 3-methylcrotonyl-CoA substrates. This has been previously described for P. citronellolis (7, 9) and could suggest that the GCCase enzyme (encoded by the atuCF genes) may also have MCCase activity.
TABLE 3.
Carboxylase activity in P. aeruginosa PAO1 cell extracts
| Carbon sourceb | Carboxylase activitya (U/mg of protein) on substrate:
|
||
|---|---|---|---|
| 3-Methylcrotonyl-CoA | Acetyl-CoA | Propionyl-CoA | |
| Glucose | 82.0 | 36.2 | 47.8 |
| Glucose + leucine | 97.0 | 143.0 | 82.0 |
| Glucose + citronellol | 45.1 | 50.0 | 44.0 |
| Leucine | 492.0 | NDc | ND |
| Citronellol | 1,312.0 | 73.8 | 66.9 |
Enzymatic quantification was carried out as described in Materials and Methods. One unit of enzyme activity catalyzed the incorporation of 1 μmol of 14C into acid-stable product/min. Data given are the averages of two quantifications in duplicate; the standard deviation was less than 10%.
Cell extracts were grown on minimal medium M9 with the carbon source indicated.
ND, not determined.
Phylogenetic analysis.
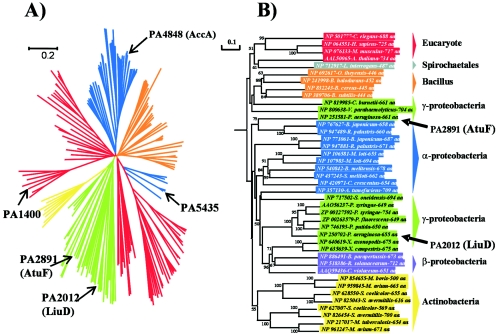
The liu cluster products from P. aeruginosa PAO1 showed more similarity to the products of the liu homologs from other Pseudomonas species (also called mcc homologs) than to the proteins encoded in the atu homologous genes from PAO1 (Fig. 2). Amino acid sequence alignments and phylogenetic analysis of the more important enzymes of the pathway, LiuD/AtuF, were done (Fig. 6). The phylogenetic tree of the LiuD/AtuF (α-MCCase/α-GCCase) proteins with their orthologs showed five phylogenetic groups of the biotinylated subunit of acyl-CoA carboxylases: ACCase, PCCase, MCCase, pyruvate carboxylases, and urea carboxylases (see the supplemental material). The LiuD and AtuF proteins from P. aeruginosa were located in the MCCase root, as shown in Fig. 6A. The LiuD protein from P. aeruginosa PAO1 was phylogenetically related to the MccA proteins of P. putida KT2440, P. fluorescens PfO-1, P. syringae pv. tomato D3000, and P. syringae pv. syringae B728a (Fig. 6A). Further analysis of the MCCase root showed that the similarities of AtuF and LiuD to their homologs from Rhizobiaceae of the Alphaproteobacteria, such as Bradyrhizobium japonicum and Rhodopseudomonas palustris, were higher than those to the LiuD protein from P. aeruginosa itself, which is classified in the Gammaproteobacteria root (Fig. 6B). In these bacteria the liu homolog cluster could be involved in leucine catabolism, while both the liu and atu clusters in P. aeruginosa are involved in the metabolism of both leucine and AMTC, as described above. These results further suggest that the current atu cluster from P. aeruginosa PAO1 probably originated by horizontal transfer from bacteria of the Rhizobiaceae (from the Alphaproteobacteria root) that contain a GC percentage (64.66 and 65.53% for B. japonicum and R. palustris, respectively) close to that of P. aeruginosa (67.14%). Also, codon usage and codon frequency among these bacteria are highly similar (codon usage database, http://www.kazusa.or.jp/codon/). Genetic transfer has been shown to occur in several examples of the evolution of catabolic pathways in bacteria (31, 34).
FIG. 6.
Phylogenetic trees of AtuF/LiuD protein orthologs. Trees were obtained by the UPGMA, NJ, and ME methods in the Mega2 package as described in Materials and Methods, and the NJ tree is shown. (A) The tree shows the five phylogenetic groups of the biotinylated subunits of acyl-CoA carboxylases aligned; the probable functions of the ortholog proteins are indicated in the phylogenetic groups, in agreement with some characterized carboxylases: acetyl-CoA carboxylases (blue), propionyl-CoA carboxylases (orange), 3-methylcrotonyl-CoA carboxylases (green), pyruvate carboxylases (red), and urea carboxylases (yellow). Acyl-CoA carboxylases from P. aeruginosa PAO1 are shown by arrows. (B) Phylogenetic tree of MCCase root constructed with the 41 proteins showing the higher phylogenetic relationship. The AtuF and LiuD proteins of P. aeruginosa are shown by arrows. Bootstrap values of higher than 50% obtained by the UPGMA, NJ, and ME methods are shown. Blue, Alphaproteobacteria; purple, Betaproteobacteria; green, Gammaproteobacteria; yellow, Actinobacteria; orange, Bacillus; gray, Spirochaeta; red, eukaryote.
In conclusion, our results suggest that both the atu and liu clusters encode the enzymes involved in the AMTC and leucine catabolic pathways, and probably both carboxylase enzymes (encoded by the atuCF and liuBD genes) have principally GCCase and MCCase activities, respectively. In addition, the atu cluster from P. aeruginosa PAO1 lacks an HMG-CoA lyase homolog (LiuE), suggesting a dual function for this enzyme, catalyzing the deacetylation of both 3-hydroxy-γ- carboxygeranyl-CoA in AMTC catabolism and 3-hydroxy-3-methylglutaryl-CoA for leucine breakdown (Fig. 1). Phylogenetic analysis suggests that the atu cluster was acquired by a horizontal transfer event, probably from Rhizobiaceae, which by a xenologous gene displacement event rendered in P. aeruginosa the heterofunctional liu and atu clusters, implicated in both the AMTC and leucine catabolic pathways.
Supplementary Material
Acknowledgments
We thank M. A. Jacobs, the Pseudomonas Genome Project, and the Pseudomonas Genetic Stock Center for mutant strain and cosmid donation and the Pseudomonas aeruginosa Community Annotation Project for use of the updated database.
This research was funded by CONACYT (J35095-B) and C.I.C./UMSNH grants.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, M. D., P. Che, J. Song, B. J. Nikolau, and E. S. Wurtele. 1998. 3-Methylcrotonyl-coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol. 118:1127-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos-García, J., and G. Soberón-Chávez. 2000. Degradation of the methyl substituted alkene, citronellol, by Pseudomonas aeruginosa, wild type and mutant strains. Biotechnol. Lett. 22:235-237. [Google Scholar]
- 3.Cantwell, S. G., E. P. Lau, D. S. Watt, and R. R. Fall. 1978. Biodegradation of acyclic isoprenoids by Pseudomonas species. J. Bacteriol. 153:324-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad, R. S., L. K. Massey, and J. R. Sokatch. 1974. d- and l-isoleucine metabolism and regulation of their pathways in Pseudomonas putida. J. Bacteriol. 11:103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Díaz-Pérez, A. L., N. A. Zavala-Hernández, C. Cervantes, and J. Campos-García. 2004. The atuRDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 70:5102-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fall, R. R., J. L. Brown, and T. L. Schaeffer. 1979. Enzyme recruitment allows the biodegradation of recalcitrant branched hydrocarbons by Pseudomonas citronellolis. Appl. Environ. Microbiol. 38:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fall, R. R., and M. L. Hector. 1977. Acyl-coenzyme A carboxylases. Homologous 3-methylcrotonyl-CoA and geranyl-CoA carboxylases from Pseudomonas citronellolis. Biochemistry 16:4000-4005. [DOI] [PubMed] [Google Scholar]
- 8.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan, X., T. Diez, T. K. Prasad, B. J. Nikolau, and E. S. Wurtele. 1999. Geranoyl-CoA carboxylase: a novel biotin-containing enzyme in plants. Arch. Biochem. Biophys. 362:12-21. [DOI] [PubMed] [Google Scholar]
- 10.Hector, M. L., and R. R. Fall. 1976. Multiple acyl-coenzyme A carboxylases in Pseudomonas citronellolis. Biochemistry 15:3465-3472. [DOI] [PubMed] [Google Scholar]
- 11.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 12.Hoschle, B., V. Gnau, and D. Jendrossek. 2005. Methylcrotonyl-CoA and geranyl-CoA carboxylases are involved in leucine/isovalerate utilization (Liu) and acyclic terpene utilization (Atu), and are encoded by liuB/liuD and atuC/atuF, in Pseudomonas aeruginosa. Microbiology 151:3649-3656. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura, Y., R. Miyake, Y. Tokumasu, and M. Sato. 2000. Molecular cloning and characterization of two genes for the biotin carboxylase and carboxyltransferase subunits of acetyl coenzyme A carboxylase in Myxococcus xanthus. J. Bacteriol. 182:5462-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo, H., K. Shiratsuchi, T. Yoshimoto, T. Masuda, A. Kitazono, D. Tsuru, M. Anai, M. Sekiguchi, and T. Tanabe. 1991. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc. Natl. Acad. Sci. USA 88:9730-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Marshall, V. P., and J. R. Sokatch. 1968. Oxidation of d-amino acids by particulate enzyme from Pseudomonas aeruginosa. J. Bacteriol. 95:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, R. R., V. D. Marshall, J. R. Sokatch, and L. Unger. 1973. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J. Bacteriol. 115:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massey, L. K., R. S. Conrad, and J. R. Sokatch. 1974. Regulation of leucine catabolism in Pseudomonas putida. J. Bacteriol. 118:112-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKean, A. L., J. Ke, J. Song, P. Che, S. Achenbach, B. J. Nikolau, and E. S. Wurtele. 2000. Molecular characterization of the non-biotin-containing subunit of 3-methylcrotonyl-CoA carboxylase. J. Biol. Chem. 275:5582-5590. [DOI] [PubMed] [Google Scholar]
- 21.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 22.Poston, J. M. 1976. Leucine 2,3-aminomutase, an enzyme of leucine catabolism. J. Biol. Chem. 251:1859-1863. [PubMed] [Google Scholar]
- 23.Preston, M. J., P. C. Seed, D. S. Toder, B. H. Iglewski, D. E. Ohman, J. K. Gustin, J. B. Goldberg, and G. B. Pier. 1997. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect. Immun. 65:3086-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert, E. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez, E., and H. Gramajo. 1999. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2). Microbiology 145:3109-3119. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez, J. M., P. Ruíz-Salas, M. Ugarte, and M. A. Peñalva. 2004. Fungal metabolic model for 3-methylcrotonyl-CoA carboxylase deficiency. J. Biol. Chem. 279:4578-4587. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schaeffer, T. L., S. G. Cantwell, J. L. Brown, D. S. Watt, and R. R. Fall. 1979. Microbial growth on hydrocarbons: terminal branching inhibits biodegradation. Appl. Environ. Microbiol. 38:742-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song, J., E. S. Wurtele, and B. J. Nikolau. 1994. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc. Natl. Acad. Sci. USA 91:5779-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stover, K. C., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1: an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 31.Van Der Meer, J. R., W. M. De Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West, S. E. H., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia coli-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 128:81-86. [DOI] [PubMed] [Google Scholar]
- 33.Wong, S. M., and J. J. Mekalanos. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:10191-10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh, W. K., and L. N. Ornston. 1980. Origins of metabolic diversity: substitution of homologous sequences into genes for enzyme with different catalytic activities. Proc. Natl. Acad. Sci. USA 77:5365-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.