Abstract
The carbazole dioxygenase genes were introduced into a dibenzothiophene degrader. The recombinant Rhodococcus erythropolis SN8 was capable of efficiently degrading dibenzothiophene and carbazole simultaneously. SN8 could also degrade various alkylated derivatives of carbazole and dibenzothiophene in FS4800 crude oil by just a one-step bioprocess.
Acid precipitation resulting from fossil fuel combustion has driven enormous efforts to remove contaminants from fossil fuels. Heteroatoms such as nitrogen and sulfur in crude oil can also poison the catalysts used in catalytic cracking and hydrotreating processes (6). Microorganisms possess the capability to metabolize sulfur- and nitrogen-containing compounds in crude oil (18). Dibenzothiophene (DBT) has been used as a model sulfur compound, and research has been focused on strains that can selectively remove sulfur by a 4S pathway, in which DBT is converted to 2-hydroxybiphenyl (2-HBP) (5, 8, 12, 14, 15). Carbazole (CA) is the most abundant nitrogen-containing compound in many petroleum samples and was therefore chosen as a model compound (7). A variety of CA-degrading microorganisms have been reported (9, 10, 17, 21, 22). These different CA degraders follow similar degradation pathways, and the first step is catalysis by CA dioxygenase, which converts CA to 2′-aminobiphenyl-2,3-diol (18, 20).
Unlike biodesulfurization, where sulfur is selectively removed from substrates, leaving the hydrocarbon portion of the molecule intact, the pathway of CA degradation only liberates nitrogen in the course of completely degrading the substrates (1). Mutants or recombinants capable of carrying out only the first step(s) of the CA degradation pathway could be used to avoid the loss of fuel value. Since the nitrogen compounds in petroleum refinery feed stocks are known to poison catalysts, the microbial products from CA degradation might be expected to cause less catalyst inhibition than their parent compounds (18). Therefore, there is much hope that the efficiency of conventional catalytic processing of crude oil can be improved by microbial modification of nitrogenous catalyst poisons. Blocked mutants also have the ability to preserve the carbon content of the fuel being treated, retaining the fuel value. However, there have been no reports concerning host engineering to investigate the partial transformation pathway of CA in crude oil as a possible alternative to its total removal.
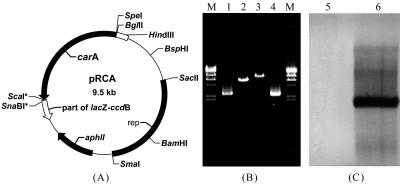
In this study, a CA dioxygenase gene was introduced into the excellent DBT degrader Rhodococcus erythropolis XP (24), which could desulfurize DBT via a 4S pathway. The resultant recombinant was designated SN8. Based on the DNA sequence of Pseudomonas sp. strain CA10 (20), the primers used were as follows: Primer1, 5′-GCCGACTAGTAAGGAGATGGACGTGGCG-3′ (SpeI restriction site underlined); Primer2, 5′-CATGCAAATTTCCTTCTAGTTCCTTCAGCCCGAAACGTGCGCTT-3′; Primer3, 5′-AAGCGCACGTTTCGGGCTGAAGGAACTAGAAGGAAATTTGCATG-3′; Primer4, 5′-GACGAGTACTGCAGCGCCGTCATACGTTGC-3′ (ScaI site underlined). The underlined bases in Primer2 matched the 5′ end of the carAc gene, while the rest matched the 3′ end of carAa. Primer2 and Primer3 were fully matched. The genes encoding CA dioxygenase (carA) containing carAacd were amplified from Pseudomonas sp. strain XLDN4-9 (13) by the recombinant PCR method. The carAa gene was amplified by Primer1 and Primer2, and the carAcd genes were amplified by Primer3 and Primer4. carAa and carAcd were then used as templates, while Primer1 and Primer4 were added to amplify the integrated gene cluster of carA. The fragment obtained was digested with SpeI and ScaI and ligated to SpeI-SnaBI-digested pRESQ (Fig. 1) (23). The resulting plasmid, pRCA, was introduced into strain XP by electroporation. No native promoter sequence of carA was amplified in this study. The expression of the carA gene cluster in R. erythropolis SN8 was driven by the lac promoter present in the vector pRESQ. Southern blot analysis was used to confirm that the carA gene cluster existed in SN8 but not in XP (Fig. 1C). The carA gene was also amplified by Primer1 and Primer4 by using the total DNA of R. erythropolis SN8 as a template. The gene cluster was sequenced, and it was 99% similar to that of strain CA10.
FIG. 1.
Scheme for plasmid pRCA construction and Southern blot analysis. (A) Map of recombinant plasmid pRCA. (B) Ethidium bromide-stained agarose gel of the genetically manipulated products. Lanes: M, HindIII-digested λ DNA molecular size markers; 1 and 4, PCR-amplified products of carA from strains XLDN4-9 and SN8, respectively; 2, native plasmid pRESQ; 3, recombinant plasmid pRCA. (C) Southern blot analysis of the total DNAs digested with HindIII from the XP (lane 5) negative control and the recombinant SN8 (lane 6) probed with the carAa gene.
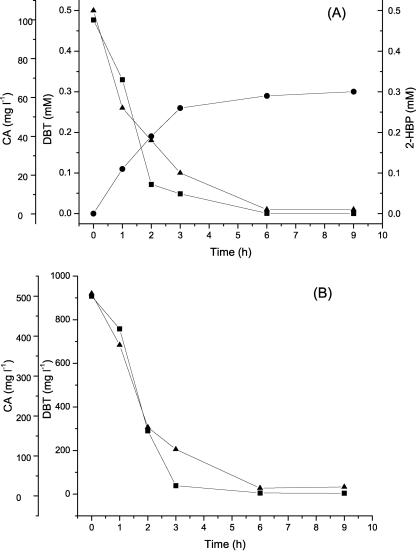
Cells of strain SN8 cultivated in basal salts medium (24) supplemented with 100-mg liter−1 kanamycin sulfate were harvested and washed twice with 100 mM potassium phosphate buffer (pH 7.0). A cell suspension of 8 g (dry cell weight)/liter was incubated at 30°C with shaking at 250 rpm. After reaction for 6 h, 98% of the 0.5 mM (92-mg liter−1) DBT present and 100% of the 100-mg liter−1 CA present were removed by the resting cells of SN8 (Fig. 2A). A resting-cell reaction mixture of host strain XP was also subjected to the same conditions as a control. After 6 h, 92-mg liter−1 DBT was degraded while only a 5% decrease in the CA concentration was detected. The rate of DBT degradation by SN8 was equivalent to that of host strain XP. Biotransformation of DBT and CA by SN8 was also investigated in n-tetradecane. Nine hundred twenty milligrams per liter of DBT and 500-mg liter−1 CA in n-tetradecane were transformed in 6 h (Fig. 2B). Riddle et al. introduced the carA gene cluster into some Pseudomonas strains and investigated the biotransformation of CA in a liquid two-phase system. About 37% of the CA present (0.8% by weight) was removed after treatment for 24 h (18). To the best of our knowledge, no publication has yet been concerned with the introduction of CA-degrading genes into a DBT-degrading strain.
FIG. 2.
Degradation of DBT and CA by resting cells of SN8 at 30°C. (A) Degradation of DBT and CA in aqueous phase. (B) Time course of degradation of DBT and CA dissolved in n-tetradecane. Symbols: ▪, removal of CA; ▴, removal of DBT; •, 2-HBP production.
CA dioxygenase has been demonstrated to be able to attack diverse aromatic compounds, including DBT and biphenyl (16). To eliminate the effects of the native dsz enzymatic system of XP, plasmid pRCA was introduced into Escherichia coli DH5α. The resting-cell reaction mixture was incubated for 48 h at 30°C in the dark to prevent photooxidation. Dibenzothiophene-5-oxide was detected as the product of DBT degradation in this study, which could be further converted to dibenzothiophene sulfone and then to 2-HBP by the native DszABC enzymes of XP (15). Although DBT might be converted to dibenzothiophene dihydrodiol and monohydroxy-dibenzothiophene by the introduced dioxygenase, the amount of these two compounds was too little to have a significant effect on DBT degradation (16). Resting cells of strain SN8 were employed to treat 0.5 mM DBT, but only 0.3 mM 2-HBP accumulated (Fig. 2A). By contrast, the relationship between the decrease in DBT and the increase in 2-HBP caused by host strain XP was stoichiometric. Further investigation showed that SN8 was also able to transform 2-HBP, and 0.28 mM 2-HBP was converted in 68 h while only 0.04 mM 2-HBP was assimilated by XP under the same conditions. Similarly, as an analog of 2-HBP, biphenyl could also be converted by CA dioxygenase to yield cis-2,3-dihydroxy-2,3-dihydrobiphenyl (16). Therefore, it is expected that, catalyzed by CA dioxygenase, 2-HBP will be converted to a more highly hydroxylated biphenyl, which also retains the carbon skeleton of the fuels.
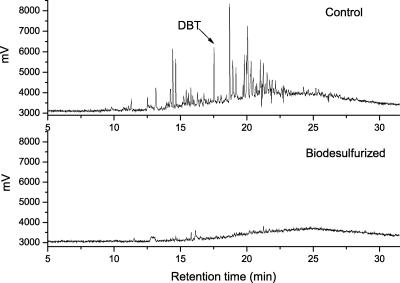
While strain SN8 was shown to transform DBT and CA in model oil experiments, it was of interest to determine if similar results could be obtained in experiments using crude oil, where a complex mixture of chemicals is present and exposure to petroleum could be potentially damaging to the biocatalysts. FS4800 crude oil was obtained from the Fushun Research Institute of Petroleum and Petrochemicals, SINOPEC, China. The concentrations of CA and its derivatives were determined by gas chromatography (GC)-mass spectrometry (3, 11). The results in Table 1 show that SN8 was capable of degrading various alkylated derivatives, including methyl-, dimethyl-, and trimethyl-CA, in FS4800 crude oil. In general, the lower the number of alkyl carbons on the CA ring, the more susceptible the compound was to biodegradation. The distribution of sulfur-containing compounds in crude oil was determined by GC with a pulsed-flame photometric detector (24). Sulfur compounds with retention times longer than that of DBT were desulfurized, and almost all of the highly alkylated sulfur compounds present at a detectable level were also removed (Fig. 3). In previous studies, mixed microbial populations were used to degrade either sulfur or nitrogen heterocycles in crude oil and the cultures were still able to degrade alkane (2, 3, 4). Sphingomonas sp. strain GTIN11, also a CA degrader, could not remove CA that was replaced by more than one methyl group from shale oil (9). Here, we used a purely sulfur-specific degrading bacterium as a host, and the recombinant was capable of degrading a wide range of alkyl CAs and dibenzothiophenes in just a one-step bioprocess. Recently, a Gordonia sp. strain was reported to be capable of DBT desulfurization and CA utilization (19). However, its desulfurization activity was very low and no data concerning selective CA degradation and oil processing were given. The study described in this report is a significant step toward the development of a bioprocess for removing recalcitrant nitrogen and sulfur compounds from petroleum. To the best of our knowledge, this is the first recombinant strain capable of selectively biodegrading DBT and CA simultaneously.
TABLE 1.
Degradation of CA and its derivatives in FS4800 crude oil by strain SN8
| Compounda | Concn (ng/g) in:
|
|
|---|---|---|
| Controlled crude oil | Treated crude oil | |
| CA | 326.46 | 9.70 |
| 1-M-CA | 7.55 | 5.10 |
| 3-M-CA | 3.16 | 1.28 |
| 2-M-CA | 3.32 | 1.42 |
| 4-M-CA | 4.24 | 1.20 |
| 1,8-DM-CA | 6.68 | 5.67 |
| 1,3-DM-CA | 6.15 | 4.66 |
| 1,6-DM-CA | 7.04 | 5.28 |
| 1,7-DM-CA | 7.56 | 5.85 |
| 1,4-DM-CA | 8.64 | 5.46 |
| 1,5-DM-CA | 8.68 | 7.48 |
| 2,6-DM-CA | 2.65 | 1.40 |
| 1,2-DM-CA | 2.06 | 0.00 |
| 2,4-DM-CA | 3.93 | 2.50 |
| 2,5-DM-CA | 3.58 | 2.78 |
| 2,3-DM-CA | 0.81 | 0.58 |
| 3,4-DM-CA | 0.99 | 0.59 |
| TM-CA (a) | 17.58 | 13.84 |
| TM-CA (b) | 20.72 | 16.72 |
| TM-CA (c) | 9.05 | 7.10 |
M, methyl; DM, dimethyl; TM, trimethyl.
FIG. 3.
Analysis of the sulfur content of FS4800 crude oil, before and after treatment, by GC with a pulsed-flame photometric detector. The control sample was an oil sample without bacterial amendment that was subjected to the same conditions as the treated sample.
Nucleotide sequence accession number.
The sequence determined in this study was submitted to the GenBank database and assigned accession number DQ060076.
Acknowledgments
We are grateful to Quan Shi at the China University of Petroleum for analytic assistance with crude oil samples. Thanks to Robert van der Geize (University of Groningen, Groningen, The Netherlands) for helpful discussion of the molecular manipulation of Rhodococcus strains.
This work was supported by the Chinese National Natural Science Foundation (grants 20590368, 20377026, and 30400008) and the Chinese National Programs for High Technology Research & Development (grant 2004AA649160).
REFERENCES
- 1.Benedik, M. J., P. R. Gibbs, R. R. Riddle, and R. C. Willson. 1998. Microbial denitrogenation of fossil fuels. Trends Biotechnol. 16:390-395. [DOI] [PubMed] [Google Scholar]
- 2.Fedorak, P. M., and D. W. S. Westlake. 1983. Microbial degradation of organic sulfur compounds in Prudhoe Bay crude oil. Can. J. Microbiol. 29:291-296. [DOI] [PubMed] [Google Scholar]
- 3.Fedorak, P. M., and D. W. S. Westlake. 1984. Microbial degradation of alkyl carbazoles in Norman Wells crude oil. Appl. Environ. Microbiol. 47:858-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedorak, P. M., and D. W. S. Westlake. 1984. Degradation of sulfur heterocycles in Prudhoe Bay crude oil by soil enrichments. Water Air Soil Pollut. 21:225-230. [Google Scholar]
- 5.Gray, K. A., O. S. Pogrebinsky, G. T. Mrachko, L. Xi, D. J. Monticello, and C. H. Squires. 1996. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat. Biotechnol. 14:1705-1709. [DOI] [PubMed] [Google Scholar]
- 6.Hegedus, L. L., and R. W. McCabe. 1981. Catalyst poisoning. Catal. Rev. 23:377-476. [Google Scholar]
- 7.Hsu, C. S., K. Qian, and W. K. Robbins. 1994. Nitrogen speciation of polar petroleum compounds by compounds class separation and on-line liquid chromatography-mass spectrometry (LC-MS). J. High Resolut. Chromatogr. 17:271-276. [Google Scholar]
- 8.Kilbane, J. J., II. 1989. Desulfurization of coal: the microbial solution. Trends Biotechnol. 7:97-101. [Google Scholar]
- 9.Kilbane, J. J., II, A. Daram, J. Abbasian, and K. J. Kayser. 2002. Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole metabolism in petroleum. Biochem. Biophys. Res. Commun. 297:242-248. [DOI] [PubMed] [Google Scholar]
- 10.Kirimura, K., H. Nakagawa, K. Tsuji, K. Matsuda, R. Kurane, and S. Usami. 1999. Selective and continuous degradation of carbazole contained in petroleum oil by resting cells of Sphingomonas sp. CDH-7. Biosci. Biotechnol. Biochem. 63:1563-1568. [DOI] [PubMed] [Google Scholar]
- 11.Li, M., S. R. Larter, and D. Stoddert. 1992. Liquid chromatographic separation schemes for pyrrole and pyridine nitrogen aromatic heterocycle fractions from crude oils suitable for rapid characterization of geochemical samples. Anal. Chem. 64:1337-1344. [DOI] [PubMed] [Google Scholar]
- 12.Li, F. L., P. Xu, C. Q. Ma, L. L. Luo, and X. S. Wang. 2003. Deep desulfurization of hydrodesulfurization-treated diesel oil by a facultative thermophilic bacterium Mycobacterium sp. X7B. FEMS Microbiol. Lett. 223:301-307. [DOI] [PubMed] [Google Scholar]
- 13.Li, L., P. Xu, and H. D. Blankerspoor. 2004. Degradation of carbazole in the presence of non-aqueous phase liquids by Pseudomonas sp. Biotechnol. Lett. 26:581-584. [DOI] [PubMed] [Google Scholar]
- 14.Li, F. L., P. Xu, J. H. Feng, L. Meng, Y. Zheng, L. L. Luo, and C. Q. Ma. 2005. Microbial desulfurization of gasoline in a Mycobacterium goodii X7B immobilized-cell system. Appl. Environ. Microbiol. 71:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monticello, D. J. 2000. Biodesulfurization and the upgrading of petroleum distillates. Curr. Opin. Biotechnol. 11:540-546. [DOI] [PubMed] [Google Scholar]
- 16.Nojiri, H., J. Nam, M. Kosaka, K. Morii, T. Takemura, K. Furihata, H. Yamane, and T. Omori. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouchiyama, N., Y. Zhang, T. Omori, and T. Kodama. 1993. Biodegradation of carbazole by Pseudomonas spp. CA06 and CA10. Biosci. Biotechnol. Biochem. 57:455-460. [DOI] [PubMed] [Google Scholar]
- 18.Riddle, R. R., P. R. Gibbs, R. C. Willson, and M. J. Benedik. 2003. Recombinant carbazole-degrading strains for enhanced petroleum processing. J. Ind. Microbiol. Biotechnol. 30:6-12. [DOI] [PubMed] [Google Scholar]
- 19.Santos, S. C. C., D. S. Alviano, C. S. Alviano, M. Pádula, A. C. Leitão, O. B. Martins, C. M. S. Ribeiro, M. Y. M. Sassaki, C. P. S. Matta, J. Bevilaqua, G. V. Sebastián, and L. Seldin. 2005. Characterization of Gordonia sp. strain F.5.25.8 capable of dibenzothiophene desulfurization and carbazole utilization. Appl. Microbiol. Biotechnol. [Epub ahead of print.] doi: 10.1007/s00253-005-0154-z. [DOI] [PubMed]
- 20.Sato, S., J. Nam, K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1997. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179:4850-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider, J., R. J. Grosser, K. Jayasimhulu, W. Xue, B. Kinkle, and D. Warshawsky. 2000. Biodegradation of carbazole by Ralstonia sp. RJGII.123 isolated from a hydrocarbon contaminated soil. Can. J. Microbiol. 46:269-277. [DOI] [PubMed] [Google Scholar]
- 22.Shotbolt-Brown, J., D. W. Hunter, and J. Aislabie. 1996. Isolation and description of carbazole-degrading bacteria. Can. J. Microbiol. 42:79-82. [DOI] [PubMed] [Google Scholar]
- 23.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2002. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45:1007-1018. [DOI] [PubMed] [Google Scholar]
- 24.Yu, B., P. Xu, Q. Shi, and C. Q. Ma. 2006. Deep desulfurization of diesel oil and crude oils by a newly isolated Rhodococcus erythropolis strain. Appl. Environ. Microbiol. 72:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]