Abstract
BBn (BioBreeding) rats were fed casein-based diets supplemented with barley flour, oatmeal flour, cellulose, or barley β-glucans of high [HV] or low viscosity [LV] in order to measure the prebiotic effects of these different sources of dietary fiber. The dietary impact on the composition of the cecal microbiota was determined by the generation of denaturing gradient gel electrophoresis (DGGE) profiles of PCR-amplified 16S rRNA gene sequences. The DGGE profiles produced from the cecal microbiota of rats within each dietary group were similar, but consensus profiles generated from pooled bacterial DNAs showed differences between rat groups. Animals fed HV glucans (HV-fed rats) had DGGE consensus profiles that were 30% dissimilar from those of the other rat groups. A 16S rRNA gene fragment that was more conspicuous in the profiles of HV-fed animals than in those of cellulose-fed rats had sequence identity with Lactobacillus acidophilus. Measurements of L. acidophilus rRNA abundance (DNA-RNA hybridization), the preparation of cloned 16S rRNA gene libraries, and the enumeration of Lactobacillus cells (fluorescent in situ hybridization) showed that lactobacilli formed a greater proportion of the cecal microbiota in HV-fed rats. In vitro experiments confirmed that some lactobacilli utilize oligosaccharides (degree of polymerization, 3 or 4) present in β-glucan hydrolysates. The results of this study have relevance to the use of purified β-glucan products as dietary supplements for human consumption.
Bacterial communities (microbiota) of considerable biodiversity populate the large bowels of animals, including humans, and influence the physiology, biochemistry, and immunology of the animal host (19, 22). It has been proposed that altering bacterial activities in the bowel could promote the general health of humans or be used in the prophylaxis or treatment of specific diseases (30). Such health-related measures would involve modification of the composition or functioning of bacterial communities, which might be achieved through the use of probiotics or prebiotics (30).
Probiotic products contain bacterial cells that are often administered in food or as dietary supplements (30). These bacteria are allochthonous to the bowel of the consumer and have a transient existence there: they are detectable in the feces of the consumer only during the period of probiotic administration (reviewed in reference 7). Prebiotics, in contrast, are dietary components or supplements that pass undigested through the small bowel. In the large bowel, they have the potential to become sources of carbon and energy for bacterial residents (autochthonous strains) and thus to boost bacterial numbers or metabolism (16). Most prebiotic research has been directed at the effects of inulin or fructo-oligosaccharide consumption, with a lesser interest in galacto-oligosaccharides (1, 11). This research has focused on members of the bacterial genus Bifidobacterium that are physiologically endowed for the uptake and well-regulated catabolism of complex carbohydrates (35). Other plant polymers, such as β-glucans (polymers of glucose residues linked by β1-3 or β1-4 glycosidic bonds), have been neglected in prebiotic research, even though they are a major component of grains such as barley and oats and are therefore common in the food of farm animals and humans (12). The consumption of β-glucans has been associated with potential health benefits for humans, such as reducing postprandial glycemia in type 2 diabetic patients and a reduction of hyperlipidemia and hypercholesterolemia (8, 9, 13, 23, 38). The amounts of β-glucans produced by cereals vary according to plant variety, growth conditions, and postharvest handling and processing (14, 24). β-Glucans can be purified and processed, however, providing the opportunity to produce products that could be added to foods to promote specific bacteriological and physiological phenomena in the bowel. Therefore, the aim of our study was to measure the impact on the gut microbiota of rats of the consumption of diets supplemented with β-glucans extracted from barley flour.
MATERIALS AND METHODS
Preparation of HV and LV β-glucans for inclusion in rat diets.
High-purity, high-viscosity (HV) β-glucan products were prepared by alkaline extraction (sodium bicarbonate at pH 9, 55°C, 1 h) of β-glucan from barley flour (grains pearled to 30% and pin milled), followed by removal of starchy residue by centrifugation, inactivation of endogenous enzymes by heat treatment of the supernatant (85°C, 1 h), and digestion of residual starch with heat-stable α-amylase (Termamyl). Proteins were then removed by first precipitating them by the addition of hydrochloric acid until the isoelectric point (pH 5.0) was reached, followed by centrifugation. Finally, precipitation of β-glucan was achieved by the addition of an equal volume of ethanol, and then the β-glucan was washed with ethanol and air dried (10). A low-viscosity (LV) β-glucan product was obtained by exposing the HV β-glucan solution to excess shear, using a Microfluidizer processor (M-110 EH; Microfluidics, Newton, MA) at a pressure between 15,000 and 20,000 lb/in2. The low-viscosity β-glucan was then recovered by ethanol precipitation and air dried.
The β-glucan contents of the fractions were determined by an enzymatic assay (28) using a kit (Megazyme Inc., Wicklow, Ireland). Nitrogen contents were determined using a nitrogen analyzer (model FP-428; Leco Instruments Ltd., Mississauga, ON), and protein contents were estimated based on the nitrogen contents, using a conversion factor of 6.25. Starch contents were determined as described by Bhatty (4). The viscosities of β-glucan solutions were determined using a UDS 200 dynamic spectrometer (rheometer; PAAR Physica, Glen Allen, VA), in control shear rate mode, equipped with a DG27 double-gap cup and bob and a Peltier temperature control unit. Viscosities were determined in duplicate at a shear rate of 129 s−1 (100 rpm). The instrument was calibrated with S3 standard oil (3.408 mPa s−1 at 25°C; Cannon Instrument Co., State College, PA). Tests were performed at 20°C.
Experimental animals and diets.
BBn (BioBreeding) rats were obtained from a colony maintained at the Department of Agricultural, Food, and Nutritional Science at the University of Alberta. Experimental procedures used in the study were approved by the Faculty of Agriculture, Forestry and Home Economics' Animal Policy and Welfare Committee (protocol 2002-19C), and the animals were maintained according to the guidelines of the Canadian Council on Animal Care. Rats were housed in temperature- and humidity-controlled rooms with a 12-h light-dark cycle. Pregnant dams were fed standard rat chow (5001 rodent diet; Lab Diet, Brentwood, MO) until their litters were born. On the day of pup birth, the dams' diet was switched to a semipurified casein-based diet containing nonnutritive cellulose (Harlan-Teklad, Madison, WI) as the fiber source (cellulose diet; Table 1). Dams remained on this diet until the rat pups were weaned. Groups of BBn weanling rats (21 days) were then fed one of five experimental diets. Six rats per dietary group were used for bacteriological analyses. The diets were semipurified and casein based, with different fiber compositions, as follows: approximately 8% (wt/wt) of the total fiber was derived from either HV β-glucan (228 mPa s−1), LV β-glucan (27 mPa s−1), barley flour, oatmeal flour, or nonnutritive cellulose. Barley and oatmeal flours were prepared from 18 to 20% pearled grains by grinding. The macronutrient and fiber compositions of the diets are shown in Table 1. At 35 days of age, rats were euthanized with halothane (MTC Pharmaceutical, Cambridge, Ontario, Canada). The cecum of each rat was removed, snap frozen in liquid nitrogen, and stored at −80°C until assayed.
TABLE 1.
Amounts and sources of macronutrients and fiber in rat dietsa
| Macronutrient | Value for indicated diet
|
||||
|---|---|---|---|---|---|
| HV | LV | Barley | Oatmeal | Cellulose | |
| Total amount of diet (g) | 1,010.4 | 1,015.1 | 997.0 | 989.2 | 999.9 |
| Total protein (g/kg) | 267.8 | 269.8 | 264.8 | 266.9 | 270 |
| Casein | 263.3 | 266.7 | 258.3 | 259 | 270 |
| β-Glucan | 4.5 | 3.1 | |||
| Barley flour | 6.5 | ||||
| Oatmeal flour | 7.9 | ||||
| Total lipids (g/kg) | 200.4 | 200.4 | 297.3 | 201.5 | 200.4 |
| Soybean stearine | 131 | 131 | 129 | 130 | 131 |
| Safflower oil | 62 | 62 | 60 | 61 | 62 |
| Linseed oil | 7.4 | 7.4 | 7.1 | 7.2 | 7.4 |
| Barley flour | 1.2 | ||||
| Oatmeal flour | 3.3 | ||||
| Total carboydrates (g/kg) | 379.2 | 382.2 | 376.2 | 372 | 378 |
| Corn starch | 378 | 381.7 | 346.7 | 330 | 378 |
| β-Glucan | 1.2 | 0.5 | |||
| Barley flour | 29.5 | ||||
| Oatmeal flour | 42 | ||||
| Total fiber (g/kg) | 78 | 78.8 | 79.2 | 77.3 | 80 |
| Cellulose | 22 | 22 | 75.3 | 75 | 80 |
| β-Glucan | 56 | 56.8 | |||
| Barley flour | 3.9 | ||||
| Oatmeal flour | 2.3 | ||||
Micronutrients were added to the diets in the form of AOAC vitamin mix (Association of Official Analytical Chemists) (10 g/kg) and Bernhart-Tomarelli mineral mix (Harlan Teklad, Madison, WI) (50 g/kg). The diets contained approximately 25, 34, and 41% of total energy derived from protein, carbohydrate, and fat, respectively.
DNA and RNA extractions from rat cecal contents.
Bacterial DNAs and RNAs were extracted from cecal contents as previously described by Walter et al. (41) and Tannock et al. (37). Briefly, bacterial cells in homogenates of cecal contents were lysed mechanically, and DNAs were purified using phenol-chloroform treatment and ethanol precipitation. RNAs were purified by phenol-chloroform treatment, isopropanol and ethanol precipitations, and the use of QIAGEN RNA/DNA columns (QIAGEN, Victoria, Australia).
PCR and RT-PCR.
PCRs were conducted with either individual or pooled bacterial DNAs or RNAs as templates. Pooled samples (consensus profiles) were prepared by combining 1 μl of each nucleic acid sample from the rats in a specific dietary group. The V3 region of the 16S rRNA gene was amplified in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA), using universal bacterial primers HDA1-GC and HDA2 described by Walter et al. (41) (HDA1-GC, 5′-CGC CCG GGG CGC GCC CCGT GGC GGG GCG GGG GCG CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′ [bold type indicates the GC clamp]; and HDA2, 5′-GTA TTA CCG CGG CTG CTG GCA C-3′). Reverse transcription-PCR (RT-PCR) was performed using a QIAGEN One-Step RT-PCR kit. Each 50-μl reaction mixture was prepared with HDA primers at a concentration of 0.6 μM and with 10 to 40 ng of template DNA. Amplification was performed as described by Tannock et al. (37). The PCR and RT-PCR products were checked by electrophoresis using a 2% agarose gel prior to analysis.
Generation of cecal microbiota profiles by DGGE.
Denaturing gradient gel electrophoresis (DGGE) analysis was performed using the Bio-Rad DCode universal mutation detection system (Hercules, CA) as described by Walter et al. (41). The gels contained a 30 to 55% gradient of 7.0 M urea and 40% (vol/vol) formamide increasing in the direction of electrophoresis. Electrophoresis was conducted with a constant voltage of 130 V at 60°C for 4 h. Gels were stained with ethidium bromide solution, washed with deionized water, and visualized by UV transillumination. DGGE profiles were compared using Dice's similarity coefficient (Dsc) with the Bionumerics software package (Applied Maths, Austin, TX). When performing Dsc analysis, profiles were only compared within the same gel, not between gels. A position tolerance of 3.0% was used for all analyses.
Bacterial origin of DNA fragments.
DNA fragments of interest were aseptically excised from the polyacrylamide gel and extracted as described by Knarreborg et al. (25). Cloning, sequencing, and other procedures have been reported previously (37). Sequencing was performed by the Agricultural, Food and Nutritional Science Biotech Core, University of Alberta. The sequences were compared with those in the NCBI database by using the BLASTN algorithm (2).
Preparation of 16S rRNA gene clone libraries.
Libraries were prepared from cDNAs generated by RT-PCR from pooled RNAs extracted from rats fed diets of cellulose and HV glucans (referred to hereafter as cellulose- and HV-fed rats). The PCR primers SacI-POmod and HDA-2 (34, 41) were used to amplify a partial sequence of the 16S rRNA gene (528 bp; nucleotides 11 to 539). PCRs were performed with a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA), using a QIAGEN Taq core kit (QIAGEN, Canada), in a total volume of 50 μl according to the manufacturer's instructions. The PCR program included preheating at 95°C for 4 min, 25 cycles of denaturation (1 min at 95°C), annealing (1 min at 58°C), and extension (2 min at 72°C), and a final extension at 72°C for 7 min. The 16S rRNA gene amplicons were ligated into vector pCR2.1-TOPO (Invitrogen, Canada), and One Shot Top10 (Invitrogen, Canada) competent Escherichia coli cells were chemically transformed as described by the manufacturer. Recombinant cells were grown on Luria-Bertani (LB) agar plates containing 100 μg of ampicillin per ml and 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml. For each clone library, 200 white colonies were picked with a sterile toothpick, checked by subculture to confirm color selection, and stored in 96-well microtiter plates containing 200 μl of LB freezing buffer (42) per well. Plates were incubated at 37°C overnight, duplicated, and stored at −80°C until further analysis. Cloned 16S rRNA gene sequences were determined using M13 primers by the Molecular Biology Services Unit at the University of Alberta, and the retrieved sequences were compared to those in the NCBI database by using the BLASTN algorithm (2).
DNA-RNA dot blot hybridizations.
The abundance of Lactobacillus acidophilus rRNAs in cecal contents of HV- and cellulose-fed rats was determined by dot blot hybridizations using 16S or 23S rRNA-targeted oligonucleotide probes (3, 29). RNA extraction from cecal contents and reference bacterial cultures, labeling of the probes, and dot blot hybridizations were achieved as described previously (6). The oligonucleotide probes and hybridization conditions used in these experiments are shown in Table 2. RNAs extracted from cecal contents and dilutions of RNA extracted from a pure culture of Lactobacillus acidophilus MB443, as well as dilutions of RNA from Escherichia coli (strain W; Sigma, St. Louis, MO), were immobilized on nylon membranes. Hybridizations were carried out overnight at appropriate temperatures (Table 2). After washing of the membranes, hybridization signals on the dot blots were detected by autoradiography at room temperature, and films were scanned and analyzed using Quantity One software (version 4.2; Bio-Rad Laboratories). The abundance of rRNA from the Lactobacillus population (Lba 23S probe) was expressed as a proportion of the total microbiota by reference to the bacterial domain probe (Eub 338).
TABLE 2.
Probes, targets, and hybridization and washing conditions used in dot blot analysis
Enumeration of lactobacilli in cecal contents by fluorescent in situ hybridization (FISH)/flow cytometry.
Cecal contents were homogenized (1/10) in phosphate-buffered saline (PBS; 130 mM NaCl, 3 mM NaH2PO4, 7 mM Na2HPO4, pH 7.2) by vortex mixing with glass beads (3-mm diameter) for 3 min. Three hundred microliters of the suspension was added to 900 μl of 4% paraformaldehyde (wt/vol). After overnight storage at 4°C, the preparation was centrifuged at 7,600 × g for 5 min at 4°C. The pellet was suspended in 1 ml of PBS and then centrifuged at 7,600 × g for 5 min at 4°C. Finally, the pellet was resuspended in 400 μl of PBS, to which 400 μl of ice-cold ethanol was added. Six preparations per sample (800 μl each) were stored at −20°C in microcentrifuge tubes. Cecal samples from six rats fed the HV diet and four rats fed the cellulose diet were analyzed.
Probe Eub 338 (5′-GCTGCCTCCCGTAGGAGT-3′) (3) was used as the positive control. Probe NON 338 (5′-ACATCCTACGGGAGGC-3′), designed by Wallner et al. (39), was used as the negative control. The control probes were covalently linked at the 5′ end either to fluorescein isothiocyanate (FITC) or to the sulfoindocyanine dye indodicarbocyanine (Cy5). Probe Lab 158 (5′-GGT ATTAGCAYCTGTTTCCA-3′) (20), specific for the Lactobacillus-Enterococcus group, was linked at the 5′ end to either Cy5 or FITC. Probe Lba 23S (used in dot blot hybridizations), targeting L. acidophilus, was linked at the 5′ end to Cy5. Validation of probe Lba 23S was performed with L. acidophilus ATCC 4356 as described previously (27). All of the probes were purchased from MWG Biotech (Bangalore, India).
For each cecal sample, three fixed (see above) 800-μl preparations were pooled as follows. Each tube was centrifuged at 14,600 × g for 5 min. The resulting bacterial pellets were then pooled in a single tube and suspended in 1 ml of PBS. The bacterial cells were harvested by centrifugation at 14,600 × g for 5 min and then resuspended in 1 ml of Tris-EDTA buffer (100 mM Tris-HCl, pH 8.0, 50 mM EDTA). After being washed in Tris-EDTA buffer, the pellets were resuspended in Tris-EDTA buffer containing 1 mg of lysozyme per ml (Serva, Heidelberg, Germany) and left for 10 min at room temperature. The cells were then washed in PBS and suspended in 1 ml of hybridization solution (900 mM NaCl, 20 mM Tris-HCl, pH 8.0, 0.01% sodium dodecyl sulfate [SDS], 30% formamide). Fifty-microliter aliquots were used for FISH with control and group-specific probes. Hybridizations were performed overnight at 35°C in microcentrifuge tubes protected from light and containing the appropriate probe at a final concentration of 4 ng per μl. Following hybridization, 150 μl of hybridization solution was added to each tube, and the bacterial cells were harvested by centrifugation at 14,600 × g for 5 min at room temperature. The cells were then resuspended and incubated at 37°C for 20 min in 200 μl of washing solution (65 mM NaCl, 20 mM Tris-HCl, pH 8.0, 5 mM EDTA, pH 8.0, 0.01% SDS). After a final centrifugation, the cells were suspended in 150 μl of PBS, and 100-μl aliquots were added to 500 μl of PBS for data acquisition by flow cytometry.
A FACSCalibur flow cytometer (Becton Dickinson) was used to determine the compositions of bacterial populations as described previously (26, 27, 32, 33). The cytometer was equipped with an air-cooled argon ion laser providing 15 mW at 488 nm combined with a 635-nm red-diode laser. All of the parameters were collected as logarithmic signals. The 488-nm laser was used to measure the forward-angle light scatter (488-nm band-pass filter), the side-angle light scatter (488-nm band-pass filter), and the green fluorescence intensity conferred by FITC-labeled probes (FL1; 530-nm band-pass filter). The red-diode laser was used to detect the red fluorescence conferred by Cy5-labeled probes (FL4; 660-nm band-pass filter). The acquisition threshold was set in the side scatter channel. The rate of events in the flow was generally below 3,000 events/s. A total of 100,000 events were stored in list mode files. Subsequent analyses were conducted using CellQuest software (Becton Dickinson, Erembodegem-Aalst, Belgium).
The enumeration of bacterial groups was performed as described by Lay et al. (26), where a group probe labeled with Cy5 was combined with the EUB 338-FITC probe. An FL1 histogram was used to evaluate the total number of bacteria hybridizing with the EUB 338-FITC probe. A region, R1, was delineated in this histogram to define all of the events considered to represent bacterial cells. Region R1 was used to gate all density plots. For each Cy5-labeled specific probe, a rectangular region was delineated within the density plot to encompass double-labeled bacteria. These regions permitted the positioning of histogram markers which defined the sections for which the area under the curve could be integrated and bacterial cells counted. These steps allowed an estimation of the proportion of the bacterial group targeted by the Cy5-labeled probe as a proportion of total bacteria (Eub 338-FITC-labeled cells). This proportion was corrected for background fluorescence using the data relating to the negative control probe NON 338. The enumeration of L. acidophilus cells within the Lactobacillus-Enterococcus group was performed by combining the Lba 23S probe labeled with Cy5 with the Lab 158-FITC probe.
Preparation of β-glucan hydrolysates for in vitro studies.
Ten grams of purified HV β-glucan (see above) was slowly added to 1 liter of 19 mM sodium phosphate buffer, pH 6.5, with continuous stirring. After 30 min at 50°C, the temperature was increased to 100°C and held for 5 min. After cooling to 40°C, lichenase (Megazyme; EC 3.2.1.73) was added (1 U per 100 mg of β-glucan for partial hydrolysis [hydrolysate 1] or 20 U per 100 mg of β-glucan for a hydrolysate enriched in oligosaccharides with a degree of polymerization [DP] of 3 or 4 [hydrolysate 2]), and the mixture was incubated at 40°C for 1.5 h with continuous stirring. The mixture was then boiled for 5 min to inactivate the enzyme. An equal volume of ethanol was added to the mixture and held at 20°C for 1 h. After centrifugation at 5,000 × g for 10 min at 20°C, the supernatant was rotary evaporated almost to dryness and then freeze-dried. The compositions of the hydrolysates were determined by high-performance liquid chromatography (HPLC)-evaporative light-scattering detection (see below).
Utilization of oligosaccharides in β-glucan hydrolysates, as measured by HPLC.
The utilization of oligosaccharides in β-glucan hydrolysates was measured by the culture of strains of lactobacilli (L. acidophilus ATCC 4356T, Lactobacillus crispatus ATCC 33820T, Lactobacillus gasseri ATCC 33323T, Lactobacillus johnsonii ATCC 33200T, Lactobacillus hamsteri ATCC 43851T, and Lactobacillus ruminis strains ATCC 27780T, ATCC 27781, and ATCC 27782) in peptone-yeast extract medium supplemented with vitamin K-hemin, Tween 80, and l-cysteine (21) and containing a 2% (wt/vol) concentration of hydrolysate. Ten milliliters of prereduced culture medium was inoculated with 100 μl of an overnight Lactobacillus culture in peptone-yeast extract medium containing 1% (wt/vol) glucose. The cultures were incubated anaerobically for 72 h. One milliliter of culture supernatant was used for HPLC-evaporative light-scattering detection using a SUPELCOSIL LC-NH2 column (25 cm by 4.6 mm by 5 μm; Supelco) equipped with a Hewlett Packard series 1050 autosampler (Agilent Technologies, Wilmington, Delaware), an Alltech 500 evaporative light-scattering detector (Alltech, Deerfield, IL), and a Varian 9010 solvent delivery system. The temperature of the detector was 125°C, the gas pressure was >10 lb/in2, and the gas flow rate was 3 liters/min. Eluents A and B were water (HPLC-grade) and acetonitrile, respectively. The solvent gradient used was as follows: 8% (vol/vol) A and 92% B for 35 min; 35% A and 65% B for 35 min; 55% A and 45% B for 5 minutes, with a hold for an additional 10 min; and a change to 8% A and 92% B over a period of 5 min, with a hold for an additional 5 min. The mobile phase had a flow rate of 1.0 ml/min. The sample injection volume was 10 μl, and the total running time was 60 min. Peak integration was performed using the Shimadzu Class-VP chromatography laboratory automated software system (Shimadzu Scientific Instruments Inc., Columbia, MD). The utilization of specific oligosaccharide fractions (DP) was determined by reference to amounts of oligosaccharides measured in uninoculated medium containing β-glucan hydrolysate.
RESULTS
Cecal microbiota DGGE profiles.
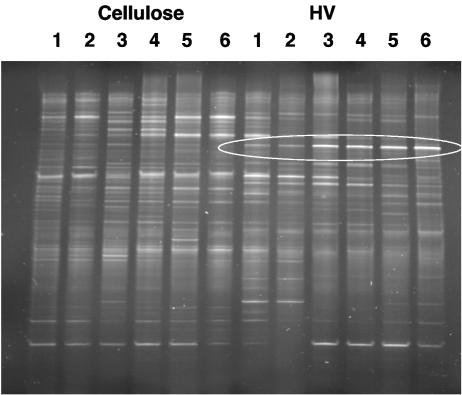
The compositions of the cecal microbiota, as reflected by DGGE profiles generated from bacterial DNAs, differed between dietary groups (Fig. 1). Consensus profiles (prepared from six animals per group) for rats fed cellulose, barley flour, or oatmeal flour were approximately 90% similar, but the profiles for LV- and HV-fed rats were about 20% and 30% dissimilar, respectively, to those of the other groups. The cecal profiles for rats in the same dietary group were similar except in the case of animals fed the LV diet, where there was considerable animal-to-animal variation (Table 3). A comparison of DGGE profiles generated from pooled DNAs or RNAs extracted from the cecal contents of cellulose- or HV-fed animals (six animals per group) showed that the profiles were similar regardless of the nucleic acid used as a template (Dsc, 88.9% and 85.7%, respectively). A 16S rRNA gene fragment that was more apparent in the profiles of HV-fed animals than in those of cellulose-fed rats was excised from the DGGE gel, cloned, and sequenced (Fig. 2). The sequence had identity with a fragment from L. acidophilus (100% identity over a total sequence length of 200 bp).
FIG. 1.
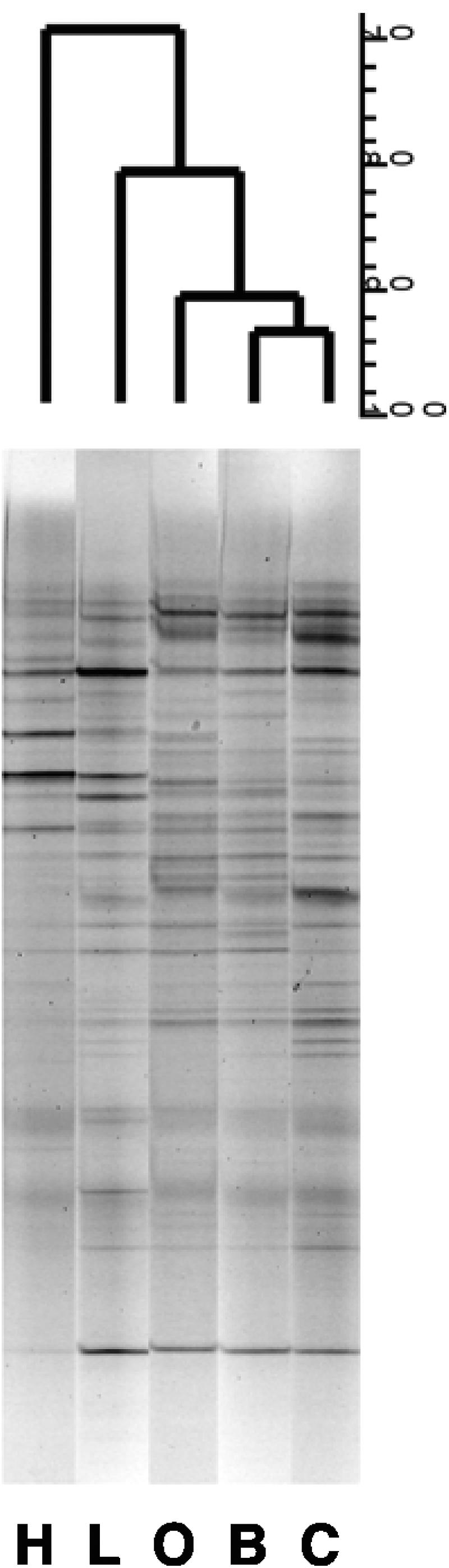
Similarity of consensus DGGE profiles generated from the cecal microbiota of rats fed HV (H), LV (L), oatmeal (O), barley (B), and cellulose (C) diets. Bar = Dice's similarity coefficient (70 to 100%).
TABLE 3.
Similarities of DGGE profiles within dietary groups (six rats per group)
| Dietary group | Dice similarity coefficient (%) |
|---|---|
| HV | 80.0 |
| LV | 66.3 |
| Barley | 85.7 |
| Oatmeal | 81.0 |
| Cellulose | 82.1 |
FIG. 2.
DGGE profiles generated from bacterial DNAs extracted from the cecal contents of rats fed a cellulose or HV diet. Six rats per group (lanes 1 to 6) were used. 16S rRNA gene sequences with greater staining intensities in HV-fed rat profiles than in cellulose-fed rat profiles are circled. These sequences were subsequently shown to originate from L. acidophilus.
16S rRNA gene clone libraries.
Comparison of the 16S rRNA gene libraries prepared from cDNAs showed that Lactobacillus sequences were more prevalent in the library prepared from HV-fed rats (51.25%) than in that prepared from rats consuming the cellulose diet (2.5%). The Lactobacillus sequences showed similarity to those of members of the L. acidophilus group (92 to 100% identity), including L. johnsonii (97.8 to 100% identity). One clone had low identity with L. hamsteri (92.6%).
Abundance of Lactobacillus 16S rRNAs, as measured by dot blot hybridizations.
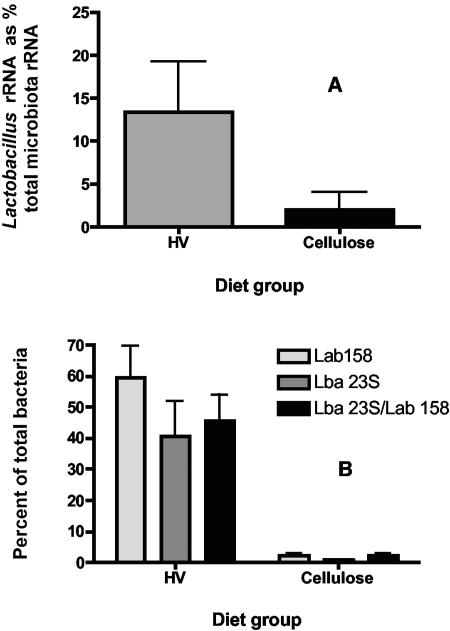
The abundance of rRNAs derived from L. acidophilus, expressed as a percentage of the total microbiota rRNA, was greater in cecal contents from HV-fed rats than in those from cellulose-fed animals (Fig. 3) (in Mann-Whitney test, P = 0.0411).
FIG. 3.
(A) Relative abundance of L. acidophilus rRNAs in cecal contents of HV- and cellulose-fed rats, as determined by dot blot DNA-RNA hybridizations. Means and standard errors of the means are shown for six rats per group. (B) Measurements of cecal bacterial populations using FISH/fluorescence-activated cell sorting. HV, rats fed the HV diet; cellulose, rats fed the cellulose diet; Lab 158, oligonucleotide probe specific for Lactobacillus-Enterococcus group; Lba 23S, probe specific for L. acidophilus; Lba 23S/Lab 158, proportion of bacterial cells detected by Lab 158 probe that also hybridized with the Lba 23S probe. Means and standard errors of the means are shown for six rats per HV diet group and four rats per cellulose diet group.
Enumeration of lactobacilli in cecal contents.
FISH probes targeting members of the Lactobacillus-Enterococcus group or L. acidophilus showed that populations of these bacteria were larger in HV-fed rats than in cellulose-fed animals (Fig. 3) (in Kruskal-Wallis nonparametric analysis of variance test, P < 0.01). L. acidophilus populations were approximately 40-fold larger in animals fed the HV diet.
Utilization of oligosaccharides in β-glucan hydrolysates.
Culture medium supplemented to a final concentration of 2% β-glucan hydrolysate 1 contained oligosaccharides with a DP ranging from 3 to 7, whereas hydrolysate 2 contained oligosaccharides with a DP of 3 to 5. L. gasseri ATCC 33323T, L. johnsonii ATCC 33200T, L. crispatus ATCC 33820T, L. hamsteri ATCC 43851T, and the L. ruminis strains utilized lower-molecular-weight oligosaccharides in hydrolysate 1 (data not shown). Maximal cell densities determined spectrophotometrically (A600) after 72 h of incubation ranged from 1.0 to 2.7. The type culture strain of L. acidophilus did not utilize the oligosaccharides (A600, 0.7; the HPLC profile was unaltered relative to that of the control). Utilization by the lactobacilli of the components of hydrolysate 2, rich in DP3 and DP4 oligosaccharides, is recorded in Table 4. Under these conditions, the DP3 oligosaccharides were used by all of the strains, but two of the L. ruminis strains also utilized DP4 molecules.
TABLE 4.
Utilization by lactobacilli of β-glucan hydrolysate enriched for DP3 and DP4 oligosaccharides (72 h of anaerobic incubation at 37°C)a
| Lactobacillus species and strain | % of oligosaccharide fraction used by bacteriumb
|
|
|---|---|---|
| DP3 | DP4 | |
| L. crispatus ATCC 33820T | 63 | 0 |
| L. gasseri ATCC 33323T | 57 | 0 |
| L. ruminis ATCC 27781 | 93 | 82 |
| L. ruminis ATCC 27782 | 63 | 22 |
| L. ruminis ATCC 27780T | 54 | 0 |
| L. hamsteri ATCC 43851T | 63 | 0 |
| L. johnsonii ATCC 33200T | 85 | 0 |
A single assay was performed with each culture.
Percentage of oligosaccharides utilized by bacterium relative to uninoculated medium (DP3, 17.3 mg/ml; DP4, 7.5 mg/ml).
DISCUSSION
A comparison of the DGGE profiles generated from the cecal microbiota of rats showed that the HV-containing diet produced a large, highly consistent difference in microbiota composition relative to the other diets. Therefore, we investigated the composition of the microbiota of HV-fed and cellulose-fed animals in more detail. The DGGE profiles generated from the cecal microbiota of HV-fed rats contained a 16S rRNA fragment that was more intensely stained than in corresponding profiles of cellulose-fed rats. This rRNA gene sequence originated from bacteria with identity to L. acidophilus. The abundance of rRNAs derived from L. acidophilus in cecal contents from HV-fed rats was greater than that in cecal contents from cellulose-fed animals. Bacterial rRNA can be used as an indicator of metabolic activity because the ribosome-to-cell ratio is roughly proportional to the growth rate of the bacteria (15). The proportion of Lactobacillus 16S rRNA sequences randomly cloned from cDNAs from the cecal microbiota of HV-fed rats was greater than that in the case of cellulose-fed rats. This supported the outcome of the DNA-RNA hybridization experiment: copies of the 16S rRNA genes of lactobacilli were more prevalent in HV-fed rats. FISH/flow cytometry analysis of the microbiota showed that the increased abundance of Lactobacillus rRNAs was in fact due to the presence of large populations of L. acidophilus in the ceca of HV-fed rats relative to those of cellulose-fed animals. Therefore, consumption of the HV diet did not just increase bacterial metabolic activity, which is sometimes the only outcome of prebiotic administration (37), but resulted in increased numbers of Lactobacillus cells.
Lactobacillus strains belonging to the L. acidophilus group (L. crispatus, L. gasseri, amd L. johnsonii) as well as L. hamsteri, cultured in medium containing β-glucan hydrolysates, utilized oligosaccharides of short chain length (DP3). The type culture strain of L. acidophilus that we tested did not utilize oligosaccharides in the hydrolysates. There may be strain-to-strain variation in this species with regard to the ability to utilize oligosaccharides, but we did not test this possibility. We presume that DP3 oligosaccharides were the substrates that produced the “lactobacillogenic” effect in the cecum and that these were released from the HV β-glucan by hydrolysis catalyzed by extracellular β-glucanases secreted by other cecal residents (40). We do not know whether any proportion of the HV β-glucan was digested in proximal regions of the gut, but evidence of an effect on the cecal microbiota suggests that this did not occur to any extent. Strains of L. ruminis were also tested for the ability to utilize oligosaccharides in β-glucan hydrolysates and were found to ferment the DP3 and DP4 fractions. Only two Lactobacillus species can presently be considered autochthonous to the digestive tract of humans, namely, L. salivarius and L. ruminis (reviewed in reference 7). The oral cavity, however, appears to be the likely habitat of L. salivarius, whereas L. ruminis seems to be a true resident of the colons of a proportion of healthy humans (7, 31, 36). The utilization by L. ruminis of oligosaccharides present in β-glucan hydrolysates as carbon and energy sources is of significance in the development of prebiotics for human consumption because these lactobacilli normally comprise approximately 0.1% of the fecal microbiota, a much greater concentration of bacteria than can be achieved by the administration of probiotic strains (36). Future work to determine the impact of defined products containing purified oligosaccharides of DP3/DP4 derived from barley β-glucans on L. ruminis populations in the human colon would clearly be worth pursuing.
The results of our study clearly demonstrate that HV β-glucan can produce a prebiotic effect in the ceca of rats, at least under the specific dietary conditions that were used. While extrapolation of our observations to humans is not yet appropriate, experimental animal studies provide an informative screening method by which products for potential use in human nutrition or medicine can be tested. Lactobacilli and other lactic acid bacteria have recently generated considerable interest as probiotic agents in the prophylaxis or treatment of chronic pouchitis and ulcerative colitis (5, 17, 18). Thus, food-derived products that stimulate autochthonous Lactobacillus populations in human colons are of major biotechnological and medical interest.
Acknowledgments
The support of Alberta Agriculture, the Alberta Barley Commission, and the Alberta Value Added Corporation is gratefully acknowledged. R.B. was supported by a grant from the Crohns and Colitis Foundation of Canada, and C.L. was supported by the Crohns and Colitis Foundation of America.
REFERENCES
- 1.Alles, M. S., R. Hartemink, S. Meyboom, J. L. Harryman, K. M. J. Van Laere, F. M. Nagengast, and J. G. A. J. Hautvast. 1999. Effect of transgalactooligosaccharides on the composition of the human intestinal microflora and on the putative risk markers for colon cancer. Am. J. Clin. Nutr. 69:980-991. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and L. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatty, R. S. 1992. β-Glucan content and viscosities of barleys and their roller-milled flour and bran products. Cereal Chem. 69:469-471. [Google Scholar]
- 5.Bibiloni, R., R. N. Fedorak, G. W. Tannock, K. L. Madsen, P. Gionchetti, M. Campieri, C. De Simone, and R. B. Sartor. 2005. VSL#3 probiotic-mixture induces remission in patients with active colitis. Am. J. Gastroenterol. 100:1539-1546. [DOI] [PubMed] [Google Scholar]
- 6.Bibiloni, R., M. A. Simon, C. Albright, B. Sartor, and G. W. Tannock. 2005. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Lett. Appl. Microbiol. 41:45-51. [DOI] [PubMed] [Google Scholar]
- 7.Bibiloni, R., J. Walter, and G. W. Tannock. 2004. The gut microflora, p. 125-143. In M. M. Nakano and P. Zuber (ed.), Strict and facultative anaerobes. Medical and environmental aspects. Horizon Bioscience, Wymondham, England.
- 8.Braaten, J. T., F. W. Scott, P. J. Wood, K. D. Riedel, M. S. Wolynetz, D. Brule, and M. W. Collins. 1994. High beta-glucan oat bran and oat gum reduce postprandial blood glucose and insulin in subjects with and without type 2 diabetes. Diabet. Med. 11:312-318. [DOI] [PubMed] [Google Scholar]
- 9.Braaten, J. T., P. J. Wood, F. W. Scott, M. S. Wolynetz, M. K. Lowe, P. Bradley-White, and M. W. Collins. 1994. Oat beta-glucan reduces blood cholesterol concentration in hypercholesterolemic subjects. Eur. J. Clin. Nutr. 48:465-474. [PubMed] [Google Scholar]
- 10.Burkus, Z., and F. Temelli. 1998. Effect of extraction conditions on the yield, composition and viscosity stability of barley β-glucan gum. Cereal Chem. 75:805-809. [Google Scholar]
- 11.Crittenden, R. G. 1999. Prebiotics, p. 141-156. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, England.
- 12.Cui, S. 2001. Cereal non-starch polysaccharides. I. (1-3)(1-4)-β-d-Glucans, p. 103-166. In S. Cui (ed.), Polysaccharide gums from agricultural products: processing, structures and functionality. Technomic Publishing Co., Lancaster, United Kingdom.
- 13.Davidson, M., L. Dugan, J. Burns, J. Bova, K. Sory, and K. Drennan. 1991. The hypocholesterolemic effects of beta-glucan in oatmeal and oat bran. A dose-controlled study. JAMA 265:1833-1839. [PubMed] [Google Scholar]
- 14.Duffus, C. M., and M. P. Cochrane. 1993. Formation of the barley grain—morphology, physiology, and biochemistry, p. 31-72. In A. W. Macgregor and R. S. Bhatty (ed.), Barley chemistry and technology. American Association of Cereal Chemists Inc., St. Paul, Minn.
- 15.Felske, A., H. Rheims, A. Wolerink, E. Stackbrandt, and A. D. L. Akkermans. 1997. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983-2989. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 17.Gionchetti, P., F. Rizzello, U. Helwig, A. Venturi, K. M. Lammers, P. Brigidi, B. Vitali, G. Poggioli, M. Miglioli, and M. Campieri. 2003. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 124:1535-1538. [DOI] [PubMed] [Google Scholar]
- 18.Gionchetti, P., F. Rizzello, A. Venturi, P. Brigidi, D. Matteuzzi, G. Bazzocchi, G. Poggioli, M. Miglioli, and M. Campieri. 2000. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 191:584-587. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host-microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1973. Anaerobe laboratory manual. VPI Anaerobe Laboratory, Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 22.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins, D. J., C. W. Kendall, and V. Vuskan. 2002. Viscous fibres, health claims, and strategies to reduce cardiovascular disease risk. Am. J. Clin. Nutr. 71:401-402. [DOI] [PubMed] [Google Scholar]
- 24.Kerckhoffs, D., G. Hornstra, and R. Mensink. 2003. Cholesterol-lowering effect of β-glucan from oat bran in mildly hypercholesterolemic subjects may decrease when a β-glucan is incorporated into bread and cookies. Am. J. Clin. Nutr. 78:221-227. [DOI] [PubMed] [Google Scholar]
- 25.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lay, C., L. Rigottier-Gois, K. Holmstrom, M. Rajilic, E. E. Vaughan, W. M. de Vos, M. D. Collins, R. Thiel, P. Namsolleck, M. Blaut, and J. Dore. 2005. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71:4153-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lay, C., M. Sutren, V. Rochet, K. Saunier, J. Dore, and L. Rigottier-Gois. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ. Microbiol. 7:933-946. [DOI] [PubMed] [Google Scholar]
- 28.McCleary, B. V., and M. Glennie-Holmes. 1985. Enzymatic quantification of (1-3)(1-4)-β-d-glucan in barley and malt. J. Inst. Brew. 91:285-295. [Google Scholar]
- 29.Pot, B., C. Hertel, W. Ludwig, P. Descheemaeker, K. Kersters, and K. H. Schleifer. 1993. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA-targeted oligonucleotide probe hybridization. J. Gen. Microbiol. 139:513-517. [DOI] [PubMed] [Google Scholar]
- 30.Reid, G. 1999. The scientific basis for probiotic strains of Lactobacillus. Appl. Environ. Microbiol. 65:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 32.Rigottier-Gois, L., A.-G. Le Bourhis, G. Gramet, V. Rochet, and J. Dore. 2003. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 43:237-245. [DOI] [PubMed] [Google Scholar]
- 33.Rigottier-Gois, L., V. Rochet, N. Garrec, A. Suau, and J. Dore. 2003. Enumeration of Bacteroides species in human faeces by fluorescent in situ hybridisation combined with flow cytometry using 16S rRNA probes. Syst. Appl. Microbiol. 26:110-118. [DOI] [PubMed] [Google Scholar]
- 34.Rodtong, S., and G. W. Tannock. 1993. Differentiation of Lactobacillus strains by ribotyping. Appl. Environ. Microbiol. 59:3480-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannock, G. W., K. Munro, R. Bibiloni, M. A. Simon, P. Hargreaves, P. Gopal, H. Harmsen, and G. Welling. 2004. Impact of the consumption of oligosaccharide-containing biscuits on the fecal microbiota of humans. Appl. Environ. Microbiol. 70:2129-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tappy, L., E. Gugolz, and P. Wursch. 1996. Effects of breakfast cereals containing various amounts of beta-glucan fibers on plasma glucose and insulin responses in NIDDM subjects. Diabetes Care 19:831-834. [DOI] [PubMed] [Google Scholar]
- 39.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 40.Walter, J., M. Mangold, and G. W. Tannock. 2005. Construction, analysis, and β-glucanase screening of a bacterial artificial chromosome library from the large-bowel microbiota of mice. Appl. Environ. Microbiol. 71:2347-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munroo, and T. Alatossava. 2000. Detection and identification of gastrointestinal species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmer, R., and A. M. V. Gibbins. 1997. Construction and characterization of a large-fragment chicken bacterial artificial chromosome library. Genomics 42:217-226. [DOI] [PubMed] [Google Scholar]