Abstract
The Bacteria community composition in an acidic Sphagnum peat bog (pH 3.9 to 4.5) was characterized by a combination of 16S rRNA gene clone library analysis, rRNA-targeted fluorescence in situ hybridization (FISH), and cultivation. Among 84 environmental 16S rRNA gene clones, a set of only 16 cloned sequences was closely related (≥95% similarity) to taxonomically described organisms. Main groups of clones were affiliated with the Acidobacteria (24 clones), Alphaproteobacteria (20), Verrucomicrobia (13), Actinobacteria (8), Deltaproteobacteria (4), Chloroflexi (3), and Planctomycetes (3). The proportion of cells that hybridized with oligonucleotide probes specific for members of the domains Bacteria (EUB338-mix) and Archaea (ARCH915 and ARC344) accounted for only 12 to 22% of the total cell counts. Up to 24% of the EUB338-positive cells could be assigned by FISH to specific bacterial phyla. Alphaproteobacteria and Planctomycetes were the most numerous bacterial groups (up to 1.3 × 107 and 1.1 × 107 cells g−1 peat, respectively). In contrast to conventional plating techniques, a novel biofilm-mediated enrichment approach allowed us to isolate some representatives of predominant Bacteria groups, such as Acidobacteria and Planctomycetes. This novel strategy has great potential to enable the isolation of a significant proportion of the peat bog bacterial diversity.
Sphagnum-dominated acidic peat bogs represent one of the most extensive wetland types in North America and Eurasia. They occupy about 3% of the Earth's terrestrial surface (18), comprising up to 80% of the area in some regions of West Siberia. The environmental and ecological significance of peat soils is immense because of the well-recognized role of the northern wetlands in the global carbon budget and emission of the greenhouse gas methane. Due to low biodegradative activity in these ecosystems, the northern peatlands harbor approximately 30% of the global reserves of soil organic carbon (12). Furthermore, these wetlands determine the hydrology of northern rivers and represent one of the largest reservoirs of freshwater in the Northern Hemisphere. Despite their major impact on the global carbon and water cycles, the microbial diversity in acidic wetlands remains poorly understood. High acidity (pH 3.5 to 5.0), low temperatures, and extremely low concentrations of mineral nutrients (5 to 50 mg per liter) make these wetlands an extreme habitat. So far, only microbial populations involved in CH4 cycling, i.e., methanotrophic bacteria (5-9, 23, 28) and methanogenic archaea (1, 13, 19, 35), have attracted considerable research interest. Other members of the microbial communities in acidic Sphagnum peatlands remain largely unknown. The molecular diversity of bacterial 16S rRNA genes in peat bogs has been addressed only once (29, 30). However, the focus of that research was restricted to a few novel actinomycete lineages (30).
In our study, we aimed to elucidate the overall bacterial community composition in an acidic Sphagnum peat soil by a combination of 16S rRNA gene clone libraries, fluorescence in situ hybridization (FISH), and cultivation.
MATERIALS AND METHODS
Sampling site.
The peat samples were collected from the depths 0 to 10, 10 to 20, 20 to 30, 30 to 40, 40 to 50, and 70 to 80 cm over the profile of the acidic (pH 3.9 to 4.5) Sphagnum peat bog Bakchar, Plotnikovo field station of the Institute of Soil Science and Agrochemistry (Siberian Branch of Russian Academy of Sciences), which is located in the Tomsk region, West Siberia (56°51′N, 82°50′E). This bog, which is 15 × 30 km2 in size, occupies the flat watershed between the rivers Bakchar and Iksa and is part of the worldwide-largest Great Vasyugan Wetland. The sampling was done under a Sphagnum-Carex plant community, with a water table at approximately 15-cm depth. The samples were transported to the laboratory, homogenized by cutting the peat material into small fragments (about 0.5 cm) with sterile scissors, and fixed for FISH or used for DNA extraction as well as for cultivation studies (see below).
FISH.
The fixation procedure was carried out as described by Dedysh et al. (8) and included (i) the separation of the peat water enriched with microbial cells from the rough (≥2- to 3-mm) Sphagnum debris by repeated stomacher treatments, (ii) recovery of the microbial cells from the peat water, and (iii) cell fixation with 4% (wt/vol) freshly prepared paraformaldehyde solution. A set of Cy3-labeled oligonucleotide probes with reported group specificity for the domains Bacteria and Archaea, as well as for some phyla within the domain Bacteria and some classes within the Proteobacteria, was used in this study (see Table S1 in the supplemental material). In addition, the oligonucleotide probes VER139 (5′-CGAGCTATTCCCCTCTTG-3′) and VER1463 (5′-CCATCCATACCTTCG GCA-3′) were developed to enumerate two subgroups of the Verrucomicrobia in Sphagnum peat. All oligonucleotide probes used in this study were purchased from Syntol (Moscow, Russia).
Hybridization was done on gelatin-coated (0.1%, wt/vol) and dried Teflon-laminated slides (MAGV, Germany) with eight wells for independent positioning of the samples. The fixed samples were applied to these wells, hybridized to the respective fluorescent probes, and stained with the universal DNA stain 4′,6′-diamidino-2-phenylindole (DAPI) (1 μM) as described earlier (8). The cell counts were determined with a Zeiss Axioplan 2 microscope (Zeiss, Jena, Germany) equipped with the Zeiss filters no. 20 and 02 for Cy3-labeled probes and for DAPI staining, respectively.
DNA extraction.
Extracts of total DNA were obtained from peat soil sampled at a depth of 10 to 20 cm. Two subsamples, each of 0.5 g (wet weight), were taken from this peat sample and processed separately. To increase the lysis efficiency, these subsamples were subjected to three repeated cycles of grinding with liquid nitrogen using a sterile pestle and a mortar. Further extraction steps were performed using an FP 120 FastPrep cell disruptor (Savant Instruments Inc., Farmingdale, NY) and a FastDNA SPIN kit for soil (Bio101, Carlsbad, CA) according to the manufacturer's instructions, with an additional step of humic acid removal by repeated washing of the matrix-bound DNA with 5.5 M guanidine isothiocyanate solution (modified as described by Yeates and Gillings [40]). Extraction of genomic DNA from newly isolated strains was performed using a sodium dodecyl sulfate-based procedure described previously (5).
PCR amplification, cloning, and sequencing.
Two environmental clone libraries were constructed, one from each DNA extract. The peat soil 16S rRNA gene pool was PCR amplified using the Bacteria-specific primers Eub9f and Eub1492r (20). In order to reduce the potential bias of separate PCRs, the amplicons of two independent PCRs were combined before cloning (26). The mixed PCR product was cloned using a TA cloning kit (Invitrogen, San Diego, Calif.) as recommended by the manufacturer. Cloned inserts were PCR amplified using the T7 promoter and the M13 reverse primers, purified using QIAquick spin columns (QIAGEN, Hilden, Germany), and sequenced on an ABI Prism 377 DNA sequencer as specified by the manufacturer (PE Applied Biosystems, Foster City, Calif.).
Conventional cultivation approach.
One gram of wet peat was suspended in 50 ml of sterile water and treated in a laboratory stomacher at 240 rpm for 5 min. The resultant suspensions were serially diluted in 10-fold steps, and the aliquots from dilutions of 10−3 to 10−5 were spread onto solid media (see below). Each dilution level consisted of five replicate plates per sample. The plates were incubated at 24°C for 4 weeks in the dark, and the number of colonies was then counted. Two slightly different basal media were used for enumeration and subcultivation of peat-inhabiting bacteria. Medium M1 contained the following (in grams per liter of distilled water): KNO3, 0.25; KH2PO4, 0.1; MgSO4, 0.1; CaCl2 · 2H2O, 0.02; yeast extract, 0.1; and Na2MoO4, 0.005. A 0.1% (vol/vol) concentration of a trace element stock solution described by Dedysh et al. (5) was added. For isolation of nitrogen-fixing bacteria, the same medium without KNO3 was used. Medium M2 contained the following (in grams per liter of distilled water): (NH4)2SO4, 0.1; KH2PO4, 0.1; MgSO4, 0.05; and CaCl2 · 2H2O, 0.02. A 0.1% (vol/vol) concentration of the same trace element stock solution was added. After sterilization, one of the following carbon sources was added to these mineral base media at a concentration of 0.05% (wt/vol): glucose, sodium gluconate, sodium acetate, methanol, cellobiose, carboxymethylcellulose, or starch. The pH range of the prepared media was 4.4 to 5.5. For preparation of peat extract medium, 400 g of wet peat was mixed with 200 ml of distilled water, homogenized, and centrifuged to sediment peat debris. Five hundred milliliters of this extract was further mixed with 500 ml of the base medium M2. The solidifying agents used for these media preparations were agar (Difco) or agarose or Gel-Gro (ICN Biomedicals Inc.). Colonies were selected randomly and restreaked on homologous media. When subcultures were grown, defined pools of cells were taken for identification by means of whole-cell hybridization with a set of Cy3-labeled oligonucleotide probes. In addition, partial 16S rRNA gene sequences (500 to 700 nucleotides) were determined for some of these isolates as described earlier (7).
Biofilm-mediated isolation approach.
The petri dish technique (34) was used to enrich microbial biofilms consisting of cells of peat-inhabiting bacteria. The bottom of a glass petri dish was covered with a 2-cm layer of sterilized water agar (1.5%, wt/vol), which contained 0.002% (wt/vol) cycloheximide to inhibit growth of fungi. Fifteen to 20 sterilized cover slides were placed vertically and partially into the agar layer. These plates were then inoculated with 10 to 15 ml of peat water and incubated in the dark at 10 and 24°C for 1 to 2 months. Every 3 weeks, a few slides from each plate were taken out and subjected to examination by FISH with oligonucleotide probes specific for the Acidobacteria, Planctomycetes, or Verrucomicrobia. When good development of the target bacteria had been observed on these cover slides, a few other slides were taken out for agar streak isolation. The following medium was used for isolation of planctomycetes (in grams per liter of distilled water except as noted): KH2PO4, 0.1; MgSO4 · 7H2O, 0.05; CaCl2 · 2H2O, 0.01; Hutner's basal salts (34), 20 ml; N-acetylglucosamine, 1.0; Na-ampicillin, 0.2; peptone, 0.1; and yeast extract, 0.1 (pH 5.8). For isolation of acidobacteria, 10-fold diluted R2A medium (Difco) at pH 5.8 was used. The colonies appearing on the plates were randomly picked for FISH-based examination. This procedure was continued until individual colonies of target organisms were ultimately identified and obtained in pure culture. Finally, nearly full-length 16S rRNA gene sequences were determined for the isolates as described by Dedysh et al. (7).
Phylogenetic analysis.
Data processing was done by using the ARB program package (21). Primary screening of the clone library for the presence of chimeric sequences was done using the Bellerophon program (14). The screening results were further verified by independently subjecting the first 5′ 600 nucleotide positions or the last 3′ 600 nucleotide positions of each sequence to phylogenetic analysis. The resulting alignments were used for analyses without making changes of possible errors in the public-domain 16S rRNA gene sequences. Phylogenetic analyses were carried out using the PHYLIP program package (10). Trees were constructed using the neighbor-joining and maximum-likelihood methods. The significance levels of interior branch points obtained in neighbor-joining analysis were determined by bootstrap analyses (1,000 data resamplings).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences retrieved from Sphagnum peat and those obtained from bacterial isolates have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession numbers AM162408 to AM162489 and AM162403 to AM162407, respectively.
RESULTS AND DISCUSSION
16S rRNA gene clone library analysis.
We anticipated that the greatest bacterial diversity would be detectable at the oxic-anoxic interface of the bog profile, corresponding to a depth of 10 to 20 cm. A total of 101 clones were selected at random, designated B (for “Bakchar”), sequenced, and analyzed phylogenetically. Seventeen cloned sequences were considered chimeric and excluded from further analyses. Among the other 84 nearly full-length 16S rRNA gene sequences, a set of only 16 peat clones was closely related (≥95% similarity) to taxonomically described organisms. Major groups of peat clones were affiliated with the Acidobacteria (24 clones), Alphaproteobacteria (20), Verrucomicrobia (13), and Actinobacteria (8). Minor groups of clones were assigned to the Deltaproteobacteria (four clones), Chloroflexi (three), Planctomycetes (three), Bacteroidetes (two), and Chlorobi (two). Four 16S rRNA gene sequences were of unknown phylogenetic affiliation.
(i) Acidobacteria.
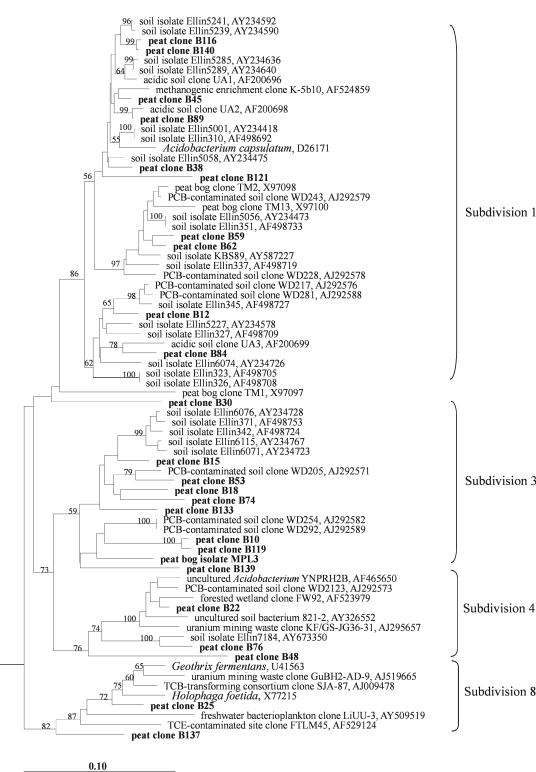
The 24 peat clones that belonged to the Acidobacteria were widely distributed among the subdivisions 1, 3, 4, and 8 (Fig. 1). Twelve clones had sequence similarities of ≥95% to 16S rRNA gene sequences from isolates named with the prefix “Ellin,” which were recently obtained from different soils by using newly developed media and extended incubation time (16, 32). Two peat clones, B59 and B62, were closely related to another soil isolate, strain KBS89 (36). However, except for 16S rRNA gene sequence data, no further information is as yet available for the “Ellin” isolates and strain KBS89, and none of them was described taxonomically. Thus, only a single peat clone (B25) was closely related to taxonomically described organisms, i.e., Geothrix fermentans and Holophaga foetida (96% and 95% sequence similarity, respectively). G. fermentans is an anaerobic chemo-organotroph that utilizes various organic acids as electron donors and Fe(III) as the electron acceptor (4). In addition to Fe(III), these bacteria are also capable of using humic acids as alternative electron acceptors, a feature which makes these organisms highly relevant for the peat bog environment.
FIG. 1.
16S rRNA gene-based dendrogram showing the phylogenetic relationship of 24 peat clones (boldface) to the peat bog isolate MPL3 and to representatives of the phylum Acidobacteria. The classification into subdivisions follows the proposal by Hugenholtz et al. (15). Bootstrap values (1,000 data resamplings) of >50% are shown. The root was determined by using the 16S rRNA gene sequence of Chlorobium limicola Y10643 as an outgroup. The scale bar represents 0.1 substitution per nucleotide position.
(ii) Proteobacteria.
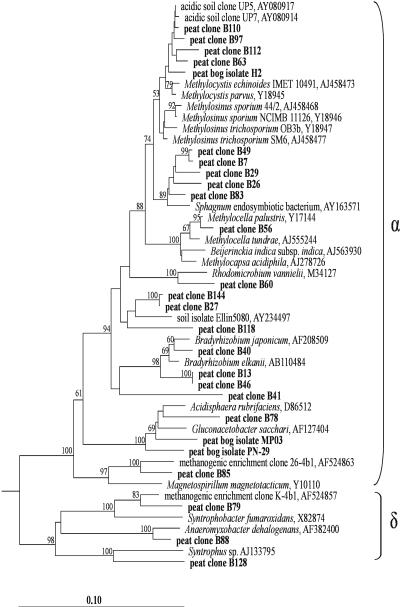
Only sequence types affiliated with the Alpha- and Deltaproteobacteria were present in the clone library (Fig. 2). Ten of the alphaproteobacterial peat clones clustered within a polyphyletic group of serine pathway methanotrophs defined by members of the genera Methylocystis, Methylosinus, Methylocella, and Methylocapsa. However, only one of the clones (B56) could be assigned on the species level (98% sequence similarity) to a taxonomically described organism, the acidophilic methanotroph Methylocella palustris (7). Four peat clones (B63, B97, B110, and B112) showed 97 to 98% sequence similarity to different strains of Methylocystis and to the environmental sequences UP4, UP5, and UP7, which had been obtained from acidic forest soil (27). An isolate representative of this group of sequences was recently obtained from the acidic Sphagnum peat lake Teufelssee in Germany and was designated strain H2 (S. N. Dedysh et al., unpublished data). This isolate is now being described as a novel, acidophilic species of the genus Methylocystis. Five other peat clones (B7, B26, B29, B49, and B83) formed another separate cluster within the phylogenetic radiation of serine pathway methanotrophs. This cluster includes a recently published sequence from an uncultured putative methanotroph, which was detected inside hyaline cells of Sphagnum cuspidatum (28). Other alphaproteobacterial sequences in the clone library were affiliated with the genera Bradyrhizobium (clones B13, B40, and B46), Acidisphaera (B78), and Magnetospirillum (B85). The deltaproteobacterial peat clone B88 was assigned at the species or genus level (97.2% sequence similarity) to Anaeromyxobacter dehalogenans (33), providing further evidence that the acidic peat bog is colonized by bacteria capable of dissimilatory reduction of Fe(III). Two additional peat clones were related to the genera Syntrophobacter (B79) and Syntrophus (B128). The peat clone B122 was only distantly related (90% similarity) to Desulfuromonas thiophila and formed a novel lineage.
FIG. 2.
16S rRNA gene-based dendrogram showing the phylogenetic relationship of 23 peat clones (boldface) to peat bog isolates MP03 and PN-29 and to representatives of the classes Alphaproteobacteria and Deltaproteobacteria. Bootstrap values (1,000 data resamplings) of >50% are shown. The root was determined by using the 16S rRNA gene sequence of Pseudomonas fluorescens AF094730 as an outgroup. The scale bar represents 0.1 substitution per nucleotide position.
(iii) Verrucomicrobia.
Four clones (B36, B68, B70, and B102) of the 13 verrucomicrobial peat clones formed a coherent cluster with Opitutus terrae, an anaerobic polysaccharide utilizing ultramicrobacterium (2) (see Fig. S1A in the supplemental material). The remaining peat clones were more diverse and spread over a broad phylogenetic sequence cluster for which cultured representatives have not yet been reported. The environmental 16S rRNA gene sequences of this cluster were retrieved from a variety of different habitats, including forested wetland, manure leakage, thermal soil, Antarctic sediments, oligotrophic lake, and river water.
(iv) Actinobacteria.
Seven of the eight actinobacterial peat clones were assigned to two clusters that also contained environmental sequences (abbreviated TM) from a peat bog near Gifhorn, Germany (30) (see Fig. S1B in the supplemental material). The first group, defined by the peat clones B2, B4, B31, TM36, TM146, and TM220, includes a single cultured representative, Conexibacter woesei (22). This aerobic bacterium is a deep-rooting member of the Actinobacteria that possesses an unusual combination of chemotaxonomic characteristics (22) and appears to be ubiquitously distributed in soils (30). The second group, defined by the peat clones B124, B145, TM56, and TM226, showed a moderate relationship (91 to 92% sequence similarity) to Acidimicrobium ferrooxidans, an acidophilic autotrophic bacterium (3). The peat clone B125 was only distantly related (≤90% similarity) to any other actinobacterial sequence and formed a novel lineage.
(v) Other bacterial phyla.
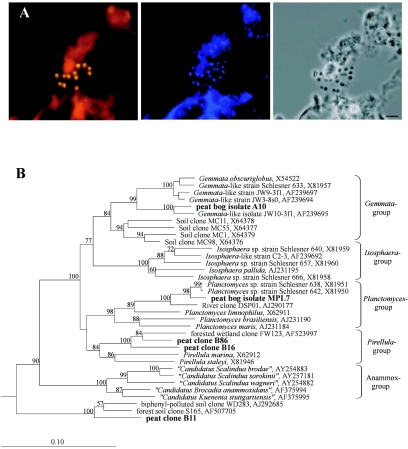
Three peat clones were affiliated with the Planctomycetes (Fig. 3B). Two of those (B16 and B86) fell within the Pirellula group, while the third (B11) belonged to a deeply branching planctomycete cluster for which no taxonomically described representatives are known. Three cloned 16S rRNA gene sequences (B14, B71, and B131) grouped within the phylum Chloroflexi and showed 88 to 90% sequence similarity to the clone K-4b6 (accession no. AF524858). This clone was obtained from an acidophilic methanogenic consortium that had been enriched from the same peat bog Bakchar (35). Only two peat clones (B9 and B114) were affiliated with the Bacteroidetes. Both exhibited a distant relationship (89 to 91%) to Cytophaga sp. strain BHI60-95B (AJ431254). Two peat clones (B44 and B81) were related to the Chlorobi. The closest relative was the uncultured anoxic soil bacterium BSV19 (AJ229184), exhibiting a 16S rRNA gene sequence similarity of 91%. Finally, four peat clones (B39, B54, B55, and B72) could not be unambiguously assigned to any of the currently recognized main lines of bacterial descent. Clone B39 had 88% similarity to a member of candidate division OP3, wastewater biofilm clone koll11 (AJ224540). Peat clone B54 exhibited 86% similarity to another cloned sequence, OPd3 (AF047562), affiliated with candidate division OP11. Clone B55 had relatively high similarity (94%) to unclassified clone RB25 (Z95718) but only a distant relationship (85% similarity) to any other sequence in the database. Peat clone B72 exhibited 85% similarity to thesequence of the Calyptogena elongata gill symbiont (AF035719).
FIG. 3.
(A) Specific detection of planctomycete microcolonies in Sphagnum peat by FISH. Epifluorescent micrographs of in situ hybridizations with Cy3-labeled probes Pla46 and Pla886 (left), DAPI staining (center), and the phase-contrast image (right) are shown. Bar, 5 μm. (B) 16S rRNA gene-based dendrogram showing the phylogenetic relationship of three peat clones (boldface) to peat bog isolates A10 and MPL7 and to representatives of the phylum Planctomycetes. Bootstrap values (1,000 data resamplings) of >50% are shown. The root was determined by using the 16S rRNA gene sequence of Chlorobium limicola Y10643 as an outgroup. The scale bar represents 0.1 substitution per nucleotide position.
FISH studies. (i) Bacterial community structure.
The number of DAPI-stained cells in peat sampled over the Bakchar bog profile increased with depth and was in the range of 0.67 × 109 to 1.45 × 109 cells per g of wet peat (Table 1). In FISH with EUB338-mix, the number of bacterial cells varied along the depth profile in a narrow range of 1.0 × 108 to 1.5 × 108 cells per g of wet peat. This comprised only 9 to 18% of the total DAPI cell counts. The number of cells targeted with archaeal probes ARCH915 and ARC344 clearly increased with depth and comprised up to 8.2% of the total cell number. Thus, the proportion of cells detectable in Sphagnum peat with both sets of domain-specific probes did not exceed 22% of the DAPI-stained cells, and the percentage of bacterial cells detectable by FISH in relation to total DAPI cell counts decreased with increasing depth (Table 1). The DAPI-stained objects which were targeted neither with EUB338-mix nor with ARCH915 plus ARC344 were represented by cells of a very small size, i.e., ≤0.5 μm in length. They accounted for up to 80% of the DAPI-stained cells and were not identified by any of the fluorescent oligonucleotide probes further applied in this study. The nature of these small objects in peat from the same Siberian wetland was recently addressed in a separate study (25). Combined application of fractionation technique, scanning electron microscopy, and respiration measurements showed that these cells are viable nano-sized microorganisms which do not form colonies on conventional media and account for only 2% of the total microbial respiration in this peat sample. Only Archaea-affiliated 16S rRNA gene sequences were retrieved from DNA extracted from a collected fraction of these nano-sized cells.
TABLE 1.
Total cell numbers over the depth profile of Sphagnum peat bog Bakchar determined by different detection methods
| Depth (cm) | No. of cells (108) per g of wet peata determined with:
|
||
|---|---|---|---|
| DAPI staining | EUB338-mix | ARCH915 + ARC344 | |
| 0-10 | 6.68 ± 1.35 | 1.22 ± 0.31 (18.2) | 0.27 ± 0.07 (4.1) |
| 10-20 | 8.42 ± 1.33 | 1.08 ± 0.31 (12.8) | 0.19 ± 0.05 (2.2) |
| 20-30 | 7.89 ± 1.55 | 0.98 ± 0.28 (12.4) | 0.37 ± 0.11 (4.7) |
| 30-40 | 12.24 ± 2.24 | 1.06 ± 0.34 (8.7) | 0.46 ± 0.15 (3.8) |
| 40-50 | 14.46 ± 2.57 | 1.50 ± 0.34 (10.4) | 0.51 ± 0.15 (3.5) |
| 70-80 | 11.39 ± 2.19 | 1.24 ± 0.25 (10.9) | 0.94 ± 0.26 (8.2) |
Data are means ± standard errors. Values in parentheses indicate the percentage of cells targeted with the probe set in relation to DAPI counts.
A set of fluorescently labeled 16S and 23S rRNA-targeted oligonucleotide probes with reported specificity for major phylogenetic groups within the domain Bacteria (see Table S1 in the supplemental material) was applied in FISH to determine the abundance of the respective groups in the same Sphagnum peat sample (depth of 10 to 20 cm) which had been used for the cultivation-independent retrieval of 16S rRNA gene sequences. The Alphaproteobacteria were detected as the most abundant bacterial population and comprised 1.1 × 107 ± 0.4 × 107 cells per g of wet peat. Beta-, Gamma-, and Deltaproteobacteria were not abundant, and their population sizes ranged from 6.9 × 105 to 8.8 × 105 cells per g of wet peat. These findings agreed well with the results of the 16S rRNA gene clone library analysis. This was also true for the low population sizes of the Firmicutes and Bacteroidetes (3.9 × 105 ± 1.9 × 105 and 2.6 × 105 ± 2.1 × 105 cells per g of wet peat, respectively), two major bacterial groups known to be key players in biopolymer degradation in various habitats. The minor role of these bacteria in the Sphagnum peat bog might be one of the reasons for the extremely low rate of biodegradation in this acidic ecosystem.
A strong disagreement of the FISH and clone library data was observed with regard to the numerical significance of the Acidobacteria and Planctomycetes. In contrast to the high proportion of Acidobacteria-affiliated sequences in the clone library, the population size of these bacteria revealed by FISH with probe HoAc1402 (17) was unexpectedly low, i.e., 1.2 × 106 ± 0.4 × 106 cells per g of wet peat. Given that most of these acidobacterial clone sequences had no mismatches in the target region of probe HoAc1402, three possible explanations for the discrepancy between the FISH and clone library data could be offered: (i) most acidobacteria were present in a dormant state, (ii) lysis of acidobacterial cells was more efficient than that for other bacterial populations, and (iii) the 16S rRNA genes of Acidobacteria were preferentially amplified by PCR. However, at present, no preference can be given to any of these hypotheses. Vise versa, the number of planctomycete cells targeted with the probes PLA46 and PLA886 (24) was 4.8 × 106 ± 1.6 × 106 cells per g of wet peat, while only three cloned sequences were affiliated with the Planctomycetes. This might be due to mismatches revealed in planctomycete full-length 16S rRNA gene sequences for the Bacteria-specific PCR primers 27f and 1492r (37). Thus, the clone libraries were most likely biased toward underrepresentation of Planctomycetes-affiliated sequences, while FISH allowed us to reveal this pitfall of the PCR-based approach and to show that planctomycetes represent one of the numerically significant populations in acidic Sphagnum peatlands.
Although much less pronounced, the same kind of disagreement between the FISH and clone library data was also observed for the numerical significance of the Actinobacteria and Verrucomicrobia. The cell number determined by FISH with the Actinobacteria-specific probe HGC69a was not particularly high and comprised 1.0 × 106 ± 0.3 × 106 cells per g of wet peat. This might be partially due to the limited detection scope of probe HGC69a, which was designed to target an insertion within domain III of actinobacterial 23S rRNA genes (31). However, this insertion was reported to be absent in Conexibacter woesei (22), suggesting that deeply branching lineages in the Actinobacteria might lack this feature. Thus, the C. woesei-related bacteria detected by the clone library analysis might not be targeted by probe HGC96a. The probes VER139 and 1463 were designed to enumerate two “peat-specific” subgroups of the Verrucomicrobia (see Fig. S1A in the supplemental material). Use of these two probes in FISH identified 1.0 × 106 ± 0.7 × 106 verrucomicrobial cells per g of wet peat. Thus, in this particular peat sample, only 20% of all EUB338-mix-positive cells could be assigned to specific bacterial phyla by using the described set of probes (see Table S1 in the supplemental material).
(ii) Depth distribution.
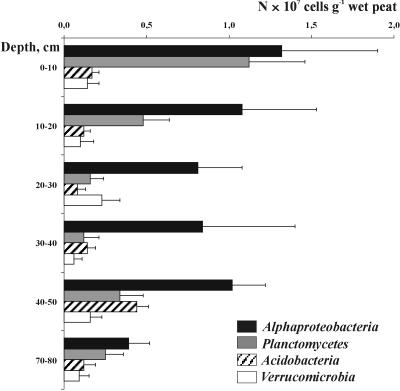
FISH was further applied to assess the depth distribution of major bacterial groups (Fig. 4). The Alphaproteobacteria remained the most abundant group over the peat bog profile and comprised 45 to 72% of all cells targeted in peat with the set of probes applied. Members of the Planctomycetes showed two population maxima. The first maximum (1.1 × 107 ± 0.3 × 107 cells per g of wet peat) was detected in the uppermost, oxic layer of the bog profile. We observed numerous microcolonies of planctomycetes being attached to particles of nondecomposed organic material (Fig. 3A). Interestingly, a sharp decline of planctomycete abundance with depth was followed by a second population maximum in an anoxic layer of the bog profile, at a depth of 40 to 50 cm. At the same depth we observed a population maximum (4.4 × 106 ± 0.7 × 106 cells per g of wet peat) of members of the Acidobacteria, while a stable population size at a lower level was observed in the upper bog layers (1.1 × 106 to 1.7 × 106 cells per g of wet peat). In summary, the proportion of EUB338-mix-positive cells that could be assigned to specific bacterial phyla by using this set of probes varied from 7% in the deeper peat layers to 24% in an uppermost peat layer.
FIG. 4.
Depth distribution of bacterial cells that were detected in Sphagnum peat bog Bakchar with probes specific for the Alphaproteobacteria, Planctomycetes, Acidobacteria, or Verrucomicrobia. Error bars indicate standard errors.
Cultivation. (i) Conventional approach.
The numbers of CFU obtained on different solid media were quite low and varied between 0.2 × 106 and 3.1 × 106 CFU per g of wet peat (see Table S2 in the supplemental material), which is in the range of values obtained for Sphagnum peat by other authors (11). This corresponded to 0.02 to 0.36% of total DAPI cell counts. One hundred colonies were randomly picked from the plates and screened by whole-cell hybridization with rRNA-targeted fluorescent probes. Only 49 of these colonies represented pure cultures of bacteria, while the other 51 colonies were the cocultures of two to four organisms. Almost half of the pure cultures were affiliated with the Betaproteobacteria. Other isolates were affiliated with the Alphaproteobacteria (20%), Actinobacteria (10%), Gammaproteobacteria (8%), Firmicutes (4%), and Bacteroidetes (2%). None of the pure cultures belonged to the Acidobacteria or Planctomycetes. Partial sequencing of the 16S rRNA genes from a representative set of isolates showed that almost all of them were most closely related to taxonomically described organisms (see Table S3 in the supplemental material). None of these 16S rRNA gene sequences was identical to the peat clones in the library. Only two alphaproteobacterial isolates, i.e., PN-29 and MP03, were affiliated with the acidophilic heterotrophic bacteria Acidisphaera and Gluconacetobacter, which had also been detected at the genus level by the clone library analysis (Fig. 2).
A somewhat different situation was observed when all isolated strains, including cocultures, were taken into account. The major difference was that the cocultures contained representatives of both the Acidobacteria and Planctomycetes. This observation gave us the idea that an isolation approach based on enrichment of mixed cultures or microbial biofilms might be more efficient for isolating bacteria indigenous to Sphagnum peat than a routine “single-colony pick-up” strategy.
(ii) Biofilm-mediated isolation.
Microbial biofilms that developed on the surfaces of the cover slides were composed of morphologically diverse organisms. Biofilm screening by FISH with the probes specific for the Acidobacteria or Planctomycetes showed that a variety of organisms from both phyla were present in high cell numbers. The cell material taken from these biofilms was further used for isolation of the target organisms. All steps of the isolation procedure were monitored by FISH. As a result, a single strain of the phylum Acidobacteria (designated MPL3) and two representatives of the phylum Planctomycetes (designated A10 and MPL7) were obtained in a pure culture.
Strain MPL3 is the first isolate of Acidobacteria from Sphagnum peatland. It belongs to subdivision 3 of this phylogenetically diverse phylum (Fig. 1) and is only distantly related (90% sequence similarity) to several acidobacterial isolates (Ellin 342, 371, 6071, 6076, and 6115) (32). Thus, strain MPL3 represents a new family-level group within the Acidobacteria.
Strains A10 and MPL7 belong to planctomycete lineages defined by the genera Gemmata and Planctomyces, respectively (Fig. 3B). The 16S rRNA gene sequence from isolate A10 showed 90% similarity to that of Gemmata obscuriglobus, the only taxonomically described organism in this group. Strain A10 was more closely related (96% sequence similarity) to the taxonomically undescribed Gemmata-like isolate JW10-3f1, which was obtained from a eutrophic lake (38). The morphology of peat bog isolate A10 was different from the morphology described for representatives of the genus Gemmata. In contrast to single, spherical cells typical for Gemmata, the ovoid-like cells of strain A10 were assembled in large rosette-like cell clusters, encompassing up to 20 or more cells. The 16S rRNA gene sequence of the other planctomycete isolate, strain MPL7, exhibited 90% similarity to that of Planctomyces limnophilus and 96 to 97% similarity to those of the taxonomically undescribed Planctomyces sp. strains Schlesner 638 and 642. The latter two organisms were obtained from a compost leakage water (39). Notably, strain MPL7 was represented by spherical cells lacking a stalk, while the presence of stalks is considered a typical genus-specific feature of Planctomyces. In this respect, isolate MPL7 is similar to strain Schlesner 642, since the latter has been, so far, the only reported example of a stalkless, spherical cell morphology found in the genus Planctomyces (39). Thus, both isolates, i.e., strains A10 and MPL7, were only distantly related to taxonomically described planctomycetes and possessed different cell morphologies. The physiology and specific adaptations of these organisms to the acidic environment remain to be investigated. So far, there has been only one previous report on the isolation of a planctomycete-like strain from acidic peat water, i.e., the bog water (pH 4.2) of the Kaltenhofer Moor near Kiel, Germany (34). This strain has not been described taxonomically, and no further information except a brief morphological characterization was given. According to the morphological characterization, it had the same cell morphotype as our isolate MPL7. Thus, Sphagnum-dominated wetlands seem to represent a novel yet unexploited source for exploring the biodiversity of the Planctomycetes.
In summary, a phylogenetically diverse and mostly unknown 16S rRNA gene sequence diversity was detected in an acidic Sphagnum peat bog. The greatest diversity was found for members of the Acidobacteria. FISH showed that Alphaproteobacteria and Planctomycetes are numerically significant groups in this environment. Due to methodological limitations, most of the peat bog Bacteria community remained inaccessible by FISH. However, we found a key to the isolation of peat-inhabiting organisms, such as members of the Acidobacteria and Planctomycetes. The biofilm-mediated approach will provide us with a set of unique organisms for a detailed study of those phenotypic and genotypic traits that enable indigenous bacterial life in this extreme environment.
Supplementary Material
Acknowledgments
This research was supported by the program “Molecular and Cell Biology” of the Russian Academy of Sciences, the Russian Fund of Basic Research (grant no. 04-04-04000), and the Deutsche Forschungsgemeinschaft (436 RUS 113/543/0-3).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Basiliko, N., J. B. Yavitt, P. M. Dees, and S. M. Merkel. 2003. Methane biogeochemistry and methanogen communities in two northern peatland ecosystems, New York State. Geomicrobiol. J. 20:563-577. [Google Scholar]
- 2.Chin, K.-J., W. Liesack, and P. H. Janssen. 2001. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int. J. Syst. Evol. Microbiol. 51:1965-1968. [DOI] [PubMed] [Google Scholar]
- 3.Clark, D. A., and P. R. Norris. 1996. Acidimicrobium ferrooxidans gen. nov., sp. nov.: mixed culture ferrous iron oxidation with Sulfobacillus species. Microbiology 142:785-790. [DOI] [PubMed] [Google Scholar]
- 4.Coates, J. D., D. J. Ellis, C. V. Gaw, and D. R. Lovley. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49:1615-1622. [DOI] [PubMed] [Google Scholar]
- 5.Dedysh, S. N., N. S. Panikov, and J. M. Tiedje. 1998. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl. Environ. Microbiol. 64:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedysh, S. N., N. S. Panikov, W. Liesack, R. Großkopf, J. Zhou, and J. M. Tiedje. 1998. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282:281-284. [DOI] [PubMed] [Google Scholar]
- 7.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 8.Dedysh, S. N., M. Derakshani, and W. Liesack. 2001. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67:4850-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedysh, S. N., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, W. Liesack, and J. M. Tiedje. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52:251-261. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 11.Golovchenko, A. V., Y. V. Sannikova, T. G. Dobrovol'skaya, and D. G. Zvyagintsev. 2005. The saprotrophic bacterial complex in the raised peat bogs of western Siberia. Mikrobiologiia 74:545-551. (In Russian). [PubMed] [Google Scholar]
- 12.Gorham, E. 1991. Northern peatlands: role in carbon cycle and probable responses to climate warming. Ecol. Appl. 1:182-195. [DOI] [PubMed] [Google Scholar]
- 13.Horn, M. A., C. Matthies, K. Küsel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 18.Kivinen, E., and P. Pakarinen. 1981. Geographical distribution of peat resource and major peatland complex types in the world. Ann. Acad. Sci. Fenn. Ser. A3 132:1-28. [Google Scholar]
- 19.Kotsyurbenko, O. R., K.-J. Chin, M. V. Glagolev, S. Stubner, M. V. Simankova, A. N. Nozhevnikova, and R. Conrad. 2004. Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6:1159-1173. [DOI] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 21.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckman, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monciardini, P., L. Cavaletti, P. Schumann, M. Rohde, and S. Donadio. 2003. Conexibacter woesei gen. nov., sp. nov., a novel representative of a deep evolutionary line of descent within the class Actinobacteria. Int. J. Syst. Evol. Microbiol. 53:569-576. [DOI] [PubMed] [Google Scholar]
- 23.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neef, A., R. Amann, H. Schlesner, and K.-H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of Planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 25.Panikov, N. S. 2005. Contribution of nanosized bacteria to the total biomass and activity of a soil microbial community. Adv. Appl. Microbiol. 57:245-296. [DOI] [PubMed] [Google Scholar]
- 26.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radajewski, S., G. Webster, D. S. Reay, S. A. Morris, P. Ineson, D. B. Nedwell, J. I. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 28.Raghoebarsing, A. A., A. J. P. Smolders, M. C. Schmid, W. I. C. Rijpstra, M. Wolters-Arts, J. Derksen, M. S. M. Jetten, S. Schouten, J. S. Sinninghe Damsté, L. P. M. Lamers, J. G. M. Roelofs, H. J. M. Op den Camp, and M. Strous. 2005. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 436:1153-1156. [DOI] [PubMed] [Google Scholar]
- 29.Rheims, H., F. A. Rainey, and E. Stackebrandt. 1996. A molecular approach to search for diversity among bacteria in the environment. J. Ind. Microbiol. 17:159-169. [Google Scholar]
- 30.Rheims, H., C. Spröer, F. A. Rainey, and E. Stackebrandt. 1996. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology 142:2863-2870. [DOI] [PubMed] [Google Scholar]
- 31.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 32.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 33.Sanford, R. A., J. R. Cole, and J. M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesner, H. 1994. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17:135-145. [Google Scholar]
- 35.Sizova, M. V., N. S. Panikov, T. P. Tourova, and P. W. Flanagan. 2003. Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a Sphagnum peat bog. FEMS Microbiol. Ecol. 45:301-315. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, J., C. Jenkins, R. I. Webb, and J. A. Fuerst. 2002. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 68:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward, N., F. A. Rainey, E. Stackebrandt, and H. Schlesner. 1995. Unraveling the extent of diversity within the order Planctomycetales. Appl. Environ. Microbiol. 61:2270-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.