Abstract
Since 1998 a lethal disease of carp and ornamental koi (Cyprinus carpio) has afflicted fisheries in North America, Europe, and Asia, causing severe economic losses to the fish farming industry. This review summarizes the isolation and identification of the disease-causing agent and describes the currently known molecular characteristics of this newly isolated virus, distinguishing it from other known large DNA viruses. In addition, we summarize the clinical and histopathological manifestations of the disease. Providing information on the immune response to this virus and evaluating the available means of diagnosis and protection should help to reduce the damage induced by this disease. This review does not discuss the economic aspects of the disease or the debate on whether the disease should be registered; both of these issues were recently reviewed in detail (O. L. M. Haenen, K. Way, S. M. Bergmann, and E. Ariel, Bull. Eur. Assoc. Fish Pathol. 24:293-307, 2004; D. Pokorova, T. Vesely, V. Piackova, S. Reschova, and J. Hulova, Vet. Med. Czech. 50:139-147, 2005).
INTRODUCTION
Common carp (Cyprinus carpio carpio) is a fish species that is widely cultivated for human consumption, with 1.5 million metric tons harvested annually, principally by China, other Asian countries, and European countries (www.fao.org). In contrast to common carp, the subspecies koi (Cyprinus carpio koi) is a beautiful and colorful fish, the focus of a hobby whose enthusiasts keep koi in backyard ponds and large display aquaria for personal pleasure or competitive showing. The hobby originated with the Romans in the first century A.D., matured into the present science and art practiced in Japan, and subsequently spread worldwide (2).
A mortal viral carp disease was detected in the United Kingdom in 1996, but the first scientific reports did not appear until 1998, when Ariav and coworkers described the disease (1) following major outbreaks in several carp farms along the Israeli coast. The disease was not restricted to the United Kingdom and Israel, and reports appeared describing a similar disease with mass mortality in Germany (5, 24). Soon afterwards, countries all over the world reported the presence of this contagious disease (7, 22).
Although the virus has been isolated, its morphology intensively studied by electron microscopy, the molecular size of the genome estimated, and about 16% of its genome sequenced and published in GenBank (see below), the International Committee on Virus Taxonomy has not yet determined the nomenclature of this virus. The virus has been designated by several names, first as koi herpesvirus (KHV), according to its morphological manifestation (23). However, this name is problematic, since the virus infects common carp and perhaps other cyprinids (S. M. Bergmann, unpublished data). By using pulsed-field gel electrophoresis, it was estimated that its genomic size is 277 kbp, larger than that of any known herpesvirus (26), and it bears sequences divergent from those of all other known herpesviruses. Moreover, this virus bears at least two genes which have not yet been described for the genomes of Herpesviridae members: the thymidylate kinase (TmpK) and serine protease inhibitor (serpin) genes (M. Ilouze, unpublished data). Based on these findings, we temporarily designated it carp interstitial nephritis and gill necrosis virus (CNGV), according to the pathogenic effects it induces in fish (36, 40). Following recognition that the viral genome contains a significant number of DNA sequences without homology to any other known viral sequences, the name koi herpes-like virus was proposed. Recent findings demonstrated that CNGV (KHV) bears several genes whose sequences resemble the DNA sequences of CyHV-1 and CyHV-2 and that the CyHV-1 genome is approximately 295 kbp, similar in size to CNGV (KHV) (26, 40, 49). These findings led to the suggestion that CNGV be designated cyprinid herpesvirus 3 (CyHV-3) in order to emphasize its unique characteristics (49). Previously, we suggested that CNGV is distinguishable from other Herpesviridae members. Now, when the genomes of CyHV-1 and CyHV-2 have been partially sequenced and have been found to be closely related to CNGV, it has become clear that it is not just CNGV that should be classified in a specific clade but that this cluster should include at least the two other CyHVs and probably the distant relatives ranid herpesvirus 1 and ictalurid herpesvirus 1 (49).
Virions of herpesviruses have complex and characteristic structures consisting of both symmetrical and nonsymmetrical components (25, 46). The spherical virion comprises the core, capsid, tegument, and envelope (32). The core consists of the viral genome, which is packaged as a single linear double-stranded DNA (dsDNA) molecule ranging from 125 to ∼240 kbp in size. Thus, the use of “herpes” in the CyHV may require either changing the definition of the herpesvirus family to include viruses with a genome much larger than 240 kbp or classifying this virus with a large genome into a specific group. In addition, mammalian and avian herpesviruses are highly adapted to their hosts, and lethal infection is usually observed only in fetuses, in immunosuppressed organisms, or following infection of an alternative host. Herpesviruses establish lifelong latent infection, a feature which is assumed to be the hallmark of all herpesviruses (47a). In contrast, infection with CNGV causes an acute disease with greater than 90% mortality of both fries and adult carp. Assuming CNGV to be truly a member of the Herpesviridae family suggests that it persists in the host as a latent virus. Solving the question of latency will contribute not only to the phylogenic classification of this virus but also to understanding its epidemiology and improving prophylactic measures to prevent the spread of the disease. In the absence of an official name for this virus, we will continue to use the term CNGV in this review, although we agree that the virus shows similarity to the unassigned CyHVs. Once the evolutionary origin of these viruses is resolved, the most appropriate name can be assigned.
IDENTIFICATION AND CHARACTERIZATION OF CNGV
Isolation and General Description
Hedrick and coworkers described the isolation of the virus causing this carp and koi disease after infection of cultured KF-1 cells with cell extracts prepared from the organs of sick fish (23). Establishment of primary cultures of koi fin cells (KFC) and carp fin cells (CFC) allowed independent isolation of the etiologic agent in Israel (35, 40). The isolated virus from sick carp was confirmed as the etiologic agent for this disease based on the following data: (i) the virus was successfully isolated from infected fish but not from naive specimens; (ii) inoculation of the virus propagated in KFC culture into naive fish induced a similar disease; (iii) cocultivation of kidney cells taken from specimens with the induced disease, but not from the mock-infected fish, yielded a similar virus, which propagated in KFC cultures; (iv) three cycles of transferring the virus from sick fish to cultured CFC (“ping-pong” technique) were successfully applied; (v) cloned virus isolated in tissue culture induced the same disease in fish; (vi) rabbit sera prepared against the purified virus interacted specifically with tissues from experimentally infected fish as well as with sick fish from ponds; and (vii) viral DNA was identified in infected KFC and in sick fish but not in naive fish (26, 35, 40). This initial identification of CNGV facilitated diagnosis of the disease by infection of KFC, PCR, and immunological methods.
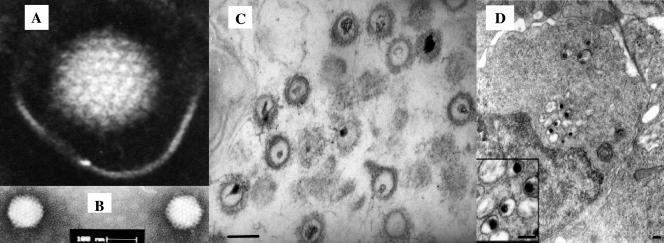
Electron microscopy of negative-stained particles showed an icosahedral morphology with an average core diameter of 100 to 110 nm, resembling the core of herpesvirus (23, 26, 39). Thin sections of purified virus pellet revealed enveloped particles with a thread-like structure (tegument) on the surface of the core. The cores bear atypically nonsymmetrical electron-dense regions, which probably contain the viral genome (26) (Fig. 1). The virus has a buoyant density of 1.16 g/ml and bands at 37 to 39% (wt/vol) sucrose following centrifugation in a sucrose gradient (26).
FIG. 1.
Electron micrographs of CNGV. (A and B) CNGV harvested from infected KFC was purified by sucrose gradient centrifugation, and virus was negatively stained with 2% phosphotungstate. The core size was in the 96- to 105-nm range, with an average diameter of 103 nm. Bar, 0.1 μm. (C) Thin sections were made for ultrastructural analysis by transmission electron microscopy. Purified virus pellets were fixed with 3% glutaraldehyde in 0.1 M sodium cacodylate and stained with uranyl acetate and lead citrate. (D) Ultrastructural appearance of CNGV particles in infected kidney at 8 days postinfection. This cell harbors several cytoplasmic viral particles with round electron-dense cores (magnified in the inset). Bars, 200 nm.
CNGV survived in pond water for 4 h at temperatures of around 22°C and probably survives for a longer period in droppings and pond mud (7, 12, 26, 35). However, as a guideline for virus inactivation, infectivity is abolished after 2 days at 35°C (35) or after 30 min at 60°C (7). The virus is inactivated at pH values of below 3 and above 11, and it is also readily inactivated in chloroform, 25% ether, or 0.1% Triton X-100 (7, 26).
Propagation of the Virus in Cultured Cells
At present, CNGV propagates well on cell lines derived from koi fin (KF-1) (23) and common carp brain (33), as well as on KFC and CFC (40), and induces plaques at 3 to 4 days postinfection (dpi) at 22°C. The titer of the virus produced under these conditions exceeds 2 × 106 PFU/ml, enabling molecular studies as well as production of virus for vaccination (34, 40).
The virus isolated from sick fish induced typical plaques in all of the cultured cells listed above: it increased cell volume and induced the formation of syncytia and abundant cytoplasmic vacuoles, and the cells became rounded before they detached from the substrate. It is noteworthy that infected cells with foamy cytoplasm were also observed in kidneys of diseased fish (36).
The number of plaques induced by CNGV is directly related to the viral dilution: serial dilution yields a single-hit curve, indicating that a single infectious unit is sufficient to produce each plaque. In addition, virus isolated from a single plaque was sufficient to induce the disease in fish, demonstrating that no helper virus or additional microorganism is required for inducing the disease (36).
The propagation of the virus in tissue cultures is temperature restricted. The optimal temperature for viral propagation in KF-1 cells is 15 to 25°C; at 30°C these cells did not produce virus (16). It would be interesting to determine whether viral and/or host factors are involved in restricting virus propagation to the permissive temperatures and to clarify the molecular machinery that represses viral function at nonpermissive temperatures.
Viral DNA
Sequencing shows that the CNGV genome is a large linear dsDNA of 295 kbp (49), similar in size to the CyHV-1 genome as determined by pulsed-field gel electrophoresis (26, 49). The CNGV genome is larger than those of vaccinia virus, herpes simplex virus type 1, and all known members of the Herpesviridae except CyHV-1. It is possible that the large genome is characteristic for the CyHVs. A large fragment or perhaps even the entire CNGV genome has been resolved (49, 50), yet at present only ∼16% of the genome sequence is available to the scientific community via GenBank.
Of the CNGV DNA sequences available in GenBank, many are highly divergent from all other DNA viruses. Small genomic fragments of 16 to 45 bp show similarity mainly to members of the Herpesviridae, Adenoviridae, Poxviridae, and Baculoviridae families (26). One example is the DNA sequence of the CNGV thymidine kinase (TK) gene (accession no. AJ535112) (4), of which only a 21-bp DNA fragment is similar to the Shope fibroma virus (a poxvirus) TK gene. However, recent studies found that the DNA sequences of the CNGV DNA polymerase, major capsid protein, helicase, and intercapsomeric triplex protein genes and of an additional open reading frame encoding an unidentified protein resemble those of CyHV-1 and CyHV-2 (49) (Table 1). Although these genes of CNGV and CyHVs are similar to each other, there was no apparent homology with other DNA viruses. Providing additional CyHV-1, CyHV-2, and CNGV DNA sequences to GenBank will assist in the phylogenic classification of CNGV, which will be instrumental in controlling the lethal disease caused by CNGV. It is conceivable that fish viruses that evolve in distinct ecological environments are genetically highly divergent from each other. In this case it would be expected that CNGV, which thrives in captivity and is restricted to limited species of Cyprinus, bears only slight similarity to other viruses.
TABLE 1.
Relationship of CNGV to fish and amphibian herpesvirus genes (Blast N)a
| Gene (accession no.) | Virus | Accession no. | Identityb (%) | E value |
|---|---|---|---|---|
| DNA polymerase gene (AY939862) | Cyprinid herpesvirus 1 | AY939868 | 927/1,121 (82) | 0 |
| Cyprinid herpesvirus 2 | AY939863 | 372/467 (79) | 1e−40 | |
| Major capsid protein gene (AY939864) | Cyprinid herpesvirus 1 | AY939865 | 293/362 (80) | 2e−38 |
| Intercapsomeric triplex protein gene (AY939859) | Cyprinid herpesvirus 1 | AY939860 | 41/45 (91) | 5e−05 |
| Cyprinid herpesvirus 2 | AY939861 | 54/65 (83) | 3.6 | |
| DNA helicase gene (AY939857) | Cyprinid herpesvirus 2 | AY939867 | 90/101 (89) | 2e−21 |
| Cyprinid herpesvirus 1 | AY939858 | 101/116 (87) | 6e−21 | |
| Unidentified open reading frame (AY208988) | Cyprinid herpesvirus 1 | AY939866 | 37/41 (90) | 0.0038 |
Five viral genes were analyzed by using the Blast N program with default parameters. Data are based on reference 49.
Number of identical nucleotides/total. The assessed fragments do not represent the sequences of complete genes.
Viral Proteins
The number of open reading frames in the viral genome and the number of proteins assembled in the virion are not yet known. Rough estimations made by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of purified virions approximated the number of proteins to be between 31 (17) and 80 (our unpublished results). However, due to the large genomic size of this virus, the number of proteins expressed by CNGV may be even larger. Recent data show that the CNGV genome allows 183 to 185 open reading frames (T. Aoki, personal communication).
In addition to these CNGV genes resembling those of CyHV-1 and CyHV-2 at the DNA level, analysis by use of the BLAST X program, which allows comparison of translation products of viral DNA sequences to viral protein databases, showed that the proteins encoded by the major capsid protein, DNA polymerase, helicase, and intercapsomeric triplex genes have some similarity to the proteins of anguillid herpesvirus 1 (AngHV-1) (28), ictalurid herpesvirus 1 (8), ranid herpesvirus 1 (10), and salmonid herpesvirus 1 (Table 2) (9). This similarity led to the suggestion that all these viruses be included in a new group of aquatic herpesvirus-like viruses (49). In agreement with this view, Rijsewijk and coworkers (38) demonstrated that CyHV (KHV) and AngHV-1 are closely related according to the sequences of their DNA polymerase proteins. Although the amino acid sequences of these two viruses are similar to those of ranid herpesvirus 1 and ictalurid herpesvirus 1, it has been suggested that they be categorized as a separate group (38).
TABLE 2.
Similarity of CNGV to fish and amphibian herpesvirus genes (Blast X)a
| Gene (accession no.) | Virus | Accession no. | Identity (%) | Similarity (%) | Gaps (%) |
|---|---|---|---|---|---|
| DNA polymerase gene (AY939862) | Cyprinid herpesvirus 1 | AAX53084 | 976/1,309 (74) | 1,121/1,309 (85) | 11/1,309 (0) |
| Cyprinid herpesvirus 2 | AAX53083 | 140/155 (90) | 147/155 (94) | ||
| Anguillid herpesvirus 1 | AAK00356 | 99/155 (63) | 122/155 (77) | 1/155 (0) | |
| Ranid herpesvirus 1 | AAD12269 | 282/1,047 (26) | 449/1,047 (41) | 140/1,047 (13) | |
| Ictalurid herpesvirus 1 | NP_041148 | 217/801 (27) | 349/801 (43) | 107/801 (13) | |
| DNA helicase gene (AY939857) | Cyprinid herpesvirus 1 | AAX53077 | 344/482 (71) | 398/482 (82) | 2/482 (0) |
| Cyprinid herpesvirus 2 | AAX53078 | 219/302 (72) | 238/302 (78) | 26/302 (0) | |
| Ictalurid herpesvirus 1 | NP_041116 | 127/493 (25) | 214/493 (42) | 64/493 (12) | |
| Intercapsomeric triplex protein gene (AY939859) | Cyprinid herpesvirus 1 | AAX53080 | 205/370 (55) | 272/370 (73) | 5/370 (1) |
| Cyprinid herpesvirus 2 | AAX53081 | 58/88 (65) | 67/88 (75) | 3/88 (3) | |
| Ictalurid herpesvirus 1 | NP_041118 | 76/333 (22) | 118/333 (34) | 85/333 (25) | |
| Major capsid protein gene (AY787402) | Cyprinid herpesvirus 1 | AAX53086 | 823/1,279 (64) | 1,008/1,279 (78) | 21/1,279 (1) |
| Ictalurid herpesvirus 1 | NP_041130 | 252/1,170 (21) | 414/1,170 (34) | 211/1,170 (18) | |
| Unidentified open reading frame (AY208988) | Cyprinid herpesvirus 1 | AAX53075 | 96/135 (71) | 118/135 (87) | 1/135 (0) |
| Ranid herpesvirus 1 | AAD12270 | 72/260 (27) | 119/260 (45) | 16/260 (6) | |
| Ictalurid herpesvirus 1 | NP_041147 | 54/186 (29) | 87/186 (46) | 13/186 (6) |
Five viral proteins were analyzed by using the standard Blast X program. Identity is the number of amino acids with identity/total number of amino acids of the protein, similarity is the number of conserved amino acids/total number of amino acids; and gaps are number of gaps/total number of amino acids. Data are based on references 38 and 49.
Importantly, the CNGV sequence encodes three enzymes involved in the processing of nucleotides for DNA synthesis, i.e., ribonucleotide reductase, TmpK, and TK, which are similar to those encoded by the poxvirus genome (Table 3). It also should be emphasized that serine protease inhibitor (serpin) encoded by CNGV is a protein found exclusively in poxviruses. Serpin and TmpK have not yet been found in herpesviruses (6, 44). It would be of great interest to determine whether CyHV-1 and CyHV-2 code for these three proteins as well. Several other CNGV polypeptides show similarity to proteins derived from ictalurid herpesvirus 1 (8), African swine fever virus, and shrimp white spot. Iyer et al. (27) showed that African swine fever virus, poxviruses, iridoviruses, and phycodnaviruses share a common evolutionary origin but have no direct evolutionary relationship to herpesviruses. Based on these varied homologies to small parts of other viruses, we speculate that either CNGV evolved by intensive horizontal incorporation (probably by recombination) of genetic fragments derived from poxviruses and other DNA viruses or the CyHVs group descended from a unique common ancestor differing from the mammalian, avian, and reptile viruses, as well as from the clade of ostreid herpesvirus (11). If the latter possibility proves to be true, it would be logical to include the CyHVs as a separate family under the umbrella of the order Herpesviralles.
TABLE 3.
Similarity of CNGV to representatives of poxviruses and other large DNA viruses (Blast X)a
| Gene (accession no.) | Family | Virus | Accession no. | Identity (%) | Similarity (%) | Gaps (%) |
|---|---|---|---|---|---|---|
| Serine protease inhibitor homologue gene (AY661550) | Poxviridae | Fowlpox virus (B22R homologue) | NP_039060 | 55/139 (39) | 81/139 (58)b | 3/139 (2) |
| Variola virus (B26R homologue) | CAA49135 | 45/112 (40) | 64/112 (57)b | 1/112 (0) | ||
| Thymidylate kinase gene (DQ118125) | Poxviridae | Canarypox virus | NP_955193 | 106/216 (49) | 143/216 (66) | 8/216 (3) |
| Nimaviridae | Shrimp white spot syndrome virus | AAK77840 | 82/191 (42) | 117/191 (61) | 1/191 (0) | |
| TK-dTMP | ||||||
| Iridoviridae | Chilo iridescent virus | AAK82112 | 72/176 (40) | 117/176 (66) | 3/176 (1) | |
| Asfarviridae | African swine fever virus | CAA79604 | 57/192 (29) | 89/192 (46) | 8/192 (4) | |
| Ribonucleotide reductase gene (AY786308) | Poxviridae | Monkeypox virus | AAU01269 | 447/803 (55) | 580/803 (72) | 35/803 (4) |
| Baculoviridae | Spodoptera litura nucleopolyhedrovirus | AAL01709 | 415/763 (54) | 555/763 (72) | 13/763 (1) | |
| Phycodnaviridae | Feldmannia irregularis virus a | AAR26844 | 394/765 (51) | 529/765 (69) | 12/765 (1) | |
| Mimivirus | Acanthamoeba polyphaga mimivirus | YP_142667 | 346/733 (47) | 489/733 (66) | 12/733 (1) | |
| Nimaviridae | Shrimp white spot syndrome virus | AAL89096 | 379/845 (44) | 538/845 (63) | 49/845 (5) | |
| Asfarviridae | African swine fever virus | P42491 | 285/737 (38) | 415/737 (56) | 64/737 (8) | |
| Herpesviridae | Ostreid herpesvirus 1 | YP_024594 | 263/741 (35) | 415/741 (56) | 58/741 (7) | |
| Thymidine kinase gene (AJ535112) | Poxviridae | Cowpox virus | AAF21104 | 71/171 (41) | 114/171 (66) | 3/171 (1) |
| Mimivirus | Acanthamoeba polyphaga mimivirus | YP_142612 | 60/181 (33) | 102/181 (56) | 14/181 (7) | |
| Nimaviridae | Shrimp white spot syndrome virus | AAG40728 | 69/178 (38) | 100/178 (56) | 8/178 (4) | |
| Asfarviridae | African swine fever virus | AAQ07945 | 52/177 (29) | 81/177 (45) | 10/177 (5) |
Four viral genes were analyzed by using the standard Blast X program. For definitions, see Table 2, footnote a.
The assessed fragments do not represent the sequences of complete proteins.
In conclusion, the morphology of CNGV is similar to that of the Herpesviridae. However, the genome of the isolated virus is composed of a ∼295-kbp linear DNA molecule, larger than that of the other Herpesviridae members and bearing highly divergent DNA sequences that encode polypeptides resembling those of other large dsDNA viruses. Analysis of the DNA and amino acid sequences of this high-molecular-weight viral genome revealed that CNGV could be a novel virus bearing a mosaic genome including genetic elements derived from phylogenetically distant DNA viruses.
THE DISEASE CAUSED BY CNGV
The disease caused by CNGV is seasonal, appearing in spring and autumn when the water temperature in open-air ponds is 17 to 28°C. It is highly contagious, spreads from infected to healthy fish sharing the same pond, and is lethal to 80 to 100% of the fish in infected open-air ponds. Mortality occurs within 6 to 22 dpi, peaking at between 8 and 12 dpi (35).
Fish exposed to the virus at 20 to 24°C for 3 days and then transferred to a nonpermissive temperature survived the disease (34, 35, 40). On the other hand, many of the fish held at 13°C for 30 days developed the disease following a temperature shift up to 22 to 24°C. Persistence of the virus in the fish is limited, since fish transferred to the permissive temperature after 64 days at 13°C did not die (16).
Disease Characteristics
Although the disease is highly contagious and extremely virulent, morbidity and mortality are restricted to koi and common carp (Cyprinus carpio) populations (5, 35, 48). The virus does not induce the disease in other cyprinid or noncyprinid species, and these virus-exposed fish do not transfer the disease to naive carps, suggesting that species other than Cyprinus carpio are not carriers of CNGV (35; A. Perelberg, unpublished results).
Clinical Signs of the Disease
Sick fish were apathetic and gathered close to the water surface, suffering from suffocation. Gill necrosis appeared as early as 3 dpi, coupled with an increase in the levels of external parasites and bacteria (1). The skin showed a lack of luster, with pale patches and increased mucus secretions (22, 23, 34, 35; Bergmann, unpublished data).
Histopathology
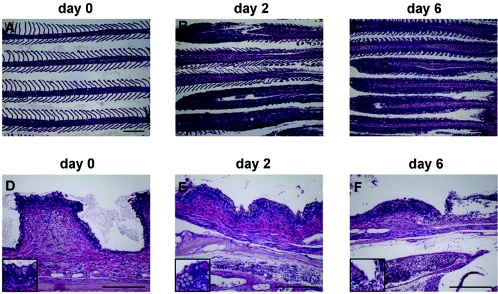
In sick fish the most prominent lesions were observed in the gill, skin, kidney, spleen, liver, and gastrointestinal systems (23). Pathological changes were noted in the gills as early as 2 dpi, as evidenced by a loss of lamellae accompanied by a mixed inflammatory cell infiltrate (Fig. 2). From 6 dpi onward, changes in the gills became more pronounced, with complete effacement of the gill architecture accompanied by severe inflammation in nearly all of the filaments. Congestion of the central venous sinus of the gill was also evident at this stage. Similar changes were evident at 8 and 10 dpi. In fish infected with CNGV, the gill rakers showed effects that were more easily recognizable than the changes observed in the filaments. These include increased subepithelial inflammation and congestion of blood vessels in the gill arch (Fig. 2D to F). The inflammatory process was accompanied by attenuation of the rakers' height. Focal sloughing of the surface epithelium was noted as early as 6 dpi.
FIG. 2.
CNGV induces gill inflammation as early as 2 dpi. Carp were infected with CNGV and harvested on the indicated dpi. Gills were collected and submitted for histological analyses. (A to C) Gill filaments. Normally, gill filaments are slender structures containing numerous lamellae (A). As early as 2 dpi, many lamellae are infiltrated by inflammatory cells (B). At 6 dpi (C) and onwards, all lamellae are heavily infiltrated. (D to F) Gill rakers. As early as 2 dpi (E), an increased inflammatory infiltrate is present in the subepithelial zone. In addition, at the bottom of the photomicrograph a congested vessel in the gill arch is seen. At 6 dpi (F), the inflammatory process is more pronounced, with sloughing of the overlying epithelium (upper right). This is accompanied by increased congestion and edema. All the sections were stained with hematoxylin and eosin. Bars, 200 μm.
While gill injury preceded all other histological changes, it is plausible, as suggested before (23), that this effect is caused by secondary infections. However, the inflammation process detected in the gills appeared to be induced by CNGV itself rather than by secondary infection, since no increase in microorganisms was detected at 2 to 6 dpi despite the marked inflammatory response in the gills, which was apparent as early as 2 dpi. The population of microorganisms did not increase until 8 to 10 dpi, when focal denudation of the gill epithelium occurs.
In addition to the gills, the most prominent pathological changes were noted in the kidneys. A mild peritubular inflammatory infiltrate was evident as early as 2 dpi. On day 6, a heavy interstitial inflammatory infiltrate was observed, along with congestion of blood vessels. At 8 dpi the infiltrates were more severe and were accompanied by a feathery degeneration of the tubular epithelium in many nephrons, together with the presence of intraepithelial lymphocytes. As early as 6 dpi, large cells with a foamy distended cytoplasm and a few intranuclear inclusion bodies were scattered among the inflammatory interstitial cells. These “foamy” cells are reminiscent of the cytopathic effect observed in infected cultured cells. Other organs are also affected: liver analysis showed mild inflammatory infiltrates located mainly in the parenchyma, while brain sections showed focal meningeal and parameningeal inflammation (36).
Detection of CNGV in Fish Tissues
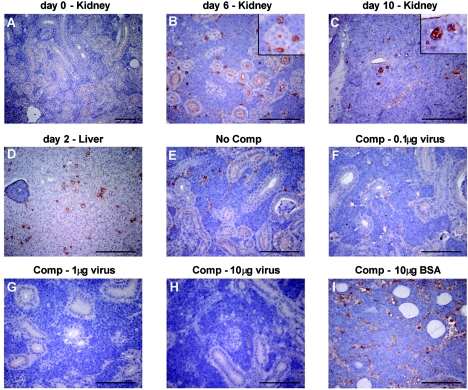
Cells expressing viral proteins were identified using rabbit anti-CNGV antiserum. As early as 2 to 3 dpi cells expressing viral proteins were detected in the interstitial cells, most of which appeared large, corresponding to the foamy cytomegalic cells described above. The number of CNGV-labeled interstitial cells was maximal at 6 dpi and remained constant through 10 dpi (Fig. 3). Interestingly, at 10 dpi, when hematoxylin and eosin staining showed maximal feathery degeneration of the tubular epithelium, viral proteins were also detected in some of the tubular epithelial cells, suggesting that the virus is not restricted to cells of the hematolymphoid lineage. Cells expressing viral proteins were also detected in brain and liver (36). In agreement with the immunohistochemical results, electron microscopy studies revealed viral particles in several fish tissues derived from sick fish, including bronchial epithelial cells (23) and kidney cells (36). Large amounts of viral particles were revealed in intestinal tissues of symptomatic common carp (35).
FIG. 3.
Immunohistochemical staining of carp tissues with antiserum against CNGV. (A to D). Tissue sections from kidney or liver of healthy carp (A) or from carp sacrificed at the indicated dpi (B to D) were incubated with antiserum against CNGV. Note that while at 6 dpi only interstitial cells are stained, at 10 dpi viral proteins are also detected in the epithelial cells. (E to I) To demonstrate the specificity of the staining reaction, the serum was incubated with increasing amounts of CNGV protein (F to H), bovine serum albumin (I), or no protein (E) prior to its application to the tissue sections. Bars, 100 μm. The insets in panels B and C show enlargements. Comp, competition.
CNGV DNA-bearing cells were found in the gill mucus and also between the secondary lamellae of the gills by using in situ hybridization (Bergmann, unpublished data). Several available PCR primers are currently in use for detecting CNGV DNA (17, 20, 26). The number of genome equivalents of CNGV in tissues from infected koi was determined using real-time TaqMan PCR. The results of these experiments (18) correlate well with the mortality observed in experimentally exposed koi at 13, 18, 23, and 28°C (16). The number of CNGV genomes per 106 host cells in the gills and kidneys of fish held at higher temperatures (23 and 28°C) increased earlier than that in fish kept at lower water temperatures. These experiments not only confirmed the relative number of CNGV genomes in fish organs but also provided information about the spread of virus in the fish body (18). By 2 to 3 dpi, viruses appeared in the blood and kidneys of carps infected by bathing or by cohabitation with sick fish, and their titer in these tissues increased up to 7 dpi (26).
Diagnosis of CNGV
The rapid dissemination of the virus in the fish body stimulated several laboratories to develop efficient diagnostic means to identify the virus. Early diagnosis of the disease is extremely important to farmers, hobbyists, and veterinary authorities, allowing them to by take appropriate precautions to prevent spread of the disease. It also allows hobbyists to save their fish by elevating the temperature and allows farmers to advance the marketing of their fish. Several diagnostic techniques are currently in use, including isolation of the virus in cultured cells, histological and immunohistochemical methods, electron microscopy, enzyme-linked immunosorbent assay (ELISA), PCR, and in situ hybridization. These methods were described in detail by Haenen et al. and Pokorova et al. (22, 37).
The early appearance (3 to 5 dpi) of viral particles in carp droppings allows noninvasive detection of CNGV by using PCR, ELISA, and infection of cultured cells (12). Although most of the diagnostic methods are in use, they are time-consuming and laborious and require specialized equipment. Recently, a “one-step ELISA kit” was developed by A. D. Thompson and her collaborators (personal communication), and a loop-mediated isothermal DNA amplification method was described by Gunimaladevi et al. and by Soliman and El-Matbouli (21, 45). Both techniques are easy to use as rapid field diagnostic tools. It would be very helpful if these two new techniques were adapted to test for the presence of viral components in fish droppings.
Detection of prevaccinated fish and/or fish surviving the disease following exposure to CNGV is crucial for veterinary authorities. Identification of viral DNA is not always successful, while detection of anti-CNGV antibodies in the fish's serum is easily done by ELISA (34, 40). Unfortunately, a suitable kit for detecting anti-CNGV antibodies is not yet available.
Dissemination of the Virus
It is still unclear how viruses spread among ponds or from farm to farm and where the virus remains between seasons. Virus harvested from tissue cultures remains infective in water for at least 4 h (35), explaining the highly contagious nature of the virus in ponds. It is not yet known how the virus enters the fish body, i.e., through the gills or through the intestine (23, 35).
Based on the detection of CNGV in gill mucus and lamellae (18, 36; Bergmann, unpublished data), it appears likely that virus infects the fish via the gills, replicates there, induces mucosal sloughing and necrosis, and is then shed into the water. This scenario could explain the rapid and efficient spread of this contagious disease. From the gills, the virus can be rapidly transferred to the kidneys, where it resides in white blood cells and induces severe interstitial nephritis. Localization of the virus within white blood cells raises the intriguing possibility that the virus is rapidly transferred to the viscera via infected white blood cells and then multiplies in the epithelial cells of the kidney and intestine. The virus is released into the water either through shedding or together with the sloughed epithelial and inflammatory cells resulting from severe local inflammation.
The ability to invade the fish through the gills, multiply there, and then be released through the water is analogous to the case for respiratory viruses in mammals that infect the respiratory epithelium, replicate there, and are spread through air droplets and aerosols. This may turn out to be the most common means of spreading of aquatic viruses. However, large amounts of viral DNA were found in the gut early after infection (18), and clusters of virus particles were detected by electron microscopy in the intestinal system (35). Thus, the possibility that the virus penetrates the fish body through the digestive system should also be considered. Recently we found that droppings of infected fish contain CNGV antigens, viral DNA, and infectious particles (12). It can be assumed, but has not yet been proven, that the virus is preserved in stool for a long period, especially during cold seasons.
PROTECTION OF FISH AGAINST CNGV
One way to reduce the threat caused by CNGV is to select strains and crossbreeds which are more resistant to viral infection. Screening of several edible carp strains and their crossbreeds revealed that the Dor-70 × wild-type Sassan fish were quite resistant to CNGV infection (60.7% survivors) (43). However, the design and development of carp strains resistant to CNGV will require the use of modern molecular genetic methodologies such as quantitative trait loci and microarrays (19). Even so, the breeding system would not be appropriate for selecting resistant ornamental koi fish. Immunization of fish against the virus may be a useful tool to overcome the CNGV threat. Unfortunately, thus far, all efforts to immunize carps with inactivated virus or with viral proteins have proved unsuccessful. To eradicate this disease from fish husbandries, two methods of fish immunization were developed in Israel: challenge of the fish with the pathogenic virus (see below) and fish immunization with attenuated CNGV. Although the use of active virus for immunization was found to be quite efficient, these methods are not yet risk free.
Fish with Naturally Acquired Immunity
Based on the observation that the disease breaks out when the water temperature is between 18 and 28°C, I. Bejerano developed a protocol for selecting carp and koi fish with naturally acquired immunity. According to this procedure, healthy fingerlings were exposed to the virus by cohabitation with sick fish for 2 to 5 days at 22 to 24°C (permissive temperature). Thereafter the water temperature was elevated above 30°C for 25 to 30 days, and the fish were then transferred to open-air ponds. This procedure was found to be quite efficient, and 60% of the immunized fingerlings survived a challenge with sick fish (40). We find that the fish with naturally acquired immunity remain resistant in ponds for a long time, as revealed by challenge infection even a year after exposure to the pathogenic virus (our unpublished results). Although it is beneficial to Israeli fisheries, this method has several disadvantages: (i) by using this method, farmers spread the pathogenic virus over many fisheries and risk spreading it into wild carp populations; (ii) the procedure involves a loss of 40% or more of the fingerlings; (iii) economically the procedure is costly, and (iv) most important, it involves a serious risk, because the pathogenic CNGV used for immunization may persist in the fish body and could reproduce following stress, inducing the disease in the infected fish themselves and/or in nonimmunized fish. So far, using specific and efficient primers and conventional PCR, we and others have been unable to detect any latent viral DNA in organs of fish with naturally acquired immunity. However, Gilad et al. found small traces of viral DNA in surviving fish at 64 dpi by using real-time TaqMan PCR (18).
Development of an Efficient Vaccine against the Virus
Immunization of carp by injection of inactivated virus has failed so far. Live, attenuated vaccines potentially have many advantages in aquaculture (3). In general, live vaccine stimulates all phases of the immune system, resulting in balanced systemic and local responses involving both humoral and cellular branches of the immune system. The advantages of using live attenuated virus vaccine are especially prominent in fish, where heat-inactivated virus is poorly immunogenic and large amounts of proteins are required for achieving an efficient and durable immune response (29, 30). However, the chance that reverted mutated virus will appear and threaten immunized populations is very small.
Experiments to achieve a nonpathogenic attenuated virus have been carried out in Israel since 2003 (34, 40). The attenuated virus was isolated following serial transfer of the Israeli CNGV isolate in KFC. Viruses harvested after 20 passages in culture induced the disease in a small percentage of naive fingerlings following injection or bathing (34, 40). It can be postulated, therefore, that the genetic alterations that accumulated in both the viral and host cell genomes facilitated the isolation of an attenuated virus. The attenuated virus was cloned in tissue culture in order to avoid undesired recombination, complementation, and reversion to a pathogenic virus. Several cloned viruses were UV irradiated and then recloned in order to insert additional mutations into the viral genome (13-15). Currently, the selected attenuated virus clone does not induce the lethal disease and efficiently protects the immunized fish against challenge infection (34, 40).
Carp are very sensitive to pathogenic and attenuated viruses, and a short immersion of fish in water containing virus is sufficient for infection. The infection of fish with pathogenic and attenuated viruses is temperature restricted; fish held at the nonpermissive temperature immediately following infection were not affected by the pathogenic virus and were not rendered resistant to the disease. The attenuated virus must propagate in the host fish in order to induce protection against the virus. Like the pathogenic virus, which induces the disease only at the permissive temperature, the attenuated virus requires the appropriate temperature to confer protection. Efficient protection is achieved by immersing the fish in water containing the attenuated virus (10 to 100 PFU/ml) for 40 min, followed by incubation at the permissive temperature for an additional 48 to 72 h (34).
Protection against CNGV is associated with elevation of specific antibodies against the virus. The CNGV-specific antibody titer rises after 7 dpi and peaks at 21 dpi (40). Similar kinetics of antibody production were found in fingerlings immunized with the attenuated virus. The levels of anti-CNGV antibodies remained high in fish injected with either the pathogenic or the attenuated virus during the entire test period of 56 days. These results point to a correlation between the survival rate and increased titers of anti-CNGV antibodies in the infected fish. The fish with naturally acquired immunity in ponds remain resistant for a long time (6 to 12 months). At present, we know that vaccination with the live attenuated virus confers resistance to a challenge infection for at least 8 months (our unpublished results).
Latency
The use of live vaccine for immunization and the distribution of fish with naturally acquired immunity to prevent disease spread are both potentially risky if CNGV persists as a latent virus in the surviving infected/immunized fish. The answer to this question about latency not only is important because of the possibility of future outbreaks but also will contribute to the classification of the virus. Herpesviridae members persist as latent viruses in their hosts. Therefore, this information would be suggestive, although not conclusive, as some nonherpesviruses also become latent in infected hosts. At present, there are no solid data supporting the assumption that CNGV persists as a latent virus in either surviving infected fish or immunized fish. It has been shown that some apparently related aquatic viruses exhibit latency in their hosts. For instance, latent CyHV-1 persists in carp that survive infection (41), and AngHV-1 persists in surviving infected eels (38, 47). In these cases, the viral genome was found in cranial nerve cells and spinal nerves by using in situ hybridization (31, 41, 42). It would be interesting to determine whether the pathogenic and attenuated CNGV and CyHV-2 also persist in nerves of survivors. An additional relevant question is whether the use of live vaccine is reasonable even if CNGV remains latent in its host fish. Live attenuated viruses are currently used for human vaccination, including viruses which generate latent infection such as herpes zoster virus. The critical questions, therefore, are whether the vaccine is safe and whether attenuated CNGV can convert into pathogenic CNGV and cause sickness.
CONCLUSION
The following characteristics of CNGV are noteworthy: (i). The virus is very contagious; (ii) it is transmitted through water; (iii) induction of the disease is restricted to 18 to 28°C, as observed in open-air ponds and under laboratory conditions; (iv) the virus has a narrow host range, as even closely related cyprinid fish were resistant to this disease; (v) the virus morphology and diameter are consistent with herpesviruses, although the virus contains a relatively small, nonsymmetrical electron-dense region in the viral core; (vi) the virus contains a very large dsDNA molecule (ca. ∼295 kbp), larger than all known Herpesviridae genomes; (vii) of the DNA fragment sequences analyzed so far, some bear significant similarity to CyHV-1 and CyHV-2; and (viii) the protein sequences derived from the sequenced DNA show some similarity to those of members of Herpesviridae, Poxviridae, and other large dsDNA viruses. Additional sequencing and characterization of the virus with regard to latency should allow better classification of this virus. Importantly, several methods for the prevention of this disease are under development, and it seems that developing an attenuated virus vaccine may be instrumental in reducing the threat of CNGV outbreaks.
Acknowledgments
We are indebted to I. Bejerano and Ayana Perelberg, Aquaculture Research Station Dor, Ministry of Agriculture and Rural Development, Israel, for helping and encouraging us during all our studies. We thank Susan Lewis and S. Amir for editing the manuscript.
REFERENCES
- 1.Ariav, R., S. Tinman, I. Paperna, and I. Bejerano. 1998. Presented at the EAFP 9th International Conference, Rhodes, Greece.
- 2.Balon, E. K. 1995. Origin and domestication of the wild carp, Cyprinus carpio: from Roman gourmets to the swimming flowers. Aquaculture 129:3-48. [Google Scholar]
- 3.Benmansour, A., and P. de Kinkelin. 1997. Live fish vaccines: history and perspectives. Dev. Biol. Stand. 90:279-289. [PubMed] [Google Scholar]
- 4.Bercovier, H., Y. Fishman, R. Nahary, S. Sinai, A. Zlotkin, M. Eyngor, O. Gilad, A. Eldar, and R. P. Hedrick. 2005. Cloning of the koi herpesvirus (KHV) gene encoding thymidine kinase and its use for a highly sensitive PCR based diagnosis. BMC Microbiol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretzinger, A., T. Fischer-Scherl, R. Oumouma, R. Hoffmann, and U. Truyen. 1999. Mass mortalities in koi, Cyprinus carpio, associated with gill and skin disease. Bull. Eur. Assoc. Fish Pathol. 19:182-185. [Google Scholar]
- 6.Brooks, M. A., A. N. Ali, P. C. Turner, and R. W. Moyer. 1995. A rabbitpox virus serpin gene controls host range by inhibiting apoptosis in restrictive cells. J. Virol. 69:7688-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane, M., M. Sano, and C. Komar. 2004. Infection with koi herpesvirus-disease card. Developed to support the NACA/FAO/OIE regional quarterly aquatic animal disease (QAAD) reporting system in the Asia-Pacific. [Online.] http://www.enaca.org/modules/mydownloads/singlefile.php?cid=23&lid=557.
- 8.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J. 1998. The genome of salmonid herpesvirus 1. J. Virol. 72:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, A. J., W. Sauerbier, A. Dolan, C. Addison, and R. G. McKinnell. 1999. Genomic studies of the Lucke tumor herpesvirus (RaHV-1). J. Cancer Res. Clin. Oncol. 125:232-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, A. J., B. L. Trus, N. Cheng, A. C. Steven, M. S. Watson, C. Cunningham, R. M. Le Deuff, and T. Renault. 2005. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 86:41-53. [DOI] [PubMed] [Google Scholar]
- 12.Dishon, A., A. Perelberg, J. Bishara-Shieban, M. Ilouze, M. Davidovich, S. Werker, and M. Kotler. 2005. Detection of carp interstitial nephritis and gill necrosis virus (CNGV) in fish droppings. Appl. Environ. Microbiol. 71:7285-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake, J. W. 1976. The biochemistry of mutagenesis. Annu. Rev. Biochem. 45:11-37. [DOI] [PubMed] [Google Scholar]
- 14.Drake, J. W. 1969. Mutagenic mechanisms. Annu. Rev. Genetics. 3:247-268. [Google Scholar]
- 15.Freese, E. 1963. Molecular mechanisms of mutations, p. 207. In J. H. Taylor (ed.), Molecular genetics. Academic Press, New York.
- 16.Gilad, O., S. Yun, M. A. Adkison, K. Way, N. H. Willits, H. Bercovier, and R. P. Hedrick. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J. Gen. Virol. 84:2661-2667. [DOI] [PubMed] [Google Scholar]
- 17.Gilad, O., S. Yun, K. B. Andree, M. A. Adkison, A. Zlotkin, H. Bercovier, A. Eldar, and R. P. Hedrick. 2002. Initial characteristics of koi herpesvirus and development of a polymerase chain reaction assay to detect the virus in koi, Cyprinus carpio koi. Dis. Aquat. Organ. 48:101-108. [DOI] [PubMed] [Google Scholar]
- 18.Gilad, O., S. Yun, F. J. Zagmutt-Vergara, C. M. Leutenegger, H. Bercovier, and R. P. Hedrick. 2004. Concentrations of a koi herpesvirus (KHV) in tissues of experimentally infected Cyprinus carpio koi as assessed by real-time TaqMan PCR. Dis. Aquat. Org. 60:179-187. [DOI] [PubMed] [Google Scholar]
- 19.Gracey, A. Y., E. J. Fraser, W. Li, Y. Fang, R. R. Taylor, J. Rogers, A. Brass, and A. R. Cossins. 2004. Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc. Natl. Acad. Sci. USA 101:16970-16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, W. L., L. Mullis, S. E. LaPatra, M. Groff, and A. Goodwin. 2002. Detection of koi herpesvirus DNA in tissues of infected fish. J. Fish Dis. 25:171-178. [Google Scholar]
- 21.Gunimaladevi, I., T. Kono, M. N. Venugopal, and M. Sakai. 2004. Detection of koi herpesvirus in common carp, Cyprinus carpio L., by loop-mediated isothermal amplification. J. Fish Dis. 27:583-589. [DOI] [PubMed] [Google Scholar]
- 22.Haenen, O. L. M., K. Way, S. M. Bergmann, and E. Ariel. 2004. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 24:293-307. [Google Scholar]
- 23.Hedrick, R. P., O. Gilad, S. Yun, J. Spangenberg, G. Marty, R. Nordhausen, M. Kebus, H. Bercovier, and A. Eldar. 2000. A herpesvirus associated with mass mortality of juvenile and adult koi, a strain of common carp. J. Aquat. Anim. Health 12:44-55. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, R. 2000. Koiseuche bedroht Karpfenteichwirtschaft. Fischer Teichwirt. 11:432. [Google Scholar]
- 25.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 26.Hutoran, M., A. Ronen, A. Perelberg, M. Ilouze, A. Dishon, I. Bejerano, N. Chen, and M. Kotler. 2005. Description of an as yet unclassified DNA virus from diseased Cyprinus carpio species. J. Virol. 79:1983-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, N. S., J. Kobayashi, and T. Miyazaki. 1999. Gill filament necrosis in farmed Japanese eels, Anguilla japonica (Temminck & Schlegel), infected with Herpevirus anguillae. J. Fish Dis. 22:457-463. [Google Scholar]
- 29.Marsden, M. J., L. M. Vaughan, R. M. Fitzpatrick, T. J. Foster, and C. J. Secombes. 1998. Potency testing of a live, genetically attenuated vaccine for salmonids. Vaccine 16:1087-1094. [DOI] [PubMed] [Google Scholar]
- 30.Marsden, M. J., L. M. Vaughan, T. J. Foster, and C. J. Secombes. 1996. A live (delta aroA) Aeromonas salmonicida vaccine for furunculosis preferentially stimulates T-cell responses relative to B-cell responses in rainbow trout (Oncorhynchus mykiss). Infect. Immun. 64:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita, N., and T. Sano. 1990. Regression effect of carp Cyprinus carpio L., peripheral blood lymphocytes on CHV-induced carp papilloma. J. Fish Dis. 13:505-511. [Google Scholar]
- 32.Nathanson, N., and F. Murphy. 1997. An atlas of viral disease pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 33.Neukirch, M., K. Bottcher, and S. Bunnajirakul. 1999. Isolation of a virus from koi with altered gills. Bull. Eur. Assoc. Fish Pathol. 19:221-224. [Google Scholar]
- 34.Perelberg, A., A. Ronen, M. Hutoran, Y. Smith, and M. Kotler. 2005. Protection of cultured Cyprinus carpio against a lethal viral disease by an attenuated virus vaccine. Vaccine 23:3396-3403. [DOI] [PubMed] [Google Scholar]
- 35.Perelberg, A., M. Smirnov, M. Hutoran, A. Diamant, I. Bejerano, and M. Kotler. 2003. Epidemiological description of a new viral disease afflicting cultured Cyprinus carpio in Israel. Isr. J. Aquaculture-Bamidgeh. 55:5-12. [Google Scholar]
- 36.Pikarsky, E., A. Ronen, J. Abramowitz, B. Levavi-Sivan, M. Hutoran, Y. Shapira, M. Steinitz, A. Perelberg, D. Soffer, and M. Kotler. 2004. Pathogenesis of acute viral disease induced in fish by carp interstitial nephritis and gill necrosis virus. J. Virol. 78:9544-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pokorova, D., T. Vesely, V. Piackova, S. Reschova, and J. Hulova. 2005. Current knowledge on koi herpesvirus (KHV): a review. Vet. Med. Czech. 50:139-147. [Google Scholar]
- 38.Rijsewijk, F., S. Pritz-Verschuren, S. Kerkhoff, A. Botter, M. Willemsen, T. van Nieuwstadt, and O. Haenen. 2005. Development of a polymerase chain reaction for the detection of Anguillid herpesvirus DNA in eels based on the herpesvirus DNA polymerase gene. J. Virol. Methods 124:87-94. [DOI] [PubMed] [Google Scholar]
- 39.Roizman, B. 1990. Whither herpesviruses? Adv. Exp. Med. Biol. 278:285-291. [DOI] [PubMed] [Google Scholar]
- 40.Ronen, A., A. Perelberg, J. Abramowitz, M. Hutoran, S. Tinman, I. Bejerano, M. Steinitz, and M. Kotler. 2003. Efficient vaccine against the virus causing a lethal disease in cultured Cyprinus carpio. Vaccine 21:4677-4684. [DOI] [PubMed] [Google Scholar]
- 41.Sano, N., M. Moriwake, R. Hondo, and T. Sano. 1993. Herpesvirus cyprini: a search for viral genome in infected fish by in situ hybridization. J. Fish Dis. 16:495-499. [Google Scholar]
- 42.Sano, N., M. Moriwake, and T. Sano. 1993. Herpes virus cyprini: thermal effects on pathogenicity and oncogenicity. Fish Pathol. 28:171-175. [Google Scholar]
- 43.Shapira, Y., Y. Magen, T. Zak, M. Kotler, G. Hulata, and B. Levavi-Sivan. 2005. Differential resistance to koi herpes virus (KHV)/carp nephritis and gill necrosis virus (CNGV) among common carp (Cyprinus carpio) strains and crossbreds. Aquaculture 245:1-11. [Google Scholar]
- 44.Smith, G. L., A. de Carlos, and Y. S. Chan. 1989. Vaccinia virus encodes a thymidylate kinase gene: sequence and transcriptional mapping. Nucleic Acids Res. 17:7581-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soliman, H., and M. El-Matbouli. 2005. An inexpensive and rapid diagnostic method of koi herpesvirus (KHV) infection by loop-mediated isothermal amplification. Virol. J. 2:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment. Oxford University Press, New York, N.Y.
- 47.Van Nieuwstadt, A. P., S. G. Dijkstra, and O. L. Haenen. 2001. Persistence of herpesvirus of eel Herpesvirus anguillae in farmed European eel Anguilla anguilla. Dis. Aquat. Org. 45:103-107. [DOI] [PubMed] [Google Scholar]
- 47a.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 48.Walster, C. 1999. Clinical observations of severe mortalities in Koi carp, Cyprinus carpio, with gill disease. Fish Vet. J. 3:54-58. [Google Scholar]
- 49.Waltzek, T. B., G. O. Kelley, D. M. Stone, K. Way, L. Hanson, H. Fukuda, I. Hirono, T. Aoki, A. J. Davison, and R. P. Hedrick. 2005. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 86:1659-1667. [DOI] [PubMed] [Google Scholar]
- 50.Way, K., R. M. Le Deuff, D. M. Stone, K. L. Denham, and S. St. Hilaire. 2004. Koi herpesvirus: diagnostics and research at CEFAS Weymouth laboratory 2000-2003, p. 9-10. In Report of International Workshop on Koi Herpesvirus, Defra, London, United Kingdom.