Abstract
High-resolution crystallographic structures of multisubunit RNA polymerases (RNAPs) have increased our understanding of transcriptional mechanisms. Based on a thorough review of the literature, we have compiled the mutations affecting the function of multisubunit RNA polymerases, many of which having been generated and studied prior to the publication of the first high-resolution structure, and highlighted the positions of the altered amino acids in the structures of both the prokaryotic and eukaryotic enzymes. The observations support many previous hypotheses on the transcriptional process, including the implication of the bridge helix and the trigger loop in the processivity of RNAP, the importance of contacts between the RNAP jaw-lobe module and the downstream DNA in the establishment of a transcription bubble and selection of the transcription start site, the destabilizing effects of ppGpp on the open promoter complex, and the link between RNAP processivity and termination. This study also revealed novel, remarkable features of the RNA polymerase catalytic mechanisms that will require additional investigation, including the putative roles of fork loop 2 in the establishment of a transcription bubble, the trigger loop in start site selection, and the uncharacterized funnel domain in RNAP processivity.
INTRODUCTION
In prokaryotes, a single DNA-dependent RNA polymerase (RNAP) synthesizes all classes of RNAs including mRNAs, rRNAs, and tRNAs. Prokaryotic core RNAPs are composed of five subunits (α2, β′, β, and ω) which associate with a σ factor to form the holoenzyme required for promoter recognition (26). In eukaryotes, RNAP I synthesizes the rRNAs and RNAP II forms the mRNA and the small nuclear RNA, while RNAP III is responsible for the synthesis of the tRNA and the 5S rRNA.
RNAPs I, II, and III contain 14, 12, and 17 subunits, respectively. These three enzymes are functionally and structurally related; five subunits are common to all three enzymes, while another four are related (32, 198). The two largest subunits of prokaryotic (bacterial RNAP β′ and β) and eukaryotic RNAPs (RNAP I Rpa190 and Rpa135, RNAP II Rpb1 and Rpb2, and RNAP III Rpc160 and Rpc128) share a high degree of sequence similarity and form the catalytic center of the enzyme. The multisubunit archaebacterial RNAP is composed of 13 polypeptides, the three largest subunits being the homologues of the two largest subunits of the eukaryotic and prokaryotic RNAPs (101). Six other archaeal subunits show sequence similarity with bacterial and eukaryotic RNAP polypeptides.
The resolution of the crystallographic structures of multisubunit RNAP has provided a framework for elucidating transcriptional mechanisms. To date, high-resolution structures are available for the bacterial RNAP and the eukaryotic RNAP II alone (7, 8, 28, 44, 116) as well as part of a complex with nucleic acids (57, 87, 118, 194, 195) or regulatory factors (11, 29, 86, 119, 185) (see Table 1 and Fig. 1A and B for models).
TABLE 1.
RNAPs
| Group and PDB no. | Reference | Resolution (Å) | Description |
|---|---|---|---|
| Eukaryotes | |||
| 1I3Q | Cramer et al. (44) | 3.10 | RNA polymerase II crystal form I |
| 1I50 | Cramer et al. (44) | 2.80 | RNA polymerase II crystal form II |
| 1I6H | Gnatt et al. (57) | 3.30 | RNA polymerase II elongation complex |
| 1R9S | Westover et al. (195) | 4.25 | RNA polymerase II strand separated elongation complex, matched nucleotide |
| 1NIK | Bushnell et al. (28) | 4.10 | Wild-type RNA polymerase II |
| 1PQV | Kettenberger et al. (86) | 3.80 | RNA polymerase II-TFIIS complex |
| 1R5U | Bushnell et al. (29) | 4.50 | RNA polymerase II TFIIB complex |
| 1R9T | Westover et al. (195) | 3.50 | RNA polymerase II strand separated elongation complex, mismatched nucleotide |
| 1K83 | Bushnell et al. (27) | 2.80 | Yeast RNA polymerase II complexed with the inhibitor amanitin |
| 1NT9 | Armache et al. (7) | 4.20 | Complete 12-subunit RNA polymerase II |
| 1SFO/1R9R | Westover et al. (194) | 3.61 | RNA polymerase II strand separated elongation complex |
| 1TWA | Westover et al. (195) | 3.20 | RNA polymerase II complexed with ATP |
| 1TWC | Westover et al. (195) | 3.00 | RNA polymerase II complexed with GTP |
| 1TWF | Westover et al. (195) | 2.30 | RNA polymerase II complexed with UTP |
| 1TWG | Westover et al. (195) | 3.30 | RNA polymerase II complexed with CTP |
| 1TWH | Westover et al. (195) | 3.40 | RNA polymerase II complexed with 2′dATP |
| 1WCM | Armache et al. (8) | 3.80 | Complete 12-subunit RNA polymerase II |
| 1YIV | Kettenberger et al. (87) | 3.80 | Refined RNA polymerase II-TFIIS complex |
| 1YIW | Kettenberger et al. (87) | 4.00 | Complete RNA polymerase II elongation complex |
| 1YIY | Kettenberger et al. (87) | 4.00 | RNA polymerase II-TFIIS-DNA/RNA complex |
| 1Y77 | Kettenberger et al. (87) | 4.50 | Complete RNA polymerase II elongation complex with substrate analogue gmpcpp |
| Prokaryotes | |||
| 1HQM | Minakhin et al. (116) | 3.30 | Thermus aquaticus core RNA polymerase, includes complete structure with side chains |
| 116V | Campbell et al. (30) | 3.30 | Thermus aquaticus core RNA polymerase-rifampin complex |
| 11W7 | Vassylyev et al. (185) | 2.60 | RNA polymerase holoenzyme from Thermus thermophilus |
| 1L9U | Murakami et al. (119) | 4.00 | Thermus aquaticus RNA polymerase holoenzyme |
| 1L9Z | Murakami et al. (118) | 6.50 | Thermus aquaticus RNA polymerase holoenzyme/fork-function promoter DNA complex |
| 1SMY | Artsimovitch et al. (11) | 2.70 | Transcription regulation by alarmone ppGpp from Thermus thermophilus |
| 1YNJ | Campbell et al. (31) | 3.20 | Thermus aquaticus RNA polymerase-sorangicin complex |
| 1YNN | Campbell et al. (31) | 3.30 | Thermus aquaticus RNA polymerase-rifampin complex |
| 1ZYR | Tuske et al. (182) | 3.00 | Thermus aquaticus RNA polymerase-streptolydigin complex |
| 2A6E | Artsimovitch et al. (12) | 2.80 | Thermus aquaticus RNA polymerase apo-holoenzyme complex |
| 2A6H | Temiakov et al. (176) | 2.40 | Thermus aquaticus RNA polymerase-streptolydigin complex |
| 2A69 | Artsimovitch et al. (12) | 2.50 | Thermus aquaticus RNA polymerase-rifapentine complex |
| 2A68 | Artsimovitch et al. (12) | 2.50 | Thermus aquaticus RNA polymerase-rifabutin complex |
FIG. 1.
Structures of eukaryotic and prokaryotic RNAPs. (A) Structure of S. cerevisiae RNAP II bound to nucleic acids (PDB accession number 1Y1W). (Left panel) The various subunits and the DNA-RNA hybrid are shown (see color code). Some main domains of the enzyme are circled in black and gray. (Right panel) Structure of T. aquaticus RNAP (PDB accession number 1I6V). The various subunits are shown (see color code), as well as the positions of the cleft, pore, and RNA exit channel. (B) Structural elements of RNAP II in the region of the DNA-RNA hybrid. The rudder, switch 2, funnel, bridge helix, trigger loop, lid, switch 1, and switch 5 features of Rpb1 and the fork loop 1, fork loop 2, switch 3, and switch 4 features of Rpb2 are indicated. The positions of metals A and B and the residues coordinating them are shown. (C) Mechanism of ribonucleotide addition to the RNA chain. Residues known to coordinate the two Mg2+ ions (pink spheres) are shown and coordination bonds are in green dotted lines. DNA and RNA are shown in gray and white, respectively. Black arrows show the nucleophilic attack.
Both the prokaryotic and eukaryotic transcription reactions involve a number of steps, starting with promoter recognition. The bacterial σ factor (60) and the eukaryotic general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (42, 62) are required for specific binding of the polymerase to promoters. In both systems, promoter binding is accompanied by bending and wrapping of the promoter DNA against the body of the polymerase (55, 100, 142-144). DNA wrapping is involved in promoter melting in the region of the transcriptional initiation site, probably through the induction of a torsional strain that generates unwinding of the DNA double helix (50, 54). On the basis of recent site-specific protein-DNA photo-cross-linking experiments (54), it was proposed that formation of a right-handed loop in the promoter DNA wrapped around a mobile portion of the enzyme, named the clamp domain, induces DNA unwinding, which can then be stabilized by some general transcription factors. The formation of this open complex allows pairing of incoming ribonucleoside triphosphates (NTPs) to the template DNA strand for phosphodiester bond formation (43, 69, 70).
It has been proposed that all polymerases catalyze phosphodiester bond formation by a two metal ion mechanism (166) (Fig. 1C). By this mechanism, a first Mg2+ ion, termed metal A, facilitates the nucleophilic attack of the 3′ oxygen on the 5′ α-phosphate. The second Mg2+ ion, metal B, facilitates the release of the pyrophosphate. In prokaryotic RNAP and eukaryotic RNAP II, metal A is coordinated by three strictly conserved aspartates of β′/Rpb1 contained in the NADFDGD motif (44, 57, 206). Metal B has a low apparent affinity for free RNAP II (44) and appears to enter the active site with the incoming NTP and is coordinated by three aspartates, two from β′/Rpb1 and one from β/Rpb2, located in a conserved ED motif (195). Formation of a phosphodiester bond is followed by translocation of the nucleic acids in order to present the next template register for a second nucleotide addition cycle.
Transcription initiation is characterized by a cycle of abortive initiation events, where RNAP synthesizes and releases small transcripts without disengaging from the DNA template (188). When the transcript reaches a length of approximately 10 to 12 nucleotides, stabilization of an early elongation complex occurs and RNAP breaks its contacts with the general transcription factors/σ and clears the promoter for elongation (43, 69).
In the elongation phase, RNAPs require the help of protein factors to bypass impediments to elongation. Replacement of the general transcription factors by various elongation factors has been demonstrated for RNAP II (134) and numerous elongation factors have been identified to date (159). These factors affect transcription in various ways from rescue of elongation complexes stalled at pause and arrest sites to phosphorylation of the C-terminal domain of the Rpb1 subunit of RNAP II and chromatin remodeling and modification.
Because mRNA processing occurs cotranscriptionally in the RNAP II system (68), the complexity of the network of factors interacting with the elongating enzyme is much greater than in the RNAP I and III systems. Recruitment of both mRNA processing and elongation factors is mainly directed through specific interactions with the phosphorylated RNAP II C-terminal domain. Nonetheless, evidence supports the existence of elongation factors during transcription by RNAP I (95) and RNAP III (113), which lack a C-terminal domain. In prokaryotes, the number of characterized elongation factors is smaller, the best-characterized being GreA, NusA, and Mfd (23).
The termination process is different for prokaryotic RNAPs than for eukaryotic RNAPs I, II, and III. Termination in prokaryotes is the better understood. Two major mechanisms have been proposed. The first is independent of termination factors but rather requires formation of stem-loop structures in the transcribed RNA that interacts with the RNAP to induce pausing and which, combined with a weak RNA-DNA hybrid, stimulates transcript release from the template (140). The second mechanism involves the activity of the rho helicase to facilitate transcript release and is therefore referred to as rho-dependent termination (140).
Eukaryotic RNAP I termination, although poorly understood, is for some genes dependent on a short repeated sequence element (93) that is recognized by a nuclear termination factor called Reb1 in Saccharomyces cerevisiae and TTF1 in the mouse (162). Termination depends on the interaction of this protein with RNAP I and could involve some protein-induced changes in DNA structure. Other types of signals have been identified but to date (85, 98, 187), no specific termination element has been unequivocally identified.
RNAP II termination is more complex because it is coupled to 3′-end processing of the transcript (39). It requires association of the 3′-end processing complexes CPSF and CstF (13, 47) with RNAP II via the phosphorylated C-terminal domain of Rpb1 as well as the polyadenylation signals at the 3′ end of the pre-mRNA (41, 102, 109).
Many cleavage and polyadenylation factors have been identified and studies of thermosensitive alleles of cleavage factors show that some of them are required for RNAP II termination (21). RNAP III termination most resembles bacterial RNAP termination and involves recognition of short termination signals rich in T stretches (136). However, in contrast to prokaryotes, RNAP III termination does not involve the formation of stem-loop structures and no auxiliary factor has been identified to date.
Crystallographic studies have revealed that the structure of RNAP is highly conserved from prokaryotes to eukaryotes, with the regions of highest homology forming the enzyme's active center (44, 206). The two largest RNAP subunits (β′ and β in bacterial RNAPs; Rpb1 and Rpb2 in RNAP II) form a positively charged cleft (also termed the main channel) that accommodates the nucleic acids during transcription (44, 206) (Fig. 1A). The two catalytic Mg2+ ions, metals A and B, are buried deep in this cleft (Fig. 1B) (195). A domain of Rpb2/β called the wall closes the upstream extremity of this cleft and is a binding site for the upstream end of the RNA-DNA hybrid. The wall domain contains the flap feature, ordered in the prokaryotic structures and disordered in the eukaryotic structures, which serves as a binding site for transcription factors and would be implicated in the obstruction of the RNA exit channel by the σ factor region 4 (123).
The nucleic acids are held in the cleft by a number of protein domains, including the upper and lower jaws, which grab the downstream DNA, and by the clamp domain, which locks the nucleic acids in the cleft (57). Comparison of the structures of the free core enzyme (44) with that of the elongating core enzyme (57) from yeast led to the hypothesis that the mobile clamp domain folds over DNA to lock it during elongation. However, the backbone models (7, 28) and refined atomic model (8) of complete RNAP containing Rpb4 and Rpb7 show the clamp domain in the same position as in the elongating core structure, suggesting that promoter DNA is first loaded on the top of the clamp as seen for bacterial RNAP (7, 28, 118). The template strand would then reach the active site only after promoter melting had occurred. A pore structure (also termed secondary channel), which has the shape of an inverted funnel that opens in the cleft near the active site, was proposed as an entry route for the incoming NTP and an exit for the released pyrophosphate (44, 57). The mRNA exit channel lies between the wall and the clamp domain (Fig. 1A).
In addition to their polymerization activity, DNA-dependent RNAPs can also catalyze the 3′ endonucleolytic cleavage of transcripts under certain circumstances (40, 158). First discovered in Escherichia coli (92, 172), this cleavage activity takes place when RNAP has backtracked at pause and arrest sites (53, 196). Weakly associated DNA-RNA hybrids can induce backtracking of the enzyme to a more stable register (125). To resume elongation from this more stable position, RNAP needs to cleave the 3′ end of the transcript that now extrudes from the catalytic site through the pore.
The 3′ cleavage activity is enhanced by the cleavage factors GreA and GreB in bacteria (24, 25) and TFIIS in eukaryotes (53, 196). The backbone structure of TFIIS in a complex with complete RNAP II (86) provided insight into the mechanism by which this factor enhances transcript cleavage. More specifically, it revealed that two conserved essential acidic residues of TFIIS (79) are located in the vicinity of the polymerase metal B and could participate in its coordination. Therefore it was suggested that the two metal ion mechanism invoked for NTP polymerization is also involved in the 3′ endonucleolytic activity of RNAP II. Similar studies on GreB, the prokaryotic homologue of TFIIS, have also reached similar conclusions for bacterial RNAP (128, 164).
Structures of yeast elongating core RNAP have revealed the presence of an 8- to 9-base-pair DNA-RNA hybrid in the cleft (57, 194). The crystallographic data have also shown that a number of loops and helices of Rpb1/β′ and Rpb2/β are located in this region close to the catalytic site (Fig. 1B). These structural motifs have been named either according to their location, aspect or presumed role in the transcription reaction. For example, the Rpb1/β′ bridge helix separates the main channel and is located near the template DNA at the +1 site.
Because crystallographic data revealed two different conformations for this structure, bent (or distorted) (206) or straight (or continuous) (44, 57, 194), it was proposed that the bridge helix might be involved in translocation of the nucleic acids during transcription. The location of the rudder, the lid and the fork loop 1 suggests that these loops are involved in DNA-RNA strand separation, in order to maintain an 8- to 9-base-pair hybrid (194). The fork loop 2 and the zipper could be involved in delineating the downstream and upstream boundaries of the transcription bubble respectively (44, 57). Five loops of Rpb1 and Rpb2 have been termed the switches and could participate in controlling the position of the clamp or, for switches 1 to 3, in forming a binding site for the DNA-RNA hybrid (7, 44, 57).
Crystallographic studies of yeast core RNAP in complex with the general transcription factor TFIIB have shown contacts of this factor with the dock domain (29) of the enzyme. This domain of Rpb1 is located between the wall and the clamp and lies on the surface of the structure (Fig. 1A). The finger domain of TFIIB enters deep into the active site after passing across the saddle, between the wall and clamp domains. This feature is reminiscent of the 3.2 linker loop of the σ factor in prokaryotic RNAP, which follows a similar path through the RNAP flap and clamp domains (185).
Superposition of the crystallographic structures of the core RNAP/TFIIB complex and the core elongation complex (57) shows that the RNA would clash with the TFIIB finger domain in the active site beyond synthesis of the fifth residue and that TFIIB, if it were not displaced, would compete with RNA for binding to the saddle after synthesis of the 10th nucleotide. These observations provide a molecular basis for abortive initiation before synthesis of the 10th nucleotide and explain why TFIIB is released from the complex beyond synthesis of this register.
Crystallographic structures of RNAPs in complexes with known catalytic inhibitors have also been resolved and have brought new insight into the catalytic mechanisms of RNAPs. Observation of these inhibitors at their binding sites allows a better understanding of their mode of action and the role of the structural features they interact with. The structures of cocrystals of α-amanitin (27), rifampin (30), sorangicin (31), and streptolydigin (176, 182) with core RNAP have all been resolved. α-Amanitin, an inhibitor of the translocation step, is seen binding to a region located between the funnel and the bridge helix (27) of yeast RNAP II (Fig. 1B).
Many observations support the hypothesis that restriction in the movement of the bridge helix is required for translocation of nucleic acids in the active site (61, 184). In prokaryotes, rifampin and sorangicin both bind in a pocket juxtaposed to fork loop 2 (30, 31). Modeling of an RNA-DNA hybrid in a bacterial RNAP structure (90) combined with positioning of rifampin to its binding site reveals that a steric clash would occur after synthesis of the second or third nucleotide (30), which is in good agreement with observations that rifampin and sorangicin specifically inhibit RNAP during initiation at a step before addition of the second or third nucleotide (90, 115).
An additional hypothesis about the mode of action of rifampin has been suggested and involves an allosteric signal transmitted through the binding site of rifampin to the catalytic aspartates in order to lower the affinity for the Mg2+ ion (12). The location of streptolydigin in the RNAP structure and its binding to the fork 2, bridge helix, and trigger loop features indicate that this drug could interfere with the translocation of nucleic acids in the active site (182). However, an alternative hypothesis suggests an allosteric mechanism trapping RNAP in a conformation inappropriate for transcript elongation (176).
Recent reports have enlightened the evolutionary connection between prokaryotic and eukaryotic RNAPs (75, 76). The presence of the catalytic double-psi β-barrel domain, which contains a signature metal-coordinating motif, in both eukaryotic RNA-dependent RNA polymerases and the universally conserved β′ subunit of DNA-dependent RNA polymerases, coupled to the absence of other common domains, suggests that they have evolved through the early divergence of a common ancestor. Interestingly, the presence of another, although highly diverged, double-psi β-barrel domain in the β subunit of DNA-dependent RNA polymerases supports this idea of a common ancestor that diverged through lineage-specific insertions of domains and motifs.
During the past two decades many mutational studies of RNAP have been performed with the aim of understanding the function and regulation of this enzyme. A great number of these studies were performed before the resolution of RNAP crystal structures. We surmised that reexamining the effect of amino acid alterations in RNAP in light of their available three-dimensional structures might provide new insight into the structure-activity relationship of these enzymes.
We therefore created an extensive catalogue of published mutations affecting the function of multisubunit RNAP and mapped them on the structures of the prokaryotic and eukaryotic enzymes. This catalogue is presented in the form of a table (see Table S2 in the supplemental material for mutations discussed in this article and additional mutations; the complete catalogue is also available in a Web format at http://www.ircm.qc.ca/microsites/mutationsaffecting/fr/index.html) indicating (i) the reference in which the mutation is described, (ii) the name of the mutant allele when available, (iii) the organism and subunit, (iv) the position of the altered amino acids on the linear amino acid sequence of the altered subunit, (v) the equivalent position on either S. cerevisiae RNAP II or Thermus aquaticus RNAP (the amino acid numbering in the text and figures refers to these specific positions), (vi) the name of the structural domain where the altered amino acid is located, (vii) the homology region, and (viii) the function affected by the altered amino acids or phenotype for 6-azauracil sensitivity or drug resistance (blanks stand for mutants with phenotypes for which no affected function could be attributed). This work, as presented in this review, has allowed us to make a series of observations concerning the functions of domains and structural elements of RNAP.
AMINO ACID SUBSTITUTIONS IN RNAP THAT LOWER THE POLYMERIZATION RATE ARE LOCATED CLOSE TO THE ACTIVE SITE
An important, albeit predictable, observation is that a great number of substitutions inducing a low polymerization rate map in proximity to the active site. RNAP mutants that have been included in this category are defective in transcription assays in vitro. In most cases, these mutants have not been characterized sufficiently to define which step of the transcription process is impaired.
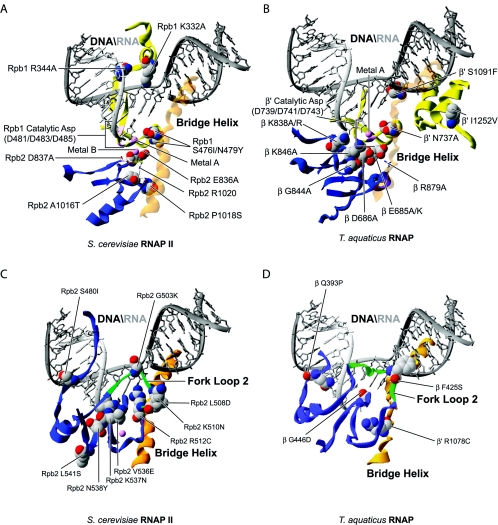
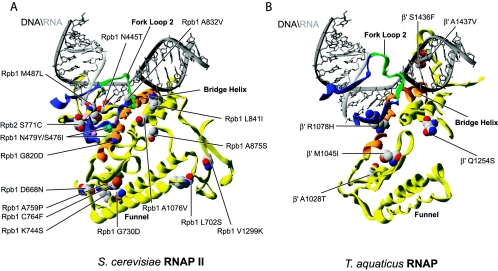
In Fig. 2, we have modeled in the structure of S. cerevisiae RNAP II (87) (Fig. 2A and C) and T. aquaticus RNAP (30) (Fig. 2B and D) a series of substitutions in both prokaryotic and eukaryotic enzymes that induce a low polymerization phenotype. Site-directed (191, 205) and random mutagenesis (48, 117) of the three strictly conserved aspartates of β′/Rpb1 binding metal A, which are shown in Fig. 1C and 2 (for β′, D739, 741, and 743; and for Rpb1, D481, 483, and 485), generates mutants that are completely inactive in transcription (191, 205) with the surprising exception of the random mutation D743G conferring microcin J25 resistance in E. coli. (The amino acid numbering in the text and figures refers to amino acid positions on either S. cerevisiae RNAP II or T. aquaticus RNAP.)
FIG. 2.
Distribution of the substitutions inducing low polymerization rates. (A) Substitutions near the active site of both archaeal and eukaryotic RNAPs have been mapped onto the structure of S. cerevisiae RNAP II. The two magnesium ions are shown in pink. (B) Substitutions near the active site of prokaryotic RNAP have been mapped onto the structure of T. aquaticus RNAP. (C) Substitutions near the fork loop 2 of archaeal and eukaryotic RNAPs have been mapped onto the structure of S. cerevisiae RNAP II. The two magnesium ions are shown in pink. (D) Substitutions near the fork loop 2 of prokaryotic RNAP have been mapped onto the structure of T. aquaticus. For all the prokaryotic models, the nucleic acids were imported from PDB 1Y1W and placed by overlaying the eukaryotic and prokaryotic structures.
Mutants arising from site-directed (99, 163, 191) and random mutagenesis (103) of residues ED of β/Rpb2 (β, E685K/A and D686A; Rpb2, E836A and D837A) present a reduced polymerization activity, especially at low NTP and Mg2+ concentrations (99, 103, 163, 191) (Fig. 2A). Interestingly, the D837A substitution in yeast Rpb2 gives rise to a dominant negative phenotype, indicating that the mutated subunit interferes with the function and/or assembly of the wild-type enzyme (99).
The substitutions that cause a low polymerization rate and map near the catalytic acidic residues are indicated in Fig. 2. Bacterial residue N737 and its eukaryotic counterpart N479 are juxtaposed to the three aspartates of β′/Rpb1. In yeast, the N479Y substitution is lethal (4, 48), but when combined with an S476I substitution results in a low-catalytic-rate phenotype in vitro (48, 66) as seen in the site-directed E. coli mutant N737A (163).
Based on the structure of RNAP, it was proposed that residue N737/N479 binds the ribose 2′-OH group and is involved in discrimination of NTP versus dNTP (87, 173). Replacement of this amino acid might allow inclusion of dNTPs in the RNA chain, a situation that may eventually lead to a block in polymerization. This block could be a consequence of the fact that the nucleic acid binding site is highly specific for a particular conformation of the DNA-RNA hybrid that is modified upon addition of a dNTP (57). Alternatively the N737/N479 replacement could impair binding of the incoming NTP to the active site. In addition, because this amino acid is located in close proximity to the catalytic aspartates of the largest subunit, it is possible that its replacement causes a distortion of the active center, leading to a defect in transcription.
In prokaryotes, the engineered β R879A replacement (Fig. 2B) lowers the polymerization rate of the enzyme (163). In the crystallographic structure of yeast RNAP II (194), the equivalent amino acid, R1020, is involved in a salt bridge with the catalytic residue D837 of Rpb2. Disruption of this bridge could cause changes in the conformation of the active site, having a negative effect on the catalytic activity of the enzyme. Two thermosensitive and 6-azauracil-sensitive replacements in the Rpb2 subunit of yeast RNAP II, A1016T (135, 150) and P1018S (105, 135, 150), are located near R1020, and could have a similar effect by modifying the position of R1020 and its interaction with D837.
The bacterial replacement S1091F (154) which confers resistance to streptolydigin is located in the bridge helix, an important helix of β′/Rpb1 proposed to be involved in nucleic acid translocation (44, 57) (Fig. 2B). In the structure of T. aquaticus RNAP (30) with a DNA-RNA hybrid modeled in the active site, S1091 could contact the template DNA strand near +2. The replacement of a small serine for a large phenylalanine probably can result in a steric clash with the template DNA and disturb its presence in the active site during polymerization.
The I1252V replacement in E. coli also alters the bridge helix function, but indirectly through an effect on the juxtaposed trigger loop. This mutant is also overreactive to regulatory signals such as terminators and pause signals (16). The site-directed β K838A and K838R replacements, identified by affinity cross-linking studies as residues being close to the active site, confer a defect at the promoter clearance step of the transcription cycle (83, 120, 148). These two K838 mutants synthesize abortive transcripts but are unable to escape the promoter and proceed into elongation.
Interestingly, the positively charged side chain of K838 is directed toward the RNA around position −2. One can propose that K838 contacts the RNA at this position, leading to its stabilization in the active site. Replacement of this important lysine with a neutral alanine residue or arginine, a residue with a less-flexible and bulkier side chain, possibly results in the inability to retain the transcript in the catalytic center, leading to abortive initiation and impairment of the transition to elongation.
The following mutants were engineered in order to investigate the properties of the conserved residues surrounding K838. K846A (148) (Fig. 2B) is similarly oriented toward the DNA-RNA hybrid. It is possible that the positively charged K846 amino acid is also involved in the interaction with the nucleic acids and their stabilization in the active site. The G844A replacement could similarly affect the polymerization rate by changing the conformation of this important hybrid binding region. Two replacements in the switch 2 region of Rpb1, K332A and R344A, confer an increase in abortive initiation (110). Both replacements are located in close proximity to the template DNA strand around positions +1 to −3. Although the R344A mutant is able to bind DNA as well as the wild type, this mutant is unable to form stable RNAP II-DNA-RNA complexes containing a 5-nucleotide RNA. This observation suggests that the observed defects are linked to an inability to stabilize the hybrid in the active site.
Among the amino acid replacements near the catalytic center that lower the polymerization rate, many are located in proximity to fork loop 2 (Fig. 2C and D). This β/Rpb2 structural element was proposed to be involved in maintaining the downstream end of the transcription bubble (44, 57, 86). In prokaryotes, the replacement F425S (64) conferring streptolydigin resistance is located directly in fork loop 2. Considering its location, the neutral phenylalanine F425 could be implicated in an interaction with the DNA helix during strand separation (Fig. 2D). We speculate that the replacement for a serine could lead to a promoter opening defect and therefore to a low polymerization rate. Similarly, replacements Q393P (81, 82, 202), conferring rifampin resistance, and G446D (174, 175), isolated based on its increased termination phenotype, both surround fork loop 2 and possibly destabilize the structure of this element resulting in the inability to maintain the transcription bubble open.
Because R1078, a residue located in the E. coli β′ bridge helix, forms a hydrogen bond with the backbone of fork loop 2 at position H431, the disruption of this bond by the R1078C (17) replacement, isolated from its ability to mimic the ppGpp effect (see below), could induce a conformational change that alters an important interaction of the bridge helix with fork loop 2. Figure 2C shows that the eukaryotic replacements S480I, G503K, G503K, L508D, and K510N (22), isolated from their altered termination properties, are localized in fork loop 2 and induce a low-polymerization phenotype.
Other replacements from these studies, including V536E, L541S, K537N, and N538Y (22), are positioned in a region that interacts with both the N-terminal and C-terminal ends of fork loop 2; they possibly induce misfolding of this element. Finally, the R512C replacement in yeast Rpb2 is also located in the fork 2 element. Isolated as a suppressor of the ssu72-2 defect (131), this mutation affects transcription. It would be interesting to assess whether all of these fork 2 mutations alter the architecture of the transcription bubble in vitro.
AMINO ACID REPLACEMENTS IN RNAP II AFFECTING SELECTION OF THE TRANSCRIPTION INITIATION SITE
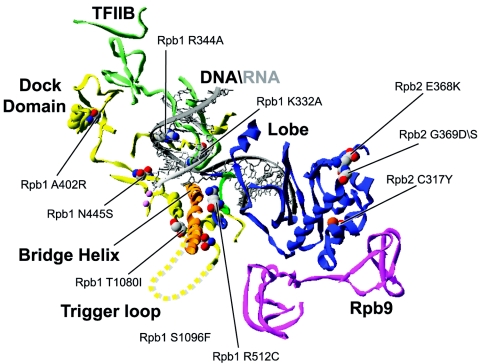
In eukaryotes, selection of the transcription initiation site is dictated by different promoter elements, such as the TATA box and the initiator (161). The general transcription factor TFIIB was also shown to play a role in initiation site selection. Indeed, many mutants of TFIIB that alter initiation site selection have been isolated (20, 132, 133). Consistently, photo-cross-linking studies have shown that a domain of TFIIB approaches the DNA template near +1 in the initiation complex (35). The Rpb9 subunit of RNAP II also plays a role in initiation site selection and many replacements affecting this process affect residues of this RNAP II subunit (71, 171). Rpb9 forms the jaw-lobe module, in combination with the jaw domain of Rpb1 and the lobe domain of Rpb2 (see Fig. 1A). This module may grip the downstream DNA during initiation, suggesting that these contacts are important for selection of the initiation site.
Figure 3 shows the locations of amino acid changes affecting initiation site selection by RNAP II. Replacements C317Y (65) and E368K (65) were isolated for their ability to restore the expression of genes rendered inactive by insertion of a Ty retrotransposon in their promoter regions. Both of these replacements map to the Rpb2 lobe domain which interacts with Rpb9. We can hypothesize that these replacements alter the lobe in a way that affects its interaction with Rpb9, thereby affecting initiation site selection. Alternatively, these replacements may alter the ability of the jaw-lobe module to firmly contact downstream DNA, impairing the accurate positioning in the +1 register of the active site.
FIG. 3.
Distribution of the substitutions affecting initiation site selection. Substitutions in eukaryotic RNAP affecting start site selection have been mapped onto the structure of S. cerevisiae RNAP II. The TFIIB structure (PDB accession number 1R5U) has been placed onto 1Y1W (see legend to Fig. 1) by overlaying the polymerase subunits.
The G369D (65) and G369S (34) replacements also affect the Rpb2 lobe domain. The G369S mutation was isolated as a suppressor of a TFIIB mutation affecting initiation site selection (34). The Rpb2 replacement R512C (131) located in the fork 2 domain also induces a downstream shift in transcription start site. This mutation has been isolated as a suppressor of the ssu72-2 temperature-sensitive defect, an essential uncharacterized protein known to interact with TFIIB. The location of this replacement within the fork 2 element suggests that this residue participates in stabilizing contacts with nucleic acids or maintaining the transcription bubble architecture. The proximity of the fork 2 element to the finger domain of TFIIB raises the possibility of an altered interaction between these two features, leading to a defect in start site selection.
The trigger loop is a structural element that was recently shown to play a role in elongation by modulating the oscillating properties of the adjacent bridge helix (16) (Fig. 3). Two initiation site selection mutants carry alterations in residues of the trigger loop (T1080I and S1096F) (65). In the available structures, a large portion of the trigger loop is missing, consistent with this element being mobile and capable of adopting different conformations relative to the +1 site. Therefore, replacing T1080 and S1096 could affect the selection of the initiation site by directly altering the structure and/or function of the trigger loop. Alternatively, replacement of T1080 and S1096 could affect the interaction of the trigger loop with the bridge helix, affecting the lateral mobility of RNAP (16) and consequently altering start site selection.
Many amino acid replacements affecting start site selection could exert their effect by altering the interaction between RNAP II and TFIIB. A402R (20) is located in the dock domain of Rpb1 (Fig. 3) and has been isolated as a suppressor of a TFIIB mutation affecting start site selection. Crystallographic structures of TFIIB bound to RNAP II revealed that TFIIB contacts the dock domain (29). Replacement N445T (4) displays a sit phenotype (see below) and this replacement affects start site selection. The N445S (20) replacement also affects start site selection and both N445T and N445S locate close to the saddle and thus could destabilize TFIIB function by altering its interaction with RNAP II.
Finally, replacements K332A and R344A in the switch 2 element of Rpb1 have been isolated from their reduced growth phenotype and exhibit an increase in abortive initiation but also a downstream shift in start site utilization (110). These two residues are close to the template DNA at positions +1 to −3 and the authors suggest that the mutant RNAPs are unable to form a stable complex with the DNA-RNA hybrid. It should be noted that K332A is within 5 angstroms of the finger domain of TFIIB, providing another basis to the observed start site selection defect.
AMINO ACID DELETIONS AFFECTING FORMATION OF THE OPEN COMPLEX
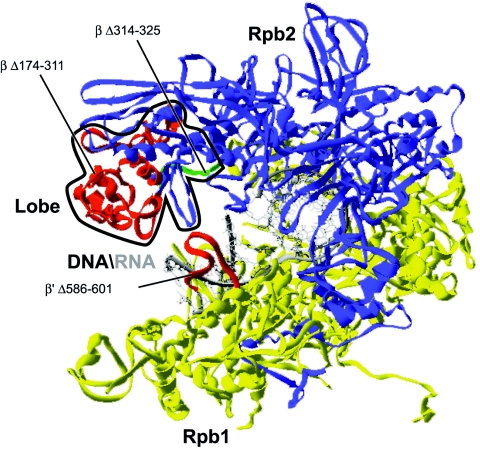
Figure 4 shows the locations of three prokaryotic RNAP mutations that affect open complex formation prior to initiation. The engineered mutant Δ174-311 (121, 152) carries an almost complete deletion of the β dispensable region which is located in the lobe domain. The transcription bubble formed by this mutant is shorter at its downstream end. A shorter deletion engineered in the lobe (Δ314-325) also leads to the formation of an open complex that is shortened in the downstream region but shows a slightly more severe phenotype in that it fails to fully protect the promoter DNA in a DNase I footprinting assay (121). Even though the deleted region is longer for Δ174-311 than Δ314-325, the N- and C-terminal ends of the deleted regions in Δ314-325 are spatially more distant than they are in Δ174-311, where they appear almost juxtaposed (Fig. 4).
FIG. 4.
Locations of the deletions affecting the architecture of the transcription bubble. Deletions in prokaryotic RNAP affecting the architecture of the transcription bubble were mapped onto the structure of T. aquaticus RNAP. The lobe domain is circled in black.
It was proposed that the movement of the lobe domain toward DNA is necessary to maintain a grip on the DNA (90). A suggested mechanism for open complex formation involves downstream promoter contacts with the lobe regions of RNAP to allow for the downstream propagation of the transcription bubble (121). We can hypothesize that the formation of the transcription bubble requires a certain degree of restriction in the axial rotation of DNA in order to generate an unwound helix at the active site.
Mutant Δ586-601 is also defective in downstream promoter opening (94). Indeed, Δ586-601 is unable to initiate transcription unless the template is premelted in the −7 to +1 region. This deletion mutant was constructed to assess the role of the β′ rudder which is an element that does not contact the template DNA in the RNAP structure. The position of the nontemplate DNA is not revealed in the different crystallographic structures of RNAP; it therefore remains possible that the rudder interacts with the nontemplate DNA strand and thereby contributes to establishing or maintaining an open promoter.
RNAP II MUTATIONS ASSOCIATED WITH 6-AZAURACIL SENSITIVITY
TFIIS was first identified as an elongation factor that assists RNAP II to elongate through arrest sites in vitro (137, 139). TFIIS, like its bacterial counterparts GreA and GreB (24, 25), greatly stimulates the 3′ transcript cleavage activity of RNAP (77, 138, 189). This intrinsic endonucleolytic cleavage activity takes place when RNAP has backtracked at pause and arrest sites (53, 196). To resume elongation, RNAP needs to cleave the 3′ end of the transcript that now extrudes from the catalytic site through the pore.
The crystal structure of TFIIS in a complex with RNAP II (87) revealed that TFIIS binds RNAP II along the floor of the cleft and repositions the RNA in the active site. An acidic hairpin of TFIIS reaches all the way into the pore in such a way that it positions two highly conserved residues, D290 and E291, essential for stimulating cleavage activity (79), near the metal B ion of the enzyme's active site. These acidic amino acids have been proposed to participate in the coordination of metal B and to directly position and activate a nucleophilic water molecule (87), thereby stimulating cleavage activity and/or elongation by RNAP.
Inhibition of the enzyme IMP dehydrogenase by 6-azauracil results in depletion of the intracellular pools of GTP and to a lesser extent that of UTP in eukaryotes (52). In yeast, mutants with a defect in transcriptional elongation often show a growth defect on 6-azauracil-containing medium (141). Not surprisingly, a number of RNAP II in-frame linker insertion mutants with 6-azauracil sensitivity have been isolated (Fig. 5). For some of these mutants, overexpression of TFIIS (by increased dosage of the PPR2 gene) was shown to suppress the 6-azauracil phenotype.
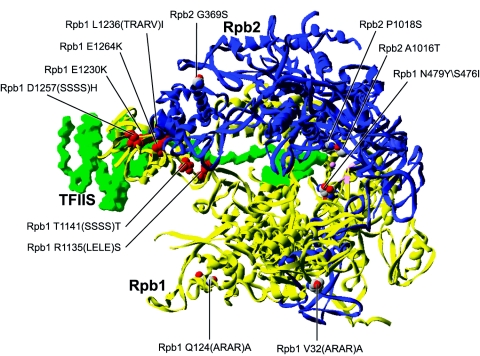
FIG. 5.
Distribution of the altered amino acids conferring 6-azauracil sensitivity. Altered amino acids in eukaryotic RNAP conferring 6-azauracil sensitivity were mapped onto the structure of S. cerevisiae RNAP II. The structure of TFIIS (PDB accession number 1PQV) has been placed onto 1Y1W (see legend to Fig. 1) by overlaying the polymerase subunits. The altered amino acids producing 6-azauracil sensitivity that can be suppressed by TFIIS overexpression are colored red.
An interesting observation is that almost all the 6-azauracil mutations suppressed by overexpression of TFIIS, R1135(LELE)S (3, 5), T1141(SSSS)T (3, 5), E1230K (5), E1230K E1264K (5), L1236(TRARV)I (5), and D1257(SSSS)H (5), affect residues in Rpb1 located along the TFIIS-RNAP II binding interface (86). Alterations of these RNAP II residues probably affect the interaction with TFIIS, conferring an elongation defect and the 6-azauracil sensitivity phenotype. In fact, it was confirmed by biochemical means for two of these RNAP II mutants, E1230K and L1236(TRARV)I, that they indeed have a lower affinity for TFIIS (199).
A series of additional replacements conferring 6-azauracil sensitivity are not suppressed by TFIIS overexpression and are scattered throughout the structure of the enzyme (Fig. 5). The V32(ARAR)A insertion (3, 5) in Rpb1 is located on the outside of the clamp domain. It is possible that this insertion alters the interaction of RNAP II with TFIIF. Indeed, TFIIF was found to localize to the clamp domain of RNAP II (38, 54). The Q124(ARAR)A insertion (3, 5) is located on the clamp domain at the opening of the cleft, where downstream DNA is located. Therefore, this amino acid is possibly important for the interaction of RNAP II with DNA. The G369S (34) replacement is located in the lobe region. Notably, this replacement was identified in a screen for suppressors of TFIIB mutations affecting initiation site selection. The G369S replacement may well alter the conformation of the lobe region, resulting in the correction of the TFIIB defect, but also conferring an elongation defect leading to 6-azauracil sensitivity.
Finally, three additional RNAP II mutants, the A1016T (135, 150), P1018S (105, 135, 150), and S476I/N479Y (147) mutants, present a 6-azauracil sensitivity phenotype because of amino acid alterations in Rpb2. Two of these replacements, A1016T and P1018S, are located in the hybrid binding region, while S476I N479Y is located near the aspartates coordinating metal A. It is therefore likely that the 6-azauracil sensitivity of these mutants results from a transcriptional elongation defect brought about by distortion of the enzyme's active site, rather than a defect in its interaction with elongation factors.
RNAP MUTANTS PRESENTING HIGHER POLYMERIZATION RATES OR SUPPRESSING POLYMERIZATION DEFECTS
Yeast sit mutants (suppressor of initiation of transcription defect) were isolated in a genetic screen for suppression of a transcriptional defect at the HIS4 gene (10) brought about by removal of three transcription factors required for HIS4 expression. In a strain carrying deletions of the BAS1, BAS2, and GCN4 genes, which encode transcription factors that act at the HIS4 promoter, the HIS4 gene is poorly transcribed, leading to auxotrophy for histidine (9, 10). Interestingly, the low level of HIS4 transcription observed in such a yeast strain can be increased by mutations in RPB1 or RPB2. It was proposed that these mutations increase the processivity of the polymerase and/or the number of initiation events at the HIS4 promoter. Unfortunately, only the sit mutations mapping in RPB1, not those in RPB2, have been sequenced. In either case the mutant RNAP II enzymes remain to be characterized in vitro in order to pinpoint the mechanistic basis for suppression.
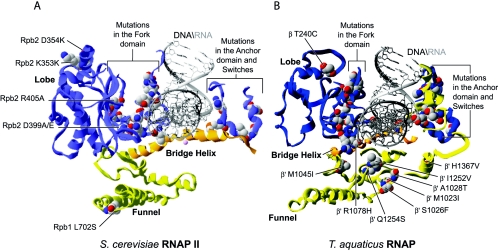
RNAP mutants having a higher polymerization rate or suppressing polymerization defects appear to affect different regions of the enzyme. A first group of amino acid replacements are located near the catalytic aspartates of Rpb1/β′ or near the active center (Fig. 6A). N445T (4) and M487L (4) are located in this region and were isolated as sit mutations. It is possible that these replacements affect the position of the catalytic aspartates, thereby affecting the polymerization rate.
FIG. 6.
Distribution of substitutions conferring a sit phenotype or a high polymerization rate, or that can suppress a low-polymerization-rate phenotype. (A) Substitutions in eukaryotic RNAP inducing sit, high polymerization rate or suppression of low polymerization rate phenotypes have been mapped onto the structure of S. cerevisiae RNAP II. (B) Substitutions in prokaryotic RNAP showing higher polymerization rate or suppression of a low polymerization rate have been mapped onto the structure of T. aquaticus RNAP.
Because N445S (20) was found to affect start site selection, and because position N445 falls near the saddle which interacts with TFIIB, it is conceivable that N445T also affects the number of initiation events. The Rpb2 S771C (36) replacement, identified as an intergenic suppressor of the synthetic lethality of the Drosophila RpII215 K1 mutation, is located in the hybrid binding region, near the RNA at position +9 and the fork loop 2 segment. Replacement with cysteine could cause an increased interaction between the hybrid region and fork loop 2. Therefore, and as replacements in the fork loop 2 region can induce lower polymerization rates (see above), it is possible that S771C has the opposite effect.
A second group of replacements are localized in the Rpb1/β′ funnel domain (Fig. 6). The role of the funnel in transcription has not yet been defined. However, it was shown that the funnel and the bridge helix form the binding site for the inhibitor α-amanitin (27). Many prokaryotic mutations, including A1028T (190) and M1045I (154, 190) isolated from altered termination and streptolydigin resistance, and eukaryotic ones, including L702S (147), a suppressor of elongation defects of RNAP III mutant rpc160-112; G730D (4), a sit mutation, K744S (147); A759P (4); and C764F D668N (4), that cause either a higher polymerization rate, a suppressed polymerization defect, or a sit phenotype are found in this region, suggesting a role for the funnel in transcript elongation. Because the funnel is juxtaposed to the bridge helix, we propose that it regulates its ability to translocate the nucleic acids during polymerization. The funnel is also exposed at the surface of the enzyme and could theoretically be contacted by certain transcriptional regulators.
The bridge helix is the site of many replacements that confer a higher polymerization activity or can suppress mutations inducing polymerization defects (Fig. 6). In eukaryotes, replacement of glycine 820 with an aspartate (G820D) (4) probably alters the conformation of the bridge helix in a way that facilitates translocation (Fig. 6A). The A832V (4, 190) replacement is located near the base moiety of the +1 nucleotide in the template DNA. It was proposed that the invariant alanine 832 helps position the +1 nucleotide through van der Waals interactions (57).
The replacement of the alanine for a valine, a bulkier hydrophobic residue, might allow a better interaction with the base of this nucleotide. This could stabilize the nucleic acids in the cleft, resulting in a more efficient polymerization. The L841I replacement (147) has its side chain oriented toward the two helices that form the N- and C-terminal ends of the trigger loop. The L841 residue could be involved in a number of interactions with neutral amino acids in these structural elements. Replacement of a leucine with an isoleucine is not a drastic modification. It may be that this region of the bridge helix is critical for the processivity of the enzyme. In prokaryotes, the R1078H (153, 190) replacement is part of the bridge helix, but is oriented toward fork loop 2 (Fig. 6B). The R1078 residue is involved in an interaction with the backbone of fork loop 2. Replacement of an arginine for a histidine could potentially stabilize an important interaction of the bridge helix with fork loop 2, leading to a higher rate of polymerization. Consistently, replacement of arginine 1078 with a cysteine (14, 17), which is too small to allow any interaction with fork loop 2, produces a low-polymerization phenotype as discussed above.
Other prokaryotic and eukaryotic RNAP mutants are affected in residues close to the bridge helix. The replacements S1436F (190) and A1437V (190) in E. coli RNAP are found in a β′ loop that could potentially interact with the bridge helix (Fig. 6B). The A1076V replacement in yeast RNAP II (4) (Fig. 6A) when combined with N479Y, a residue implicated in NTP versus dNTP selection (87, 173), confers a sit phenotype. The N479Y replacement responsible for the sit phenotype is lethal when combined with a PPR2 deletion and this effect is suppressed by A1076V. These effects might be explained if the N479Y replacement positively affects the initiation of transcription but has a negative effect on transcription elongation, which then requires TFIIS. Similar to the RNAP III replacement N479Y that is lethal unless compensated for by S476I, the requirement for TFIIS imposed by the RNAP II replacement N479Y can be overcome by the A1076V replacement.
A1076V reconfigures the active site for stimulation of the intrinsic cleavage activity of RNAP to promote pause escape, or, because A1076 is located in a helix bordering the trigger loop, by stimulating the processivity of RNAP through altered interactions of this element with the bridge helix. A875S (147) is another replacement suppressing the slow elongation phenotype imposed by the double amino acid replacement of Rpc160-112 N479Y/S476I (48, 66) (see above). A875S might be involved in interactions with a region of the enzyme located close to the trigger loop, maybe correcting the N479Y/S476I induced defect by a mechanism similar to that brought about by A1076V.
The V1299K (147) replacement is located at the center of a β-sheet surrounded by two β-strands. One of these β-strands is located in the region adjacent to the trigger loop and in proximity to the bridge helix. Therefore it is possible that this replacement also increases the enzyme's polymerization rate by affecting the functions of the trigger loop or the bridge helix. Finally, a replacement in E. coli, Q1254S (16), which is located in the trigger loop, produces a fast-elongating RNAP in vitro and is thought to affect the oscillating properties of the bridge helix (16).
AMINO ACID REPLACEMENTS AFFECTING TERMINATION
A great number of replacements in E. coli and eukaryotic RNAPs were reported to affect the termination step of the transcription reaction (Fig. 7) (16, 22, 78, 82, 97, 147, 154, 157, 165, 174, 190). These mutants were isolated from their altered ability to read through terminators located in specific genes. Many terminators have been used, including SUP4, T1, ρ-dependent and ρ-independent terminators, trp attenuator, thr attenuator, rrnT1, T7, his, Tr2, P14, T3, lambda Tr2, and ops, in various contexts.
FIG. 7.
Distribution of substitutions affecting termination. (A) Substitutions in eukaryotic RNAP affecting termination and located near the lobe, funnel, and fork domains and switches have been mapped onto the structure of S. cerevisiae RNAP II. (B) Substitutions in prokaryotic RNAP affecting termination and located near the lobe, funnel, and fork domains and switches have been mapped onto the structure of T. aquaticus RNAP.
In some cases the ability to read through a terminator leads to the normal expression of a gene required for survival, allowing isolation of RNAP mutants with decreased termination ability. In contrast, expression of toxic genes from readthrough of terminators by wild-type RNAP allows for screening of mutants with increased termination properties. Most of these replacements affect the lobe, including G338E (22, 157), T339T (22, 157), T339T K353K (22, 157), A340K (22, 78, 157), L341I (22), D354K (22, 157), D399E (22), D399A (22), R405A (22), and T240C (97); the cleft, including I1252V (16), S1091F (154), Q1254S (16), R1078H (154, 190), and H1367Y (190); the fork, including S480I G503K (22, 157), V536E L541S (22, 157), K537N (22, 157), N538Y (22, 157), K510N (22, 157), G503K (22, 157), L508D (22, 157), S480I (22, 157), L483H (22, 157), H494I G503Q (157), P501R (22, 157), M542I (22, 157), C544P (22, 157), G446D (22, 157), Q393P (81, 82), P440S T443I (97), P440L (175), and S454F S591F (175); and the funnel, including L702S (147), M1023I (190), S1026F (154, 190), A1028T (190), and M1045I (190), regions of the enzyme or are into or close to switches, including R1129K E1149D (22, 157), I1139L R1129S (22, 157), M1133L (22, 157), S1436F (190), A1437V (190), G609D (190), S626F (190), S1439L (190), G1011S (175), P1020L A1045V (97), and G1028D (175).
These domains also regroup replacements carrying particular phenotypes such as altered polymerization rates and unstable open promoter complexes. For example, the funnel and the fork domains are altered in mutants showing various polymerization defects. It is tempting to speculate that the termination phenotypes observed in this case derive from an altered elongation phenotype. The lobe seems important for opening and maintaining the transcription bubble. We can hypothesize that a transcription complex in which the transcription bubble has a tendency to collapse is more prone to arrest at a termination site.
Many termination mutants also show altered switch domains. The switches were proposed to form a portion of the binding site for the DNA-RNA hybrid (7). Therefore, it is possible that replacements in these structural elements affect the stability of the RNA and its retention in the active site, leading to increased or reduced ability to terminate transcription.
DRUG-RESISTANT RNAP MUTANTS
Over the past years, numerous drug-resistant RNAP mutants have been isolated and characterized. α-Amanitin, rifampin, streptolydigin, sorangicin, and microcin J25 resistances have all been studied thoroughly allowing new insights regarding the transcriptional mechanisms of RNAPs to be gained. Moreover, understanding the mode of action of these and other inhibitors is pivotal for the design of novel and potent antipathogen drugs.
α-Amanitin is a bicyclic octapeptide used for a long time as a specific RNAP II inhibitor (106). The crystallographic structures of RNAP II in a complex with α-amanitin were resolved by Bushnell and colleagues in 2002 (27) and show binding of this drug to the funnel and the bridge helix features. Known replacements conferring α-amanitin resistance, including L722F (18), R726H (37)/P (19), I756F (19), C764Y (19), S769D (18, 19), and G772E (19), have been mapped onto the crystal structure of RNAP II in Fig. 8A to show that they all cluster around this binding site.
FIG.8.
(A) Distribution of RNAP II substitutions conferring α-amanitin resistance and mapped onto the S. cerevisiae RNAP II structure 1K83. Residues are colored differently for clarity. (B) Distribution of RNAP substitutions conferring rifampin resistance and mapped onto the T. aquaticus RNAP structure 1I6V. Substitutions inducing low levels of rifampin resistance are shown in pink. (C) Distribution of RNAP substitutions and insertions conferring streptolydigin resistance and mapped onto the T. aquaticus RNAP structure 1ZYR. Red, pink, and magenta residues are located in the bridge helix, fork domain, and trigger loop, respectively. (D) Distribution of RNAP substitutions conferring sorangicin resistance and mapped onto the T. aquaticus RNAP structure 1YNJ. (E) Distribution of RNAP substitutions conferring microcin J25 resistance and mapped onto the T. aquaticus RNAP structure 1I6V.
Because the bridge helix feature has been proposed to be involved in nucleic acid translocation, α-amanitin most probably impairs the translocation of nucleic acids by interfering with the function of the bridge (58). This is further supported by studies showing that after addition of α-amanitin to a transcribing RNAP II complex, synthesis of a single phosphodiester bond can still occur (61, 184).
Rifampin, a member of the rifamycin family, is an efficient antibiotic against bacterial pathogens and binds with high affinity to bacterial RNAPs (63). Crystals of RNAP in a complex with rifampin were obtained by Campbell and colleagues in 2001 (30) and permitted the identification of the binding pocket and the description of a mechanism of action for this drug. Direct interactions of rifampin with RNAP are mostly made with a region of the β subunit forming the fork domain.
In accordance with these findings, most of the rifampin-resistant mutations isolated so far map directly in or adjacent to the fork domain (Fig. 8B). More specifically, they are found in the protrusion, fork and external 1 and 2 domains of β (14, 31, 81, 82, 97, 108, 120, 127, 129, 130, 156, 160, 174, 178, 186, 201, 202, 207). Thus, these replacements most probably affect the conformation of the binding pocket and lower its affinity for rifampin. Other mutations conferring lower levels of rifampin resistance are mostly located away from this binding pocket. Many of them are found in the lobe region and suggest that binding of rifampin to the fork region is somehow linked to the lobe's conformation when holding the downstream DNA. Thus, it appears that binding of the lobe domain to DNA is necessary, either directly or indirectly, for the formation of the rifampin-binding pocket.
Numerous rifampin-resistant mutations have been isolated through the years and their locations overlap a small region where some streptolydigin-resistant mutations (74) cluster. Nonetheless, the biochemical effects of these two drugs on transcription are different. Rifampin inhibits initiating RNAPs at the promoter and has no effects once RNAP has elongated past the promoter (183). Consistently, rifampin's mechanism of action based on the crystallographic studies implies a steric clash between the drug and the RNA hybrid when it reaches 2 or 3 nucleotides in length (30). In contrast, streptolydigin can block transcription of initiating as well as elongating RNAPs (114).
Nonetheless, this steric mechanism alone cannot explain why some rifampin-resistant mutations can only confer resistance to certain members of the rifamycin family, without impeding the antibiotic binding to RNAP. Why, for example, can an elevated Mg2+ concentration confer a certain Rif resistance phenotype and why is rifambutin, a member of the Rif family, able to inhibit formation of the first phosphodiester bond? From the study of cocrystals of RNAP structures with different members of the Rif family (rifampin and rifambutin), the authors proposed two allosteric mechanisms, the σ pathway, which implicates an interaction between the drug and the σ factor, and the β pathway, which implicates specific residues for transmission of a signal from the Rif binding site to the catalytic aspartates, altering binding of Mg2+ in the active site and leading to inhibition of transcription. The σ pathway was evidenced by the finding that rifambutin, but not rifampin, can bind a portion of σ70, an interaction that could account for its specific effect on the formation of the first phosphodiester bond. The β pathway was defined by the engineering of specific RNAP mutations lying in the signal transmission path, far from the Rif binding pocket, which induce resistance without altering Rif binding (12).
Streptolydigin is an antibiotic that also specifically inhibits bacterial RNAPs (114). Mapping of amino acid replacements conferring streptolydigin resistance onto the RNAP structure, as shown in Fig. 8C, shows clustering in the fork loop 2 region, including A423V/P/T (64, 182), G424R (64, 182), G424D-F425S (107), F425S/C/I (64, 182), D426V (64), and 6His tag insertion at 420 to 421 (156); the fork domain, including G450C (182), and L451R (182); the bridge helix, including R1078H (156), L1086M/V (182), A1089G (182), S1091F (156, 182), and R1096I; the trigger loop, including P1232L, L1236M, T1237I/N, V1246V, Q1254R, Q1254S P1257A, G1255D, L1256V P1257T, and P1257R/L/S (182); the funnel domain (M1045I) (156); and a region adjacent to the trigger loop (V1361G) (182).
This location for the streptolydigin binding pocket strongly suggests that the mechanism of action of this drug involves an impaired fork loop 2 function and a restriction of the bridge helix and trigger loop movement, these being required for translocation of nucleic acids in the active site, reminiscent of the α-amanitin mode of action (176). However, some recent results show that streptolydigin blocks neither translocation nor phosphodiester bond formation (182). These findings raise important concerns about the respective implication of the bridge helix/trigger loop and of the complementary NTP binding in the process of translocation. The authors rather propose that binding of streptolydigin traps the RNAP active site in an inactive intermediate state (182).
Sorangicin is a macrolide polyether antibiotic that inhibits transcription initiation in a way very similar to rifampin (72). It has also been observed that some rifampin-resistant mutations can confer sorangicin resistance, suggesting largely overlapping binding sites for these two drugs (31, 127, 145). However, the structures of these drugs are different and observation that some rifampin-resistant mutants are sorangicin sensitive, while most sorangicin-resistant mutations confer rifampin resistance, suggests that subtle differences exist with regard to their modes of action (145).
In order to better understand the basis of similarities and differences between these drugs, crystallographic studies of sorangicin in a complex with T. aquaticus RNAP have been performed which demonstrated that both drugs binds to the same RNAP region (31). The mapping of mutations conferring sorangicin resistance is shown in Fig. 8D. All (31, 127) cluster to the protrusion and fork domains, suggesting that the modes of action of these two drugs are the same, and that the higher conformational flexibility of sorangicin renders it more potent to adapt to structural alterations of its binding pocket.
Microcin J25 is an antibacterial peptide inhibiting transcription by RNAP (149). Recent studies by Adelman and colleagues and Mukhopadhyay and colleagues (1, 117), including saturation mutagenesis and mapping of mutations conferring microcin J25 resistance, have led to the identification of the microcin J25 binding site made of more than 50 amino acids. These replacements and those isolated in previous reports have been mapped in Fig. 8E. The defined region surrounds the secondary channel which is proposed to be the entry site of NTPs. Thus, based on these studies, the suggested mode of action of microcin J25 consists in the occlusion of the secondary channel, interfering with NTP entry to the active site and leading to inhibition of transcription.
RNAP MUTANTS MIMICKING THE EFFECTS OF ALARMONE ppGpp
In nutriment-limiting conditions, such as amino acid starvation, bacterial cells rapidly accumulate a guanine derivative, ppGpp. The production of this metabolite is dependent on the proteins RelA and SpoT. The presence of ppGpp leads to the inhibition of the expression of the translational machinery and to an increase in the synthesis of many enzymes involved in the biosynthesis of amino acids and many other products (33) in a process called the stringent response. ΔrelA ΔspoT strains are auxotrophic for many amino acids (200) and this phenotype, due to the absence of ppGpp, can be suppressed by changes in the β, β′, and σ subunits of RNAP.
Although recent crystallographic studies of T. thermophilus RNAP in a complex with alarmone ppGpp have been resolved (11), the precise mechanism of inhibition and activation of respective promoters by this metabolite is still not completely understood. Studies of promoters like rrnB P1, rrnD P1, and tyrT have showed that open promoter complexes at these growth rate-regulated promoters are unstable (56, 59, 104). In addition to the short-lived open complexes, other determinants for regulation by ppGpp have been identified in promoters, including a C-rich sequence called discriminator located between the −10 box and +1 site on the nontemplate DNA strand (179), a shortened 16-bp spacer between the −35 and −10 elements, and a noncanonical sequence in the −35 region (14, 15, 33).
Based in part on the crystallographic structures showing the ppGpp molecule bound to RNAP in the pore near the active site, it has been proposed that alarmone could make sequence-specific contacts with the nontemplate strand (11), this interaction being possibly responsible for the reduced stability of open complexes at these promoters.
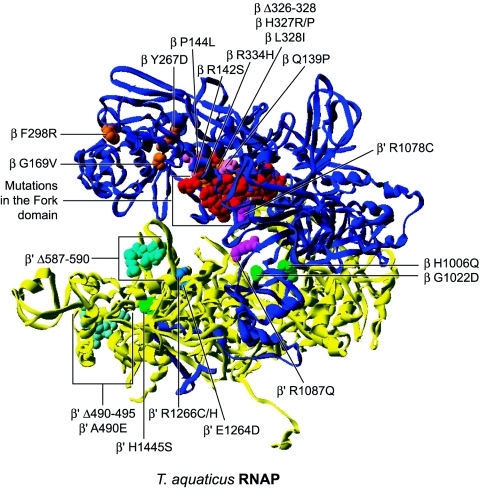
The mapping of the RNAP replacements mimicking ppGpp effect on the T. aquaticus RNAP structure is shown in Fig. 9. A great number of β replacements cluster in the fork domain (14, 178, 201, 202, 207) and some in the protrusion domain, including Q139P (178), P144L (178), R142S (178), and R334H (14, 17). Interestingly, many of them also confer rifampin resistance (178, 201, 207). The fork loop 2 element in the fork domain protrudes in the cleft near the DNA-RNA hybrid and is proposed to be involved in maintaining the architecture of the transcription bubble (44). Replacements falling in or near this element are thus likely to destabilize the open complex in a manner similar to that brought about by ppGpp.
FIG. 9.
Distribution of substitutions and deletions mimicking the alarmone ppGpp effect on transcription mapped onto the T. aquaticus RNAP structure 1I6V. ppGpp has been placed by overlaying the T. thermophilus RNAP structure 1SMY. Substitutions located in the lobe and protrusion domain are shown in orange and pink, respectively. The many substitutions in the fork domain are shown in red and are not labeled. Substitutions in the cleft, switches, and jaw domains are shown in magenta, green, and blue, respectively. Cyan-colored altered amino acids are located in the clamp core and clamp head region.
It is not clear whether replacements located more or less at a distance from the fork loop 2 element simply destabilize the fork domain or alter the flexibility of this domain linked to the lobe region. It is conceivable that the later replacements also alter the lobe domain function. Indeed, a set of replacements in the β subunit are located directly in the lobe region, including G169V (178), Y267D (178), F298R (178), Δ326-328 (14), H327R/P (178), and L328I (178). This domain is suggested to grip tightly the downstream DNA. Deletion in this domain have also been isolated that confer defects in open promoter complexes. Thus, mutational alteration of this region is another way to destabilize open complexes and mimic the effect of ppGpp in the stringent response.
Two other replacements, H1006Q (14, 178) and G1022D (178), in the β subunit are localized in the C-terminal domain in the switch 3 element. The switches are at the base of the clamp and interact with the DNA-RNA hybrid (57). It is conceivable that these mutated residues make important contacts with the hybrid and that breaking these interactions lead to unstable open promoter complexes.
Many of the β′ subunit replacements map within the clamp domain. H1445S (178) lies in the switch 1 element and, because this element is also at the base of the clamp and interacts with the template strand, it is conceivable that the replacement destabilizes the open complex. Another deletion in the clamp, the Δ587-590 deletion (178), affects the rudder element. This element is thought to help separate the RNA transcript from DNA by preventing reannealing of the two strands (94). Moreover, the complete deletion of this element (Δ586-601) leads to an altered transcription bubble in the downstream part (94), and thus the Δ587-590 replacement is consistent with the mode of action of ppGpp.
The A490E (178) replacement and the Δ490-495 (17) deletion are located in the clamp head near the downstream part of the DNA in the cleft. It is possible that this region of the clamp helps hold DNA in a fashion similar to the lobe and jaw regions. Thus, effects on the stability of the open complex arise from an altered capacity of RNAP to grip the downstream DNA and maintain a stable transcription bubble. Two other replacements, E1264D (178) and R1266C (14), are located in the cleft region and may act similarly to replacements in the lobe and clamp head regions because this part of the cleft also participates in holding the downstream DNA.
Finally, two replacements are located in the bridge helix. This element has been proposed to be involved in translocation of nucleic acids and to maintain a specific conformation in the template strand in the active site. Because this element interacts with nucleic acids, replacements could easily destabilize the open complex. The mutated residue R1078C (14) is oriented toward the fork loop 2 element and may affect open promoter stability by disrupting an important interaction between this element and the bridge helix. The other replacement in the bridge helix, R1087Q (178), is oriented toward the nucleic acids in the prokaryotic structure, but in the opposite direction, toward the trigger loop, in eukaryotic structures. This could be attributable to conformational changes between the bent and straight conformations observed in these respective structures. Because this replacement is near the point of passage of nucleic acids over the bridge helix, replacement to glutamine could interfere with the movement of nucleic acids in the cleft. All the replacements discussed in this section support a destabilizing effect of ppGpp on open promoter complexes.
RNAP MUTATIONS CONFERRING ASSEMBLY DEFECTS
This section focuses on mutations affecting the correct assembly of RNAPs. Many of the assembly mutations most probably alter the correct folding of the subunit either locally or globally, rather than abolish specific key interactions that disrupt the contacts between subunits.
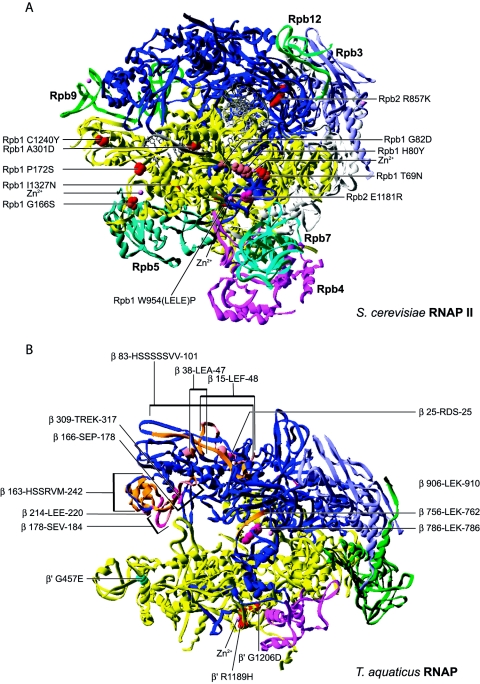
Many zinc atoms are known to bind and stabilize the RNAP structures (44). The eukaryotic mutation T69N (192), isolated in the largest subunit of RNAP II by random mutagenesis, and temperature-sensitive mutations G166S (89, 150) and P172S (89, 150), isolated by random hydroxylamine mutagenesis, are located near or in Zn2+ binding regions and show altered RNAP composition after chromatography (Fig. 10).
FIG. 10.
(A) Distribution of substitutions and insertions affecting assembly of eukaryotic RNAP mapped onto the S. cerevisiae RNAP II structure. (B) Distribution of substitutions and insertions affecting assembly of prokaryotic RNAP mapped onto the T. aquaticus structure.
The RNAP I temperature-sensitive mutations H80Y (197, 203) and G82D (197, 203), isolated by hydroxylamine mutagenesis, also fall near a Zn2+ binding domain. It is thus tempting to speculate that the observed assembly defects come from a destabilized RNAP structure altered in its ability to bind zinc. Isolation of E1181R (203) in the second largest subunit of RNAP I, an extragenic spontaneous revertant suppressing Rpa190 replacements H80Y and G82D, is also localized in a Zn2+ binding region. These two metal binding regions stabilize the clamp domain and interact with each other. It is on the basis of this extragenic suppression that G82D and H80Y have been proposed to induce assembly defects. Rpb4 and Rpb7 also associate with the RNAP in this region.
Finally, the temperature-sensitive prokaryotic β′ subunit mutations R1189H (122) and G1206D (122) also map to a Zn2+ region and these subunits fail to reassemble after denaturation and renaturation. This occurrence of assembly defective mutations in the Zn2+ binding regions suggests an important contribution of these regions and of zinc to the correct folding of subunits and the assembly of RNAP.
The next group of replacements localize in regions more or less distant from the interface of subunit-subunit interactions. These replacements most probably affect RNAP assembly by altering either globally or locally the folding of the given subunit. The temperature-sensitive eukaryotic Rpb1 mutation A301D (169) is located in a helix of the clamp domain. An assembly defect has been proposed based on the altered cellular distribution of this Rpb1 mutant, which localizes in the cytoplasm instead of the nucleus at the nonpermissive temperature. The replacement could disturb the correct folding of the clamp domain known to interact with the C-terminal part of Rpb2.
In the second largest subunit of eukaryotic RNAP, the R857K (89, 150) replacement maps to the wall domain. The replacement of R857 with a lysine may also disturb the folding of this domain bound by Rpb3, Rpb10, and Rpb12. Finally, replacements in the largest Rpb1 subunit, I1327N (89, 150) and C1240Y (89, 150), affect residues located in regions interacting with other subunits, Rpb5 and Rpb9, respectively. This observation suggests an alteration of the interaction domains of Rpb1 with these subunits. However, these mutants were isolated based upon their ability to assemble stably with Rpb3, which is quite distant from these Rpb1 regions interacting with Rpb5 and Rpb9. In the crystal structure, Rpb3 interacts mostly with Rpb2 and Rpb11. It is then more probable that these replacements alter the stability and/or folding of Rpb1 which no longer associates stably with Rpb3, consequently altering interaction with either Rpb2 or Rpb11.
The W954(LELE)P insertion (3, 5, 6, 51) is positioned in the foot domain of Rpb1 between the interaction domains of TFIIS and Rpb6. The slow-growth, temperature-sensitive, and inositol auxotrophy phenotypes exhibited by this mutant can be suppressed by overexpression of Rpb6, suggesting that the observed defects arise from an altered ability of W954(LELE)P to bind Rpb6. Observed in the crystal structure, the W954(LELE)P insertion in the foot domain of Rpb1 is located too far away from Rpb6 to interfere directly with its association. Thus, this insertion may lead to instability of this Rpb1 region, which can be overcome by formation of the complete RNAP complex, this later step being favored by overexpression of Rpb6.
Finally, the prokaryotic β′ replacement G457E (122) is located in the N-terminal part of a helix of the clamp and replacement for a glutamate may alter the correct folding in this region and, consequently, its association with the β subunit.
The final set of mutants discussed here come from deletion or insertion mutagenesis. Even though large deletions of dispensable regions in the β subunit have been reported (155), these mutations are generally more prone to severely affect the folding of the subunits, given the complexity of the RNAP structure. A number of XhoI linker insertions have been identified (96) in the β subunit (Xho-42, 25-RDS-25; Xho-5, 178-SEV-184; Xho-6, 38-LEA-47; Xho-16, 214-LEE-220; Xho-29, 786-LEK-786; Xho-31, 83-HSSSSSVV-101; Xho-1, 309-TREK-317; Xho-4, 166-SEP-178; Xho-19, 756-LEK-762; Xho-36, 15-LEF-48; Xho-54, 906-LEK-910; and Xho-73, 163-HSSRVM-242) (Fig. 10). Assembly defects have been confirmed by measuring the amount of rifampin-sensitive (Rifs) RNAP activity arising from association of these Rifs mutant β subunits into RNAP when overexpressed in a strain carrying a rifampin-resistant chromosomal copy of β. A number of these mutants show detectable (Xho-42, Xho-5, Xho-6, Xho-29, and Xho-31), inhibitory (Xho-1, Xho-4, and Xho-19) or no catalytic activity (Xho-16, Xho-36, Xho-54, and Xho-73). These insertions most probably induce more or less severe misfolding of the respective subunits or domains.
CONCLUSIONS AND PERSPECTIVES
Mapping the amino acid changes reported to affect the function of multisubunit RNAP on the structure of the prokaryotic and eukaryotic enzymes revealed interesting features on the role of many structural elements during the transcription reaction. Many of these changes have not been fully characterized and knowing their position within the structure now allows us to make novel predictions that can be tested experimentally. For example, fork loop 2, which, based on its location, was proposed to be involved in setting the downstream boundary of the transcription bubble, is the site of many replacements that negatively regulate the polymerization rate. It would be interesting to further characterize these mutant RNAPs in vitro in order to determine whether or not they are altered in their ability to form and maintain a transcription bubble; these experiments would either confirm the implication of fork loop 2 in promoter opening or reveal another role for this element in the transcription reaction.
The trigger loop is targeted in a number of mutants affected in initiation site selection; these mutations could impair either direct contact of nucleic acids around +1 or the movement of the bridge helix. A functional analysis of mutants with an altered trigger loop would be required to discriminate between these two possibilities. Yet another example is the funnel domain, which is the site of many replacements that increase the polymerization rate or suppress polymerization defects. In vitro characterization of amino acid changes in this domain should allow us to clarify the role of the funnel in transcription.
It is important to note that a significant number of mutations listed in the catalogue (see Table S2 in the supplemental material) have not been described in this article (2, 45, 46, 49, 66, 67, 73, 78, 80, 84, 88, 91, 97, 108, 111, 112, 124, 126, 129, 130, 146, 151, 157, 160, 165, 167, 168, 170, 177, 180, 181, 186, 193, 204). For most of them, only in vivo phenotypes have been described and no in vitro characterization has yet been performed. Other mutations listed in Table S2 in the supplemental material that have not been discussed in this article are those whose effect on the enzyme has been characterized in vitro but whose location in the RNAP structure does not suggest a mechanistic basis for their effect. Some of these replacements may act indirectly by affecting known functional elements of the enzyme or may define new functional regions. As a whole, all these mutations are of great interest and have been included in our table in the supplemental material. Their positions within the structure and the domain in which they lie are indicated.
We hope that the structural perspective on mutations affecting the function of multisubunit RNA polymerases provided herein will stimulate the design of new mutations to be characterized and provide novel information on transcriptional mechanisms.
Supplementary Material
Acknowledgments
We apologize to our colleagues whose work on RNAP could not be incorporated in this review due to space limitation. We are grateful to the members of our laboratory for helpful discussions and comments on this paper. We thank Diane Bourque for artwork and Julie Edwards for critical reading of the manuscript.
This work is supported by grants from the Canadian Institutes for Health Research (to B.C. and J.A.), Genome Canada (to B.C.), and Genome Québec (to B.C.). M.F.L. holds a studentship from the Fonds Québécois de la Recherche sur la Nature et les Technologies and V.T. from the Fonds de la Recherche en Santé du Québec. J.A. is a senior scholar from the Fonds de la Recherche en Santé du Québec.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Adelman, K., J. Yuzenkova, A. La Porta, N. Zenkin, J. Lee, J. T. Lis, S. Borukhov, M. D. Wang, and K. Severinov. 2004. Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol. Cell 14:753-762. [DOI] [PubMed] [Google Scholar]
- 2.Allison, L. A., and C. J. Ingles. 1989. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc. Natl. Acad. Sci. USA 86:2794-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault, J., M. A. Drebot, J. C. Stone, and J. D. Friesen. 1992. Isolation and phenotypic analysis of conditional-lethal, linker-insertion mutations in the gene encoding the largest subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Gen. Genet. 232:408-414. [DOI] [PubMed] [Google Scholar]
- 4.Archambault, J., D. B. Jansma, J. H. Kawasoe, K. T. Arndt, J. Greenblatt, and J. D. Friesen. 1998. Stimulation of transcription by mutations affecting conserved regions of RNA polymerase II. J. Bacteriol. 180:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archambault, J., F. Lacroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archambault, J., K. T. Schappert, and J. D. Friesen. 1990. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol. Cell. Biol. 10:6123-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armache, K. J., H. Kettenberger, and P. Cramer. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100:6964-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armache, K. J., S. Mitterweger, A. Meinhart, and P. Cramer. 2005. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280:7131-7134. [DOI] [PubMed] [Google Scholar]
- 9.Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874-880. [DOI] [PubMed] [Google Scholar]
- 10.Arndt, K. T., C. A. Styles, and G. R. Fink. 1989. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell 56:527-537. [DOI] [PubMed] [Google Scholar]
- 11.Artsimovitch, I., V. Patlan, S. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299-310. [DOI] [PubMed] [Google Scholar]
- 12.Artsimovitch, I., M. N. Vassylyeva, D. Svetlov, V. Svetlov, A. Perederina, N. Igarashi, N. Matsugaki, S. Wakatsuki, T. H. Tahirov, and D. G. Vassylyev. 2005. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell 122:351-363. [DOI] [PubMed] [Google Scholar]
- 13.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 15.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 16.Bar-Nahum, G., V. Epshtein, A. E. Ruckenstein, R. Rafikov, A. Mustaev, and E. Nudler. 2005. A ratchet mechanism of transcription elongation and its control. Cell 120:183-193. [DOI] [PubMed] [Google Scholar]
- 17.Bartlett, M. S., T. Gaal, W. Ross, and R. L. Gourse. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:331-345. [DOI] [PubMed] [Google Scholar]
- 18.Bartolomei, M. S., and J. L. Corden. 1987. Localization of an α-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 7:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomei, M. S., and J. L. Corden. 1995. Clustered alpha-amanitin resistance mutations in mouse. Mol. Gen. Genet. 246:778-782. [DOI] [PubMed] [Google Scholar]
- 20.Berroteran, R. W., D. E. Ware, and M. Hampsey. 1994. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol. Cell. Biol. 14:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. [DOI] [PubMed] [Google Scholar]
- 22.Bobkova, E. V., N. Habib, G. Alexander, and B. D. Hall. 1999. Mutational analysis of the hydrolytic activity of yeast RNA polymerase III. J. Biol. Chem. 274:21342-21348. [DOI] [PubMed] [Google Scholar]
- 23.Borukhov, S., J. Lee, and O. Laptenko. 2005. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55:1315-1324. [DOI] [PubMed] [Google Scholar]
- 24.Borukhov, S., A. Polyakov, V. Nikiforov, and A. Goldfarb. 1992. GreA protein: a transcription elongation factor from Escherichia coli. Proc. Natl. Acad. Sci. USA 89:8899-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borukhov, S., V. Sagitov, and A. Goldfarb. 1993. Transcript cleavage factors from E. coli. Cell 72:459-466. [DOI] [PubMed] [Google Scholar]
- 26.Burgess, R. R., and A. A. Travers. 1970. Escherichia coli RNA polymerase: purification, subunit structure, and factor requirements. Fed. Proc. 29:1164-1169. [PubMed] [Google Scholar]
- 27.Bushnell, D. A., P. Cramer, and R. D. Kornberg. 2002. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. USA 99:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushnell, D. A., and R. D. Kornberg. 2003. Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA 100:6969-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bushnell, D. A., K. D. Westover, R. E. Davis, and R. D. Kornberg. 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303:983-988. [DOI] [PubMed] [Google Scholar]
- 30.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 31.Campbell, E. A., O. Pavlova, N. Zenkin, F. Leon, H. Irschik, R. Jansen, K. Severinov, and S. A. Darst. 2005. Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO J. 24:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carles, C., I. Treich, F. Bouet, M. Riva, and A. Sentenac. 1991. Two additional common subunits, ABC10 alpha and ABC10 beta, are shared by yeast RNA polymerases. J. Biol. Chem. 266:24092-24096. [PubMed] [Google Scholar]
- 33.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 34.Chen, B. S., and M. Hampsey. 2004. Functional interaction between TFIIB and the Rpb2 subunit of RNA polymerase II: implications for the mechanism of transcription initiation. Mol. Cell. Biol. 24:3983-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, H. T., and S. Hahn. 2004. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119:169-180. [DOI] [PubMed] [Google Scholar]
- 36.Chen, Y., D. Chafin, D. H. Price, and A. L. Greenleaf. 1996. Drosophila RNA polymerase II mutants that affect transcription elongation. J. Biol. Chem. 271:5993-5999. [PubMed] [Google Scholar]
- 37.Chen, Y., J. Weeks, M. A. Mortin, and A. L. Greenleaf. 1993. Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol. Cell. Biol. 13:4214-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung, W. H., J. L. Craighead, W. H. Chang, C. Ezeokonkwo, A. Bareket-Samish, R. D. Kornberg, and F. J. Asturias. 2003. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell 12:1003-1013. [DOI] [PubMed] [Google Scholar]
- 39.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 40.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 41.Connelly, S., and J. L. Manley. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 2:440-452. [DOI] [PubMed] [Google Scholar]
- 42.Coulombe, B., and Z. F. Burton. 1999. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev. 63:457-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig, M. L., O. V. Tsodikov, K. L. McQuade, P. E. Schlax, Jr., M. W. Capp, R. M. Saecker, and M. T. Record, Jr. 1998. DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J. Mol. Biol. 283:741-756. [DOI] [PubMed] [Google Scholar]
- 44.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876. [DOI] [PubMed] [Google Scholar]
- 45.de la Mata, M., C. R. Alonso, S. Kadener, J. P. Fededa, M. Blaustein, F. Pelisch, P. Cramer, D. Bentley, and A. R. Kornblihtt. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12:525-532. [DOI] [PubMed] [Google Scholar]
- 46.Delgado, M. A., M. R. Rintoul, R. N. Farias, and R. A. Salomon. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dichtl, B., D. Blank, M. Sadowski, W. Hubner, S. Weiser, and W. Keller. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dieci, G., S. Hermann-Le Denmat, E. Lukhtanov, P. Thuriaux, M. Werner, and A. Sentenac. 1995. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 14:3766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donaldson, I. M., and J. D. Friesen. 2000. Zinc stoichiometry of yeast RNA polymerase II and characterization of mutations in the zinc-binding domain of the largest subunit. J. Biol. Chem. 275:13780-13788. [DOI] [PubMed] [Google Scholar]
- 50.Douziech, M., F. Coin, J. M. Chipoulet, Y. Arai, Y. Ohkuma, J. M. Egly, and B. Coulombe. 2000. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol. Cell. Biol. 20:8168-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drebot, M. A., G. C. Johnston, J. D. Friesen, and R. A. Singer. 1993. An impaired RNA polymerase II activity in Saccharomyces cerevisiae causes cell-cycle inhibition at START. Mol. Gen. Genet. 241:327-334. [DOI] [PubMed] [Google Scholar]
- 52.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of Gtp biosynthesis in Saccharomyces-Cerevisiae. Curr. Genet. 22:9-11. [DOI] [PubMed] [Google Scholar]
- 53.Fish, R. N., and C. M. Kane. 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577:287-307. [DOI] [PubMed] [Google Scholar]
- 54.Forget, D., M. F. Langelier, C. Therien, V. Trinh, and B. Coulombe. 2004. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol. Cell. Biol. 24:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forget, D., F. Robert, G. Grondin, Z. F. Burton, J. Greenblatt, and B. Coulombe. 1997. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc. Natl. Acad. Sci. USA 94:7150-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 57.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 58.Gong, X. Q., Y. A. Nedialkov, and Z. F. Burton. 2004. Alpha-amanitin blocks translocation by human RNA polymerase II. J. Biol. Chem. 279:27422-27427. [DOI] [PubMed] [Google Scholar]
- 59.Gourse, R. L. 1988. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 16:9789-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 61.Gu, W., W. Powell, J. Mote, Jr., and D. Reines. 1993. Nascent RNA cleavage by arrested RNA polymerase II does not require upstream translocation of the elongation complex on DNA. J. Biol. Chem. 268:25604-25616. [PMC free article] [PubMed] [Google Scholar]
- 62.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hartmann, G., K. O. Honikel, F. Knusel, and J. Nuesch. 1967. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta 145:843-844. [DOI] [PubMed] [Google Scholar]
- 64.Heisler, L. M., H. Suzuki, R. Landick, and C. A. Gross. 1993. Four contiguous amino acids define the target for streptolydigin resistance in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:25369-25375. [PubMed] [Google Scholar]
- 65.Hekmatpanah, D. S., and R. A. Young. 1991. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol. Cell. Biol. 11:5781-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hermann-Le Denmat, S., M. Werner, A. Sentenac, and P. Thuriaux. 1994. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol. 14:2905-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Himmelfarb, H. J., E. M. Simpson, and J. D. Friesen. 1987. Isolation and characterization of temperature-sensitive RNA polymerase II mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2155-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 69.Holstege, F. C., U. Fiedler, and H. T. Timmers. 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16:7468-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holstege, F. C., P. C. van der Vliet, and H. T. Timmers. 1996. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15:1666-1677. [PMC free article] [PubMed] [Google Scholar]
- 71.Hull, M. W., K. McKune, and N. A. Woychik. 1995. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 9:481-490. [DOI] [PubMed] [Google Scholar]
- 72.Irschik, H., R. Jansen, K. Gerth, G. Hofle, and H. Reichenbach. 1987. The sorangicins, novel and powerful inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J. Antibiot. (Tokyo) 40:7-13. [DOI] [PubMed] [Google Scholar]
- 73.Ito, K., and Y. Nakamura. 1993. Pleiotropic effects of the rpoC10 mutation affecting the RNA polymerase beta′ subunit of Escherichia coli on factor-dependent transcription termination and antitermination. Mol. Microbiol. 9:285-293. [DOI] [PubMed] [Google Scholar]
- 74.Iwakura, Y., A. Ishihama, and T. Yura. 1973. RNA polymerase mutants of Escherichia coli. Streptolydigin resistance and its relation to rifampicin resistance. Mol. Gen. Genet. 121:181-196. [DOI] [PubMed] [Google Scholar]
- 75.Iyer, L. M., E. V. Koonin, and L. Aravind. 2003. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iyer, L. M., E. V. Koonin, and L. Aravind. 2004. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene 335:73-88. [DOI] [PubMed] [Google Scholar]
- 77.Izban, M. G., and D. S. Luse. 1992. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′-5′ direction in the presence of elongation factor SII. Genes Dev. 6:1342-1356. [DOI] [PubMed] [Google Scholar]
- 78.James, P., S. Whelen, and B. D. Hall. 1991. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. Structural analysis of the wild-type and ret1-1 mutant alleles. J. Biol. Chem. 266:5616-5624. [PubMed] [Google Scholar]
- 79.Jeon, C., H. Yoon, and K. Agarwal. 1994. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl. Acad. Sci. USA 91:9106-9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeronimo, C., M. F. Langelier, M. Zeghouf, M. Cojocaru, D. Bergeron, D. Baali, D. Forget, S. Mnaimneh, A. P. Davierwala, J. Pootoolal, M. Chandy, V. Canadien, B. K. Beattie, D. P. Richards, J. L. Workman, T. R. Hughes, J. Greenblatt, and B. Coulombe. 2004. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell. Biol. 24:7043-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 82.Jin, D. J., and C. A. Gross. 1991. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J. Biol. Chem. 266:14478-14485. [PubMed] [Google Scholar]
- 83.Kashlev, M., J. Lee, K. Zalenskaya, V. Nikiforov, and A. Goldfarb. 1990. Blocking of the initiation-to-elongation transition by a transdominant RNA polymerase mutation. Science 248:1006-1009. [DOI] [PubMed] [Google Scholar]
- 84.Kawagishi-Kobayashi, M., M. Yamamoto, and A. Ishihama. 1996. Mutational analysis of the RNase-like domain in subunit 2 of fission yeast RNA polymerase II. Mol. Gen. Genet. 250:1-6. [DOI] [PubMed] [Google Scholar]
- 85.Kempers-Veenstra, A. E., J. Oliemans, H. Offenberg, A. F. Dekker, P. W. Piper, R. J. Planta, and J. Klootwijk. 1986. 3′-end formation of transcripts from the yeast rRNA operon. EMBO J. 5:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kettenberger, H., K. J. Armache, and P. Cramer. 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114:347-357. [DOI] [PubMed] [Google Scholar]
- 87.Kettenberger, H., K. J. Armache, and P. Cramer. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16:955-965. [DOI] [PubMed] [Google Scholar]
- 88.Kim, W. J., L. P. Burke, and M. A. Mortin. 1994. Molecular modeling of RNA polymerase II mutations onto DNA polymerase I. J. Mol. Biol. 244:13-22. [DOI] [PubMed] [Google Scholar]
- 89.Kolodziej, P. A., and R. A. Young. 1991. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol. Cell. Biol. 11:4669-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619-625. [DOI] [PubMed] [Google Scholar]
- 91.Krasnoselskaya, I., J. Huang, T. Jones, C. Dezan, and M. A. Mortin. 1998. Selection and analysis of rare second-site suppressors of Drosophila RNA polymerase II mutations. Mol. Gen. Genet. 258:457-465. [DOI] [PubMed] [Google Scholar]
- 92.Krummel, B., and M. J. Chamberlin. 1989. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry 28:7829-7842. [DOI] [PubMed] [Google Scholar]
- 93.Kuhn, A., A. Normann, I. Bartsch, and I. Grummt. 1988. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 7:1497-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuznedelov, K., N. Korzheva, A. Mustaev, and K. Severinov. 2002. Structure-based analysis of RNA polymerase function: the largest subunit's rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J. 21:1369-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Labhart, P. 1997. Transcript cleavage in an RNA polymerase I elongation complex. Evidence for a dissociable activity similar to but distinct from TFIIS. J. Biol. Chem. 272:9055-9061. [DOI] [PubMed] [Google Scholar]
- 96.Landick, R., A. Colwell, and J. Stewart. 1990. Insertional mutagenesis of a plasmid-borne Escherichia coli rpoB gene reveals alterations that inhibit beta-subunit assembly into RNA polymerase. J. Bacteriol. 172:2844-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Landick, R., J. Stewart, and D. N. Lee. 1990. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 4:1623-1636. [DOI] [PubMed] [Google Scholar]
- 98.Lang, W. H., and R. H. Reeder. 1993. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Langelier, M. F., D. Baali, V. Trinh, J. Greenblatt, J. Archambault, and B. Coulombe. 2005. The highly conserved glutaminic acid 791 of Rpb2 is involved in binding of NTP and Mg(B) in the active center of human RNA polymerase II. Nucleic Acids Res. 33:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Langelier, M. F., D. Forget, A. Rojas, Y. Porlier, Z. F. Burton, and B. Coulombe. 2001. Structural and functional interactions of transcription factor (TF) IIA with TFIIE and TFIIF in transcription initiation by RNA polymerase II. J. Biol. Chem. 276:38652-38657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langer, D., J. Hain, P. Thuriaux, and W. Zillig. 1995. Transcription in archaea: similarity to that in eucarya. Proc. Natl. Acad. Sci. USA 92:5768-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lanoix, J., and N. H. Acheson. 1988. A rabbit beta-globin polyadenylation signal directs efficient termination of transcription of polyomavirus DNA. EMBO J. 7:2515-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee, J., M. Kashlev, S. Borukhov, and A. Goldfarb. 1991. A β subunit mutation disrupting the catalytic function of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 88:6018-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leirmo, S., and R. L. Gourse. 1991. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J. Mol. Biol. 220:555-568. [DOI] [PubMed] [Google Scholar]
- 105.Lennon, J. C., III, M. Wind, L. Saunders, M. B. Hock, and D. Reines. 1998. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lindell, T. J., F. Weinberg, P. W. Morris, R. G. Roeder, and W. J. Rutter. 1970. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science 170:447-449. [DOI] [PubMed] [Google Scholar]
- 107.Lisitsyn, N. A., E. D. Sverdlov, E. P. Moiseeva, and V. G. Nikiforov. 1985. Localization of mutation leading to resistance of E. coli RNA polymerase to the antibiotic streptolydigin in the gene rpoB coding for the beta-subunit of the enzyme. Bioorg. Khim. 11:132-134. (In Russian.) [PubMed] [Google Scholar]
- 108.Lisitsyn, N. A., E. D. Sverdlov, E. P. Moiseyeva, O. N. Danilevskaya, and V. G. Nikiforov. 1984. Mutation to rifampicin resistance at the beginning of the RNA polymerase beta subunit gene in Escherichia coli. Mol. Gen. Genet. 196:173-174. [DOI] [PubMed] [Google Scholar]
- 109.Logan, J., E. Falck-Pedersen, J. E. Darnell, Jr., and T. Shenk. 1987. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl. Acad. Sci. USA 84:8306-8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Majovski, R. C., D. A. Khaperskyy, M. A. Ghazy, and A. S. Ponticelli. 2005. A functional role for the switch 2 region of yeast RNAPII in transcription start site utilization and abortive initiation. J. Biol. Chem. 280:34917-34923. [DOI] [PubMed]
- 111.Martin, C., S. Okamura, and R. Young. 1990. Genetic exploration of interactive domains in RNA polymerase II subunits. Mol. Cell. Biol. 10:1908-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martin, E., V. Sagitov, E. Burova, V. Nikiforov, and A. Goldfarb. 1992. Genetic dissection of the transcription cycle. A mutant RNA polymerase that cannot hold onto a promoter. J. Biol. Chem. 267:20175-20180. [PubMed] [Google Scholar]
- 113.Matsuzaki, H., G. A. Kassavetis, and E. P. Geiduschek. 1994. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol. 235:1173-1192. [DOI] [PubMed] [Google Scholar]
- 114.McClure, W. R. 1980. On the mechanism of streptolydigin inhibition of Escherichia coli RNA polymerase. J. Biol. Chem. 255:1610-1616. [PubMed] [Google Scholar]
- 115.McClure, W. R., and C. L. Cech. 1978. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253:8949-8956. [PubMed] [Google Scholar]
- 116.Minakhin, L., S. Bhagat, A. Brunning, E. A. Campbell, S. A. Darst, R. H. Ebright, and K. Severinov. 2001. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 98:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mukhopadhyay, J., E. Sineva, J. Knight, R. M. Levy, and R. H. Ebright. 2004. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 14:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 119.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 120.Mustaev, A., M. Kashlev, J. Y. Lee, A. Polyakov, A. Lebedev, K. Zalenskaya, M. Grachev, A. Goldfarb, and V. Nikiforov. 1991. Mapping of the priming substrate contacts in the active center of Escherichia coli RNA polymerase. J. Biol. Chem. 266:23927-23931. [PubMed] [Google Scholar]
- 121.Nechaev, S., M. Chlenov, and K. Severinov. 2000. Dissection of two hallmarks of the open promoter complex by mutation in an RNA polymerase core subunit. J. Biol. Chem. 275:25516-25522. [DOI] [PubMed] [Google Scholar]
- 122.Nedea, E. C., D. Markov, T. Naryshkina, and K. Severinov. 1999. Localization of Escherichia coli rpoC mutations that affect RNA polymerase assembly and activity at high temperature. J. Bacteriol. 181:2663-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nickels, B. E., S. J. Garrity, V. Mekler, L. Minakhin, K. Severinov, R. H. Ebright, and A. Hochschild. 2005. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl. Acad. Sci. USA 102:4488-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nonet, M. L., and R. A. Young. 1989. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123:715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nudler, E., A. Mustaev, E. Lukhtanov, and A. Goldfarb. 1997. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89:33-41. [DOI] [PubMed] [Google Scholar]
- 126.Onai, K., S. Katagiri, M. Akiyama, and H. Nakashima. 1998. Mutation of the gene for the second-largest subunit of RNA polymerase I prolongs the period length of the circadian conidiation rhythm in Neurospora crassa. Mol. Gen. Genet. 259:264-271. [DOI] [PubMed] [Google Scholar]
- 127.O'Neill, A., B. Oliva, C. Storey, A. Hoyle, C. Fishwick, and I. Chopra. 2000. RNA polymerase inhibitors with activity against rifampin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 44:3163-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Opalka, N., M. Chlenov, P. Chacon, W. J. Rice, W. Wriggers, and S. A. Darst. 2003. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114:335-345. [DOI] [PubMed] [Google Scholar]
- 129.Ovchinnikov, Y. A., G. S. Monastyrskaya, S. O. Guriev, N. F. Kalinina, E. D. Sverdlov, A. I. Gragerov, I. A. Bass, I. F. Kiver, E. P. Moiseyeva, V. N. Igumnov, S. Z. Mindlin, V. G. Nikiforov, and R. B. Khesin. 1983. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol. Gen. Genet. 190:344-348. [DOI] [PubMed] [Google Scholar]
- 130.Ovchinnikov, Y., G. S. Monastyrskaya, V. V. Gubanov, V. M. Lipkin, E. D. Sverdlov, I. F. Kiver, I. A. Bass, S. Z. Mindlin, O. N. Danilevskaya, and R. B. Khesin. 1981. Primary structure of Escherichia coli RNA polymerase nucleotide substitution in the beta subunit gene of the rifampicin resistant rpoB255 mutant. Mol. Gen. Genet. 184:536-538. [DOI] [PubMed] [Google Scholar]
- 131.Pappas, D. L., Jr., and M. Hampsey. 2000. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:8343-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pinto, I., D. E. Ware, and M. Hampsey. 1992. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68:977-988. [DOI] [PubMed] [Google Scholar]
- 133.Pinto, I., W. H. Wu, J. G. Na, and M. Hampsey. 1994. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 269:30569-30573. [PubMed] [Google Scholar]
- 134.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 135.Powell, W., and D. Reines. 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reeder, R. H., and W. H. Lang. 1997. Terminating transcription in eukaryotes: lessons learned from RNA polymerase I. Trends Biochem. Sci. 22:473-477. [DOI] [PubMed] [Google Scholar]
- 137.Reinberg, D., and R. G. Roeder. 1987. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J. Biol. Chem. 262:3331-3337. [PubMed] [Google Scholar]
- 138.Reines, D. 1992. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem. 267:3795-3800. [PMC free article] [PubMed] [Google Scholar]
- 139.Reines, D., M. J. Chamberlin, and C. M. Kane. 1989. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J. Biol. Chem. 264:10799-10809. [PubMed] [Google Scholar]
- 140.Richardson, J. P., and J. Greenblatt. 1996. Control of RNA chain elongation and termination, p. 822-848. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 141.Riles, L., R. J. Shaw, M. Johnston, and D. Reines. 2004. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rivetti, C., S. Codeluppi, G. Dieci, and C. Bustamante. 2003. Visualizing RNA extrusion and DNA wrapping in transcription elongation complexes of bacterial and eukaryotic RNA polymerases. J. Mol. Biol. 326:1413-1426. [DOI] [PubMed] [Google Scholar]
- 143.Rivetti, C., M. Guthold, and C. Bustamante. 1999. Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J. 18:4464-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Robert, F., M. Douziech, D. Forget, J. M. Egly, J. Greenblatt, Z. F. Burton, and B. Coulombe. 1998. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol. Cell 2:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rommele, G., G. Wirz, R. Solf, K. Vosbeck, J. Gruner, and W. Wehrli. 1990. Resistance of Escherichia-Coli to rifampicin and sorangicin-A—a comparison. J. Antibiot. 43:88-91. [DOI] [PubMed] [Google Scholar]
- 146.Rowland, G. C., P. P. Lim, and R. E. Glass. 1995. In vivo cloning of a carboxy-terminal rpoB allele which confers altered transcriptional properties. Folia Microbiol. (Praha) 40:588-594. [DOI] [PubMed] [Google Scholar]
- 147.Rozenfeld, S., and P. Thuriaux. 2001. A genetic look at the active site of RNA polymerase III. EMBO Rep. 2:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sagitov, V., V. Nikiforov, and A. Goldfarb. 1993. Dominant lethal mutations near the 5′ substrate binding site affect RNA polymerase propagation. J. Biol. Chem. 268:2195-2202. [PubMed] [Google Scholar]
- 149.Salomon, R. A., and R. N. Farias. 1992. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Scafe, C., C. Martin, M. Nonet, S. Podos, S. Okamura, and R. A. Young. 1990. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 10:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Scafe, C., M. Nonet, and R. A. Young. 1990. RNA polymerase II mutants defective in transcription of a subset of genes. Mol. Cell. Biol. 10:1010-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Severinov, K., and S. A. Darst. 1997. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl. Acad. Sci. USA 94:13481-13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Severinov, K., D. Fenyo, E. Severinova, A. Mustaev, B. T. Chait, A. Goldfarb, and S. A. Darst. 1994. The sigma subunit conserved region 3 is part of “5′-face” of active center of Escherichia coli RNA polymerase. J. Biol. Chem. 269:20826-20828. [PubMed] [Google Scholar]
- 154.Severinov, K., D. Markov, E. Severinova, V. Nikiforov, R. Landick, S. A. Darst, and A. Goldfarb. 1995. Streptolydigin-resistant mutants in an evolutionarily conserved region of the beta′ subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 270:23926-23929. [DOI] [PubMed] [Google Scholar]
- 155.Severinov, K., A. Mustaev, M. Kashlev, S. Borukhov, V. Nikiforov, and A. Goldfarb. 1992. Dissection of the beta subunit in the Escherichia coli RNA polymerase into domains by proteolytic cleavage. J. Biol. Chem. 267:12813-12819. [PubMed] [Google Scholar]
- 156.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 157.Shaaban, S. A., B. M. Krupp, and B. D. Hall. 1995. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol. Cell. Biol. 15:1467-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shilatifard, A., R. C. Conaway, and J. W. Conaway. 2003. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72:693-715. [DOI] [PubMed] [Google Scholar]
- 159.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 160.Singer, M., D. J. Jin, W. A. Walter, and C. A. Gross. 1993. Genetic evidence for the interaction between cluster I and cluster III rifampicin resistant mutations. J. Mol. Biol. 231:1-5. [DOI] [PubMed] [Google Scholar]
- 161.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 162.Smid, A., M. Finsterer, and I. Grummt. 1992. Limited proteolysis unmasks specific DNA-binding of the murine RNA polymerase I-specific transcription termination factor TTFI. J. Mol. Biol. 227:635-647. [DOI] [PubMed] [Google Scholar]
- 163.Sosunov, V., E. Sosunova, A. Mustaev, I. Bass, V. Nikiforov, and A. Goldfarb. 2003. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 22:2234-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sosunova, E., V. Sosunov, M. Kozlov, V. Nikiforov, A. Goldfarb, and A. Mustaev. 2003. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc. Natl. Acad. Sci. USA 100:15469-15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sparkowski, J., and A. Das. 1992. Simultaneous gain and loss of functions caused by a single amino acid substitution in the beta subunit of Escherichia coli RNA polymerase: suppression of nusA and rho mutations and conditional lethality. Genetics 130:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Steitz, T. A. 1998. A mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 167.Sugaya, K. 2003. Amino acid substitution of the largest subunit of yeast RNA polymerase II: effect of a temperature-sensitive mutation related to G1 cell cycle arrest. Curr. Microbiol. 47:159-162. [DOI] [PubMed] [Google Scholar]
- 168.Sugaya, K., M. Ajimura, H. Tsuji, M. Morimyo, and K. Mita. 1998. Alteration of the largest subunit of RNA polymerase II and its effect on chromosome stability in Schizosaccharomyces pombe. Mol. Gen. Genet. 258:279-287. [DOI] [PubMed] [Google Scholar]
- 169.Sugaya, K., S. Sasanuma, P. R. Cook, and K. Mita. 2001. A mutation in the largest (catalytic) subunit of RNA polymerase II and its relation to the arrest of the cell cycle in G1 phase. Gene 274:77-81. [DOI] [PubMed] [Google Scholar]
- 170.Sujatha, S., A. Ishihama, and D. Chatterji. 2001. Functional complementation between mutations at two distant positions in Escherichia coli RNA polymerase as revealed by second-site suppression. Mol. Gen. Genet. 264:531-538. [DOI] [PubMed] [Google Scholar]
- 171.Sun, Z. W., A. Tessmer, and M. Hampsey. 1996. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 24:2560-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Surratt, C. K., S. C. Milan, and M. J. Chamberlin. 1991. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc. Natl. Acad. Sci. USA 88:7983-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Svetlov, V., D. G. Vassylyev, and I. Artsimovitch. 2004. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 279:38087-38090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tavormina, P. L., R. Landick, and C. A. Gross. 1996. Isolation, purification, and in vitro characterization of recessive-lethal-mutant RNA polymerases from Escherichia coli. J. Bacteriol. 178:5263-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tavormina, P. L., W. S. Reznikoff, and C. A. Gross. 1996. Identifying interacting regions in the beta subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 258:213-223. [DOI] [PubMed] [Google Scholar]
- 176.Temiakov, D., N. Zenkin, M. N. Vassylyeva, A. Perederina, T. H. Tahirov, E. Kashkina, M. Savkina, S. Zorov, V. Nikiforov, N. Igarashi, N. Matsugaki, S. Wakatsuki, K. Severinov, and D. G. Vassylyev. 2005. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell 19:655-666. [DOI] [PubMed] [Google Scholar]
- 177.Thuillier, V., I. Brun, A. Sentenac, and M. Werner. 1996. Mutations in the alpha-amanitin conserved domain of the largest subunit of yeast RNA polymerase III affect pausing, RNA cleavage and transcriptional transitions. EMBO J. 15:618-629. [PMC free article] [PubMed] [Google Scholar]
- 178.Trautinger, B. W., and R. G. Lloyd. 2002. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 21:6944-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Travers, A. A. 1984. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 12:2605-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Treich, I., C. Carles, A. Sentenac, and M. Riva. 1992. Determination of lysine residues affinity labeled in the active site of yeast RNA polymerase II(B) by mutagenesis. Nucleic Acids Res. 20:4721-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Treich, I., M. Riva, and A. Sentenac. 1991. Zinc-binding subunits of yeast RNA polymerases. J. Biol. Chem. 266:21971-21976. [PubMed] [Google Scholar]
- 182.Tuske, S., S. G. Sarafianos, X. Wang, B. Hudson, E. Sineva, J. Mukhopadhyay, J. J. Birktoft, O. Leroy, S. Ismail, A. D. Clark, Jr., C. Dharia, A. Napoli, O. Laptenko, J. Lee, S. Borukhov, R. H. Ebright, and E. Arnold. 2005. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell 122:541-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Umezawa, H., S. Mizuno, H. Yamazaki, and K. Nitta. 1968. Inhibition of DNA-dependent RNA synthesis by rifamycins. J. Antibiot. (Tokyo) 21:234-236. [DOI] [PubMed] [Google Scholar]
- 184.Vaisius, A. C., and T. Wieland. 1982. Formation of a single phosphodiester bond by RNA polymerase B from calf thymus is not inhibited by alpha-amanitin. Biochemistry 21:3097-3101. [DOI] [PubMed] [Google Scholar]
- 185.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 186.Vattanaviboon, P., R. Sukchawalit, P. Jearanaikoon, C. Chuchottaworn, and M. Ponglikitmongkol. 1995. Analysis of RNA polymerase gene mutation in three isolates of rifampicin resistant Mycobacterium tuberculosis. Southeast Asian J. Trop. Med. Public Health 26(Suppl. 1):333-336. [PubMed] [Google Scholar]
- 187.Veldman, G. M., R. C. Brand, J. Klootwijk, and R. Planta. 1980. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 8:2907-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vo, N. V., L. M. Hsu, C. M. Kane, and M. J. Chamberlin. 2003. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry 42:3798-3811. [DOI] [PubMed] [Google Scholar]
- 189.Wang, D., and D. K. Hawley. 1993. Identification of a 3′→5′ exonuclease activity associated with human RNA polymerase II. Proc. Natl. Acad. Sci. USA 90:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Weilbaecher, R., C. Hebron, G. Feng, and R. Landick. 1994. Termination-altering amino acid substitutions in the beta′ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 8:2913-2927. [DOI] [PubMed] [Google Scholar]
- 191.Werner, F., and R. O. Weinzierl. 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10:635-646. [DOI] [PubMed] [Google Scholar]
- 192.Werner, M., S. Hermann-Le Denmat, I. Treich, A. Sentenac, and P. Thuriaux. 1992. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol. Cell. Biol. 12:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.West, M. L., and J. L. Corden. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Westover, K. D., D. A. Bushnell, and R. D. Kornberg. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303:1014-1016. [DOI] [PubMed] [Google Scholar]
- 195.Westover, K. D., D. A. Bushnell, and R. D. Kornberg. 2004. Structural basis of transcription; nucleotide selection by rotation in the RNA polymerase II active center. Cell 119:481-489. [DOI] [PubMed] [Google Scholar]
- 196.Wind, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wittekind, M., J. Dodd, L. Vu, J. M. Kolb, J. M. Buhler, A. Sentenac, and M. Nomura. 1988. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol. Cell. Biol. 8:3997-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Woychik, N. A., and R. A. Young. 1990. RNA polymerase II: subunit structure and function. Trends Biochem. Sci. 15:347-351. [DOI] [PubMed] [Google Scholar]
- 199.Wu, J., D. E. Awrey, A. M. Edwards, J. Archambault, and J. D. Friesen. 1996. In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc. Natl. Acad. Sci. USA 93:11552-11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 201.Xu, J., Y. Tozawa, C. Lai, H. Hayashi, and K. Ochi. 2002. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol. Genet. Genomics 268:179-189. [DOI] [PubMed] [Google Scholar]
- 202.Yang, X., and E. E. Ishiguro. 2003. Temperature-sensitive growth and decreased thermotolerance associated with relA mutations in Escherichia coli. J. Bacteriol. 185:5765-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yano, R., and M. Nomura. 1991. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol. Cell. Biol. 11:754-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yuzenkova, J., M. Delgado, S. Nechaev, D. Savalia, V. Epshtein, I. Artsimovitch, R. A. Mooney, R. Landick, R. N. Farias, R. Salomon, and K. Severinov. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin j25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]
- 205.Zaychikov, E., E. Martin, L. Denissova, M. Kozlov, V. Markovtsov, M. Kashlev, H. Heumann, V. Nikiforov, A. Goldfarb, and A. Mustaev. 1996. Mapping of catalytic residues in the RNA polymerase active center. Science 273:107-109. [DOI] [PubMed] [Google Scholar]
- 206.Zhang, G., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98:811-824. [DOI] [PubMed] [Google Scholar]
- 207.Zhou, Y. N., and D. J. Jin. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like stringent RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2908-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.