Abstract
Lactic acid bacteria (LAB) employ sucrase-type enzymes to convert sucrose into homopolysaccharides consisting of either glucosyl units (glucans) or fructosyl units (fructans). The enzymes involved are labeled glucansucrases (GS) and fructansucrases (FS), respectively. The available molecular, biochemical, and structural information on sucrase genes and enzymes from various LAB and their fructan and α-glucan products is reviewed. The GS and FS enzymes are both glycoside hydrolase enzymes that act on the same substrate (sucrose) and catalyze (retaining) transglycosylation reactions that result in polysaccharide formation, but they possess completely different protein structures. GS enzymes (family GH70) are large multidomain proteins that occur exclusively in LAB. Their catalytic domain displays clear secondary-structure similarity with α-amylase enzymes (family GH13), with a predicted permuted (β/α)8 barrel structure for which detailed structural and mechanistic information is available. Emphasis now is on identification of residues and regions important for GS enzyme activity and product specificity (synthesis of α-glucans differing in glycosidic linkage type, degree and type of branching, glucan molecular mass, and solubility). FS enzymes (family GH68) occur in both gram-negative and gram-positive bacteria and synthesize β-fructan polymers with either β-(2→6) (inulin) or β-(2→1) (levan) glycosidic bonds. Recently, the first high-resolution three-dimensional structures have become available for FS (levansucrase) proteins, revealing a rare five-bladed β-propeller structure with a deep, negatively charged central pocket. Although these structures have provided detailed mechanistic insights, the structural features in FS enzymes dictating the synthesis of either β-(2→6) or β-(2→1) linkages, degree and type of branching, and fructan molecular mass remain to be identified.
INTRODUCTION
Extracellular polysaccharides (exopolysaccharides) (EPS) are commonly found in bacteria and microalgae and less frequently in yeasts and fungi (39, 142, 160, 168, 217). Several lactic acid bacteria (LAB), including species of Lactobacillus, are known to produce EPS. Depending on their composition and mechanism of biosynthesis, EPS are divided in two classes: heteropolysaccharides and homopolysaccharides. Heteropolysaccharides consist of multiple sugar types and are synthesized by the combined action of multiple different types of glycosyltransferase enzymes (39). In contrast, homopolysaccharides are synthesized from the sole substrate sucrose by the action of one sucrase enzyme. Sucrase-type enzymes synthesize polysaccharides consisting of either glucose sugar residues (glucans) or fructose residues (fructans).
Homopolysaccharide synthesis in LAB has mainly been studied in oral streptococci and Leuconostoc spp. (124, 126, 149, 153). Because of their clearly established role in formation of dental caries (7) Streptococcus mutans and Streptococcus sanguis strains have been subject to a number of studies (18, 100, 109, 157, 159). Interestingly, there is increasing evidence that a number of Lactobacillus species are also associated with advanced stages of dental caries (26). Both glucans and fructans (see below) formed by oral streptococci (and lactobacilli) apparently have major influences on the formation of dental plaque. They are involved in adherence of bacteria to each other and to the tooth surface, modulating diffusion of substances through plaque, and occasionally serving as extracellular energy reserves (29, 41, 141, 162). Alternatively, these polymers may protect microbial cells against desiccation, phagocytosis and phage attack, antibiotics or toxic compounds, predation by protozoans, and osmotic stress (20).
In general, glucans and/or fructans can be used as viscosifying, stabilizing, emulsifying, sweetening, gelling, or water-binding agents, in the food as well as in the nonfood industries (40, 51, 66, 190, 217, 218). Certain oligosaccharides (e.g., fructo-oligosaccharides, isomaltooligosaccharides, and lactulose) and polysaccharides (e.g., fructans) are used as prebiotic food additives (14, 15, 50, 84, 151, 164). Additionally, oligosaccharides containing α-(1→2) glucosidic bonds are in some cases used as feed additives (127).
Over the years a large number of glucansucrase and fructansucrase genes and enzymes have been identified by cloning, reverse genetics, and various enzyme activity assays. Enzymes synthesizing α-glucan polymers, glucansucrases (GS), are limited to LAB while enzymes synthesizing fructans, fructansucrases (FS), are present in gram-positive and gram-negative bacteria (33; http://afmb.cnrs-mrs.fr/CAZY/). Fructan biosynthesis also is known to occur in plants and fungi and involves a set of enzymes which are evolutionarily related to sucrose-hydrolyzing enzymes (invertases). They are clearly different from their bacterial counterparts (75, 106, 205, 216). Although the GS and FS enzymes perform very similar reactions on the same substrates (see below), they do not share a high amino acid sequence similarity, and differ strongly in protein structures.
The properties of GS of Streptococcus and Leuconostoc spp. (124, 126, 149, 154) and FS of LAB (126) have been reviewed previously. In view of the many recent developments in the understanding of the structure-function relationships of these sucrase enzymes, including GS and FS enzymes from lactobacilli, an overview of current knowledge of the sucrase field of research is presented here, with a focus on sucrase enzymes from LAB.
NOMENCLATURE AND CLASSIFICATION OF SUCRASE ENZYMES
According to the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology, the following GS enzymes are classified based on the reaction catalyzed and the product specificity: dextransucrase (sucrose:1,6-α-d-glucan-6-α-d-glucosyltransferase, EC 2.4.1.5) and alternansucrase [sucrose:1,6(1,3)-α-d-glucan-6(3)-α-d-glucosyltransferase, EC 2.4.1.140]. At present, the mutan-(sucrose:1,3-α-d-glucan-3-α-d-glucosyltransferase) and reuteransucrase [sucrose:1,4(6)-α-d-glucan-4(6)-α-d-glucosyltransferase] enzymes mentioned are classified together with dextransucrase enzymes in EC 2.4.1.5. Also, two FS enzymes are distinguished now, based on the different products synthesized: inulosucrase (sucrose:2,1-β-d-fructan-1-β-d-fructosyltransferase, EC 2.4.1.9) and levansucrase (sucrose:2,6-β-d-fructan-6-β-d-fructosyltransferase, EC 2.4.1.10).
As described above, glucan- and fructan-synthesizing enzymes have been referred to as glucansucrase/glucosyltransferase and fructansucrase/fructosyltransferase, respectively (33; http://afmb.cnrs-mrs.fr/CAZY/). Since glucosyltransferase and fructosyltransferase refer to enzyme activities that are widely found in nature for enzymes that catalyze the transfer of, for instance, glucosyl and fructosyl sugar units, they are not descriptive for the substrate and product specificity of sucrase-type enzymes. Therefore, in this review we only use glucansucrases and fructansucrases for enzymes synthesizing homopolysaccharides from sucrose.
In another classification system, glycoside hydrolase (GH) enzymes have been divided into 100 different families based on their amino acid sequences (33; http://afmb.cnrs-mrs.fr/CAZY/). In view of their (different) sequence similarities, GS and FS have been included in the families GH70 and GH68, respectively. Evolutionarily, structurally, and mechanistically related families are further grouped into clans. Enzymes from the families GH32 (consisting mainly of plant and fungal fructosyltransferases) and GH68 comprise clan GH-J. The members of clan GH-J possess a five-bladed β-propeller structure (33; http://afmb.cnrs-mrs.fr/CAZY/), with three identical catalytic (Asp, Glu, and Asp) residues, and use a retaining reaction mechanism. Enzymes from families GH13 (mainly starch modifying enzymes) and GH70 and GH77 (4-α-glucanotransferases) constitute clan GH-H (also known as α-amylase superfamily) containing an α-amylase-type catalytic (β/α)8-barrel, a catalytic machinery with the catalytic Asp, Glu, and Asp residues at strands β4, β5, and β7, respectively, and a retaining reaction mechanism (see below).
The nomenclature for enzymes that synthesize polymers containing predominantly one linkage type is relatively straightforward, for instance, dextransucrase and levansucrase. The nomenclature for an enzyme synthesizing comparable numbers of different glycosidic linkages is a dilemma, since it could be assigned to multiple classes. An example is the GtfL enzyme from Streptococcus salivarius ATCC 25975 (Table 1). Similar problems might arise for FS, with several enzymes already synthesizing both β-(2→6) and β-(2→1) glycosidic bonds (Table 2). This nomenclature problem is the more pressing since recent GS enzyme engineering work has revealed that a limited number of amino acid substitutions may cause large changes in glucosidic linkages in products synthesized by (mutant) sucrase enzymes (96) (see below). In view of their similar structures and reaction mechanisms, we propose that sucrase enzymes synthesizing multiple glycosidic linkages in their products in future be labeled glucansucrases (GS) and fructansucrases (FS), respectively.
TABLE 1.
Linkage type distribution in glucans synthesized by GS enzymes from LAB produced in recombinant hosts
| Glucan and strain | Glucan linkage type distributionb | Method useda | Glucan size (Da) | Gene | Protein size (residues) | Reference(s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reuteran | Terminal | α-(1→4) | α-(1→6) | α-(1→4,6) | ||||||||
| L. reuteri 121 | 11 | 46 | 26 | 17 | Met/1H | 80 × 106 | gtfA | 1,781 | 97 | |||
| L. reuteri ATCC 55730 | 9 | 69 | 11 | 13 | Met/1H | 45 × 106 | gtfO | 1,781 | 94 | |||
| Dextran | Terminal | α-(1→3) | α-(1→6) | α-(1→3,6) | ||||||||
| L. reuteri 180 | 10 | 26 | 51 | 13 | Met | gtf180 | 1,772 | 95 | ||||
| L. sakei Kg15 | 4 | 9 | 86 | 9 | Met | gtfKg15 | 1,561 | 95 | ||||
| L. fermentum Kg3 | 3 | 7 | 89 | 7 | Met | gtfKg3 | 1,595 | 95 | ||||
| L. parabuchneri 33 | 6 | 9 | 75 | 9 | Met | gtf33 | 1,463 | 95 | ||||
| L. mesenteroides NRRL B-1299 | 15 | 85 | 13C | dsrA | 1,290 | 125 | ||||||
| L. mesenteroides NRRL B-1299 | 5 | 95 | 13C | dsrB | 1,508 | 118 | ||||||
| L. mesenteroides NRRL B-512F | 5 | 95 | 13C | dsrS | 1,527 | 119, 156 | ||||||
| L. mesenteroides NRRL B-512F | 40 | 50 | 13C | dsrT5 | 1,499 | 55, 56 | ||||||
| L. mesenteroides Lcc4 | Mainly | 13C | dsrD | 1,527 | 135 | |||||||
| S. mutans GS-5 | 15 | 70 | 15 | Met | gtfD | 1,430 | 69, 76, 173 | |||||
| S. downei MFe28 | 90 | 13C | gtfS | 1,365 | 61 | |||||||
| S. sobrinus OMZ176 | 6 | 16 | 73 | 5 | Met | gtfT | 1,468 | 68 | ||||
| S. salivarius ATCC 25975 | 100 | 13C | gtfK | 1,599 | 179 | |||||||
| S. salivarius ATCC 25975 | 95 | 13C | gtfM | 1,577 | 179 | |||||||
| S. gordonii CH1 | 25 | 75 | 13C | gtfG | 1,577 | 64, 214, 215 | ||||||
| Mutan | Terminal | α-(1→3) | α-(1→6) | α-(1→3,6) | ||||||||
| L. reuteri ML1 | 18 | 47 | 26 | 13 | Met | gtfML1 | 1,772 | 95 | ||||
| S. mutans GS-5 | 88 | 7 | 5 | Met | gtfB | 1,475 | 54, 175 | |||||
| S. mutans GS-5 | 86 | 8 | 7 | Met | gtfC | 1,375 | 54, 203 | |||||
| S. downei MFe28 | 0.5 | 88 | 2 | 0.5 | 13C | gtfI | 1,597 | 163 | ||||
| S. sobrinus 6715 | Mainly | 13C | gtfIa | 1,592 | 90 | |||||||
| S. salivarius ATCC 25975 | 90 | 13C | gtfJ | 1,517 | 179 | |||||||
| Glucan | Terminal | α-(1→6) | α-(1→3,6) | |||||||||
| S. sobrinus B13N | 44 | 25 | 31 | 13C | gtfU | 1,554 | 67 | |||||
| L. mesenteroides NRRL B-1355 | Alternating α-(1→3) and α-(1→6) linkages (alternan)
|
Met/13C | asr | 2,057 | 4, 32 | |||||||
| L. mesenteroides NRRL B-1299 | α-(1→2,6) and α-(1→3,6) linkages
|
Met/13C | dsrE | 2,835 | 17, 170 | |||||||
| S. salivarius ATCC 25975 | Equal numbers of α-(1→3) and α-(1→6) linkages | 13C | gtfL | 1,449 | 179 | |||||||
Indicates method used in linkage type analysis: Met, methylation; 13C, 13C-NMR; 1H, 1H-NMR.
Boldface type indicates main linkage.
TABLE 2.
Characteristics of FS enzymes of LAB and their fructan products
| Fructan and strain | Fructan linkage type distributionc | Method useda | Fructan size (Da) | Oligonucleotideb | Gene | Protein size (residues) | Reference(s) | ||
|---|---|---|---|---|---|---|---|---|---|
| Levan | β-(2→6) | β-(2→1) | β-(2→1→6) | ||||||
| L. reuteri 121 | 98 | 2 | 13C/Met | 97% 2 × 104; 3% 3 × 106 | None | lev | 804 | 210, 211 | |
| L. sanfranciscensis | Levan | Enzymic digest (unpublished) | K, N | ftfA | 879 | 92, 93, 199 | |||
| L. mesenteroides B-512 FMC | Levan | 13C/TLC | K, N, P | mlft | 424 | 81 | |||
| S. sobrinus OMZ176 | Levan | TLC | 30 | ||||||
| S. salivarius ATCC 13419 | Levan | 20 × 106 | 136 | ||||||
| S. salivarius ATCC 25975 | 70 | 30 | TLC/Met | (20-100) × 106 | None | ftf | 969 | 45, 185, 186 | |
| S. salivarius HHT | 90 | 10 | Met | 45 | |||||
| S. salivarius 51 | 86 | 14 | Met | None | 70 | ||||
| S. salivarius SS2 | 70 | 30 | Met | 178 | |||||
| Inulin | β-(2→6) | β-(2→1) | β-(2→1→6) | ||||||
| L. reuteri 121 | 93 | 7 | 13C/Met | >107 | K, N | inu | 798 | 213 | |
| L. citreum CW28 | Mainly | 13C | islA | 1,490 | 137, 138 | ||||
| S. criceti AHT | Inulin | Met | 45 | ||||||
| S. ratti BHT | 90 | 10 | Met | 45 | |||||
| S. mutans GS-5 | Inulin | Concavalin A | (60-90) × 106 | None | ftf | 795 | 74, 166, 174 | ||
| S. mutans Ingbritt | Inulin | Mouse-AB | 2 | ||||||
| S. mutans JC1 | 95 | 5 | Met | 45 | |||||
| S. mutans JC2 | 94 | 6 | >2 × 106 | 19, 158 | |||||
| Fructan | |||||||||
| L. frumenti (5 strains) | Fructan | >106 | 198 | ||||||
| L. pontis 1.1115 and 1.675 | Fructan | >106 | 198 | ||||||
| L. panis 1.649 | Fructan | >106 | 198 | ||||||
| W. confusa 1.934 | Fructan | >106 | 198 | ||||||
Method used in linkage analysis: Met, methylation analysis; TLC, thin-layer chromatography with inulin and levan standards; concavalin A, lectin-type fructan binding protein specific for inulin; mouse-AB, mouse antibodies against inulin structures; 13C, 13C-NMR.
K, 1-kestose; N, 1,1-nystose; P, 1,1,1-kestopentaose.
Boldface type indicates main linkage.
GLUCANSUCRASES
Microbial glucansucrase enzymes (GH70) exclusively synthesize α-glucan polymers. Synthesis of these polymers has been observed in four different genera of LAB: Streptococcus, Leuconostoc, Weissella, and Lactobacillus (95, 98, 124, 176, 197, 198, 209).
GS are large, extracellular proteins with average molecular masses of 160,000 Da. A large number of GS genes have been identified; for an overview consult the CAZY website (http://afmb.cnrs-mrs.fr/CAZY/). With the release of newly sequenced LAB genomes the list of known or putative LAB GS genes requires updates. An example is a putative GS gene similar to gtfD from S. mutans GS-5 present in the genome sequence of Oenococcus oeni (http://genome.jgi-psf.org/mic_home.html). It should be emphasized that GS genes and enzymes have not been reported outside LAB. The reason for this limited distribution of GS genes and enzymes in LAB is unknown.
GS enzymes synthesize various glucans differing in the type of glucosidic linkages, degree and type of branching, length of the glucan chains, molecular mass, and conformation of the polymers. All these properties strongly contribute to specific polysaccharide properties such as solubility, rheology, and other physical characteristics (124). Many factors, including growth medium, temperature, incubation time, sucrose concentration used, and the presence of polysaccharide-degrading enzymes influence the molecular mass, structure, and physical characteristics of the polymers synthesized by a specific organism (85, 124).
Nevertheless, the information for glucosidic bond specificity (and other characteristics) must be encoded somewhere in the enzyme amino acid sequence and protein structure. Cracking the code for these product specificities is one of the clear challenges in GS enzyme research (see below).
Reactions Catalyzed and Glucan Product Synthesis
GS enzymes cleave the glycosidic bond of their substrate sucrose and couple a glucose unit to a growing glucan (polyglucose) chain (transglucosylation), water (hydrolysis), or other acceptor substrate (acceptor reaction). The energy released by cleavage of the energy-rich glycosidic bond in sucrose is used for synthesis of new glucosidic bonds. Dextran is synthesized according to the following reaction:
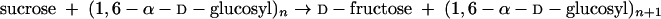 |
There are many examples of GS enzymes that synthesize linear glucans with α-(1→6) glucosidic linkages (dextran). GS enzymes introducing other glucosidic linkages are discussed below. There are also many examples of GS enzymes that catalyze the formation of two different glucosidic linkages. This results either in a (highly) branched glucan product, based on a branching type of reaction, or in a linear glucan with two different (alternating) glucosidic bonds. Unfortunately, detailed structural information for both the GS proteins and their glucan products is still lacking.
Glucan synthesis.
Depending on the main glucosidic linkages present in their glucan, four different types of α-glucans synthesized by LAB are recognized: dextran, mutan, alternan, and reuteran.
Pasteur (143) discovered the microbial origin of the jellification of cane sugar syrups. The product causing the jellification was named dextran due to its positive rotatory power. The corresponding extracellular enzyme was named dextransucrase (73). A common feature of all dextrans is the abundance of α-(1→6) linkages with some branching points at position 2, 3, or 4. Dextrans are produced by, for instance, Leuconostoc mesenteroides strains (20).
Guggenheim (63) showed that water-insoluble glucan from S. mutans OMZ176 contained a high proportion (up to 90%) of α-(1→3) glucosidic linkages. He proposed the name mutan for this polymer. The corresponding GS enzyme became named accordingly as mutansucrase. Mutan polymers are mainly produced by various streptococci (65).
Côté and Robyt isolated an α-glucan polymer from L. mesenteroides NRRL B-1355 composed of alternating α-(1→6) and α-(1→3) glucosidic linkages. To distinguish between the dextran [95% α-(1→6)] also synthesized by this strain, they named this polymer alternan and the corresponding GS enzyme alternansucrase (32).
Recently, a new type of glucan was identified from Lactobacillus reuteri 121 containing mainly α-(1→4) glucosidic linkages (208). The glucan product was named reuteran and the corresponding GS enzyme reuteransucrase (99). Reuteran, synthesized by L. reuteri 121, is a glucan with α-(1→4), α-(1→6) glucosidic bonds and α-(1→4,6) branching points (97).
Finally, an α-glucan containing, besides large numbers of α-(1→6) linkages, a substantial number of α-(1→2) linkages is produced by two different L. mesenteroides strains: NRRL B-1299 and a mutant strain (R510) of NRRL B-1355 (17, 184).
A few LAB strains carry multiple GS genes (gtf) in their genome sequence (Table 1). In order to elucidate which enzyme activities are encoded, these genes were cloned and heterologously expressed. The distribution of glucosidic linkages has been elucidated for the glucans synthesized from sucrose by heterologously produced (mostly using Escherichia coli as the host) GS enzymes from (i) seven Streptococcus strains (13 GS enzymes, synthesizing either dextran or mutan polymers) (67, 90, 124), (ii) four Leuconostoc strains (seven GS enzymes, synthesizing mainly dextran polymers, but also one alternan, and a GS synthesizing large numbers of α-(1→2,6) branch points) (4, 17, 55, 124, 135), and (iii) seven Lactobacillus strains (seven GS enzymes, synthesizing mainly dextrans, but also reuteran, and a highly branched mutan) (94, 95, 97) (Table 1).
Acceptor reaction.
Koepsell et al. (89) observed that in the presence of sucrose and saccharide acceptor substrates such as maltose, isomaltose, and O-α-methylglucoside, GS enzymes shift from glucan synthesis towards the synthesis of oligosaccharides (the acceptor reaction). Most acceptor reaction studies have been performed using saccharides (5, 42, 43, 53, 94, 99) or saccharide derivatives as substrates (31, 36, 150). However, aromatic compounds (e.g., catechine) and salicyl alcohol have also been shown to act as acceptor substrates (114, 221). GS enzymes cannot use sucrose itself as an acceptor substrate.
Where studied, glucosidic bond specificity observed in polymers is also retained in the oligosaccharides synthesized by these GS enzymes in their acceptor reaction (32, 42, 99).
Structural and Functional Organization of Glucansucrase Enzymes
Based on the deduced amino acid sequences, GS enzymes are composed of four distinct structural domains from the N to the C terminus (Fig. 1): (i) a signal peptide, (ii) an N-terminal stretch of highly variable amino acids, (iii) a highly conserved catalytic and/or sucrose binding domain of about 1,000 amino acids, and (iv) a C-terminal domain that is composed of a series of tandem repeats which is thought to be involved in glucan binding (61, 161). Characterization of several heterologously produced and purified GS enzymes has clearly shown that they are able to introduce multiple glucosidic bonds in their products, (largely) based on the single catalytic domain present (Table 1). Where studied, this was corroborated by the the composition of the polymer synthesized by the LAB host (Table 1). An exception to this rule is the GS from L. mesenteroides NRRL B-1299 (DSRE), which carries an additional C-terminally located catalytic domain (17) (Table 1). Evidence has been provided that this additional C-terminal domain is responsible for introduction of the branch points in the product synthesized (47).
FIG. 1.
Schematic representation of GS from LAB. The deduced amino acid sequence of the L. reuteri 121 glucansucrase was used as the template (97). The four different regions shown are (i) the N-terminal signal sequence; (ii) the N-terminal variable region; (iii) the catalytic core; and (iv) the C-terminal GBD. Alignments (SequenceLogo, http://weblogo.berkeley.edu/) are shown of short regions in GS enzymes (GH70) with conserved amino acid residues for which mutant information is available in the literature. The GS protein sequences used are listed in Table 1. The four conserved regions (I to IV) first identified in members of the α-amylase family (GH13) (191) which can also be found in GS enzymes (family GH70) are indicated (see also Fig. 2). The seven residues that are fully conserved in family GH13 are also present and fully conserved in the GS family (GH70), except the histidine residue in region I which is present in virtually all family GH13 enzymes but is replaced by a conserved glutamine (Gln1514) in all GS enzymes (110, 121). The seven conserved residues are indicated with arrows, and black arrows indicate the three catalytic residues. (A) Tyr residues, at positions 169 to 172 in GTFB of S. mutans GS5; mutation of these residues into Ala only changed the adhesiveness of the glucan products (202). (B) Thr344Leu, Glu349Leu, and His355Val of GTF-I, causing drops in enzyme activities of 30-, 4-, and 7-fold, respectively (122). (C) Asp511Asn and Asp513Asn, Asp411 and Asp413, and Asp437 and Asp439 of DSRS from L. mesenteroides NRRL B-512F and GTFB and GTFC from S. mutans GS-5, respectively, resulting in complete loss or strongly decreased activities (28, 119). GTFB Val412Ile and GTFC Val438Ile, resulting in enhanced insoluble glucan synthesis of about 10 to 20%, whereas soluble glucan synthesis by these enzymes was significantly lower than for the wild type (28), and GTFB (Glu422Gln) and GTFC (Glu448Gln), resulting in 40% reduced glucan synthesizing activity (28). (D) Catalytic nucleophile in region II, Asp415Asn, and Asp1024Asn (resulting in complete loss of enzyme activity) of GTFI from S. mutans (38) and GTFA from L. reuteri 121 (99). (E) Acid/base catalyst in region III, Glu453Gln, and Glu1061Gln (resulting in complete loss of enzyme activity) of GTFI (38) and GTFA (99), respectively. (F) Transition state stabilizer in region IV, Asp526Asn and Asp1133Asn (resulting in complete loss of enzyme activity) of GTFI (38) and GTFA (99). Other important residues targeted in regions 2D to 2F are discussed in detail in the text. (G) Gln937His in GTFI of Streptococcus downei, resulting in drastic but not complete loss of activity (121).
Signal peptide and N-terminal variable domain.
All GS of LAB are extracellular enzymes and the N termini of these enzymes contain a signal peptide for protein secretion (36 to 40 amino acids) (Fig. 1). The stretch of amino acids between the signal peptide and the core region of GS is highly variable both in composition and in length (200 to 700 amino acids).
Different repeating units have been identified in the variable domains of GS enzymes from various LAB organisms (17, 56, 79, 95, 97) (Table 1). The functions of the N-terminal variable domain (and repeats) have remained unclear. Deletion studies of the complete N-terminal variable domain in GTFI of Streptococcus downei MFe28 demonstrated that it does not play a significant role in glucan structure determination (116). Additional N-terminal deletions resulted in drastic loss of enzyme activity, also indicating that the N-terminal part of the catalytic core had been deleted (122). The relatively large N-terminal variable domain of GTFA of L. reuteri 121 is important for activity with sucrose, but its deletion has only minor effects on glucan product characteristics (99).
Catalytic domain.
Detailed structural data for GS proteins of family GH70 is lacking at present. However, secondary-structure prediction studies of the catalytic domain, corroborated by circular dichroism experiments, have shown that GS proteins possess a (β/α)8 barrel structure as found in glycosidases from family GH13, e.g., with α-amylase, cyclodextrin glucanotransferase (CGTase) and amylosucrase (38, 111, 117). The (β/α)8-barrel structure motif of GS is presumably circularly permuted and is characterized by the presence of eight β-sheets (E1 to E8) located in the core of the protein, alternating with eight α-helices (H1 to H8) located at the surface of the protein (Fig. 2) (111).
FIG. 2.
Topology diagrams of members of α-amylase family GH13 (A) and GS proteins of family GH70 (B). The catalytic domain of α-amylases has a (β/α)8 barrel structure, starting with β-strand 1 and ending with α-helix 8. The B domain is located between β-strand 3 and α-helix 3. GS have a putative circularly permuted (β/α)8 barrel structure (111), which starts with α-helix 3 (α-amylase numbering) and ends with β-strand 3. Between α-helix 8 and β-strand 1, a large stretch of unknown function is located. The locations of the four conserved regions (I to IV) in family GH13 (and family GH70) are indicated with dashed boxes. Amylosucrase has a domain loop (B′ domain; important for polymerizing activity; indicated with a dashed line and circle) consisting of approximately 60 amino acid residues which is located after β-strand 7 (182, 183), immediately after the two catalytically important His392 and Asp393 residues located in conserved region IV (165) (Fig. 1). GS proteins also contain an additional “loop” (about 45 amino acids; indicated with a dashed line) compared to α-amylase enzymes, which is located between β-strand 7 and α-helix 7. Conceivably, this loop is also important for polymerizing activity. The approximate sites of the three catalytic residues (D, E, and D) are indicated. B, B domain; C, C domain; GBD, glucan binding domain; VR, variable region. (Adapted from reference 96 with permission of the publisher. Copyright 2005 American Chemical Society.)
The four conserved regions (I to IV) of amino acids identified in the members of family GH13 (191) are also present in GS proteins. However, as a consequence of the circular permutation, region I in GS enzymes is located C-terminally of regions II to IV (Fig. 1 and 2). The seven amino acid residues that are fully conserved in family GH13 are also present and fully conserved in the GH70 family, except His122 (Taka-amylase A numbering), which is replaced by Gln in all GS enzymes (110) (Gln1514, GTFA L. reuteri 121 numbering; Fig. 1 and 2) (111, 121).
(i) Catalytic residues.
Since GS enzymes and the enzymes of family GH13 share a related structural fold and a similar catalytic triad of amino acid residues (see below), the wealth of structure-function information uncovered for family GH13 enzymes may serve as a template to deepen our understanding of the mode of action of GS enzymes. Most members of family GH13 act on starch, e.g., α-amylase and CGTase. Whereas α-amylases generally hydrolyze α-(1→4) glucosidic bonds, CGTases mainly catalyze transglycosylation reactions, synthesizing unique circular α-(1→4)-linked oligosaccharides from starch (cyclodextrins) (146). Amylosucrase is the only enzyme from family GH13 that uses sucrose as a substrate to synthesize α-glucan polymers containing α-(1→4) glucosidic linkages (3, 182). For family GH13 enzymes, high-resolution three-dimensional structures are available, often in complex with substrates, inhibitors, or products (207). The three-dimensional structures of amylosucrase (also in complex with sucrose, oligosaccharides, and with a covalently bound glucopyranosyl moiety) resemble those of other proteins of family GH13 (80, 115, 181, 183).
Sequence alignments, mutagenesis studies, and three-dimensional structure analyses within family GH13 have resulted in identification of the amino acid residues involved in catalysis (204). Enzymes of family GH13 contain three amino acids with Bacillus carboxyl groups crucial for catalysis, Asp229/Asp286, Glu257/Glu328, and Asp328/Asp393 (CGTase of B. circulans 251 numbering and amylosucrase of Neisseria polysaccharea numbering) present at or near C-terminal β-strands 4, 5, and 7 (using the structure element numbering of family GH13 proteins) (Fig. 1 and 2) (204). Three corresponding residues, present in similar locations in the secondary structure, are also essential for activity in family GH70 (GS) enzymes (38, 99).
Within both families GH13 and GH70 Asp1024 (catalytic nucleophile, GTFA L. reuteri 121 numbering) (Fig. 1D) is involved in the formation of the covalent glucosyl-enzyme complex (80, 111, 128, 129, 204). Its mutagenesis in family GH13 and family GH70 enzymes resulted in drastic loss of enzyme activity (38, 83, 88, 99, 119, 165). Site-directed mutagenesis studies in family GH13 and family GH70 enzymes have also provided evidence for the crucial roles of the other two invariable residues in catalysis (acid/base catalyst, Glu1061) (Fig. 1E) and, transition state stabilizer, Asp1133 (Fig. 1F) (GTFA Lactobacillus reuteri 121 numbering) (Fig. 1 and 2) (38, 88, 99, 111, 165).
(ii) Catalytic site.
Besides these three catalytic residues, the active site of members of the α-amylase superfamily contains two His amino acid residues important for activity and proposed to be involved in stabilization of the transition state (111, 131, 165). One of these His residues (His327 in B. circulans 251 CGTase and His392 in N. polysaccharea amylosucrase) is also conserved in GS enzymes (His1132 in L. reuteri 121 GTFA; Fig. 1F). Its mutation (His661Arg in L. mesenteroides NRRL B-512F DSRS and His561Gly in S. mutans GS-5 GTFB) resulted in very low residual enzyme activities (119, 202). This residue thus may play a similar role in GS enzymes.
The other His residue (His140 in B. circulans 251 CGTase and His187 in N. polysaccharea amylosucrase), involved in transition state stabilization (131, 165), is replaced by Gln in all GS enzymes known (Gln1514 in L. reuteri 121 GTFA; Fig. 1G) (111, 121). Its mutation, Gln937His, in GTFI of S. downei MFe28 resulted in drastic but not complete loss of activity (121). The authors concluded that Gln937 plays no direct role in cleavage of sucrose and in formation of the covalent glucosyl-enzyme intermediate, but may be important for transition state stabilization (121). Mutagenesis of Gln937Asn in GTFI also resulted in reduced activity and modified distribution of oligodextran and nigero-oligosaccharide products (121). Analogously, in CGTase, mutation of the corresponding His residue (His140) resulted in lower activity and altered product formation (131).
No GS enzyme mutagenesis data are available for the other two of the seven fully conserved residues, Arg1022 (Fig. 1D) and Asp1509 (Fig. 1G). However, mutagenesis of the corresponding residues in CGTase (Arg227 and Asp135, CGTase B. circulans 251 numbering) resulted in drastically decreased activities (103). Analysis of mutants in these conserved GS amino acid residues is required to elucidate their true function.
(iii) Amino acid substitutions.
The structural similarities between GS and the enzymes of family GH13 (see above) allow rational identification of regions important for enzyme activity and product specificity. Based on a comparison of sugar-binding acceptor subsites in family GH13 enzymes (111), the locations of three regions putatively involved in acceptor substrate binding in GS enzymes were identified. These were C-terminal of the catalytic residues Asp1024 (GTFA numbering, region II, Fig. 1D) and Glu1061 (region III, Fig. 1E). A third acceptor substrate binding region was identified on the basis of mutagenesis studies with different GS enzymes, involving amino acid residues 1134, 1135, 1136, 1138, and 1142 (GTFA L. reuteri 121 numbering). This region is located C-terminal of the catalytic residue Asp1133 (region IV, Fig. 1F), determining the solubility of the glucan products and the ratio of α-(1→3) versus α-(1→6) and α-(1→4) versus α-(1→6) glucosidic linkages in the polymer (96, 123, 149, 173).
For CGTase, amylosucrase, neopullulanase, acarviosyltransferase, and the branching enzyme of family GH13 (3, 13, 96, 101, 102, 104, 105, 206) and GS enzymes of family GH70, these regions have been proven to be important for product formation, giving further insights into the structural relatedness of GS and family GH13 enzymes (see below).
The effects of different amino acid substitutions in the (putative) acceptor substrate binding regions and other conserved regions in the catalytic core of LAB GS enzymes have been investigated in the past years. In many cases the mutant enzyme products and activities have been thoroughly characterized. Below, we present a structured summary of the effects of these mutations.
(a) Affecting glucosidic linkage type. Site-directed mutagenesis allowed identification of several GS regions influencing glucosidic linkage specificity in glucan and oligosaccharide products, apart from Gln937 (see above). Below, the results for a number of mutations done in amino acids in regions II and IV, by analogy to family GH13 most likely representing acceptor substrate binding subsites, are summarized (Fig. 1). Derivatives of the L. reuteri 121 GTFA protein containing mutation Pro1026Val (Fig. 1D) showed a clear change in the oligosaccharide and glucan products synthesized (96). Mutations in the tripeptide immediately following Asp1133, the putative transition state-stabilizing residue in GTFA of L. reuteri 121 (Asn1134Ser:Asn1135Glu:Ser1136Val; Fig. 1F), resulted in a drastic increase in α-(1→6) glucosidic linkages (∼40%) and a drastic decrease in α-(1→4) linkages (∼40%) in the polymer synthesized compared to the wild type (96). Also, a quintuple mutant was constructed, by combination of this triple amino acid mutant (Asn1134Ser:Asn1135Glu:Ser1136Val) with a double mutant (Pro1026Val:Ile1029Val) located in and near region II (Fig. 1D), resulting in an even further enhanced ratio of α-(1→6) to α-(1→4) glucosidic linkages in its glucan (and also oligosaccharide) products (96). Mutation Thr667Arg (Fig. 1F) in DSRS of L. mesenteroides NRRL B-512F resulted in 8% more α-(1→3) linkages in the dextran product (149).
(b) Affecting glucan solubility. Besides the glucosidic linkage type, polymers may be classified based on their solubility properties. This characteristic is undoubtly related to the structure of the polymers (linkage type, branching, and size) and thus reflects intrinsic enzyme properties.
In different GS enzymes, mutations in the residue located five amino acids behind the catalytic Asp1133 (Asp1138 GTFA of L. reuteri 121 numbering and equivalent to Thr667Arg in DSRS of L. mesenteroides NRRL B-512F) resulted in a shift in glucan solubility (Fig. 1F). Mutation Thr589Glu in GTFD of S. mutans GS-5 lowered the amount of soluble glucan synthesized from 86 to 2%, and the number of α-(1→3) linkages in the insoluble mutan product synthesized also decreased from 76 to 38% (173). The reverse shift, from a completely insoluble glucan to more soluble glucan synthesis, was observed when the similar amino acid residue was mutated in GTFB (mutant Asp567Thr) of S. mutans GS-5 (increase in soluble glucan from 0 to 24%) and in GTFI (mutant Asp569Thr) of S. downei MFe28, analysis of the soluble fraction showed production of an α-(1→6)-linked glucan (123, 173).
The effects of substitutions in a number of other amino acids on glucan solubility have also been reported: Asp457Asn (C-terminal of conserved region II), Asp571Lys (Fig. 1F), Lys779Gln and Lys1014Thr (not shown), all in GTFB of S. mutans GS-5, and Asn471Asp in GTFD of S. mutans GS-5 (173). These mutations resulted in an increase of soluble glucan synthesis from 0 to 37%, from 0 to 18%, from 0 to 3%, and from 0 to 14%, and insoluble glucan synthesis from 14 to 38%, respectively (173). The most marked effect was achieved (from 0 to 73% soluble glucan) when the six single GTFB mutants were combined (173). Mutagenesis of GTFB Val412Ile and GTFC Val438Ile resulted in enhanced insoluble glucan synthesis of about 10 to 20%, whereas soluble glucan synthesis by these enzymes was significantly lower than for the wild type (28) (Fig. 1C).
It is known that water-insoluble glucans contain mainly α-(1→3) linkages and soluble glucans mainly contain α-(1→6) glucosidic linkages (173). Unfortunately, the linkage types, degrees of branching, and sizes of the glucans synthesized by these GTFB and GTFD site-directed mutants have not been reported; conceivably, changes in either may have an effect on glucan solubility (173). Detailed analysis of these polymers may provide further insights on the structure-function relationships of GS enzymes.
(c) Affecting enzyme activity. Within the four conserved regions (I to IV) present in the catalytic core of GS enzymes, apart from the catalytic residues, other amino acid residues important for enzyme activity have been identified. Mutation of the highly conserved Trp491Gly in GTFB of S. mutans GS-5 (Fig. 1E) resulted in complete loss of enzyme activity (202) (Fig. 1). A double mutation in GTFA of L. reuteri 121, His1065Ser:Ala1066Ser, showed a very strong change in enzyme activity, but no influence on glucosidic linkage specificity (96). It was concluded that these residues are located further away from the (putative) −1 and +1 sugar binding subsites (96).
The GS-5 catalytic core commonly starts with two to four conserved Tyr residues (Fig. 1A). Conversion of one to four of these Tyr residues, at positions 169 to 172 in GTFB of S. mutans GS-5, into Ala residues had little effect on overall activity. Only the adhesiveness of the glucan products synthesized was altered by these mutations, suggesting that changes had been introduced in the glucan structure (202).
Using antibodies and chemical modification two regions, separate from the four conserved (I to IV) regions and upstream of conserved region II, that were important for activity were identified. The first of these regions extends from amino acids 368 to 382 in GTFC of S. mutans GS-5 (Fig. 1B), and the second region corresponds to GTFC residues 435 to 453 (27, 37, 57) (Fig. 1C). Subsequent mutational analysis of the first region in GTFI allowed identification of several residues (mutants Trp344Leu, Glu349Leu, and His355Val) that are important for activity (122) (Fig. 1B). Important amino acid residues in the second region have also been identified. Substitution of Asp511 and Asp513 of DSRS from L. mesenteroides NRRL B-512F in Asn residues resulted in complete loss and in a strong decrease in glucan- and oligosaccharide-synthesizing activities, respectively (119). Similar residues in GTFB from S. mutans GS-5 (Asp411 and Asp413) and GTFC from S. mutans GS-5 (Asp437 and Asp439) have been mutated into Asn residues, also affecting glucan-synthesizing activity (28). Site-directed mutagenesis of GTFB (Glu422Gln) and GTFC (Glu448Gln) resulted in 40% reduced glucan-synthesizing activity (28, 119) (Fig. 1C).
C-terminal domain.
Several studies have shown that the C-terminal domain of GS is involved in glucan binding. Therefore, it has been designated the glucan binding domain (GBD) (1, 82, 87, 99, 108, 120, 172). For a few GS enzymes, evidence has been presented that the C-terminal domain is also involved in determining the structure of the synthesized glucan (99, 214). In addition, the C-terminal GBD appears to be necessary for GS activity (99, 132). However, deletion of a large part of the C-terminal GBD had little effect on enzyme activity in, e.g., GTFI of S. downei MFe28 (116). Some GS enzymes with deletions at the C-terminal end retained hydrolytic activity, but glucan binding and synthesizing properties had become lost (56).
The precise role of the GBD domain in enzyme catalysis remains largely unknown. The GBD may be of importance for polymer chain growth. It has been suggested that the C-terminal domain plays a facilitating role in the transfer of products from the catalytic site (120).
The C-terminal domain of all reported GS is composed of a series of repeating units which have been divided into four classes, A, B, C, and D (Table 3). Site-directed mutagenesis studies of conserved amino acid residues in different A repeats of GFTI of S. downei MFe28 suggested that they do not contribute equally to the overall binding of dextran or that corresponding residues in different repeats may differ in their contributions (171).
TABLE 3.
Consensus sequences of repeating units present in the N- and/or C-terminal domain of GS enzymes from LABa
| Repeats | Strains | Gene(s) | Consensus sequence | Designation | Reference(s) |
|---|---|---|---|---|---|
| N-terminal | L. mesenteroides NRRL B-1355 | asr | WYYFDNNGYAVTLGLQTINGQHLYFDANGVQVKG | Partial A repeats | 79 |
| L. mesenteroides NRRL B-512F | dsrT | TDDKA(A/T)TTA(A/D)TS | Motif T | 56 | |
| L. mesenteroides NRRL B-1299 | dsrE | PA(A/T)DKAVDTTP(A/T)T | Motif S | 17 | |
| L. reuteri 121 | gtfA | R(P/N)DV-X12-SGF-X19-22-R(Y/F)S | RDV repeats | 97 | |
| L. sakei Kg15, L. fermentum Kg3 | gtfKg15, gtfKg3 | NDGYYFXXXGXXH°X(G/N)H°H°H° | YG repeat | 95 | |
| L. parabuchneri 33 | gtf33 | TTTQN(A/T)(P/A)NN(S/G)N(D/G)PQS | TTQ repeat | 95 | |
| C-terminal | S. downei MFe28 | gtfS | WYYFNXDGQAATGLQTIDGQTVFDDNGXQVKXG | A repeats | 8, 61, 171 |
| S. downei MFe28 | gtfI | VNGKTYYFGSDGTAQTQANPKGQTFKDGSVLR FYNLEGQYVSGSGWY | B repeats | 48 | |
| S. downei MFe28 | gtfS | DGKIFFDPDSGEVVKNRFV | C repeats | 8, 61 | |
| S. salivarius ATCC 25975 | gtfJ | GGVKNADGTYSKY | D repeats | 60 | |
| S. mutans GS-5, S. downei MFe28, S. salivarius ATCC 25975, S. sobrinus 6715 | 8 genes | NDGYYFXXXGXXH°X(G/N)H°H°H° | YG repeat | 58, 87 | |
| L. parabuchneri 33 | gtf33 | AVK(T/A)A(K/Q)(A/T)(Q/K)(L/V)(A/N)K(T/A)KAQ(I/V) (A/T)KYQKALKKAKTTKAK(A/T)QARK(S/N) LKKA(E/N(T/S)S(F/L)(S/T)KA) | KYQ repeat | 95 |
Boldface indicates conserved amino acids; RDV, KYQ, and TTQ residues are underlined; X, nonconserved amino acid residue.
Within these different repeats, a common conserved stretch of amino acids, designated the YG repeat, can be distinguished (58) (Table 3). The number, class, and distribution of these repeats are specific for each enzyme (124). L. mesenteroides NRRL B-512F dextransucrase contains, besides A and C repeats, N repeats, which have not been identified in streptococcal GS. These N repeats are not highly conserved but possess the main characteristics of YG repeats (120). Alternansucrase from L. mesenteroides NRRL B-1355 contains a single A repeat and distinct short repeats, DG(X)4APY (4).
Compared to the GBDs of other GS enzymes (average length, 400 to 500 amino acids), GTFA and GTFB from L. reuteri 121, GTFML1 from L. reuteri ML1, GTFO from L. reuteri ATCC 55730, and GTF180 from L. reuteri 180 possess relatively short GBDs of 134 to 263 amino acids (Fig. 1) (95, 97). The GBDs of L. reuteri GS enzymes lack A, B, C, and D repeats and consist of several (less well) conserved YG repeats (94, 95, 97). Also the putative GBDs of GTFKg15 from Lactobacillus sakei Kg15, GTFKg3 from Lactobacillus fermentum Kg3, and GTF33 from Lactobacillus parabuchneri 33 lack A, B, C, and D repeats and consist of various numbers of conserved and less-well-conserved YG repeating units. GTF33 contains, besides the 17 YG repeats, two unique repeating units designated KYQ that show no significant similarity to any protein motif present in the databases (Table 3). GTFKg15 possesses an additional stretch of amino acids at the end of its putative GBD showing similarity to part of a putative extracellular matrix binding protein from Streptococcus pyogenes M1 (95).
Reaction Mechanism of Glucansucrase Enzymes
The catalytic mechanism of GS enzymes is complicated and not yet fully understood. There are several aspects complicating the elucidation of the reaction mechanism: various glucans as well as oligosaccharides are synthesized and a sucrose hydrolysis reaction may also occur. Furthermore, many GS enzymes also catalyze branching reactions, resulting in synthesis of different types of glucosidic linkages.
Two alternative mechanisms have been proposed for the glucan chain growth. Nonreducing end elongation involves the presence of one amino acid residue (an Asp or Glu) acting as a nucleophilic group and another residue acting as a proton donor. The glucan chain grows by successive insertions of glucose units between the catalytic site of the enzyme and the reducing end of the glucan polymer. Reducing end elongation occurs in two steps involving two sucrose binding sites (nucleophiles): (i) the nucleophilic sites attack two sucrose molecules to give two covalent glucosyl-enzyme intermediates and (ii) the C-6 hydroxyl of one of the glucosyl intermediates makes a nucleophilic attack onto C-1 of the other glucosyl intermediate to form an α-(1→6) glucosidic linkage and an isomaltosyl intermediate. The newly released nucleophilic site attacks another sucrose molecule to give a new glucosyl-enzyme intermediate. This symmetrical and alternative role of the two sucrose binding sites results in growth of the glucan chain by its reducing end (124, 153, 155). This mechanism, however, appears more and more unlikely for GS enzymes in view of the single covalent intermediate identified (128) and mutational identification of a single catalytic triad (38, 99). Moreover, only one active site has been identified in amylosucrase (GH13) (3, 183).
The amylosucrase enzyme (from family GH13) from N. polysaccharea, synthesizing a (short) glucan polymer from sucrose, uses a double displacement mechanism, similar to that of other family GH13 enzymes. This mechanism involves the formation of a covalent glucosyl-enzyme intermediate (confirmed by three-dimensional structural data) (3, 80, 204). In a subsequent step this glucose moiety is transferred onto a water molecule (overall resulting in sucrose hydrolysis) or onto a hydroxyl group of a sugar acceptor (transglucosylation reaction). As for amylosucrase, again only one site capable of making a covalent bond with the glucose moiety, originating from the breakdown of sucrose, has clearly been identified in GS enzymes (high performance liquid chromatography analysis of tryptic digests of a trapped covalent glucosyl-GS enzyme intermediate) (80, 128).
At first a processive mechanism (polymer chain remains bound to the enzyme) was suggested for polymer formation by amylosucrase (145). A more detailed analysis showed that amylosucrase polymer formation is nonprocessive (release of the polymer chain after each glucose residue transfer and presence of intermediate oligosaccharides) (3). Early evidence indicates that GS glucan polymer synthesis proceeds in a processive manner, since oligosaccharide reaction intermediates cannot be detected and high-molecular-mass polysaccharide products are obtained at early reaction times (16, 44, 201). However, it cannot be excluded that the use of more sensitive analytical techniques of GS reaction products could reveal the presence of oligosaccharide reaction intermediates (indicative of a nonprocessive mechanism) (3).
Analysis of the three-dimensional structural information for amylosucrase proteins (GH13) with bound sucrose and oligosaccharide substrates and products provided convincing evidence for the use of a non-reducing-end elongation mechanism (3). Various studies of product formation by GS enzymes incubated with sucrose and acceptor substrates showed (e.g., conversion of maltose into panose by GTFA of L. reuteri [121]) that GS also use sucrose to elongate their oligosaccharide acceptor substrates at the nonreducing end (5, 42, 95, 99, 121, 130). Mutant data for GTFA of L. reuteri 121 showed similar changes in glucosidic bond specificity with both oligosaccharide and polysaccharide products (96). Both amylosucrase and GS enzymes thus appear to employ a non-reducing-end elongation mechanism for oligosaccharide and polymer synthesis.
Further studies need to focus on elucidation of the three-dimensional structures of GS enzymes, ideally with substrate-enzyme and/or product-enzyme complexes. This may allow elucidation of the exact reaction mechanism of family GH70 enzymes and identification of structural features determining differences in product specificity such as (i) number and position of sugar binding donor and acceptor subsites, (ii) residues involved in substrate/product binding, (iii) glucosidic bond specificity, and (iv) degree of branching.
FRUCTANSUCRASES
Bacterial FS synthesize either levan (levansucrase) or inulin (inulosucrase). Inulosucrase enzymes are exclusively present in LAB, while levansucrase enzymes are widely distributed in both gram-positive and gram-negative bacteria. At the amino acid level, the levansucrases of gram-positive and those of gram-negative bacteria show low similarity (about 20%). In general, FS enzymes of LAB origin are larger than their non-LAB counterparts. The levansucrase of S. salivarius ATCC 13419 is a particularly large enzyme with a molecular mass of 140 kDa (136). Most of the research has been performed on levansucrases, in particular on enzymes from Bacillus spp. (12, 107, 189, 195) and Zymomonas spp. (71, 187, 219, 220), and, to a lesser extent, Lactobacillus spp. (196, 211, 213), Streptococcus spp. (59, 174), and Gluconacetobacter spp. (6) species. In order to provide a state-of-the-art view of the FS field of research, work performed on non-LAB FS is also discussed.
Reactions Catalyzed and Fructan Product Synthesis
Bacterial FS are extracellular enzymes that cleave the glycosidic bond of their substrate sucrose (and in some cases also raffinose) and use the energy released to couple a fructose unit to (i) a growing fructan (either levan or inulin) chain (transfructosylation), (ii) to sucrose, (iii) to water (hydrolysis), or (iv) to another acceptor (such as raffinose). Because sucrose is used as the acceptor in the initial priming reaction, bacterial fructans contain a nonreducing glucose unit at the end of the chain (52). In the initial reaction of FS, the fructose of a sucrose molecule is coupled by the enzyme to another nonreducing fructose with a free primary alcohol at position C-2, acting as an acceptor substrate, e.g., sucrose, raffinose, or a fructan molecule (35, 152). This is also referred to as the priming reaction. In subsequent steps, the enzyme elongates the primer. A clear difference between FS and GS enzymes is the fact that GS enzymes cannot use sucrose as an acceptor but rather the cleaved glucose residue (see above).
Fructan synthesis.
In general, LAB produce two types of fructans using FS (see also Table 2): levans, consisting mainly of β-(2→6)-linked fructose residues, occasionally containing β-(2→1)-linked branches, and inulin-type fructans, with β-(2→1)-linked fructose residues, with β-(2→6)-linked branches. Levan production has been reported for streptococci (19, 70, 178), L. mesenteroides (156), L. reuteri 121 (208, 210), and Lactobacillus sanfranciscensis (92, 93). Lactobacillus frumenti, Lactobacillus pontis, Lactobacillus panis and Weissella confusa were also found to produce fructans, but their fructan binding types have not been determined (92, 93, 198). Inulin production by LAB has been observed in some cariogenic S. mutans and S. salivarius strains (45, 158, 174), Leuconostoc citreum CW28 (138), and L. reuteri 121 (212, 213).
The molecular masses of the fructans produced (if determined) show a large variation, from 2 × 104 to 50 × 106 Da (Table 2). There are some reports that the molecular mass of the fructan produced is dependent on growth and incubation conditions, e.g., the temperature, salinity, and sucrose concentration used (10, 192, 193).
Fructo-oligosaccharide synthesis.
All known bacterial FS catalyze fructose transfer from sucrose (or raffinose) to a number of acceptors other than the fructan polymer. Examples of possible acceptors are water (hydrolysis of sucrose) and sucrose and raffinose (yielding a tri- or tetrasaccharide, respectively), short-chain acylalcohols, various mono- to tetrasaccharides (22, 86, 194), and sorbitol (144). The ability of L. sanfranciscensis levansucrase to use raffinose, maltotriose, maltose, xylose, or raffinose as fructosyl acceptors, leading to the formation of a range of heterooligosaccharides, has been reported recently by Tieking et al. (199). Additionally, high performance liquid chromatography analysis of the reaction products of this levansucrase with 0.4 M raffinose as the fructose donor and acceptor revealed the presence of tetra-, penta-, and hexasaccharides (GalGF2 to GalGF4; Gal is galactose) in addition to melibiose (GalG), raffinose, kestose, and nystose. This proves the ability of the enzyme to use raffinose not only as an acceptor but also as a donor of fructose moieties.
Structural and Functional Organization of Fructansucrase Enzymes
Although the reactions performed by FS and GS are similar with respect to the use of sucrose as the substrate, the proteins involved do not share sequence similarity. Based on deduced amino acid sequences, the overall FS structure is divided into four regions (Fig. 3): (i) a signal peptide, (ii) an N-terminal stretch that varies in length, (iii) a conserved catalytic core of about 500 amino acids that is shared between all family GH68 members (carbohydrate-active enzyme website: http://afmb.cnrs-mrs.fr/CAZY) (33), and (iv) a C-terminal stretch of various lengths, in some cases with a cell wall binding domain (LPXTG; see below).
FIG. 3.
Schematic representation of FS proteins from LAB. The L. reuteri 121 inulosucrase (Inu) deduced amino acid sequence was used as the template (AF459437). The four different regions shown are (i) the N-terminal signal sequence; (ii) the N-terminal variable region; (iii) the catalytic core; and (iv) the C-terminal variable region (which in some cases contain an LPXTG cell wall anchor). Alignments (SequenceLogo, http://weblogo.berkeley.edu/) are shown of short regions in LAB FS protein amino acid sequences (Table 2) with conserved amino acid residues for which mutant information is available in literature. (A) Asp86 in B. subtilis SacB identified as the nucleophile based on crystal structure data for (inactive) mutant Glu342Ala with a bound sucrose (113) and biochemical characterization of the L. reuteri 121 Inu Asp272Asn mutant (140). (B) Mutant Glu117Gln in Z. mobilis SacB, resulting in a higher transglycosylation activity (220); SGSA, sucrose binding box 1, a region conserved between sucrose-utilizing enzymes (167). (C) Mutant Asp312Ser in S. salivarius ATCC 25975 FS (possibly involved in acceptor recognition and/or stabilizing a beta turn in the protein) (185). (D) “Sucrose binding box 2,” a region conserved between sucrose-utilizing enzymes (167). (E) RDP motif, with mutant Asp397Ser in S. salivarius ATCC 25975 FS resulting in complete loss of sucrose hydrolysis and polymerization activities (185), mutant Asp309Asn in G. diazotrophicus LsdA, with a 75-fold reduced catalytic activity (9), mutant Asp194Asn in Z. mobilis SacB, with a 3,400-fold decrease in catalytic activity (220), and Asp247 in B. subtilis SacB and Asp272 in L. reuteri 121 Inu (stabilizer of the oxocarbenium-like transition state) (113, 140). (F) Mutant Glu211Gln in Z. mobilis SacB, with sucrose hydrolysis reduced to 28%, and highly reduced transfructosylation activity. (G) Mutant Glu278Asp (30-fold lower catalytic activity), mutant Glu278His (virtually inactive enzyme) in Z. mobilis levansucrase SacB (220), and Glu342 in B. subtilis SacB and Glu523 in L. reuteri 121 Inu (acid/base catalyst) (113, 140). (H) Arg331His (higher oligosaccharide formation) in levansucrase from B. subtilis SacB (22).
Signal peptide and N-terminal variable domain.
Since bacterial FS are extracellular enzymes, they contain an N-terminal signal sequence that targets these enzymes for secretion (Fig. 3). The signal peptide-containing precursor is cleaved upon secretion of FS by gram-positive bacteria (11, 107, 147, 189, 195, 211). The N-terminal domain (Fig. 3) varies in size between the FS and no function has yet been assigned to this domain.
Catalytic domain.
Most work on structure-function relationships of the core region has been performed on non-LAB FS enzymes. The results obtained for non-LAB FS enzymes have (in part) been corroborated by work performed on LAB FS (see below). Therefore, the results for non-LAB FS are also discussed below.
(i) Catalytic residues.
Recently, high-resolution crystal structures of the non-LAB Bacillus subtilis SacB levansucrase (at 1.5 Å) and a sucrose-bound inactive mutant of the same enzyme (at 2.1 Å) have been described. Both structures show a rare five-fold β-propeller topology with a deep, negatively charged central pocket (113). This topology differs strongly from the (β/α)8 barrel identified in family GH13 enzymes and putatively assigned to GS enzymes of family GH70 (see above). The central pocket is composed mostly of residues belonging to highly conserved sequence motifs, including invariant acidic residues Asp86 (Fig. 3A), Asp247 (Fig. 3E), and Glu342 (Fig. 3G). As with GS and the enzymes of family GH13, these residues form the catalytic triad (nucleophile, transition state stabilizer, and acid-base catalyst, respectively). This has been proven by the mutational analysis of these residues and their equivalents in L. reuteri 121, the Inu and Lev enzymes (140).
Recently, a three-dimensional structure has been reported for the levansucrase enzyme of the non-LAB Gluconacetobacter diazotrophicus (112). The three-dimensional structure of this enzyme displays the same five-bladed β-propeller architecture as the B. subtilis levansucrase enzyme. The three-dimensional positions of the three catalytic residues of both levansucrases are superimposable, indicating strong structural relatedness of these enzymes.
(ii) Catalytic site.
Based on the B. subtilis SacB (113) and G. diazotrophicus LsdA (112) three-dimensional structures, residues directly involved in binding of sucrose in the active site and constituting the −1 and +1 sugar binding subsites have been identified (nomenclature according to Davies et al. [34]). In short, cleavage of the substrate (e.g., sucrose) takes place between subsites −1 and +1, and the enzyme forms a covalent intermediate with the cleaved substrate (i.e., fructose) at subsite −1. Subsequently, the cleaved substrate is coupled to the acceptor molecule (e.g., fructan).
Figure 4 shows the main amino acids involved in FS catalysis. Based on the available three-dimensional structures and sequence alignment of family GH68 proteins, residues creating subsite −1 have been identified in B. subtilis SacB, G. diazotrophicus LsdA, and L. reuteri 121 inulosucrase (Inu) and levansucrase (Lev) (Fig. 4). Strikingly, subsite +1 differs among the enzymes from family GH68. These differences at the +1 subsite distinguish FS enzymes from gram-positive (SacB and Inu) and gram-negative (LsdA) bacteria. They do not, however, distinguish between polymerizing (SacB) and oligomerizing (LsdA and Inu) or inulin (Inu)/levan (SacB, LsdA) synthesizing enzymes (72, 193, 213).
FIG. 4.
Close-up view of the active site of mutant Glu342Ala B. subtilis SacB levansucrase with a bound sucrose molecule (structure accession code 1PT2). The model was created using SwissPdb Viewer (62). Hydrogen bonds are shown by dashed lines (based on reference 113). Numbering of amino acid residues is based on B. subtilis SacB, with the numbering of L. reuteri 121 Inu (Table 2) in parentheses. Based on structural information about the SacB (113) and G. diazotrophicus LsdA (112) three-dimensional structures and alignments of FS amino acid sequences the −1 (underlined) and +1 (italics) subsites of family GH68 enzymes were identified (this work). The −1 subsite is constituted of the catalytic nucleophile (Asp86 in SacB, Asp135 in LsdA, and Asp272 in Inu) (Fig. 3A), the neighboring Trp residue (Trp85 in SacB, Trp134 in LsdA, and Trp271 in Inu) (Fig. 3A), two residues located in the RDP motif that is conserved in most members of family GH68 (Arg246, Asp247 in SacB, Arg308, Asp309 in LsdA, and Arg423, Asp424 in Inu) (Fig. 3E), and a Trp residue bordering the sucrose binding pocket and located very close to the fructose moiety at −1 (Trp163 in SacB, Trp224 in LsdA, and Trp340 in Inu) (Fig. 3B). The +1 subsite is constituted of Arg/His (Arg360 in SacB, His419 in LsdA, and Arg541 in Inu) (Fig. 3H), Glu/Gln located two residues upstream of the acid-base catalyst (Glu340 in SacB, Gln399 in LsdA, and Glu521 in Inu) (Fig. 3G), and Arg from the RDP motif (also involved in formation of the −1 subsite) (Arg246 in SacB, Arg308 in LsdA and Arg423 in Inu) (Fig. 3E).
(iii) Mutations affecting product formation.
The three-dimensional structures of the B. subtilis and G. diazotrophicus levansucrases have provided important insights into the functional roles of several conserved amino acid residues in this core region (112, 113). Two regions that are highly conserved among FS and sucrose-hydrolyzing enzymes, invertases, have been designated sucrose binding boxes (SBB) (Fig. 3B and D). Asp312, located between SBB1 and SBB2 (Fig. 3C) of S. salivarius ATCC 25975 levansucrase, is most likely involved in determining acceptor recognition or stabilizing of a β-turn in the protein (185). Analysis of the three-dimensional structure of B. subtilis SacB revealed that Asp312 indeed forms a 180° reverse β-turn between SBB1 and SBB2 and is located on the surface of the protein, far from the active site (113).
Several independent studies revealed that transglycosylation and hydrolysis reactions could be modulated separately by mutagenesis or even by changing reaction conditions. Selective inhibition of transglycosylation activity of Zymomonas mobilis levansucrase has been reported by Senthilcumar et al. (169). Point mutations in the “sucrose binding box” in levansucrase of Z. mobilis caused significant changes in the transglycosylation efficiency of the enzyme (220). The transglycosylation versus hydrolysis ratio could be also be altered by immobilization of the enzyme on hydroxyapatite, thereby mimicking in vivo conditions, where levansucrases are attached to the cell wall of bacteria or to the tooth surface. The activity of immobilized B. subtilis levansucrase was directed mainly towards its polymerizing activity (23).
Mutation of Arg331 in the B. subtilis levansucrase (Fig. 3H) yielded enzymes that were differently affected in transfructosylation activity depending on the substitution chosen; three variants (Arg331Lys, Arg331Ser, and Arg331Leu) lost the ability to synthesize levan and were only able to produce the trisaccharide kestose. It has been suggested that the side chain of Arg331 could act as a proton donor in the bifunctional catalysis (22). Analysis of the three-dimensional structure of the B. subtilis levansucrase showed, however, that this Arg331 residue is structurally involved in the acceptor binding site (113). Similar conclusions were drawn from a His296-mutagenized Z. mobilis levansucrase enzyme (220).
A pressing question is what structural features in FS enzymes determine their specificity for synthesis of either β-(2→6) or β-(2→1) linkages and the sizes of the fructans produced. At present, no three-dimensional structural information has been published for inulosucrase enzymes. The G. diazotrophicus and B. subtilis levansucrase enzymes synthesize fructo-oligosaccharides and levan polymers, respectively. Further investigation of both three-dimensional structures might give clues to the difference in product profiles of these enzymes. The lack of structural data for complexes between FS and their fructosyl acceptors makes it difficult to understand what determines the polymer versus short oligosaccharide synthesis ratio.
(iv) Calcium binding site.
Analysis of the three-dimensional structure of B. subtilis levansucrase has provided evidence for a presence of a bound metal ion, most likely Ca2+ (113). Asp339 of B. subtilis levansucrase was identified as one of the residues coordinating Ca2+ ions in the enzyme structure. The functional role of this residue has been studied by mutating equivalent amino acid residues in the inulosucrase and levansucrase from L. reuteri, Asp520Asn and Asp500Asn, respectively (139). Both mutants showed a decreased optimal temperature and the apparent affinity for Ca2+ binding was reduced significantly, 1,600-fold and 35-fold, respectively. Also, the S. salivarius ATCC 27975 levansucrase enzyme is dependent on calcium ions for enzyme activity (78).
Sequence alignment of family GH68 members revealed that residues involved in binding of the calcium are conserved in most enzymes from gram-positive bacteria, but are absent in proteins of gram-negative origin (139). Most likely a disulfide bridge present in the three-dimensional structure of G. diazotrophicus (gram-negative) plays a similar role to the calcium binding site identified in FS enzymes from gram-positive bacteria (112).
C-terminal domain.
As with GS, the C-terminal domain of FS enzymes might affect product size and/or enzyme specificity. The only evidence for such a claim is the observation that a C-terminal enlargement of the FS enzyme from the non-LAB B. subtilis was shown to produce a larger fructan polymer which was mainly due to an increase in branches in the levan (24).
A different function for the C terminus of FS is the attachment of these enzymes to the cell wall of its producing organism. A common C-terminal LPXTG cell wall-anchoring motif (49, 133, 134) (Fig. 3) is found in both inulosucrase (213) and levansucrase (211) from L. reuteri 121. A motif resembling the LPXTG cell wall-anchoring motif is present in the cell wall-associated S. salivarius ATCC 25975 levansucrase (148). Proteins displayed on the bacterial surface may have various functions for the bacterial cell. For Staphylococcus aureus, surface proteins are thought to play a major role in the infection process in humans (200). Surface proteins from urogenital Lactobacillus spp. mediate adhesion to tissue cells (77, 177) and play a role in the maintenance of a healthy urogenital microbiota. Cell-associated homopolysaccharides, produced by sucrase enzymes anchored to the cell surface, may also be involved in adherence of the organism to a surface, such as teeth and the intestinal mucosa (159).
Reaction Mechanism of Fructansucrase Enzymes
A detailed biochemical characterization of FS enzyme reactions is complicated by the fact that FS generate new fructan molecules, which in turn can be used as acceptor substrates. Accordingly, a multiple chain elongation mechanism in which the fructose residues are added randomly to all fructan acceptor molecules has been proposed (25). The nature of the fructosyl acceptor, except water, thus changes as the reaction proceeds. Kinetic and chemical studies of the levansucrase of B. subtilis further suggest that each fructose unit is added one at a time onto an acceptor molecule (25).
Most FS enzymes follow Michaelis-Menten kinetics for the hydrolysis and transferase reactions. Exceptions to this rule are the L. reuteri 121 inulosucrase and levansucrase enzymes and L. sanfranciscensis levansucrase, which cannot be saturated by their substrate, sucrose (in the case of the L. reuteri 121 levansucrase, only at 50°C) (196, 211, 212). This phenomenon has also been observed for some plant enzymes that synthesize inulin polymers (91). Low-molecular-mass fructans accelerate the rate of polysaccharide formation and increase the fructan-to-free fructose ratio (46, 212). This may partly explain the unusual reaction kinetics observed for the above-described enzymes.
A two-step mechanism has been proposed for catalysis by FS enzymes. They contain an acidic group and a nucleophilic group essential for transfructosylation (180). The first identification of a nucleophilic group responsible for binding to the fructose moiety, determined by a covalent intermediate of substrate and enzyme, was in 1976 by Chambert and Gonzy-Treboul for the B. subtilis levansucrase (21). Information about the position and identity of this Asp residue has been determined from the three-dimensional crystal structure of the B. subtilis levansucrase (113). An Asp amino acid residue in the conserved RDP motif across families GH68 and GH32 (Fig. 3E) has been shown to be involved in the stabilization of the transition state by analysis of the B. subtilis levansucrase three-dimensional structure (113). Mutation of this Asp residue resulted in dramatic decreases of catalytic activities in both L. reuteri 121 FS (140) and the S. salivarius levansucrase enzymes (185).
Based on the observation that the polymerizing activity of the enzyme could be modulated separately from the oligomerization activity, a reaction mechanism involving two active sites has been proposed for the S. mutans FS (188). However, both levansucrase three-dimensional structures provide evidence for only a single active site (112, 113).
Putative Fructansucrases in Lactic Acid Bacteria
Several putative FS-encoding genes have been identified in the genome sequences of Streptococcus, Leuconostoc, and Lactobacillus strains (Table 4). Although their sizes are quite divergent, their core regions are all around 450 amino acids long. Since the characteristic residues of the sucrose binding boxes and the catalytic triad are present, these genes may very well encode active FS enzymes. The putative Lactobacillus johnsonii and Lactobacillus gasserii FS genes (Table 4) show strong amino acid sequence similarity (62% identitity and 75% similarity over 709 amino acids and 61% identitity and 74% similarity over 663 amino acids, respectively) to the L. reuteri 121 inulosucrase, which might indicate their inulosucrase identity.
TABLE 4.
Characteristics and sequence motifs of putative levansucrase (LS) and inulosucrase (IS) enzymes identified in LAB genome sequencesa
| Organism | Accession no. | Size (amino acids) | Sequence
|
Positions of core region | |||||
|---|---|---|---|---|---|---|---|---|---|
| Sucrose box 1 | Sucrose box 2 | RDP | E211 Z. mobilis | Acid-base catalyst | R331 B. subtilis | ||||
| S. mutans UA159 putative IS | AE015025 | 795 | EWSGS | LFYTKVD | LRDPH | VFEAST | VSDELER | SRLNH | 173-636 |
| L. johnsonii NCC533 putative IS | RLJO00913 | 797 | QWSGS | LYYTKVD | MRDAH | VFEAST | VSDEIER | TRLNR | 200-663 |
| L. gasseri ATCC 33323 putative IS | RLGA00517 | 630 | QWSGS | LYYTKVD | MRDAH | VFEAST | VSDEIER | TRLNR | 173-630 |
| L. mesenteroides ATCC 8293 putative LS | RLME01775 | 1,015 | QWSGS | LYYTKVD | LRDPH | AFEANT | ITDEIER | TRLSK | 171-622 |
| L. mesenteroides ATCC 8293 putative LS | RLME01776 | 782 | QWSGS | LFYTKTD | LRDPH | TFESNT | ITDEIER | TRLSK | 166-618 |
| L. mesenteroides ATCC 8293 putative LS | RLME01780 | 1,002 | QWSGS | LFYTQVD | LRDPH | TFEGST | VTDEIER | ARLDR | 167-611 |
| L. reuteri 121 Inu EC 2.4.1.9 | AAN05575 | 798 | EWSGS | LFYTRVD | MRDAH | VFEAST | VSDEIER | TRLNR | 200-662 |
| L. reuteri 121 Lev EC 2.4.1.10 | AAO14618 | 804 | EWSGS | LFFTSND | LRDPH | VFEANT | ASDEVER | TRVSR | 177-642 |
As references, L. reuteri (121) Inu and Lev are shown (boldface). The nucleophile and acid-base catalysts have been identified in B. subtilis SacB and L. reuteri (121) Lev and Inu proteins (113, 140). Residues or motifs indicated in the top row: the RDP motif (Fig. 3E), initially identified in the G. diazotrophicus levansucrase (9); E211 residue, identified in the Z. mobilis levansucrase (220); and R331 residue, identified in the B. subtilis levansucrase (22). Sucrose binding boxes 1 and 2 are further discussed in the legend to Fig. 3.
Mining newly released genome sequences might reveal new types of sucrase genes. An example of such a new type of sucrase gene is islA of Leuconostoc citreum CW28 (Table 2), a hybrid between a GS and an FS gene (137). Its N-terminal region is similar to the variable region of alternansucrase from L. mesenteroides NRRL B-1355, its catalytic domain is similar to the core region of FS from various bacteria, and its C-terminal domain displays similarity to the GBD from alternansucrase (see above). Other exciting possibilities are enzymes that synthesize new types of polymers, such as a fructan with alternating β-(2→1)- and β-(2→6)-linked fructose residues or even a polymer containing combinations of glucose and fructose residues.
CONCLUSIONS AND FUTURE DIRECTIONS
Lactic acid bacteria are a promising source of polysaccharides and oligosaccharides for health, food, and nutritional applications. LAB synthesize a large diversity of glucans and fructans, homopolysaccharides that vary, for instance, in molecular mass, glycosidic linkages, solubility, and degree ofbranching. These parameters determine to a large extent the functional properties of both glucan and fructan polymers. As reviewed in this paper, information about structure-function relationships in the sucrase-type enzymes that synthesize these polymers from sucrose is still limited but increasing rapidly.
Most progress has been made with respect to FS enzymes. Recently, two high-resolution three-dimensional structures of the B. subtilis and G. diazotrophicus levansucrases (family GH68) have been reported (112, 113), providing strong promise for rapid further progress. The levansucrase and inulosucrase enzymes characterized are highly similar in their primary amino acid sequences and the chemical reactions they catalyze but synthesize rather different fructan polysaccharides (levan and inulin). Elucidation of an inulosucrase three-dimensional structure and further detailed biochemical comparisons of (mutant) inulosucrase and levansucrase enzymes may provide clear insights into the structure-function relationships of these FS enzymes. Questions to be answered are (i) which factors determine the fructan binding type specificity, (ii) what determines the fructan molecular mass, and (iii) what is the underlying reaction mechanism. Subsequently, mutant enzymes may be constructed, synthesizing (hybrid) fructans with specific sizes and/or containing a specific distribution of glycosidic binding types.
No three-dimensional crystal structures are available yet for glucansucrase enzymes (family GH70). Their overall sequence and secondary-structure similarity with α-amylase-type enzymes (family GH13), however, has already allowed identification of putative acceptor substrate binding subsites and rational construction of mutant GS enzymes synthesizing glucans with clearly different glucosidic bond profiles (96) and solubilities (123, 173).
Further breakthroughs in this field are expected in the years ahead, with enzyme engineering approaches increasingly allowing construction of mutant GS and FS enzymes and the discovery of new types of sucrase enzymes from genome sequences providing exciting possibilities for the synthesis of tailor-made glucan, fructan polysaccharides, and oligosaccharides for a range of applications.
Acknowledgments
We thank Thijs Kaper for assistance in creating Fig. 4 and the referees for critically reading the manuscript.
We were financially supported by the University of Groningen, TNO Quality of Life, Centre for Carbohydrate Bioprocessing, and by the EET program of the Dutch government (project number KT 97029).
REFERENCES
- 1.Abo, H., T. Matsumura, T. Kodama, H. Ohta, K. Fukui, K. Kato, and H. Kagawa. 1991. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J. Bacteriol. 173:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aduse-Opoku, J., M. L. Gilpin, and R. R. B. Russell. 1989. Genetic and antigenic comparison of Streptococcus mutans fructosyltransferase and glucan-binding protein. FEMS Microbiol. Lett. 50:279-282. [DOI] [PubMed] [Google Scholar]
- 3.Albenne, C., L. K. Skov, O. Mirza, M. Gajhede, G. Feller, S. D'Amico, G. Andre, G. Potocki-Veronese, B. A. Van Der Veen, P. F. Monsan, and M. Remaud-Simeon. 2004. Molecular basis of the amylose-like polymer formation catalyzed by Neisseria polysaccharea amylosucrase. J. Biol. Chem. 279:726-734. [DOI] [PubMed] [Google Scholar]
- 4.Arguello-Morales, M. A., M. Remaud-Simeon, S. Pizzut, P. Sarcabal, and P. F. Monsan. 2000. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 182:81-85. [DOI] [PubMed] [Google Scholar]
- 5.Arguello-Morales, M. A., M. Remaud-Simeon, R. M. Willemot, M. R. Vignon, and P. F. Monsan. 2001. Novel oligosaccharides synthesized from sucrose donor and cellobiose acceptor by alternansucrase. Carbohydr. Res. 331:403-411. [DOI] [PubMed] [Google Scholar]
- 6.Arrieta, J. G., L. Hernández, A. Coego, V. Suarez, E. Bamori, C. Menéndez, M. F. Petit-Glatron, R. Chambert, and G. Selman-Housein. 1996. Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiology 142:1077-1085. [DOI] [PubMed] [Google Scholar]
- 7.Balakrishnan, M., R. S. Simmonds, and J. R. Tagg. 2000. Dental caries is a preventable infectious disease. Aust. Dent. J. 45:235-245. [DOI] [PubMed] [Google Scholar]
- 8.Banas, J. A., R. R. B. Russell, and J. J. Ferretti. 1990. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect. Immun. 58:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batista, F. R., L. Hernández, J. R. Fernandez, J. Arrieta, C. Menéndez, R. Gomez, Y. Tambara, and T. Pons. 1999. Substitution of Asp-309 by Asn in the Arg-Asp-Pro (RDP) motif of Acetobacter diazotrophicus levansucrase affects sucrose hydrolysis, but not enzyme specificity. Biochem. J. 337:503-506. [PMC free article] [PubMed] [Google Scholar]
- 10.Ben Ammar, Y., T. Matsubara, K. Ito, M. Iizuka, T. Limpaseni, P. Pongsawasdi, and N. Minamiura. 2002. Characterization of a thermostable levansucrase from Bacillus sp. TH4-2 capable of producing high molecular weight levan at high temperature. J. Biotechnol. 99:111-119. [DOI] [PubMed] [Google Scholar]
- 11.Bezzate, S., S. Aymerich, R. Chambert, S. Czarnes, O. Berge, and T. Heulin. 2000. Disruption of the Paenibacillus polymyxa levansucrase gene impairs its ability to aggregate soil in the wheat rhizosphere. Environ. Microbiol. 2:333-342. [DOI] [PubMed] [Google Scholar]
- 12.Bezzate, S., M. Steinmetz, and S. Aymerich. 1994. Cloning, sequencing, and disruption of a levanase gene of Bacillus polymyxa CF43. J. Bacteriol. 176:2177-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binderup, K., and J. Preiss. 1998. Glutamate-459 is important for Escherichia coli branching enzyme activity. Biochemistry 37:9033-9037. [DOI] [PubMed] [Google Scholar]
- 14.Boeckner, L. S., M. I. Schnepf, and B. C. Tungland. 2001. Inulin: a review of nutritional and health implications. Adv. Food Nutr. Res. 43:1-63. [DOI] [PubMed] [Google Scholar]
- 15.Bornet, F. 2001. Fructo-oligosaccharides and other fructans: chemistry, structure and nutritional effects, p. 480-493. In B. V. McCleary and L. Prosky (ed.), Advanced dietary fibre technology. Blackwell Science, Oxford, England.
- 16.Bovey, F. 1959. Enzymatic polymerisation I: molecular weight and branching during the formation of dextran. J. Polym. Sci. 35:167-182. [Google Scholar]
- 17.Bozonnet, S., M. Dols-Laffargue, E. Fabre, S. Pizzut, M. Remaud-Simeon, P. F. Monsan, and R. M. Willemot. 2002. Molecular characterization of DSR-E, an α-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 184:5753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byun, R., M. A. Nadkarni, K. L. Chhour, F. E. Martin, N. A. Jacques, and N. Hunter. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42:3128-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlsson, J. 1970. A levansucrase from Streptococcus mutans. Caries Res. 4:97-113. [DOI] [PubMed] [Google Scholar]
- 20.Cerning, J. 1990. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol. Rev. 87:113-130. [DOI] [PubMed] [Google Scholar]
- 21.Chambert, R., and G. Gonzy-Treboul. 1976. Levansucrase of Bacillus subtilis. Characterization of a stabilized fructosyl-enzyme complex and identification of an aspartyl residue as the binding site of the fructosyl group. Eur. J. Biochem. 71:493-508. [DOI] [PubMed] [Google Scholar]
- 22.Chambert, R., and M. F. Petit-Glatron. 1991. Polymerase and hydrolase activities of Bacillus subtilis levansucrase can be separately modulated by site-directed mutagenesis. Biochem. J. 279:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambert, R., and M. F. Petit-Glatron. 1993. Immobilisation of levansucrase on calcium phosphate gel strongly increases its polymerase activity. Carbohydr. Res. 244:129-136. [DOI] [PubMed] [Google Scholar]
- 24.Chambert, R., M. C. Rain-Guion, and M. F. Petit-Glatron. 1992. Readthrough of the Bacillus subtilis stop codon produces an extended enzyme displaying a higher polymerase activity. Biochim. Biophys. Acta 1132:145-153. [DOI] [PubMed] [Google Scholar]
- 25.Chambert, R., G. Treboul, and R. Dedonder. 1974. Kinetic studies of levansucrase of Bacillus subtilis. Eur. J. Biochem. 41:285-300. [DOI] [PubMed] [Google Scholar]
- 26.Chhour, K. L., M. A. Nadkarni, R. Byun, F. E. Martin, N. A. Jacques, and N. Hunter. 2005. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 43:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chia, J. S., R. H. Lin, S. W. Lin, J. Y. Chen, and C. S. Yang. 1993. Inhibition of glucosyltransferase activities of Streptococcus mutans by a monoclonal antibody to a subsequence peptide. Infect. Immun. 61:4689-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chia, J. S., C. S. Yang, and J. Y. Chen. 1998. Functional analyses of a conserved region in glucosyltransferases of Streptococcus mutans. Infect. Immun. 66:4797-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colby, S. M., G. C. Withing, L. Tao, and R. R. B. Russell. 1995. Insertional inactivation of the Streptococcus mutans dexA (dextranase) gene results in altered adherence and dextran catabolism. Microbiology 141:2929-2936. [DOI] [PubMed] [Google Scholar]
- 30.Corrigan, A. J., and J. F. Robyt. 1979. Nature of the fructan of Streptococcus mutans OMZ 176. Infect. Immun. 26:387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cote, G. L., and C. A. Dunlap. 2003. Alternansucrase acceptor reactions with methyl hexopyranosides. Carbohydr. Res. 338:1961-1967. [DOI] [PubMed] [Google Scholar]
- 32.Côté, G. L., and J. F. Robyt. 1982. Isolation and partial characterization of an extracellular glucansucrase from Leuconostoc mesenteroides NRRL B-1355 that synthesizes an alternating (1→6), (1→3)-α-d-glucan. Carbohydr. Res. 101:57-74. [DOI] [PubMed] [Google Scholar]
- 33.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. J. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 34.Davies, G. J., K. S. Wilson, and B. Henrissat. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321:557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dedonder, R. 1966. Levansucrase from Bacillus subtilis, p. 500-505. In E. F. Neufeld and V. Ginsburg (ed.), Methods in enzymology. Academic Press, New York, N.Y.
- 36.Demuth, K., H. J. Jordening, and K. Buchholz. 2002. Oligosaccharide synthesis by dextransucrase: new unconventional acceptors. Carbohydr. Res. 337:1811-1820. [DOI] [PubMed] [Google Scholar]
- 37.Dertzbaugh, M. T., and F. L. Macrina. 1990. Inhibition of Streptococcus mutans glucosyltransferase activity by antiserum to a subsequence peptide. Infect. Immun. 58:1509-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devulapalle, K. S., S. D. Goodman, Q. Gao, A. Hemsley, and G. Mooser. 1997. Knowledge-based model of a glucosyltransferase from the oral bacterial group of mutans streptococci. Protein Sci. 6:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Vuyst, L., and B. Degeest. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 23:153-177. [DOI] [PubMed] [Google Scholar]
- 40.De Vuyst, L., and F. Vaningelgem. 2003. Developing new polysaccharides, p. 275-320. In B. M. McKenna (ed.), Texture in food, volume 1: semi-solid foods. Woodhead Publishing, Cambridge, England. [Google Scholar]
- 41.Dewar, M. D., and G. J. Walker. 1975. Metabolism of the polysaccharides of human dental plaque. I. Dextranase activity of streptococci, and the extracellular polysaccharides synthesized from sucrose. Caries Res. 9:21-35. [DOI] [PubMed] [Google Scholar]
- 42.Dols, M., M. R. Simeon, R. M. Willemot, M. R. Vignon, and P. F. Monsan. 1997. Structural characterization of the maltose acceptor-products synthesized by Leuconostoc mesenteroides NRRL B-1299 dextransucrase. Carbohydr. Res. 305:549-559. [DOI] [PubMed] [Google Scholar]
- 43.Dols-Lafargue, M., R. M. Willemot, P. F. Monsan, and M. Remaud-Simeon. 2001. Factors affecting α,-1,2 glucooligosaccharide synthesis by Leuconostoc mesenteroides NRRL B-1299 dextransucrase. Biotechnol. Bioeng. 74:498-504. [DOI] [PubMed] [Google Scholar]
- 44.Ebert, K. H., and G. Schenk. 1968. Mechanisms of biopolymer growth: the formation of dextran and levan. Adv. Enzymol. Relat. Areas Mol. Biol. 30:179-221. [DOI] [PubMed] [Google Scholar]
- 45.Ebisu, S., K. Kato, S. Kotani, and A. Misaki. 1975. Structural differences in fructans elaborated by Streptococcus mutans and Streptococcus salivarius. J. Biochem. (Tokyo) 78:879-887. [DOI] [PubMed] [Google Scholar]
- 46.Elishashvili, V. I. 1980. Purification and properties of levansucrase of Gluconobacter oxydans L-1. Biokhimiia. 45:20-27. [PubMed] [Google Scholar]
- 47.Fabre, E., S. Bozonnet, A. Arcache, R. M. Willemot, M. R. Vignon, P. F. Monsan, and M. Remaud-Simeon. 2005. Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly α-1,2 branched dextran. J. Bacteriol. 187:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferretti, J. J., M. L. Gilpin, and R. R. B. Russell. 1987. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J. Bacteriol. 169:4271-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 50.Flamm, G., W. Glinsmann, D. Kritchevsky, L. Prosky, and M. Roberfroid. 2001. Inulin and oligofructose as dietary fiber: a review of the evidence. Crit. Rev. Food Sci. Nutr. 41:353-362. [DOI] [PubMed] [Google Scholar]
- 51.Franck, A. 2002. Technological functionality of inulin and oligofructose. Br. J. Nutr. 87(Suppl. 2):S287-S291. [DOI] [PubMed] [Google Scholar]
- 52.French, A. D., and A. L. Waterhouse. 1993. Chemical structure and characteristics, p. 41-82. In M. Suzuki and N. J. Chatterton (ed.), Science and technology of fructans. CRC Press Inc., Boca Raton, Fla.
- 53.Fu, D. T., and J. F. Robyt. 1991. Maltodextrin acceptor reactions of Streptococcus mutans 6715 glucosyltransferases. Carbohydr. Res. 217:201-211:201-211. [DOI] [PubMed] [Google Scholar]
- 54.Fukushima, K., T. Ikeda, and H. K. Kuramitsu. 1992. Expression of Streptococcus mutans gtf genes in Streptococcus milleri. Infect. Immun. 60:2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Funane, K., T. Ishii, M. Matsushita, K. Hori, K. Mizuno, H. Takahara, Y. Kitamura, and M. Kobayashi. 2001. Water-soluble and water-insoluble glucans produced by Escherichia coli recombinant dextransucrases from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 334:19-25. [DOI] [PubMed] [Google Scholar]
- 56.Funane, K., K. Mizuno, H. Takahara, and M. Kobayashi. 2000. Gene encoding a dextransucrase-like protein in Leuconostoc mesenteroides NRRL B-512F. Biosci. Biotechnol. Biochem. 64:29-38. [DOI] [PubMed] [Google Scholar]
- 57.Funane, K., M. Shiraiwa, K. Hashimoto, E. Ichishima, and M. Kobayashi. 1993. An active-site peptide containing the second essential carboxyl group of dextransucrase from Leuconostoc mesenteroides by chemical modifications. Biochemistry 32:13696-13702. [DOI] [PubMed] [Google Scholar]
- 58.Giffard, P. M., and N. A. Jacques. 1994. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73:1133-1141. [DOI] [PubMed] [Google Scholar]
- 59.Giffard, P. M., C. Rathsam, E. Kwan, D. W. Kwan, K. L. Bunny, S. P. Koo, and N. A. Jacques. 1993. The ftf gene encoding the cell-bound fructosyltransferase of Streptococcus salivarius ATCC 25975 is preceded by an insertion sequence and followed by FUR1 and clpP homologues. J. Gen. Microbiol. 139:913-920. [DOI] [PubMed] [Google Scholar]
- 60.Giffard, P. M., C. L. Simpson, C. P. Milward, and N. A. Jacques. 1991. Molecular characterization of a cluster of at least two glucosyltransferase genes in Streptococcus salivarius ATCC 25975. J. Gen. Microbiol. 137:2577-2593. [DOI] [PubMed] [Google Scholar]
- 61.Gilmore, K. S., R. R. B. Russell, and J. J. Ferretti. 1990. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect. Immun. 58:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 63.Guggenheim, B. 1970. Enzymatic hydrolysis and structure of water-insoluble glucan produced by glycosyltransferases from a strain of Streptococcus mutans. Helv. Odontol. Acta 14:89-109. [PubMed] [Google Scholar]
- 64.Haisman, R. J., and H. F. Jenkinson. 1991. Mutants of Streptococcus gordonii Challis over-producing glucosyltransferase. J. Gen. Microbiol. 137:483-489. [DOI] [PubMed] [Google Scholar]
- 65.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han, Y. W. 1990. Microbial levan. Adv. Appl. Microbiol. 35:171-194. [DOI] [PubMed] [Google Scholar]
- 67.Hanada, N., K. Fukushima, Y. Nomura, H. Senpuku, M. Hayakawa, H. Mukasa, T. Shiroza, and Y. Abiko. 2002. Cloning and nucleotide sequence analysis of the Streptococcus sobrinus gtfU gene that produces a highly branched water-soluble glucan. Biochim. Biophys. Acta 1570:75-79. [DOI] [PubMed] [Google Scholar]
- 68.Hanada, N., Y. Isobe, Y. Aizawa, T. Katayama, S. Sato, and M. Inoue. 1993. Nucleotide sequence analysis of the gtfT gene from Streptococcus sobrinus OMZ176. Infect. Immun. 61:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanada, N., and H. K. Kuramitsu. 1989. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect. Immun. 57:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hancock, R. A., K. Marshall, and H. Weigel. 1976. Structure of the levan elaborated by Streptococcus salivarius strain 51: an application of chemical-ionisation mass-spectrometry. Carbohydr. Res. 49:351-360. [DOI] [PubMed] [Google Scholar]
- 71.Hartmeier, W., M. Reiss, M. Heidel, and S. P. Marx. 1994. Biochemical and economical aspects of levan synthesis by Zymomonas mobilis. Biocatalysis 10:131-136. [Google Scholar]
- 72.Hernández, L., J. Arrieta, C. Menéndez, R. Vazquez, A. Coego, V. Suarez, G. Selman, M. F. Petit-Glatron, and R. Chambert. 1995. Isolation and enzymic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem. J. 309:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hestrin, S., S. Averini-Shapiro, and M. Aschner. 1943. The enzymic production of levan. Biochem. J. 37:450-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heyer, A. G., B. Schroeer, S. Radosta, D. Wolff, S. Czapla, and J. Springer. 1998. Structure of the enzymatically synthesized fructan inulin. Carbohydr. Res. 313:165-174. [DOI] [PubMed] [Google Scholar]
- 75.Heyer, A. G., and R. Wendenburg. 2001. Gene cloning and functional characterization by heterologous expression of the fructosyltransferase of Aspergillus sydowi IAM 2544. Appl. Environ. Microbiol. 67:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Honda, O., C. Kato, and H. K. Kuramitsu. 1990. Nucleotide sequence of the Streptococcus mutans gtfD gene encoding the glucosyltransferase-S enzyme. J. Gen. Microbiol. 136:2099-2105. [DOI] [PubMed] [Google Scholar]
- 77.Howard, J. C., C. Heinemann, B. J. Thatcher, B. Martin, B. S. Gan, and G. Reid. 2000. Identification of collagen-binding proteins in Lactobacillus spp. with surface-enhanced laser desorption/ionization-time of flight ProteinChip technology. Appl. Environ. Microbiol. 66:4396-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacques, N. A. 1984. Calcium dependence of the cell-associated fructosyltransferase of Streptococcus salivarius. Carbohydr. Res. 127:349-355. [DOI] [PubMed] [Google Scholar]
- 79.Janecek, S., B. Svensson, and R. R. B. Russell. 2000. Location of repeat elements in glucansucrases of Leuconostoc and Streptococcus species. FEMS Microbiol. Lett. 192:53-57. [DOI] [PubMed] [Google Scholar]
- 80.Jensen, M. H., O. Mirza, C. Albenne, M. Remaud-Simeon, P. F. Monsan, M. Gajhede, and L. K. Skov. 2004. Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea. Biochemistry 43:3104-3110. [DOI] [PubMed] [Google Scholar]
- 81.Kang, H. K., M. Y. Seo, E. S. Seo, D. S. Kim, S. Y. Chung, A. Kimura, D. F. Day, and J. F. Robyt. 2005. Cloning and expression of levansucrase from Leuconostoc mesenteroides B-512 FMC in Escherichia coli. Biochim. Biophys. Acta 1727:5-15. [DOI] [PubMed] [Google Scholar]
- 82.Kato, C., and H. K. Kuramitsu. 1990. Carboxyl-terminal deletion analysis of the Streptococcus mutans glucosyltransferase-I enzyme. FEMS Microbiol. Lett. 72:299-302. [DOI] [PubMed] [Google Scholar]
- 83.Kato, C., Y. J. Nakano, M. Lis, and H. K. Kuramitsu. 1992. Molecular genetic analysis of the catalytic site of Streptococcus mutans glucosyltransferases. Biochem. Biophys. Res. Commun. 189:1184-1188. [DOI] [PubMed] [Google Scholar]
- 84.Kaur, N., and A. K. Gupta. 2002. Applications of inulin and oligofructose in health and nutrition. J. Biosci. 27:703-714. [DOI] [PubMed] [Google Scholar]
- 85.Kim, D. S., J. F. Robyt, S. Y. Lee, J. H. Lee, and Y. M. Kim. 2003. Dextran molecular size and degree of branching as a function of sucrose concentration, pH, and temperature of reaction of Leuconostoc mesenteroides B-512FMCM dextransucrase. Carbohydr. Res. 338:1183-1189. [DOI] [PubMed] [Google Scholar]
- 86.Kim, Y. M., J. P. Park, J. Sinha, K. H. Lim, and J. W. Yun. 2001. Acceptor reactions of a novel transfructosylating enzyme from Bacillus sp. Biotechnol. Lett. 23:13-16. [Google Scholar]
- 87.Kingston, K. B., D. M. Allen, and N. A. Jacques. 2002. Role of the C-terminal YG repeats of the primer-dependent streptococcal glucosyltransferase, GtfJ, in binding to dextran and mutan. Microbiology 148:549-558. [DOI] [PubMed] [Google Scholar]
- 88.Knegtel, R. M. A., B. Strokopytov, D. Penninga, O. G. Faber, H. J. Rozeboom, K. H. Kalk, L. Dijkhuizen, and B. W. Dijkstra. 1995. Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products. J. Biol. Chem. 270:29256-29264. [DOI] [PubMed] [Google Scholar]
- 89.Koepsell, H. J., H. M. Tsuchiya, N. N. Hellmann, A. Kazenko, C. A. Hoffman, E. S. Sharpe, and R. W. Jackson. 1953. Enzymatic synthesis of dextran acceptor specificity and chain initiation. J. Biol. Chem. 200:793-801. [PubMed] [Google Scholar]
- 90.Konishi, N., Y. Torii, T. Yamamoto, A. Miyagi, H. Ohta, K. Fukui, S. Hanamoto, H. Matsuno, H. Komatsu, T. Kodama, and E. Katayama. 1999. Structure and enzymatic properties of genetically truncated forms of the water-insoluble glucan-synthesizing glucosyltransferase from Streptococcus sobrinus. J. Biochem. (Tokyo) 126:287-295. [DOI] [PubMed] [Google Scholar]
- 91.Koops, A. J., and H. H. Jonker. 1994. Purification and characterization of the enzymes of fructan biosynthesis in tubers of Helianthus tuberosus ‘Colombia’. J. Exp. Bot. 45:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Korakli, M., M. G. Ganzle, and R. F. Vogel. 2002. Metabolism by bifidobacteria and lactic acid bacteria of polysaccharides from wheat and rye, and exopolysaccharides produced by Lactobacillus sanfranciscensis. J. Appl. Microbiol. 92:958-965. [DOI] [PubMed] [Google Scholar]
- 93.Korakli, M., A. Rossmann, M. G. Ganzle, and R. F. Vogel. 2001. Sucrose metabolism and exopolysaccharide production in wheat and rye sourdoughs by Lactobacillus sanfranciscensis. J. Agric. Food Chem. 49:5194-5200. [DOI] [PubMed] [Google Scholar]
- 94.Kralj, S., E. Stripling, G. H. Van Geel-Schutten, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2005. Highly hydrolytic reuteransucrase from probiotic Lactobacillus reuteri strain ATCC 55730. Appl. Environ. Microbiol. 71:3942-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kralj, S., G. H. Van Geel-Schutten, M. M. G. Dondorff, S. Kirsanovs, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2004. Glucan synthesis in the genus Lactobacillus: isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 150:3681-3690. [DOI] [PubMed] [Google Scholar]
- 96.Kralj, S., G. H. Van Geel-Schutten, E. J. Faber, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2005. Rational transformation of Lactobacillus reuteri 121 reuteransucrase into a dextransucrase. Biochemistry 44:9206-9216. [DOI] [PubMed] [Google Scholar]
- 97.Kralj, S., G. H. Van Geel-Schutten, H. Rahaoui, R. J. Leer, E. J. Faber, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2002. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with α-(1->4) and α-(1->6) glucosidic bonds. Appl. Environ. Microbiol. 68:4283-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kralj, S., G. H. Van Geel-Schutten, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2003. Efficient screening methods for glucosyltransferase genes in Lactobacillus strains. Biocatal. Biotransform. 21:181-187. [Google Scholar]
- 99.Kralj, S., G. H. Van Geel-Schutten, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2004. Biochemical and molecular characterization of Lactobacillus reuteri 121 reuteransucrase. Microbiology 150:2099-2112. [DOI] [PubMed] [Google Scholar]
- 100.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuriki, T., H. Takata, S. Okada, and T. Imanaka. 1991. Analysis of the active center of Bacillus stearothermophilus neopullulanase. J. Bacteriol. 173:6147-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leemhuis, H., B. W. Dijkstra, and L. Dijkhuizen. 2002. Mutations converting cyclodextrin glycosyltransferase from a transglycosylase into a starch hydrolase. FEBS Lett. 514:189-192. [DOI] [PubMed] [Google Scholar]
- 103.Leemhuis, H., H. J. Rozeboom, B. W. Dijkstra, and L. Dijkhuizen. 2003. The fully conserved Asp residue in conserved sequence region I of the alpha-amylase family is crucial for the catalytic site architecture and activity. FEBS Lett. 541:47-51. [DOI] [PubMed] [Google Scholar]
- 104.Leemhuis, H., H. J. Rozeboom, M. Wilbrink, G. J. Euverink, B. W. Dijkstra, and L. Dijkhuizen. 2003. Conversion of cyclodextrin glycosyltransferase into a starch hydrolase by directed evolution: the role of alanine 230 in acceptor subsite +1. Biochemistry 42:7518-7526. [DOI] [PubMed] [Google Scholar]
- 105.Leemhuis, H., U. F. Wehmeier, and L. Dijkhuizen. 2004. Single amino acid mutations interchange the reaction specificities of cyclodextrin glycosyltransferase and the acarbose modifying enzyme acarviosyl transferase. Biochemistry 43:13204-13213. [DOI] [PubMed] [Google Scholar]
- 106.L'Hocine, L., Z. Wang, B. Jiang, and S. Xu. 2000. Purification and partial characterization of fructosyltransferase and invertase from Aspergillus niger AS0023. J. Biotechnol. 81:73-84. [DOI] [PubMed] [Google Scholar]
- 107.Li, Y., J. A. Triccas, and T. Ferenci. 1997. A novel levansucrase-levanase gene cluster in Bacillus stearothermophilus ATCC12980. Biochim. Biophys. Acta 1353:203-208. [DOI] [PubMed] [Google Scholar]
- 108.Lis, M., T. Shiroza, and H. K. Kuramitsu. 1995. Role of C-terminal direct repeating units of the Streptococcus mutans glucosyltransferase-S in glucan binding. Appl. Environ. Microbiol. 61:2040-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 111.MacGregor, E. A., H. M. Jespersen, and B. Svensson. 1996. A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 378:263-266. [DOI] [PubMed] [Google Scholar]
- 112.Martinez-Fleites, C., M. Ortiz-Lombardia, T. Pons, N. Tarbouriech, E. J. Taylor, J. G. Arrieta, L. Hernández, and G. J. Davies. 2005. Crystal structure of levansucrase from the gram-negative bacterium Gluconacetobacter diazotrophicus. Biochem. J. 390:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meng, G., and K. Futterer. 2003. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 10:935-941. [DOI] [PubMed] [Google Scholar]
- 114.Meulenbeld, G. H., and S. Hartmans. 2000. Transglycosylation by Streptococcus mutans GS-5 glucosyltransferase-D: acceptor specificity and engineering of reaction conditions. Biotechnol. Bioeng. 70:363-369. [DOI] [PubMed] [Google Scholar]
- 115.Mirza, O., L. K. Skov, M. Remaud-Simeon, G. Potocki de Montalk, C. Albenne, P. F. Monsan, and M. Gajhede. 2001. Crystal structures of amylosucrase from Neisseria polysaccharea in complex with d-glucose and the active site mutant Glu328Gln in complex with the natural substrate sucrose. Biochemistry 40:9032-9039. [DOI] [PubMed] [Google Scholar]
- 116.Monchois, V., M. A. Arguello-Morales, and R. R. B. Russell. 1999. Isolation of an active catalytic core of Streptococcus downei MFe28 GTF-I glucosyltransferase. J. Bacteriol. 181:2290-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Monchois, V., J. H. Lakey, and R. R. B. Russell. 1999. Secondary structure of Streptococcus downei GTF-1 glucansucrase. FEMS Microbiol. Lett. 177:243-248. [DOI] [PubMed] [Google Scholar]
- 118.Monchois, V., M. Remaud-Simeon, P. F. Monsan, and R. M. Willemot. 1998. Cloning and sequencing of a gene coding for an extracellular dextransucrase (DSRB) from Leuconostoc mesenteroides NRRL B-1299 synthesizing only a α(1-6) glucan. FEMS Microbiol. Lett. 159:307-315. [DOI] [PubMed] [Google Scholar]
- 119.Monchois, V., M. Remaud-Simeon, R. R. B. Russell, P. F. Monsan, and R. M. Willemot. 1997. Characterization of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSRS) and identification of amino-acid residues playing a key role in enzyme activity. Appl. Microbiol. Biotechnol. 48:465-472. [DOI] [PubMed] [Google Scholar]
- 120.Monchois, V., A. Reverte, M. Remaud-Simeon, P. F. Monsan, and R. M. Willemot. 1998. Effect of Leuconostoc mesenteroides NRRL B-512F dextransucrase carboxy-terminal deletions on dextran and oligosaccharide synthesis. Appl. Environ. Microbiol. 64:1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Monchois, V., M. R. Vignon, P. C. Escalier, B. Svensson, and R. R. B. Russell. 2000. Involvement of Gln937 of Streptococcus downei GTF-I glucansucrase in transition-state stabilization. Eur. J. Biochem. 267:4127-4136. [DOI] [PubMed] [Google Scholar]
- 122.Monchois, V., M. R. Vignon, and R. R. B. Russell. 1999. Isolation of key amino acid residues at the N-terminal end of the core region Streptococcus downei glucansucrase, GTF-I. Appl. Microbiol. Biotechnol. 52:660-665. [DOI] [PubMed] [Google Scholar]
- 123.Monchois, V., M. R. Vignon, and R. R. B. Russell. 2000. Mutagenesis of Asp-569 of glucosyltransferase I glucansucrase modulates glucan and oligosaccharide synthesis. Appl. Environ. Microbiol. 66:1923-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Monchois, V., R. M. Willemot, and P. F. Monsan. 1999. Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol. Rev. 23:131-151. [DOI] [PubMed] [Google Scholar]
- 125.Monchois, V., R. M. Willemot, M. Remaud-Simeon, C. Croux, and P. F. Monsan. 1996. Cloning and sequencing of a gene coding for a novel dextransucrase from Leuconostoc mesenteroides NRRL B-1299 synthesizing only α(1-6) and α(1-3) linkages. Gene 182:23-32. [DOI] [PubMed] [Google Scholar]
- 126.Monsan, P. F., S. Bozonnet, C. Albenne, G. Joucla, R. M. Willemot, and M. Remaud-Simeon. 2001. Homopolysaccharides from lactic acid bacteria. Int. Dairy J. 11:675-685. [Google Scholar]
- 127.Monsan, P. F., and F. Paul. 1995. Oligosaccharide feed additives, p. 233-245. In R. J. Wallace and A. Chesson (ed.), Biotechnology in animal feeds and animal feeding. VCH, Weinheim, Germany.
- 128.Mooser, G., S. A. Hefta, R. J. Paxton, J. E. Shively, and T. D. Lee. 1991. Isolation and sequence of an active-site peptide containing a catalytic aspartic acid from two Streptococcus sobrinus α-glucosyltransferases. J. Biol. Chem. 266:8916-8922. [PubMed] [Google Scholar]
- 129.Mooser, G., and K. R. Iwaoka. 1989. Sucrose 6-α-d-glucosyltransferase from Streptococcus sobrinus: characterization of a glucosyl-enzyme complex. Biochemistry 28:443-449. [DOI] [PubMed] [Google Scholar]
- 130.Mukasa, H., A. Shimamura, and H. Tsumori. 2000. Nigerooligosaccharide acceptor reaction of Streptococcus sobrinus glucosyltransferase GTF-I. Carbohydr. Res. 326:98-103. [DOI] [PubMed] [Google Scholar]
- 131.Nakamura, A., K. Haga, and K. Yamane. 1993. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilization. Biochemistry 32:6624-6631. [DOI] [PubMed] [Google Scholar]
- 132.Nakano, Y. J., and H. K. Kuramitsu. 1992. Mechanism of Streptococcus mutans glucosyltransferases: hybrid-enzyme analysis. J. Bacteriol. 174:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 135.Neubauer, H., A. Bauche, and B. Mollet. 2003. Molecular characterization and expression analysis of the dextransucrase DsrD of Leuconostoc mesenteroides Lcc4 in homologous and heterologous Lactococcus lactis cultures. Microbiology 149:973-982. [DOI] [PubMed] [Google Scholar]
- 136.Newbrun, E., and S. Baker. 1968. Physico-chemical characteristics of the levan produced by Streptococcus salivarius. Carbohydr. Res. 6:165-170. [Google Scholar]
- 137.Olivares-Illana, V., A. Lopez-Munguia, and C. Olvera. 2003. Molecular characterization of inulosucrase from Leuconostoc citreum: a fructosyltransferase within a glucosyltransferase. J. Bacteriol. 185:3606-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Olivares-Illana, V., C. Wacher-Rodarte, S. Le Borgne, and A. López-Munguía. 2002. Characterization of a cell-associated inulosucrase from a novel source: A Leuconostoc citreum strain isolated from Pozol, a fermented corn beverage from Mayan origin. J. Ind. Microbiol. Biotechnol. 28:112-117. [DOI] [PubMed] [Google Scholar]
- 139.Ozimek, L. K., G. J. Euverink, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2005. Mutational analysis of the role of calcium ions in the Lactobacillus reuteri strain 121 fructosyltransferase (levansucrase and inulosucrase) enzymes. FEBS Lett. 579:1124-1128. [DOI] [PubMed] [Google Scholar]
- 140.Ozimek, L. K., S. A. F. T. Van Hijum, G. A. Van Koningsveld, M. J. E. C. Van Der Maarel, G. H. Van Geel-Schutten, and L. Dijkhuizen. 2004. Site-directed mutagenesis study of the three catalytic residues of the fructosyltransferases of Lactobacillus reuteri 121. FEBS Lett. 560:131-133. [DOI] [PubMed] [Google Scholar]
- 141.Parker, R. B., and H. R. Creamer. 1971. Contribution of plaque polysaccharides to growth of cariogenic microorganisms. Arch. Oral Biol. 16:855-862. [DOI] [PubMed] [Google Scholar]
- 142.Parolis, L. A., H. Parolis, L. Kenne, M. Meldal, and K. Bock. 1998. The extracellular polysaccharide of Pichia (Hansenula) holstii NRRL Y-2448: the phosphorylated side chains. Carbohydr. Res. 309:77-87. [DOI] [PubMed] [Google Scholar]
- 143.Pasteur, L. 1861. Sur la fermentation visqueuse et la fermentation butyrique. Bull. Soc. Chim. 11:30-31. [Google Scholar]
- 144.Perez-Oseguera, M. A., L. Guereca, and A. Lopez-Munguia. 1996. Properties of levansucrase from Bacillus circulans. Appl. Microbiol. Biotechnol. 45:465-471. [Google Scholar]
- 145.Potocki de Montalk, G., M. Remaud-Simeon, R. M. Willemot, P. Sarcabal, V. Planchot, and P. F. Monsan. 2000. Amylosucrase from Neisseria polysaccharea: novel catalytic properties. FEBS Lett. 471:219-223. [DOI] [PubMed] [Google Scholar]
- 146.Qi, Q., and W. Zimmermann. 2005. Cyclodextrin glucanotransferase: from gene to applications. Appl. Microbiol. Biotechnol. 66:475-485. [DOI] [PubMed] [Google Scholar]
- 147.Rathsam, C., P. M. Giffard, and N. A. Jacques. 1993. The cell-bound fructosyltransferase of Streptococcus salivarius: the carboxyl terminus specifies attachment in a Streptococcus gordonii model system. J. Bacteriol. 175:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rathsam, C., and N. A. Jacques. 1998. Role of C-terminal domains in surface attachment of the fructosyltransferase of Streptococcus salivarius ATCC 25975. J. Bacteriol. 180:6400-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Remaud-Simeon, M., R. M. Willemot, P. Sarcabal, G. Potocki de Montalk, and P. F. Monsan. 2000. Glucansucrases: molecular engineering and oligosaccharide synthesis. J. Mol. Catal. B Enzym. 10:177-198. [Google Scholar]
- 150.Richard, G., S. Morel, R. M. Willemot, P. F. Monsan, and M. Remaud-Simeon. 2003. Glucosylation of α-butyl- and α-octyl-d-glucopyranosides by dextransucrase and alternansucrase from Leuconostoc mesenteroides. Carbohydr. Res. 338:855-864. [DOI] [PubMed] [Google Scholar]
- 151.Roberfroid, M. B. 1999. Concepts in functional foods: the case of inulin and oligofructose. J. Nutr. 129:1398S-1401S. [DOI] [PubMed] [Google Scholar]
- 152.Robyt, J. F. 1998. Essentials of carbohydrate chemistry. Springer-Verlag, New York, N.Y.
- 153.Robyt, J. F. 1996. Mechanism and action of glucansucrase, p. 1-22. In K. H. Park, J. F. Robyt, and Y. D. Choi (ed.), Enzymes for carbohydrate engineering. Elsevier Science B.V., Amsterdam, The Netherlands.
- 154.Robyt, J. F. 1996. Mechanism and action of glucansucrases. Carbohydr. Bioeng. 10:295-312. [Google Scholar]
- 155.Robyt, J. F., B. K. Kimble, and T. F. Walseth. 1974. The mechanism of dextransucrase action. Direction of dextran biosynthesis. Arch. Biochem. Biophys. 165:634-640. [DOI] [PubMed] [Google Scholar]
- 156.Robyt, J. F., and T. F. Walseth. 1979. Production, purification, and properties of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 68:95-111. [DOI] [PubMed] [Google Scholar]
- 157.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 158.Rosell, K. G., and D. Birkhed. 1974. An inulin-like fructan produced by Streptococcus mutans strain JC2. Acta Chem. Scand. B 28:589. [DOI] [PubMed] [Google Scholar]
- 159.Rozen, R., G. Bachrach, M. Bronshteyn, I. Gedalia, and D. Steinberg. 2001. The role of fructans on dental biofilm formation by Streptococcus sobrinus, Streptococcus mutans, Streptococcus gordonii and Actinomyces viscosus. FEMS Microbiol. Lett. 195:205-210. [DOI] [PubMed] [Google Scholar]
- 160.Ruiz-Herrera, J. 1991. Biosynthesis of beta-glucans in fungi. Antonie van Leeuwenhoek 60:72-81. [DOI] [PubMed] [Google Scholar]
- 161.Russell, R. R. B. 1990. Molecular genetics of glucan metabolism in oral streptococci. Arch. Oral Biol. 35:53-58. [DOI] [PubMed] [Google Scholar]
- 162.Russell, R. R. B. 1994. The application of molecular genetics to the microbiology of dental caries. Caries Res. 28:69-82. [DOI] [PubMed] [Google Scholar]
- 163.Russell, R. R. B., M. L. Gilpin, H. Mukasa, and G. Dougan. 1987. Characterization of glucosyltransferase expressed from a Streptococcus sobrinus gene cloned in Escherichia coli. J. Gen. Microbiol. 133:935-944. [DOI] [PubMed] [Google Scholar]
- 164.Rycroft, C. E., M. R. Jones, G. R. Gibson, and R. A. Rastall. 2001. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91:878-887. [DOI] [PubMed] [Google Scholar]
- 165.Sarcabal, P., M. Remaud-Simeon, R. M. Willemot, G. Potocki de Montalk, B. Svensson, and P. F. Monsan. 2000. Identification of key amino acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 474:33-37. [DOI] [PubMed] [Google Scholar]
- 166.Sato, S., and H. K. Kuramitsu. 1986. Isolation and characterization of a fructosyltransferase gene from Streptococcus mutans GS-5. Infect. Immun. 52:166-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sato, Y., and H. K. Kuramitsu. 1988. Sequence analysis of the Streptococcus mutans scrB gene. Infect. Immun. 56:1956-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Selbmann, L., F. Stingele, and M. Petruccioli. 2003. Exopolysaccharide production by filamentous fungi: the example of Botryosphaeria rhodina. Antonie van Leeuwenhoek 84:135-145. [DOI] [PubMed] [Google Scholar]
- 169.Senthilkumar, V., S. J. Busby, and P. Gunasekaran. 2003. Serine substitution for cysteine residues in levansucrase selectively abolishes levan forming activity. Biotechnol. Lett. 25:1653-1656. [DOI] [PubMed] [Google Scholar]
- 170.Seymour, F. R., E. C. M. Chen, and S. H. Bishop. 1979. Methylation structural analysis of unusual dextrans by combined gas-liquid chromatography-mass spectrometry. Carbohydr. Res. 68:113-121. [Google Scholar]
- 171.Shah, D. S. H., G. Joucla, M. Remaud-Simeon, and R. R. B. Russell. 2004. Conserved repeat motifs and glucan binding by glucansucrases of oral streptococci and Leuconostoc mesenteroides. J. Bacteriol. 186:8301-8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shah, D. S. H., and R. R. B. Russell. 2002. Glucan binding domain of streptococcal glucosyltransferases. Biologia 57:129-136. [Google Scholar]
- 173.Shimamura, A., Y. J. Nakano, H. Mukasa, and H. K. Kuramitsu. 1994. Identification of amino acid residues in Streptococcus mutans. Glucosyltransferases influencing the structure of the glucan product. J. Bacteriol. 176:4845-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shiroza, T., and H. K. Kuramitsu. 1988. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J. Bacteriol. 170:810-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shiroza, T., S. Ueda, and H. K. Kuramitsu. 1987. Sequence analysis of the gtfB gene from Streptococcus mutans. J. Bacteriol. 169:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sidebotham, R. L. 1974. Dextrans. Adv. Carbohydr. Chem. Biochem. 30:371-444. [DOI] [PubMed] [Google Scholar]
- 177.Sillanpaa, J., B. Martinez, J. Antikainen, T. Toba, N. Kalkkinen, S. Tankka, K. Lounatmaa, J. Keranen, M. Hook, B. Westerlund-Wikstrom, P. H. Pouwels, and T. K. Korhonen. 2000. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182:6440-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Simms, P. J., W. J. Boyko, and J. R. Edwards. 1990. The structural analysis of a levan produced by Streptococcus salivarius SS2. Carbohydr. Res. 208:193-198. [DOI] [PubMed] [Google Scholar]
- 179.Simpson, C. L., N. W. H. Cheetham, P. M. Giffard, and N. A. Jacques. 1995. Four glucosyltransferases, GtfJ, GtfK, GtfL and GtfM, from Streptococcus salivarius ATCC 25975. Microbiology 141:1451-1460. [DOI] [PubMed] [Google Scholar]
- 180.Sinnott, M. L. 1987. Glycosyl group transfer, p. 259-297. In M. I. Page and A. Williams (ed.), Enzyme mechanisms. Royal Society of Chemistry, London, England.
- 181.Skov, L. K., O. Mirza, A. Henriksen, G. Potocki de Montalk, M. Remaud-Simeon, P. Sarcabal, R. M. Willemot, P. F. Monsan, and M. Gajhede. 2000. Crystallization and preliminary X-ray studies of recombinant amylosucrase from Neisseria polysaccharea. Acta Crystallogr. D Biol. Crystallogr. 56:203-205. [DOI] [PubMed] [Google Scholar]
- 182.Skov, L. K., O. Mirza, A. Henriksen, G. Potocki de Montalk, M. Remaud-Simeon, P. Sarcabal, R. M. Willemot, P. F. Monsan, and M. Gajhede. 2001. Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J. Biol. Chem. 276:25273-25278. [DOI] [PubMed] [Google Scholar]
- 183.Skov, L. K., O. Mirza, D. Sprogoe, I. Dar, M. Remaud-Simeon, C. Albenne, P. F. Monsan, and M. Gajhede. 2002. Oligosaccharide and sucrose complexes of amylosucrase. Structural implications for the polymerase activity. J. Biol. Chem. 277:47741-47747. [DOI] [PubMed] [Google Scholar]
- 184.Smith, M. R., R. Y. Wong, R. E. Lundin, and J. A. Ahlgren. 1998. A mutant strain of Leuconostoc mesenteroides B-1355 producing a glucosyltransferase synthesizing α(1-2) glucosidic linkages. J. Ind. Microbiol. Biotechnol. 21:37-45. [Google Scholar]
- 185.Song, D. D., and N. A. Jacques. 1999. Mutation of aspartic acid residues in the fructosyltransferase of Streptococcus salivarius ATCC 25975. Biochem. J. 344:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Song, D. D., and N. A. Jacques. 1999. Purification and enzymic properties of the fructosyltransferase of Streptococcus salivarius ATCC 25975. Biochem. J. 341:285-291. [PMC free article] [PubMed] [Google Scholar]
- 187.Song, K. B., H. K. Joo, and S. K. Rhee. 1993. Nucleotide sequence of levansucrase gene (levU) of Zymomonas mobilis ZM1 (ATCC10988). Biochim. Biophys. Acta 1173:320-324. [DOI] [PubMed] [Google Scholar]
- 188.Steinberg, D., R. Rozen, M. Bromshteym, B. Zaks, I. Gedalia, and G. Bachrach. 2002. Regulation of fructosyltransferase activity by carbohydrates, in solution and immobilized on hydroxyapatite surfaces. Carbohydr. Res. 337:701-710. [DOI] [PubMed] [Google Scholar]
- 189.Steinmetz, M., D. Le Coq, S. Aymerich, G. Gonzy-Treboul, and P. Gay. 1985. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol. Gen. Genet. 200:220-228. [DOI] [PubMed] [Google Scholar]
- 190.Sutherland, I. W. 1972. Bacterial exopolysaccharides. Adv. Microbiol. Physiol. 8:143-212. [DOI] [PubMed] [Google Scholar]
- 191.Svensson, B. 1994. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25:141-157. [DOI] [PubMed] [Google Scholar]
- 192.Tanaka, T., S. Oi, and T. Yamamoto. 1980. The molecular structure of low and high molecular weight levans synthesized by levansucrase. J. Biochem. (Tokyo) 87:297-303. [DOI] [PubMed] [Google Scholar]
- 193.Tanaka, T., S. Oi, and T. Yamamoto. 1979. Synthesis of levan by levansucrase. Some factors affecting the rate of synthesis and degree of polymerization of levan. J. Biochem. (Tokyo) 85:287-293. [DOI] [PubMed] [Google Scholar]
- 194.Tanaka, T., S. Yamamoto, S. Oi, and T. Yamamoto. 1981. Structures of heterooligosaccharides synthesized by levansucrase. J. Biochem. (Tokyo) 90:521-526. [DOI] [PubMed] [Google Scholar]
- 195.Tang, L. B., R. Lenstra, T. B. Borchert, and V. Nagarajan. 1990. Isolation and characterization of levansucrase encoding gene from Bacillus amyloliquefaciens. Gene 96:89-93. [DOI] [PubMed] [Google Scholar]
- 196.Tieking, M., M. A. Ehrmann, R. F. Vogel, and M. G. Ganzle. 2005. Molecular and functional characterization of a levansucrase from the sourdough isolate Lactobacillus sanfranciscensis TMW 1.392. Appl. Microbiol. Biotechnol. 66:655-663. [DOI] [PubMed] [Google Scholar]
- 197.Tieking, M., S. Kaditzky, R. Valcheva, M. Korakli, R. F. Vogel, and M. G. Ganzle. 2005. Extracellular homopolysaccharides and oligosaccharides from intestinal lactobacilli. J. Appl. Microbiol. 99:692-702. [DOI] [PubMed] [Google Scholar]
- 198.Tieking, M., M. Korakli, M. A. Ehrmann, M. G. Ganzle, and R. F. Vogel. 2003. In situ production of exopolysaccharides during sourdough fermentation by cereal and intestinal isolates of lactic acid bacteria. Appl. Environ. Microbiol. 69:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tieking, M., W. Kuhnl, and M. G. Ganzle. 2005. Evidence for formation of heterooligosaccharides by Lactobacillus sanfranciscensis during growth in wheat sourdough. J. Agric. Food Chem. 53:2456-2461. [DOI] [PubMed] [Google Scholar]
- 200.Ton-That, H., K. F. Faull, and O. Schneewind. 1997. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272:22285-22292. [DOI] [PubMed] [Google Scholar]
- 201.Tsuchiya, H. M., N. N. Hellmann, and H. J. Koepsell. 1953. Factors affecting molecular weight of enzymatically synthesized dextran. J. Am. Chem. Soc. 75:757-758. [Google Scholar]
- 202.Tsumori, H., T. Minami, and H. K. Kuramitsu. 1997. Identification of essential amino acids in the Streptococcus mutans glucosyltransferases. J. Bacteriol. 179:3391-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ueda, S., T. Shiroza, and H. K. Kuramitsu. 1988. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene 69:101-109. [DOI] [PubMed] [Google Scholar]
- 204.Uitdehaag, J. C., R. Mosi, K. H. Kalk, B. A. Van Der Veen, L. Dijkhuizen, S. G. Withers, and B. W. Dijkstra. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 6:432-436. [DOI] [PubMed] [Google Scholar]
- 205.Van Den Ende, W., A. Michiels, J. De Roover, and A. J. Van Laere. 2002. Fructan biosynthetic and breakdown enzymes in dicots evolved from different invertases. Expression of fructan genes throughout chicory development. Sci. World J. 2:1273-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Van Der Veen, B. A., H. Leemhuis, S. Kralj, J. C. Uitdehaag, B. W. Dijkstra, and L. Dijkhuizen. 2001. Hydrophobic amino acid residues in the acceptor binding site are main determinants for reaction mechanism and specificity of cyclodextrin-glycosyltransferase. J. Biol. Chem. 276:44557-44562. [DOI] [PubMed] [Google Scholar]
- 207.Van Der Veen, B. A., J. C. Uitdehaag, B. W. Dijkstra, and L. Dijkhuizen. 2000. Engineering of cyclodextrin glycosyltransferase reaction and product specificity. Biochim. Biophys. Acta 1543:336-360. [DOI] [PubMed] [Google Scholar]
- 208.Van Geel-Schutten, G. H., E. J. Faber, E. Smit, K. Bonting, M. R. Smith, B. Ten Brink, J. P. Kamerling, J. F. G. Vliegenthart, and L. Dijkhuizen. 1999. Biochemical and structural characterization of the glucan and fructan exopolysaccharides synthesized by the Lactobacillus reuteri wild-type strain and by mutant strains. Appl. Environ. Microbiol. 65:3008-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Van Geel-Schutten, G. H., F. Flesch, B. Ten Brink, M. R. Smith, and L. Dijkhuizen. 1998. Screening and characterization of Lactobacillus strains producing large amounts of exopolysaccharides. Appl. Microbiol. Biotechnol. 50:697-703. [Google Scholar]
- 210.Van Hijum, S. A. F. T., K. Bonting, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2001. Purification of a novel fructosyltransferase from Lactobacillus reuteri strain 121 and characterization of the levan produced. FEMS Microbiol. Lett. 205:323-328. [DOI] [PubMed] [Google Scholar]
- 211.Van Hijum, S. A. F. T., E. Szalowska, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2004. Biochemical and molecular characterization of a levansucrase from Lactobacillus reuteri. Microbiology 150:621-630. [DOI] [PubMed] [Google Scholar]
- 212.Van Hijum, S. A. F. T., M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2003. Kinetic properties of an inulosucrase from Lactobacillus reuteri 121. FEBS Lett. 534:207-210. [DOI] [PubMed] [Google Scholar]
- 213.Van Hijum, S. A. F. T., G. H. Van Geel-Schutten, H. Rahaoui, M. J. E. C. Van Der Maarel, and L. Dijkhuizen. 2002. Characterization of a novel fructosyltransferase from Lactobacillus reuteri that synthesizes high-molecular-weight inulin and inulin oligosaccharides. Appl. Environ. Microbiol. 68:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vickerman, M. M., M. C. Sulavik, P. E. Minick, and D. B. Clewell. 1996. Changes in the carboxyl-terminal repeat region affect extracellular activity and glucan products of Streptococcus gordonii glucosyltransferase. Infect. Immun. 64:5117-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vickerman, M. M., M. C. Sulavik, J. D. Nowak, N. M. Gardner, G. W. Jones, and D. B. Clewell. 1997. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene, gtfG. DNA Seq. 7:83-95. [DOI] [PubMed] [Google Scholar]
- 216.Vijn, I., and S. Smeekens. 1999. Fructan: more than a reserve carbohydrate? Plant Physiol. 120:351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Welman, A. D., and I. S. Maddox. 2003. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. Trends Biotechnol. 21:269-274. [DOI] [PubMed] [Google Scholar]
- 218.Whitfield, C. 1988. Bacterial extracellular polysaccharides. Can. J. Microbiol. 34:415-420. [DOI] [PubMed] [Google Scholar]
- 219.Yanase, H., M. Iwata, R. Nakahigashi, K. Kita, N. Kato, and K. Tonomura. 1992. Purification, crystallization and properties of the extracellular levansucrase from Zymomonas mobilis. Biosci. Biotechnol. Biochem. 56:1335-1337. [Google Scholar]
- 220.Yanase, H., M. Maeda, E. Hagiwara, H. Yagi, K. Taniguchi, and K. Okamoto. 2002. Identification of functionally important amino acid residues in Zymomonas mobilis levansucrase. J. Biochem. (Tokyo) 132:565-572. [DOI] [PubMed] [Google Scholar]
- 221.Yoon, S. H., D. B. Fulton, and J. F. Robyt. 2004. Enzymatic synthesis of two salicin analogues by reaction of salicyl alcohol with Bacillus macerans cyclomaltodextrin glucanyltransferase and Leuconostoc mesenteroides B-742CB dextransucrase. Carbohydr. Res. 339:1517-1529. [DOI] [PubMed] [Google Scholar]






