Abstract
The cell wall envelopes of gram-positive bacteria represent a surface organelle that not only functions as a cytoskeletal element but also promotes interactions between bacteria and their environment. Cell wall peptidoglycan is covalently and noncovalently decorated with teichoic acids, polysaccharides, and proteins. The sum of these molecular decorations provides bacterial envelopes with species- and strain-specific properties that are ultimately responsible for bacterial virulence, interactions with host immune systems, and the development of disease symptoms or successful outcomes of infections. Surface proteins typically carry two topogenic sequences, i.e., N-terminal signal peptides and C-terminal sorting signals. Sortases catalyze a transpeptidation reaction by first cleaving a surface protein substrate at the cell wall sorting signal. The resulting acyl enzyme intermediates between sortases and their substrates are then resolved by the nucleophilic attack of amino groups, typically provided by the cell wall cross bridges of peptidoglycan precursors. The surface protein linked to peptidoglycan is then incorporated into the envelope and displayed on the microbial surface. This review focuses on the mechanisms of surface protein anchoring to the cell wall envelope by sortases and the role that these enzymes play in bacterial physiology and pathogenesis.
INTRODUCTION
The cell wall envelopes of gram-positive bacteria represent a surface organelle that not only functions as a cytoskeletal element for the physical integrity of microbes but also promotes interactions between bacteria and their environment (60). Most importantly for bacterial pathogens, as environments are subject to change, microbes respond with alterations in envelope structure and function. Thus, one should consider the cell wall envelope a dynamic organelle, one that is continuously assembled from precursor molecules and disassembled into individual constituents.
Bacterial cell wall assembly requires peptidoglycan precursors that together form a single large macromolecule, the murein sacculus, encircling the microbial cell with a 20- to 100-nm-thick wall structure (61). Cell wall peptidoglycan is covalently and noncovalently decorated with teichoic acids, polysaccharides, and proteins. The sum of these molecular decorations provide bacterial envelopes with species- and strain-specific properties that, for pathogens, contribute greatly to bacterial virulence, interactions with host immune systems, and the development of disease symptoms or successful outcomes of infections. This review focuses on the mechanisms of surface protein anchoring to the cell wall envelope by sortases and the roles that these enzymes play in bacterial physiology and pathogenesis. Interested readers are referred to other excellent reviews that have examined in depth the structure and assembly of peptidoglycan, teichoic acids, and polysaccharides or proteins that are noncovalently associated with the cell wall envelope (136, 139, 144, 187).
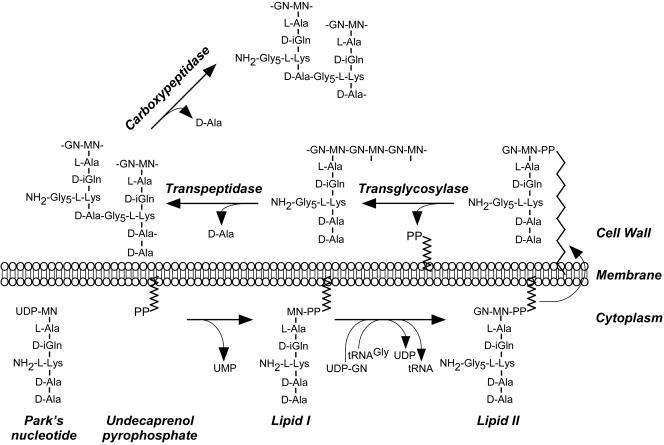
In Staphylococcus aureus, peptidoglycan precursor molecules are fabricated from N-acetylmuramic acid (MurNAc) and l- as well as d-stereoisomer amino acids in the bacterial cytoplasm to yield a soluble intermediate, Park's nucleotide (UDP-MurNAc-l-Ala-d-isoGln-l-Lys-d-Ala-d-Ala) (24) (Fig. 1). The precursor is tethered via phosphodiester linkage to a bactoprenol carrier, generating lipid I (C55-PP-MurNAc-l-Ala-d-isoGln-l-Lys-d-Ala-d-Ala) in the membrane (24, 117, 118). Further modification with N-acetylglucosamine (GlcNAc) and cross bridge decoration at the ɛ-amino of l-Lys (pentaglycine or Gly5 in staphylococci) generates lipid II {C55-PP-MurNAc-[l-Ala-d-isoGln-l-Lys(Gly5)-d-Ala-d-Ala]-β(1-4)-GlcNAc)}. Lipid II is translocated across the cell membrane (133), where it becomes a substrate for penicillin binding proteins (PBPs) that catalyze transglycosylation and transpeptidation reactions. Transglycosylation polymerizes MurNAc-GlcNAc subunits into repeating disaccharide chains, also called glycan strands (194). Transpeptidation involves first cleavage of the pentapeptide precursor [l-Ala-d-isoGln-l-Lys(Gly5)-d-Ala-d-Ala] at the terminal d-Ala and then formation of an amide bond between the carboxyl group of d-Ala at position four and the amino groups of pentaglycine cross bridges in other wall peptides (85). PBPs use these two reactions together to form a single large macromolecule that displays rigid exoskeletal functions and that serves as a scaffold for the incorporation of other molecules that can be attached to cross bridges, wall peptides, or glycan strands. Peptidoglycan biosynthesis in other bacteria follows a similar scheme, with two exceptions. First, d-isoGlu at position two of wall peptides is typically not amidated. Second, l-Lys, the diamino acid at position three of wall peptides, can be substituted with m-diaminopimelic acid, and the attached cell wall cross bridges can vary in chemical nature between different bacterial species (170).
FIG. 1.
Peptidoglycan synthesis in S. aureus. Park's nucleotide, a soluble nucleotide precursor, originates in the bacterial cytoplasm by successive addition of l-stereoisomer amino acids (l-Ala and l-Lys) as well as d-stereoisomer amino acids (d-isoglutamine [d-iGln] and d-Ala) to UDP-N-acetylmuramic acid (UDP-NM). Precursor transfer to undecaprenol pyrophosphate, a bacterial membrane carrier, generates lipid I and removes UMP nucleotide. Lipid I modification with N-acetylglucosamine (GN) and pentaglycine cross bridge formation at the ɛ-amino of l-Lys with tRNAGly substrate generates lipid II. Following translocation across the cytoplasmic membrane, lipid II serves as substrate for PBPs that catalyze three reactions: transglycosylation, transpeptidation, and carboxypeptidation. Transglycosylases polymerize MN-GN subunits into repeating disaccharide chains, the glycan strands. Transpeptidases cleave the amide bond of the terminal d-Ala in pentapeptide precursors and generate an amide bond between the carboxyl group of d-Ala at position four and the amino group of pentaglycine cross bridges in wall peptides. Carboxypeptidases hydrolyze the C-terminal d-Ala of most non-cross-linked pentapeptides to yield mature peptidoglycan.
Sortases promote the covalent anchoring of surface proteins to the cell wall envelope (120). These enzymes catalyze a transpeptidation reaction by first cleaving a surface protein substrate at the cell wall sorting signal. The resulting acyl enzyme intermediates between sortases and their substrates are then resolved by the nucleophilic attack of amino groups, typically provided by the cell wall cross bridges of peptidoglycan precursors. The product of the sortase reaction, a surface protein linked to peptidoglycan, is then incorporated into the envelope and displayed on the microbial surface. Surface proteins typically carry two topogenic sequences, N-terminal signal peptides and C-terminal sorting signals. Cell wall sorting signals span approximately 30 to 40 residues and comprise a short pentapeptide motif followed by a stretch of hydrophobic side chains and finally a mostly positively charged tail at the C-terminal end of the polypeptide (174). Sortase is a central factor in the so-called “sorting pathway.” This pathway begins with the synthesis of a surface protein precursor in the cytoplasm. The N-terminal signal peptide then directs the precursor to the membrane for translocation (1). Once the signal peptide has been cleaved and the polypeptide is moved across the plasma membrane, the cell wall sorting signal functions to retain the polypeptide within the secretory pathway. Membrane-anchored sortases cleave sorting signals at their pentapeptide motif and promote anchoring to the cell wall (120).
Recent discoveries have shown that sortases catalyze diverse transpeptidation reactions using specific polypeptide or peptidoglycan substrates. Further, sortases can target unique domains of the bacterial cell wall envelope and can even promote the assembly of pili in gram-positive bacteria. These discoveries are discussed here in the context of current research frontiers. The underlying contributions of surface proteins and sortases to the pathogenesis of bacterial infections have been revealed in animal models of disease, and these findings may be exploited for the implementation of new therapeutic strategies.
SURFACE PROTEINS, THE SUBSTRATES OF SORTASE
Staphylococcus aureus Surface Proteins and Their Functions
Staphylococcus aureus is a human and animal pathogen that causes diverse infections. As a resident of the human skin, nails, and nares, this microbe has the unique ability to penetrate deeper layers of host barriers, generating suppurative lesions in virtually all organ systems. Staphylococci lack pili or fimbrial structures and instead rely on surface protein-mediated adhesion to host cells or invasion of tissues as a strategy for escape from immune defenses (53). Furthermore, S. aureus utilizes surface proteins to sequester iron from the host during infection (182). The majority of surface proteins involved in these aspects of staphylococcal disease are sortase substrates; i.e., they are covalently linked to the cell wall by sortase (Fig. 2).
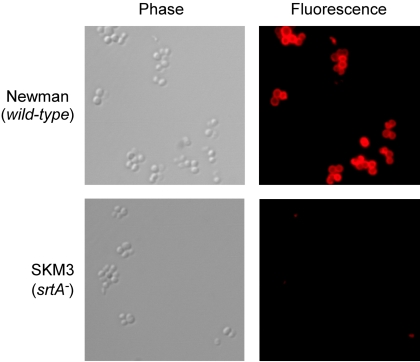
FIG. 2.
Sortase A-dependent surface display of staphylococcal proteins. Sortase is responsible for the anchoring of 20 different surface proteins to the cell wall of S. aureus strain Newman. One of these surface proteins, protein A, binds to the Fc terminus of mammalian immunoglobulins in a nonimmune fashion, causing decoration of the staphylococcal surface with antibody. Using Cy3-conjugated immunoglobulin and S. aureus strain Newman, protein A display on the bacterial surface was revealed with phase-contrast microscopy and fluorescence microscopy. Protein A display on the staphylococcal surface is abrogated in the srtA mutant strain (SKM3).
Sequence comparison of cloned surface proteins of gram-positive bacteria provided the first insight for the existence of a signal involved in anchoring these polypeptides within the envelope (51). These studies first identified six surface proteins with a common motif sequence, now referred to as LPXTG motif-type sorting signals. The sequencing of microbial genomes has greatly expanded our knowledge of the repertoire of surface proteins. Recent analyses of available sequences indicated that 732 surface protein genes carry C-terminal cell wall sorting signals in 49 microbial genome sequences (12). Here we provide a brief synopsis of what is known about surface proteins of S. aureus, molecules that have been studied for more than 50 years.
Using cell wall sorting signals as queries in bioinformatic searches, 18 to 22 genes encoding putative sortase-anchored surface proteins were identified in the genomes of S. aureus, varying with the strain under investigation (see Table 1 for a listing of 22 surface proteins) (62, 122, 123, 162). Microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) are bacterial elements of tissue adhesion and immune evasion (53). The study of several staphylococcal proteins has helped lay the foundation of our current understanding of these molecules, and these include the fibronectin binding proteins FnbpA and FnbpB (52, 89, 166, 178). Both proteins encompass a large N-terminal domain (about 500 amino acid residues) followed by four or five 50-residue repeat domains responsible for binding the N-terminal domain of fibronectin. FnbpA/FnbpB interactions with fibronectin involve structural rearrangements that lead to the ordering of the Fnbp repeat domains upon ligand binding (89, 162, 212). As fibronectin is found in extracellular matrices of most tissues as well as in soluble form within body fluids, staphylococci can adhere to virtually all tissues or serum-coated foreign bodies (151). With such widespread binding potential, one important aspect of staphylococcal binding to fibronectin is the invasion of host cells and subsequent intracellular replication (8, 44).
TABLE 1.
Staphylococcus aureus cell wall-anchored surface proteins
| Surface protein | aaa | Ligand(s)b | Motifc | Sortased | Reference(s) |
|---|---|---|---|---|---|
| Protein A (Spa) | 508 | Immunoglobulin, von Willebrand Factor, TNFRe | LPETG | A | 64, 70, 206 |
| Fibronectin binding protein A (FnbpA) | 1,018 | Fibronectin, fibrinogen, elastin | LPETG | A | 178 |
| Fibronectin binding protein B (FnbpB) | 914 | Fibronectin, fibrinogen, elastin | LPETG | A | 89 |
| Clumping factor A (ClfA) | 933 | Fibrinogen | LPDTG | A | 124 |
| Clumping factor B (ClfB) | 913 | Fibrinogen, keratin | LPETG | A | 141 |
| Collagen adhesion (Cna) | 1,183 | Collagen | LPKTG | A | 152 |
| SdrC | 947 | Unknown | LPETG | A | 90 |
| SdrD | 1,315 | Unknown | LPETG | A | 90, 91 |
| SdrE | 1,166 | Unknown | LPETG | A | 90 |
| Pls | 1,637 | Unknown | LPDTG | A | 122, 123 |
| SasA | 2,261 | Unknown | LPDTG | A | 122, 123 |
| SasB | 937 | Unknown | LPDTG | A | 122, 123 |
| SasC | 2,186 | Unknown | LPNTG | A | 122, 123 |
| SasD | 241 | Unknown | LPAAG | A | 122, 123 |
| SasE/IsdA | 354 | Heme | LPKTG | A | 121-123 |
| SasF | 637 | Unknown | LPKAG | A | 122, 123 |
| SasG/Aap | 1,117 | Unknown | LPKTG | A | 78, 122, 123 |
| SasH | 308 | Unknown | LPKTG | A | 122, 123 |
| SasI/HarA/IsdH | 895 | Haptoglobin | LPKTG | A | 42, 121-123 |
| SasJ/IsdB | 645 | Hemoglobin, heme | LPQTG | A | 121-123 |
| SasK | 211 | Unknown | LPKTG | A | 122, 123 |
| IsdC | 227 | Heme | NPQTN | B | 110, 121 |
aa, protein length in amino acids.
Molecular component(s) recognized and bound by protein.
Consensus motif recognized by sortase and present in C-terminal cell wall sorting signal.
Sortase for which cell wall surface protein is substrate.
TNFR, tumor necrosis factor receptor.
Staphylococcal strains causing connective tissue infections or osteomyelitis regularly express the collagen adhesion protein (Cna) (152, 190). A large N-terminal domain encompasses the binding site for collagen, the A domain, which assembles with a jellyroll fold (161). A molecular trench within this fold can accommodate the collagen triple helices.
S. aureus strains clump in the presence of plasma. This phenomenon, which has been exploited for diagnostic purposes, is the product of a molecular interaction between two MSCRAMMs, clumping factors A and B (ClfA and ClfB), and fibrinogen (54, 124, 141). ClfA and ClfB are structurally related and comprise a large N-terminal A domain and a repeat domain (R domain) which is composed exclusively of serine-aspartate repeats (69, 90). The ligand binding sites of ClfA and ClfB have been mapped to residues 220 to 559 (125), which assume an immunoglobulin G (IgG)-like fold (37, 125, 153, 209). An elegant molecular mechanism of fibrinogen substrate binding, coined “dock, lock, and latch,” has recently been demonstrated for SdrG, a fibrinogen binding Staphylococcus epidermidis MSCRAMM that also encompasses repeat domains (156). A cleft of 30 Å in length between two IgG-like folds of SdrG constitutes the fibrinogen binding site, with at least 62 contacts between the two molecules that occlude the cleavage sites for thrombin. Both S. aureus and S. epidermidis strains encode multiple cell wall-anchored surface proteins with large serine-aspartate repeat (Sdr) domains (69, 90, 156). Other surface proteins containing Sdr domains include the aforementioned ClfA and ClfB but also SdrC, SdrD, and SdrE. The B domains of Sdr proteins contain high-affinity calcium binding sites which adopt an EF hand fold, a common structure observed in other calcium binding proteins (91, 205). Although it seems likely that these proteins are involved in binding host factors, such interactions have thus far not been demonstrated for the majority of the Sdr proteins.
S. aureus protein A (Spa) binds to the Fc termini of mammalian immunoglobulins in a nonimmune fashion, resulting in the uniform coating of staphylococci with antibodies (86). The protein A amino acid sequence, gene sequence, and three-dimensional nuclear magnetic resonance and X-ray diffraction structures revealed a molecule comprised of five nearly identical immunoglobulin binding domains (36, 65, 179, 206). Mutations in the protein A gene (spa) cause significant defects in the pathogenesis of S. aureus infections. For example, reduced bacterial survival in blood or in the presence of macrophages is likely due to the inability of these variants to sequester immunoglobulin via Fc binding (149). However, the observed phenotypes may also be attributed to defects in the binding of protein A to von Willebrand factor, a serum polypeptide that promotes physiological homeostasis of human or animal blood, or to protein A binding to tumor necrosis factor receptor 1, a signaling molecule involved in proinflammatory cytokine responses and innate immunity (64, 70).
Four Isd proteins (iron-regulated surface determinants) are involved in binding heme or hemoproteins and appear to play a role in iron scavenging during staphylococcal host infection. HarA/IsdH is encoded by a gene outside the isd locus (see below) and has been shown to bind haptoglobin/hemoglobin complexes (42). IsdB, on the other hand, binds to hemoglobin, and four proteins, i.e., IsdA, IsdB, IsdC, and IsdH/HarA, bind heme (121, 182). It has been proposed that these proteins are involved in capturing hemoproteins on the bacterial surface, liberating heme, and promoting heme transport across the bacterial cell wall envelope (182). The functions of twelve S. aureus surface proteins with C-terminal sorting signals, i.e., SasA, SasB, SasC, SasD, SasF, SasG, SasH, SasK, SdrC, SdrD, SdrE, and Pls, are not yet known. Table 1 summarizes the current knowledge about S. aureus surface proteins.
Signal Peptides and Cell Wall Sorting Signals
All cell wall-anchored surface proteins of staphylococci or other gram-positive bacteria encode at least two topogenic sequences, an N-terminal signal peptide and a C-terminal cell wall sorting signal. For example, the N-terminal signal peptide of protein A is necessary for the secretion of precursor proteins via the Sec pathway of staphylococci and is sufficient to promote the secretion of other signal peptide-less reporter proteins (1, 4). Signal peptidase cleaves the protein A signal peptide between residues 36 and 37 (174). Following translocation across the plasma membrane, the N-terminal portion of protein A is displayed on the bacterial surface, whereas the C-terminal end is buried in the cell wall peptidoglycan and protected from extracellular protease (67). The protein A signal peptide is a member of the YSIRK-G/S family of signal peptides, which can be found in some but not all surface proteins of gram-positive bacteria and in a few secreted polypeptides (164, 192). Removal of the YSIRK-G/S motif does not abrogate the cell wall anchoring and surface display of mutant protein A; however, the rate of surface protein anchoring to the cell wall envelope is somewhat diminished (4). Clearly, signal peptides of other surface proteins or secreted polypeptides and even type II signal peptides triggering diacyl-glycerol decoration do not interfere with the function of cell wall sorting signals (134).
The C-terminal cell wall sorting signal of staphylococcal protein A encompasses a 35-residue peptide with an LPXTG motif, followed by a hydrophobic domain and a positively charged tail (173). Mutations that truncate the sorting signal cause the secretion of mutant protein A into the extracellular medium. In contrast, mutations that delete or substitute residues within the LPXTG motif abolish sortase-mediated cell wall linkage without secretion of mutant protein A (174). The cell wall sorting signal alone is sufficient to cause cell wall anchoring of other polypeptides that are initiated into the secretory pathway of S. aureus via an N-terminal signal peptide (38, 134, 135, 195). Moreover, sorting signals from one species can be functional in another microorganism (173). When the sorting function fails, mutations that either alter the distance between the LPXTG motif and the charged tail or affect residues within the two parts of the sorting signal repair the lack of function of the heterologous cell wall sorting signal (173).
Cell wall sorting signals are functional even if they do not reside at the C-terminal end of the polypeptide chain (135). Nevertheless, sorting signal function absolutely requires an upstream signal peptide. Positioning the cell wall sorting signal in the middle of an engineered polypeptide, flanked at its N-terminal side by the signal peptide-bearing reporter staphylococcal enterotoxin B (Seb) and at its C-terminal border with the mature domain of β-lactamase (BlaZ), generates a hybrid precursor that is cleaved at the N-terminal signal peptide and initiated into the secretory pathway (135). The precursor is then cleaved between the threonine and the glycine of the LPXTG motif, and the N-terminal portion of the precursor is tethered to the cell wall envelope. In contrast, the C-terminal portion of the precursor with the remainder of the cleaved cell wall sorting signal resides in the bacterial cytoplasm.
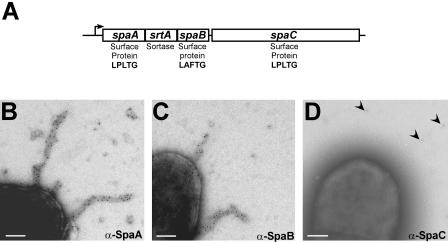
Sorting signals have been observed in a plethora of predicted gene products, most of which were identified via genome sequencing of gram-positive bacteria (12, 31, 51, 122, 136, 148). While the great majority of these sorting signals carry the LPXTG motif, others harbor variations of this sequence (Table 2) (see below). If a surface protein gene that contains such variation resides in the same transcriptional unit with a sortase gene, it is generally presumed that the two genes encode an enzyme-substrate pair, i.e., that the sortase specifically recognizes and cleaves the sorting signal of the cotranscribed substrate. This conjecture has been experimentally confirmed for Corynebacterium diphtheriae spa loci (204), S. aureus isd-srtB (123), and Listeria monocytogenes svpA-srtB (10) (see below).
TABLE 2.
Sortase classifications
| Sortase class (subfamily)a | Cleavage siteb | Membrane anchor domainc | Bacterial taxad | References |
|---|---|---|---|---|
| A (1) | LPkT-Ge* | N terminus | Bacillus, Listeria, Staphylococcus, Enterococcus, Lactobacillaceae, Streptococcaceae | 31, 41, 171, 197 |
| B (2) | NPqt-nd* | N terminus | Bacillus, Listeria, Staphylococcus, Streptococcaceae, Clostridia | 31, 41, 115 |
| C (3) | 1PkT-GG | C terminus | Actinobacteria, Bacillus, Enterococcus, Leuconostocaceae, Streptococcaceae, Clostridia | 31, 41 |
| D (4) | LPnT-At | N terminus | Bacillus | 31, 41 |
| D (5) | LAeT-Ga | N terminus | Actinobacteria | 31, 41 |
Sortase subfamily and class assignments are based on sequence, membrane topology, genomic positioning, and preference for specific amino acids within the cell wall sorting signal pentapeptide motif region of their cognate substrates (31, 41).
Cell wall sorting signal pentapeptide motif. Uppercase letters represent amino acids that are absolutely conserved. Asterisks indicate that the cleavage site has been verified experimentally.
Membrane anchor region based on transmembrane predictions and regions of high hydrophobicity.
Bacterial taxa harboring one or more sortase genes belonging to the respective sortase clasification.
Anchor Structure of Staphylococcal Surface Proteins
Sjöquist and colleagues solubilized protein A from the bacterial envelope by treatment of peptidoglycan with lysostaphin, a glycyl-glycine endopeptidase that cleaves the pentaglycine of staphylococcal cell wall cross bridges (180). Initially a protein A domain known as region X was thought to promote binding to the cell wall envelope. This domain consists of a disordered structure composed of a variable number of 8-amino-acid repeats (67). However, region X alone cannot retain protein A or other polypeptides in the envelope, and deletion of this domain does not abolish protein A anchoring or surface display (173).
Muramidases cleave the glycan strands of staphylococcal peptidoglycan and release protein A as a spectrum of molecules with different masses. In contrast, lysostaphin releases protein A species with smaller masses (173). C-terminal anchor structures of protein A were deduced by analyzing engineered surface protein sortase substrates. The protein A cell wall sorting signal was fused to the C-terminal end of Escherichia coli maltose binding protein (MalE) (171). Cell wall-anchored MalE was released with lysostaphin from the staphylococcal envelope, purified, and cleaved with trypsin, and C-terminal peptides were analyzed by Edman degradation and mass spectrometry, which revealed the sequence LPET-Gly4, LPET-Gly3, and LPET-Gly2 (171). As the cell wall sorting signal of protein A is cleaved between the threonine and glycine residues of the LPXTG motif, addition of glycine residues to the carboxyl-terminal end of protein A must be due to amide linkage of surface protein to the cell wall cross bridge of staphylococci, and this pentaglycine is cleaved by lysostaphin at positions 2, 3, and 4.
The complete anchor structure of surface proteins in staphylococci was determined after solubilization of peptidoglycan with muramidase, amidase, d-Ala-Gly endopeptidase, and lysostaphin (137, 138, 195). Seb-MHis6-Cws, an engineered reporter comprised of Seb fused to the protein A cell wall sorting signal (Cws) via a methionyl-six-histidyl linker (MHis6), can be solubilized from the peptidoglycan via cleavage with muralytic enzymes, purified by affinity chromatography on nickel-nitrilotriacetic acid resin, and then cleaved with cyanogen bromide at methionyl residues. C-terminal anchor peptides are purified by a second round of affinity chromatography and analyzed by mass spectrometry and Edman degradation. Using this technology, surface proteins were found to be linked to the cell wall cross bridges of cross-linked peptidoglycan units, comprised predominantly of murein tetrapeptides {MurNAc-[l-Ala-d-isoGln-l-Lys-(Gly5)-d-Ala-]-GlcNAc}, and only rarely to murein-pentapeptides {MurNAc-[l-Ala-d-isoGln-l-Lys-(Gly5)-d-Ala-d-Ala]-GlcNAc} that were released by muramidase cleavage of glycan strands or amidase cleavage of cell wall peptides. The overall picture that emerged from these studies indicates that surface proteins are embedded in peptidoglycan and occupy any position along glycan strands that are comprised of 2 to 11 disaccharide units and at any position along tetrapeptide cross-links with 1 to 15 wall peptide units (Fig. 3).
FIG. 3.
Cell wall anchor structure of staphylococcal surface proteins. The C-terminal threonine of surface proteins, generated by sortase A-mediated cleavage between the threonine and the glycine of the LPXTG motif, is amide linked to the pentaglycine cross bridge of S. aureus cell wall peptidoglycan. Treatment of the staphylococcal peptidoglycan with lysostaphin (glycyl-glycine endopeptidase), mutanolysin [N-acetylmuramidase that cleaves the β(1-4) O-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine (GN)], amidase (N-acetylmuramoyl-l-Ala amidase), or Φ11 hydrolase (N-acetylmuramoyl-l-Ala amidase and d-Ala-Gly endopeptidase) releases surface protein with the predicted C-terminal cell wall anchor structures.
S. AUREUS SORTASE A
Molecular Genetic Analysis of Sortase A (srtA) Function
One thousand temperature-sensitive S. aureus mutants, generated by chemical mutagenesis, were transformed with a reporter plasmid providing for the expression of Seb-SpaCWS, a hybrid between enterotoxin B and the protein A cell wall sorting signal (120). Sortase-mediated cleavage of Seb-SpaCWS was monitored by pulse-labeling experiments, and the anchoring efficiency of each mutant was scored. One variant, SKM317, with a significantly reduced rate of sorting signal cleavage was isolated. Using plasmid libraries for complementation studies in SKM317, the srtA gene was isolated and sequenced. Deletion of the srtA gene by homologous recombination results in a mutant that displays no defect in staphylococcal growth on agar or laboratory media. Pulse-labeling experiments showed that srtA mutants synthesize and secrete surface protein precursors but fail to cleave these polypeptides at their C-terminal cell wall sorting signals. As a consequence, srtA mutants do not display protein A, fibronectin binding proteins, or clumping factors on the bacterial surface, a phenotype that can be rescued by plasmid-encoded expression of the wild-type srtA gene (119). When analyzed with Seb reporter fusions to various sorting signals, srtA mutants are found to be defective in the cleavage of all staphylococcal sorting signals carrying LPXTG motif sequences (123).
Expression of several surface protein genes appears to be dramatically reduced in S. aureus srtA mutants, and the molecular mechanisms underlying this regulatory phenomenon have not yet been explored (S. K. Mazmanian and O. Schneewind, unpublished observation). Overexpression of plasmid-encoded surface protein genes or reporter genes encoding secreted proteins with C-terminal sorting signals greatly reduces the viability of staphylococci carrying srtA deletions (123). It seems plausible that srtA mutations cause the accumulation of surface proteins within the secretory pathway, which has recently been dubbed the ex-portal for Streptococcus pyogenes (163), a pathogen that is closely related to staphylococci. As these polypeptides cannot be cleaved in the absence of sortase and therefore cannot advance along the sorting pathway, it seems likely that they may block the ex-portal.
The contribution of S. aureus srtA to the pathogenesis of staphylococcal disease was examined in several different animal model systems of infection. S. aureus strain Newman, a human clinical isolate, was used as a parent, and the srtA gene was replaced with the erythromycin resistance cassette (119). Compared to the wild-type parent, sortase mutants displayed a 1.5-log-unit increase in the 50% lethal dose (LD50) measured after intraperitoneal injection of staphylococci into mice, indicating a reduction in the virulence of the srtA strain. This defect may not seem large, especially compared to virulence genes in microbes that are particularly prone to causing lethal infections in mice, such as Yersinia pestis (155). However, the LD50 for S. aureus strain Newman is already high, requires about 107 CFU (119). Any reduction in virulence of staphylococci beyond 1 to 2 log units is concealed by an experimental ceiling with a lethal dose of about 108 to 109 CFU for any bacterial organism (dead or alive), because massive induction of innate inflammatory responses by bacterial extracts is rapidly fatal.
An organ abscess model has provided greater insight into the contribution of sortase A to the pathogenesis of staphylococcal disease. Following injection of a sublethal dose of 106 CFU of S. aureus strain Newman into the bloodstream, about 1 to 2 log units of staphylococci are rapidly killed by phagocytic cells (112). Those microbes that escape phagocytosis by adherence to specific tissues or invasion of cells can seed abscesses in virtually all organ tissues of mice (104). Abscesses mature within 4 to 5 days and harbor several log units of viable staphylococci, which are then cleared over a period of 5 to 10 days (3). Removal of organ tissue from infected animals and anatomical analysis or enumeration of viable staphylococci can be used as a measure of virulence and pathogenesis. Compared to the wild-type parent strain Newman, srtA mutants display a 3-log-unit reduction in bacterial growth within abscesses in multiple different organs, consistent with the notion that surface proteins of staphylococci are required to resist phagocytic clearance and to escape innate immune responses by directing bacteria to various organ tissues (119).
The septic arthritis model was developed by Bremell et al. (15, 16). Following intravenous injection, staphylococci replicate in joints, causing infectious arthritis, bone destruction, and deformation during wound healing in addition to weight loss. The severity of the infectious arthritis can be quantified by analyzing pathological anatomical lesions after excision of joints. Again sortase A mutants displayed a large reduction in virulence in this animal model system (87, 88).
Staphylococcal endocarditis occurs mainly as infectious foci on heart valves, and damaged valve tissue with fibrin-covered lesions represent a risk factor. This important clinical infection can be recapitulated in rats by first introducing valve tissue lesions with fibrin and platelet deposits via an intravenous polyethylene catheter (130). After the catheter is implanted, animals are challenged with staphylococcal infection, which causes formation of infectious thrombi and deposits of staphylococci on valve lesions followed by tissue destruction. Two days after infection, the hearts are aseptically removed and bacterial titers are determined as CFU. In this experiment, srtA mutants displayed a 2-log-unit reduction in virulence compared with the wild-type parent strain S. aureus Newman (213).
The complete spectrum of molecular mechanisms whereby surface proteins contribute to the pathogenesis of S. aureus infectious diseases cannot yet be appreciated. In fact, only recently have we learned about the contribution of these few surface proteins to pathogenesis, and much work is required to gain a better understanding. Nevertheless, the overall contribution of these surface molecules to staphylococcal pathogenesis can be measured by comparing wild-type and srtA mutant strains in infectious disease models. As is reviewed in detail above, srtA is a key virulence factor of staphylococci. In light of the rising number of antibiotic-resistant S. aureus strains (13), the sortase enzyme has become an important target for the treatment of staphylococcal disease. Additionally, surface proteins must be considered for therapeutic and preventive strategies to combat the tide of infections with this microbe.
Sortase A Structure
Sortase A harbors an N-terminal hydrophobic segment that functions as a signal peptide for secretion and as a stop transfer signal for membrane anchoring. Membrane localization of sortase was confirmed experimentally after immunoblot analysis of S. aureus subcellular fractions (119). The enzyme adopts a type II membrane topology, with the N terminus inside the cytoplasm and the C-terminal enzymatic portion located across the plasma membrane. Sortase A is a founding member of this family of sortases (84, 199). A second group of sortase-like gene products (see below) harbor an N-terminal signal peptide and a C-terminal membrane anchor, and these enzymes are thought to assume a type I membrane topology, with the N-terminal enzymatic portion projecting towards the bacterial surface and the C-terminal end residing in the cytoplasm.
In order to obtain soluble enzyme for in vitro activity assays and structural analysis, the N-terminal signal peptide/membrane anchor of sortase A was replaced with a six-histidyl tag and recombinant protein was purified (84, 197). Preliminary examination of the NOESY (nuclear Overhauser effect spectroscopy) signals of sortase nuclear magnetic resonance (NMR) spectra suggested that the enzyme folds into a predominantly β-strand structure (83). This conjecture was corroborated by determining the three-dimensional structure of sortase by NMR spectroscopy (84) and X-ray crystallography (227). The enzyme assumes a unique fold, consisting of an eight-stranded β-barrel that includes one or two helices and several loops (Fig. 4). Strands β7 and β8 form the floor of a hydrophobic depression where the active site is located. The NMR structure showed that the absolutely conserved Cys184 and His120 residues of sortases reside within the active site (84). While Cys184 is anchored in β7, His120 is located within a helical region that connects β2 and β3, with its imidazole group in the vicinity of the sulfhydryl side chain of Cys184. The NMR structure showed Asn98 anchored at the C-terminal end of β4 and also protruding near the active site. Asn98 is only poorly conserved among sortases. Further, all three aforementioned residues were positioned in a configuration similar to that of the Cys25-His159-Asn175 triad of cysteine proteases in the papain family (84, 210). X-ray crystallography data suggest, however, that Asn98 and His120 are not in the same close proximity as is observed for papain-type proteases and that sortase-mediated catalysis at Cys184 may occur by another mechanism (227). Arg197, anchored in β8, is located in close proximity and parallel to the active-site cysteine (227) (see below). The significance of these structural observations was addressed by measuring the activity of mutant enzymes bearing alanine substitutions of critical residues (see below). Replacement of either Cys184 or His120 completely abolished sortase activity both in vivo and in vitro (197, 200, 201), and replacement of Arg197 greatly reduced the enzymatic activity (116). In contrast, replacement of Asn98 with alanine had no effect on sortase activity (116).
FIG. 4.
Structure of S. aureus sortase A bound to the LPETG substrate. Sortase folds into an eight-stranded β-barrel structure. The active site resides in a depression formed by β7 and β8 strands. The side chains of His120, Cys184, and Arg197, all of which are absolutely conserved among sortases and are required for activity, as well as the LPETG substrate are drawn with ball-and-stick structures. Cys184 performs a nucleophilic attack on the peptide bond between the threonine and the glycine residues of the substrate, resulting in the formation of an acyl intermediate with the carboxyl group of the C-terminal threonine thioester linked to the sulfur of Cys184. This intermediate is resolved by a second nucleophilic attack on the thioester bond, which results in the release of the reaction products (the structure was generated from atomic coordinates deposited in Protein Data Bank, PDB ID 1T2P) (227).
High-resolution X-ray structure data of sortase bound to LPETG peptide provided insight into the molecular interaction between the enzyme and its bound substrate (227). The substrate binding site resides in a concave plane molded by the β7 and β8 strands, and the scissile peptide bond between threonine and glycine is positioned between the side chains of Cys184 and Arg197 (Fig. 4). It seems plausible that sortase employs a cysteine-arginine dyad; i.e., arginine may function as a base for thiol ionization during catalysis (225, 228). Leucine and proline residues of the LPETG peptide are bound in the C-terminal region of β7, surrounded by several highly hydrophobic residues (228). NMR analysis of the 1H-15N chemical shifts of sortase in the presence or absence of ligand allowed identification of residues that comprise the LPXTG binding surface (108). Residues perturbed after ligand binding also mapped to the C-terminal region of the β7 strand (Thr180 and Ile182) and to the vicinity of the loop connecting strands β3 and β4 (Ala118). Importantly, Thr180 and Ala118 are absolutely conserved and Ile182 is partially conserved among sortases. Mutation of these residues significantly impaired sortase activity in vitro (108).
In the NMR structure, the β3-β4 and β6-β7 loops contain a set of acidic residues involved in calcium binding (84, 131). This cation, present in millimolar amounts in host tissues, activates sortase activity eightfold (84). Analysis of the sortase NMR spectra in the presence and absence of calcium revealed that Glu105, Glu108, and Asp108 side chains of the β3-β4 loop interact with the cation. In contrast, the β6-β7 loop forms a flap that is disordered in the absence of calcium (131, 227). As a result of metal binding, slow-motion conformational changes were detected by which Glu171, positioned in the β6-β7 loop, transiently interacts with calcium and drives the flap to a closed state (131). This motion primarily affects the wall of the groove that forms the active site, which adopts a conformation better suited for the binding of the LPXTG peptide. Therefore, the binding of calcium ions activates sortase by a mechanism that may facilitate substrate binding (84, 131).
Biochemistry of the Sortase A Reaction
Purified recombinant sortase with a six-histidyl affinity tag replacement of the N-terminal membrane anchor, SrtAΔN, cleaves LPETG peptide in vitro between the threonine and the glycine residues. Fluorescence resonance emission transmission (FRET) substrates, with fluorophore/quencher pairs 2-aminobenzoyl/2,4-dinitrophenyl or 5-[(2-aminoethyl)amino]naphtalene-1-sulfonyl/4-(4-dimethylaminophenyl-azo)benzoyl groups tethered to LPETG peptide, permit measurements of the sortase reaction as an increase in fluorescence due to substrate cleavage separating the fluorophore from the quencher (197, 201). Longer LPETG peptides would most likely improve substrate cleavage. However, the concomitant decrease in FRET due to the physical separation of functional groups diminishes the usefulness of such substrates. The addition of peptidoglycan substrates to the sortase reaction mixture stimulates cleavage of LPETG peptide and results in amide bond formation between the carboxyl group of threonine and the amino group of glycine in peptidoglycan cross bridges. Glycine, Gly2, Gly3, Gly4, and Gly5 all function as in vitro substrates; however, longer cross bridges display better substrate properties for the sortase-catalyzed transpeptidation reaction (197). Consistent with the notion that sortase functions as a transpeptidase in vivo, the velocity of the in vitro transpeptidation reaction with peptidoglycan is greater than the velocity of the hydrolysis reaction in the absence of cell wall substrate.
Sortase activity can be assessed in vivo by following the maturation of pulse-labeled surface protein, for example, the Seb-SpaCWS reporter (202). Three species can be distinguished after labeling with [35S]methionine: the full-length precursor (P1); the P2 intermediate, with cleaved a N-terminal signal peptide but still harboring the C-terminal sorting signal; and the mature (M) anchored polypeptide, in which the N-terminal signal peptide and the C-terminal sorting signal have been removed (see below and Fig. 5). The P2/M ratio is a measure of in vivo sortase activity. Using a srtA mutant strain and plasmids encoding sortase variants with amino acid substitutions, the contributions of individual amino acids to in vivo catalysis can be determined (116, 201).
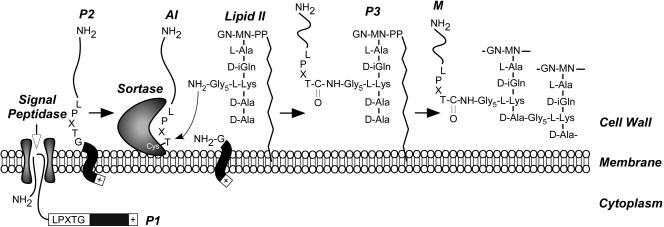
FIG. 5.
Cell wall sorting pathway of surface proteins in gram-positive bacteria. Surface proteins are first synthesized in the bacterial cytoplasm as full-length precursors (P1) containing an N-terminal signal sequence and a C-terminal sorting signal. The signal sequence directs the cellular export of the polypeptide through the Sec system and, upon translocation, is cleaved by signal peptidase. The product of this reaction, the P2 precursor harboring only the C-terminal sorting signal, is retained within the secretory pathway via its C-terminal hydrophobic domain (black box) and positively charged tail (+). Sortase, a membrane-anchored transpeptidase with active-site cysteine, cleaves the peptide bond between the threonine (T) and the glycine (G) of the LPXTG motif, generating an acyl intermediate (AI). Lipid II, the peptidoglycan biosynthesis precursor, and its pentaglycine cross bridge (Gly5) amino group attack the acyl intermediate, linking the C-terminal threonine of the surface protein to lipid II (P3 precursor) and regenerating the active site of sortase. The P3 precursor functions as a substrate for penicillin binding proteins and is incorporated into the cell wall envelope to generate mature anchored surface protein (M), which is also displayed on the bacterial surface. This pathway is universal in many gram-positive bacteria, and the functional elements of cell wall cross bridges, LPXTG motif, sortase, and penicillin binding proteins are conserved.
Even before sortase had been purified, the in vivo assay was used to demonstrate that the enzyme forms an acyl intermediate with cleaved surface protein. Surface protein anchoring can be inhibited with [2-(trimethylammonium)ethyl]methanethiosulfonate (MTSET) and p-hydroxymercuribenzoic acid (202). This suggested that the enzyme requires a cysteine residue to catalyze the transpeptidation reaction, as methane-thiosulfonate and organic mercurials react with sulfhydryl groups (2). In fact, MTSET inhibition can be rescued with dithiothreitol (DTT), which reduces the disulfide between the active-site cysteine and MTSET, thereby regenerating enzyme sulfhydryl (197). S. aureus sortase A harbors only one cysteine residue, Cys184, which is absolutely conserved in all sortases. Replacement of Cys184 with alanine completely abolishes all sortase activity both in vivo and in vitro (197, 200, 201). Addition of the strong nucleophile hydroxylamine to staphylococci results in the release of surface protein into the extracellular medium. Purification and biochemical characterization of such released products revealed threonine hydroxamate at the C-terminal end of surface proteins. Hydroxylaminolysis of surface protein occurs only in the presence of sortase and absolutely requires its active-site cysteine residue. The most likely explanation for these findings is that hydroxylamine attacks the thioester between the C-terminal threonine of cleaved surface proteins and the active-site cysteine of sortase. This acyl enzyme intermediate could indeed be detected in vitro (77). Sortase was incubated with LPETG peptide and catalysis quenched by the addition of trifluoroacetic acid. Electrospray ionization mass spectrometry revealed the presence of species in which LPET peptide was tethered to the active site cysteine. These data support a mechanistic model in which Cys184 performs nucleophilic attack on the scissile peptide bond between threonine and glycine of the LPXTG motif (acylation step) (197). The acyl intermediate is then resolved by the nucleophilic attack of the amino group of the pentaglycine cross bridge, thereby regenerating the enzyme active site and tethering surface protein to cell wall fragments (deacylation step). These reactions are as follows: R1-LPXT(CO-NH)-G-R2 + E-SH↔ R1-LPXT(CO-S)-E + NH2-G-R2 (acylation step) and R1-LPXT(CO-S)-E + NH2-Gly5-R3→R1-LPXT(CO-NH)-Gly5-R3 + E-SH (deacylation step).
Analysis of the kinetic parameters of the transpeptidation reaction indicates that it may resemble a ping-pong mechanism, whereby the binding and cleavage of the LPXTG is followed by the incorporation of the pentaglycine substrate into the active site for the separation of the acyl intermediate (77, 200). Each of these reactions appears to harbor a distinct limiting step, with that of the acylation step during transpeptidation and that of the deacylation step during hydrolysis (77).
The mechanism whereby Cys184 performs nucleophilic attack at the scissile peptide bond is not yet clear. Reagents that specifically react with sulfhydryl but not with thiolate groups such as iodoacetamide and iodoacetic acid do not inhibit sortase (202), consistent with the notion that the sortase sulfhydryl must be ionized. The NMR structure of the enzyme showed the presence of a histidine residue (His120) (see above) located in the active site of the enzyme (84). The residue is absolutely conserved and essential for sortase activity, both in vivo and in vitro (201). This result prompted the hypothesis that sortase would form an imidazolium-thiolate ion pair, mimicking active-site ionization of cysteine proteases (186). In this model, the positively charged imidazol group of His120 stabilizes the formation of a thiolate in Cys184 and acts as a proton donor/acceptor during acylation and deacylation steps (201). In papain, the Cys25-His159-Asn175 triad comprises the active site (210). While cysteine and histidine form a thiolate-imidazolium ion pair that is fundamental for papain catalysis, the asparagine side chain positions His159 in a favorable orientation towards Cys25 through hydrogen bonding. In sortase, two residues, Trp194 and Asn98, that could play a role similar to that of Asn175 are positioned near His120; however, these amino acids are not conserved among sortases. While the replacement of Asn98 with alanine or glutamine does not affect sortase activity (116), mutation of Trp194 to alanine reduced the enzyme's activity both in vitro and in vivo (201). Thus, Trp194 could play a role in positioning His120 in the proper orientation to achieve catalysis.
The observed pKas for the side chains of both Cys184 and His120 preclude the possibility of a thiolate-imidazolium ion pair within the sortase active site (32). Using an inhibitor of sortase obtained after the replacement of the T-G peptide bond with a vinyl sulfone, which reacts with cysteine thiolate, the investigators examined inhibition as a function of proton concentration. While the Ki, a value that reflects the binding of the inhibitor to the enzyme, remained constant, the ki, a measure of the effectiveness of the inhibitor, increased only beyond pH 9.4 (32). This argues in favor of the presence of a thiol group in the sortase active site at physiological pH. The pKa for the imidazol group of His120 was determined by NMR following the chemical shifts of 1H-ɛ1 and 1H-δ1 atoms of this residue as a function of pH. The titration suggested a pKa of approximately 7.0 (32). Again, this indicates that at pH 7.5 the imidazol group of His120 would be only partially protonated. Moreover, the observed pKa is independent of Cys184, as the titration curve for a sortase Cys184Ala mutant did not change (32). Together these experiments suggest that sortase catalysis cannot occur via a mechanism involving the thiolate-imidazolium ion pair, as originally proposed (84, 201).
Analysis of the X-ray crystallographic structure of sortase A with LPETG peptide led to the formulation of a new hypothesis. As is pointed out above, this structure revealed the presence of Arg197 in the active site (227). This residue is absolutely conserved among sortases and is positioned in front of and parallel to Cys184. Replacement of Arg197 with alanine, lysine, or histidine greatly impaired sortase activity, both in vivo and in vitro (116). Because the guanidinium group of Arg197 interacts with the carbonyl group of the scissile bond in the X-ray structure, it was proposed that Arg197 forms an oxyanion hole that may stabilize the acylated adduct (227). This hypothesis was corroborated by an experiment in which hydroxylamine was unable to resolve the acyl intermediate when Arg197 was replaced by alanine or lysine, indicating that in the absence of the guanidinium group, the thioacyl intermediate is not formed (116). These results suggest that the sortase active site may comprise a cysteine-arginine dyad (225, 228). It is important to note that sortases display absolute conservation of several residues. Two of these, Leu97 and Tyr153, have been replaced by alanine in order to assess their importance for the enzyme's activity. Despite their conservation, these residues were not required for sortase activity either in vitro or in vivo (201). The contribution of other conserved amino acids to sortase catalysis remains unknown.
The specificity of sortase A for different pentapeptide motifs was studied by determining the in vitro activity of the enzyme towards a peptide library with 18 amino acid substitutions in every position (99). This study confirmed bioinformatic analysis of sortase substrates, which indicate that the enzyme recognizes LPXTG sequences. Not surprisingly, initial-velocity analysis showed that only leucine is tolerated in position 1 in XPETG peptides and only proline is tolerated in position 2 in LXETG peptides, whereas any residue is tolerated in position 3 in LPXTG peptides. Only threonine in position 4 in LPEXG peptides is accepted as a substrate, and only glycine is accepted in position 5 in the LPETX peptide library. The enzyme's residues involved in this specificity were detected by comparing NMR signals of bound versus unbound sortase (see above) (108). Besides those in Cys184 and Arg197, chemical shift changes in Thr180 and Ala118 (absolutely conserved residues) and Ile182 (partially conserved) were also detected. Mutation of these residues significantly impaired sortase activity in vitro (108). Whether these residues contribute to the substrate specificity of sortase remains to be assessed, and it would be interesting to screen the peptide library and determine whether peptides with sequences differing from LPXTG can be substrates of these mutants.
Another important aspect of the sortase reaction is the interaction of the enzyme with its cell wall substrate, the pentaglycine cross bridge. In vivo, sortase can catalyze the transpeptidation of surface proteins to cell wall cross bridges containing one, three, and five glycine residues, but not to the ɛ-NH2 group of the l-lysine residue of wall peptides. This conclusion was reached following analysis of the anchor structure of surface proteins generated by S. aureus fem mutants defective in cross bridge biosynthesis (196). At least three Fem factors (factors essential for methicillin resistance) are required for the addition of glycine residues to the cross bridge of S. aureus peptidoglycan (101). FemX is responsible for the addition of the first glycine residue to the l-lysine of the wall peptide, while FemA adds the second and third glycine residues and FemB completes the cross bridge by incorporating the fourth and fifth glycine. Therefore, femB mutants synthesize Gly3 cross bridges, femA mutants synthesize Gly1 cross bridges, and a partial femAX mutant either carries Gly3 cross bridges or completely lacks cross bridge (97). The cell wall anchor structure of Φ11-hydrolase-released Seb-SpaCWS, which is expressed in each of these fem mutants (see above), revealed that sortase can link surface protein to Gly5, Gly3, and Gly1 cross bridges in wild-type, femB, femA, and femAX strains but failed to anchor protein to the ɛ-amino of l-Lys (196). Nevertheless, the velocity of the sorting reaction is diminished in fem mutants compared to the wild type (196), indicating that sortase prefers pentaglycine as a cell wall substrate. This conjecture was corroborated in vitro by the observation that Gly, Gly2, and Gly3 can be used as nucleophiles by the enzyme and are linked to the threonine of LPETG peptides (77, 200). Diglycyl-histidine and diglycyl-leucine can also be used in the in vitro transpeptidation reaction, although the binding is decreased (as deduced from the apparent Km values). Glycyl-alanine and glycyl-valine also retain substrate properties, but their binding is reduced by 10-fold. In contrast, alanyl-glycine and valyl-alanine cannot be used as substrates for the transpeptidation reaction (77). Thus, cell wall substrate recognition of sortase tolerates only glycine as the N-terminal residue and strongly prefers another glycine at the second position. While the enzyme's constraints for the selection of a cell wall substrate are being delineated, the actual binding site for peptidoglycan substrate remains unknown. Gly3 substrate was modeled into the crystal structure of sortase (227). It was speculated that Gly3 may be positioned in the loop that connects β7 and β8, replacing a water molecule that otherwise contacts the backbone atoms of this loop. Nevertheless, experimental data are needed to reveal the peptidoglycan binding site of sortase.
Lipid II, the Peptidoglycan Substrate of Sortase A
Cell wall active-antibiotics have been employed to probe the peptidoglycan substrate requirements for the sortase reaction (197). Vancomycin binds d-Ala-d-Ala within lipid II and inhibits the transglycosylation and transpeptidation reactions that assemble peptidoglycan from this precursor (193, 211). Moenomycin is a lipid II analog that interferes with the transglycosylation reaction of peptidoglycan biosynthesis (168, 207). Lastly, penicillin inhibits only the transpeptidation reaction by occupying the corresponding active sites of PBPs without affecting transglycosylation or lipid II concentrations (188, 193). By measuring cell wall anchoring of pulse-labeled reporter proteins, it was shown that both vancomycin and moenomycin, but not penicillin G, interfered with the cell wall sorting pathway (202). As the inhibition of surface protein anchoring increased during prolonged incubation of staphylococci with vancomycin or moenomycin, a plausible explanation for these results is that antibiotics reduce the availability of lipid II, which serves also as the peptidoglycan substrate of sortase A.
Additional evidence for lipid II as the peptidoglycan substrate for surface protein anchoring was garnered with in vitro reactions. LPXTG peptide is linked to lipid II by purified sortase A, and vancomycin can block this reaction (165). Analysis of surface protein anchoring in protoplasts promoted the notion that the sorting reaction does not require mature, assembled peptidoglycan (202). The cell wall envelope of S. aureus was removed by digestion with muralytic enzyme, protoplasts were pulse-labeled with [35S]methionine, and the radiolabeled surface protein was immunoprecipitated. Protoplasts catalyzed surface protein precursor cleavage at the LPXTG motif at a rate similar to that for staphylococci with intact cell wall envelopes. A unique surface protein sorting intermediate was detected in protoplast membranes. Further evidence for a linkage between surface proteins and lipid II in vivo was obtained by labeling staphylococci with [32P]phosphoric acid, which is incorporated into lipid II molecules (154). Following removal of the cell wall envelope with muramidase, which cannot cleave lipid II, labeled polypeptides were immunoprecipitated and detected by autoradiography. 32P-labeled surface protein species were identified, and their synthesis required sortase A activity. Radiolabeled lipid II could be removed from surface protein by lysostaphin cleavage at pentaglycine cross bridges, whereas muramidase, which cannot cleave lipid II, displayed no effect. Treatment of staphylococci with tunicamycin, an inhibitor of phosphor-N-acetylmuramyl-pentapeptide translocase (the enzyme required for formation of lipid I and lipid II [191]) abolished sortase A-dependent biosynthesis of 32P-labeled surface protein. The C-terminal anchor of immunoprecipitated 32P-labeled surface protein was analyzed by thin-layer chromatography and observed to bind nisin (154), an antibiotic that specifically interacts with lipid II (214). Thus, the cell wall sorting intermediate P3 is comprised of surface protein linked to lipid II (154). A model that emerged from these studies suggests that P3 not only is the product of the sortase reaction but also serves as a substrate for the transglycosylation and transpeptidation reactions of cell wall biosynthesis, similar to the case for lipid II (Fig. 5). Obviously, the amino group of the pentaglycine cross bridge of P3 is already engaged in an amide bond and cannot perform the nucleophilic attack at PBP acyl enzyme intermediates with cleaved wall peptides. Nevertheless, the pentapeptide structure permits PBP cleavage at the d-Ala-d-Ala of P3 and attachment of other pentaglycine cross bridges from neighboring wall peptides at this site. In this manner, the P3 sorting intermediate can be fully incorporated into the three-dimensional network of staphylococcal peptidoglycan.
Sortase A Inhibitors
Inhibitors of sortase should be useful for the characterization of this fascinating enzyme. However, can such inhibitors affect the outcome of human or animal infection with S. aureus? If virulence studies with srtA mutants provide a correlate for the contribution of sortase A to disease, we can be hopeful that inhibitors of the sortase reaction may display therapeutic effects. Moreover, as sortase is a universal virulence factor of gram-positive pathogens, compounds that inhibit the enzyme's activity could constitute antimicrobial agents for the treatment of many diseases, such as enterococcal and pneumococcal infections. Until such specific sortase inhibitors have been isolated and tested, it is impossible to say whether this anti-infective strategy will be inferior or equal to that of conventional antibiotic therapy. Certainly, there is no precedent for use of clinically relevant anti-infectives, i.e., inhibitors of bacterial virulence factors, as a therapeutic strategy for human infectious diseases. These thoughts should not distract us from the pressing need for the development of new therapeutic agents, as staphylococci have developed mechanisms of resistance to all known antibiotics, including methicillin and vancomycin (14).
The first search for sortase inhibitors occurred even before the enzyme was identified (202). Methane-thiosulfonates such as MTSET and (2-sulfonatoethyl)methane-thiosulfonate inhibit sortase in vivo and in vitro, with MTSET achieving complete inhibition. The mercurial p-hydroxymercuribenzoic acid could also inhibit sortase. All of these compounds react with the catalytic Cys184 and prevent formation of acyl intermediates. In contrast, sulfhydryl alkylating agents such as iodoacetamide, N-ethylmaleimide, or iodoacetic acid do not inhibit sortase. While these reagents proved useful to elucidate the catalytic mechanism of the enzyme, nondiscriminate interactions of thiol-reactive molecules renders these compounds useless for therapeutic studies because of their associated toxicity in mammalian organisms.
Several recent efforts have examined natural or chemical compounds for the property of inhibiting sortase A in vitro. For example, extracts from 80 medicinal plants were tested and those obtained from Cocculus trilobus, Fritillaria verticillata, Liriope platyphylla, and Rhus verniciflua displayed inhibitory activity (95). The extract from Fritillaria verticillata bulbs was subjected to silica gel chromatography, and a fraction with potent inhibitory effects on sortase was isolated. The constituent of this fraction was identified by NMR as glucosylsterol β-sitosterol-3-O-glucopyranol (93). As sitosterol alone does not inhibit sortase, it was concluded that the inhibitory effect must reside within the glucopyranoside moiety of the molecule. A similar experimental approach for extracts of Coptis chinensis identified the isoquinoline alkaloid berberine chloride as a sortase inhibitor (94). Both compounds exhibit a lower MIC than p-hydroxymercuribenzoic acid (see above) and were able to inhibit binding of S. aureus to fibronectin-coated surfaces (143), an interaction mediated by the sortase A substrates fibronectin binding proteins A and B (FnbpA and FnbpB) (see above). However, the ki values for these inhibitors have not been obtained, precluding their comparison with other known sortase inhibitors.
Another strategy for the development of inhibitors employed modifications to the scissile bond of LPXTG peptides. In the first of these studies, the threonine-glycine peptide bond was substituted by moieties known to alkylate the active-site thiol of cysteine proteases. These included peptidyl-diazomethane (LPAT-CHN2) and peptidyl-chloromethane (LPAT-CH2Cl) (176). Both compounds successfully inhibited sortase activity in vitro, with a ki/Ki of 2.2 ×104 M−1 · min−1 (ki = 5.8 × 10−3 min−1) for LPAT-CHN2 and a ki/Ki of 2.1 ×104 M−1 · min−1 (ki = 1.1 × 10−2 min−1) for LPAT-CH2Cl. In a second study, the scissile bond was replaced with vinyl sulfone [LPAT-SO2(Ph)], a moiety known to covalently modify the active-site thiolate of cysteine proteases via formation of a thioether adduct (32). Due to the requirement for ionization of the thiol group of Cys184, this modified peptide achieved maximal inhibition at pHs greater than 8.0. As expected, inhibition was irreversible, and at pH 7.0 the ki/Ki was measured to be 44.4 M−1 · min−1 (ki = 4 × 10−4 min−1). Different types of vinyl sulfones, i.e., di-, ethyl-, methyl-, and phenyl vinyl sulfones, all inhibited sortase A. Phenyl vinyl sulfone (PVS) displayed the greatest effect, with a ki/Ki of 20.1 M−1 · min−1 (55). Interestingly, PVS-treated S. aureus cells failed to bind to a fibronectin-coated surface, suggesting that PVS can inhibit the sortase-dependent surface display of fibronectin binding proteins in vivo. However, additional studies documenting the effects of PVS on mammalian cell viability and on other steps of the sorting reaction are required for a clearer understanding and confirmation of this inhibition.
Substrate peptides have been generated with the expectation of mimicking the transition state for the formation of sortase acyl intermediates. In order to obtain such an inhibitor, the threonine residue of an LPETG peptide was replaced by a phosphinate group (LPEΨ{PO2H-CH2}G) (98), a peptide modification that has been successfully used for the design of zinc protease inhibitors. As the tetravalent coordination of the phosphorous atom imitates the acyl intermediate transition state, this modified peptide should compete with LPETG substrate for the sortase active site. Inhibition was achieved with the phosphinate compound and was therefore exploited to determine different kinetic parameters of the sortase reaction (98).
Another strategy for the discovery of sortase inhibitors has been to screen libraries of small-molecule compounds (142). One thousand compounds were tested for their ability to inhibit sortase in vitro, and the initial hits were subjected to successive structural and chemical modifications with the goal of achieving more pronounced inhibitory effects on sortase activity. This resulted in the isolation of a set of substituted (Z)-diarylacrylonitriles that exhibit potent inhibition towards sortase. Most of the compounds described here were tested only in vitro and typically require micromolar or low millimolar concentrations for inhibition of sortase. Much work still needs to be done before one can analyze compounds with Ki at low micromolar or nanomolar concentrations and with inhibitory specificity that permits testing in animal models of S. aureus pathogenesis.
Applications of the Sortase A Reaction
Sortase-catalyzed transpeptidation is an attractive protein engineering tool for the incorporation of nonpeptide moieties into polypeptides tagged with an LPXTG motif. Several established modification systems make use of recombinant proteins conjugated to peptide analogs, unnatural amino acids, fluorophores, and other biochemical and biophysical probes. One strategy to achieve such modification is subtilisin-based peptide ligation (23). However, this technology involves several biosynthetic steps and is not efficient. Sortase transpeptidation, on the other hand, offers a simple and efficient tool for the incorporation of chemicals containing glycine residues with a free amino group to the LPXTG motif of recombinant proteins. As a proof of concept, triglycyl-lysine-folate was synthesized and incubated with purified recombinant green fluorescent protein (GFP)-LPETG-His6 (i.e., GFP containing a C-terminal LPETG-six-histidyl group) in the presence of sortase (114). The products of the reaction were separated by reverse-phase high-pressure liquid chromatography and analyzed by matrix-assisted laser desorption ionization-time-of-flight analysis, revealing that the GFP-LPET-G3K(folate) adduct was produced with high efficiency. Another biotechnological application is the incorporation of the branched peptide AT-P-022 into polypeptides. AT-P-022 possesses strong protein transduction activity; i.e., it promotes the uptake of linked proteins by eukaryotic cells. Due to its branched structure, however, it is difficult to incorporate AT-P-022 into proteins. Using sortase-mediated peptide ligation, it was possible to generate a GFP-LPET-G2(AT-P-022) conjugate in a single-step reaction. Fluorescence analysis demonstrated that the reverse-phase high-pressure liquid chromatography-purified product was taken up by NIH 3T3 cells with high efficiency (114).
Another application of the sortase reaction is the generation of self-cleavable chimeras for one-step purification of recombinant proteins (113). The concept relies on the expression and purification of a recombinant His6-sortase-LPETG-target protein fusion that cleaves itself once the enzyme has been activated by the addition of calcium and triglycine. The transpeptidation product, i.e., nontagged target protein, can be eluted in a single chromatography step, with glycine as the only modification introduced by the purification procedure. The sortase strategy differs from other systems employing N-terminal carriers that are cleaved off from the target protein by the addition of a protease, in which the separation of the target protein from the protease requires additional chromatography steps. The approach was tested for the purification of GFP, Cre, and p27 proteins (113). In all cases, the presence of an N-terminal sortase carrier increased the expression and solubility of the recombinant protein. Importantly, neither autocleavage nor transpeptidation with E. coli proteins containing an N-terminal glycine was observed during expression. Following affinity chromatography of cleared cell lysate on Ni-nitrilotriacetic acid Sepharose and several washes, charged resin was incubated in buffer containing calcium and Gly3. Concentrated and 98% pure target protein was recovered from the supernatants, indicating that sortase-based protein purification provides a simple and effective method that may be generally applicable to many proteins.
S. AUREUS SORTASE B
Sortase homologs have been revealed in every gram-positive bacterium for which genome sequences are available, and most species encode more than one sortase (148). S. aureus encodes two sortases, and the second enzyme has been named sortase B (122, 123). The structural gene for sortase B (srtB) is part of the isd (iron-regulated surface determinant) locus, which is comprised of three transcriptional units, isdA, isdB, and isdCDEF-srtB-isdG (Fig. 6A) (123). IsdA, IsdB, and IsdC are cell wall-anchored proteins. IsdD is thought to be inserted into the plasma membrane. IsdE lipoprotein and the IsdF ATP binding cassette (ABC) transporter presumably function as heme-iron transporters in the plasma membrane. IsdG is located in the cytoplasm, and it cleaves the heme tetrapyrrol ring and liberates iron for staphylococcal growth. IsdA, IsdB, IsdC, IsdD, IsdE, and IsdG all bind heme-iron (121). Additionally, IsdB (but not IsdA or IsdC) binds hemoglobin (121). Two additional Isd proteins are encoded elsewhere in the genome of S. aureus: IsdH (HarA), a haptoglobin binding, cell wall-anchored protein (42), and IsdI, an IsdG homolog with heme oxygenase activity (181). A fur box (46), i.e., a DNA sequence to which the ferric uptake repressor binds and inhibits transcription when staphylococci grow in iron-replete conditions (76, 215), is present in the promoter regions of all of these genes. Thus, the Isd proteins and sortase B are expressed only under conditions when iron is limiting (121, 123). However, fur mutant staphylococci express the isd locus and srtB in a constitutive fashion (123).
FIG. 6.
Isd-mediated heme-iron uptake in S. aureus. A. The isd locus is comprised of isdA, isdB, and isdC, which encode cell wall-anchored proteins carrying LPKTG, LPQTG, and NPQTN motifs in their respective sorting signals. Located elsewhere in the S. aureus genome, isdH and isdI encode a fourth LPKTG surface protein and a heme oxygenase, respectively. All isd genes are regulated by the ferric uptake repressor (Fur), which represses transcription under iron-replete conditions by binding to fur boxes present in promoter regions (shaded boxes). Arrows indicate the direction of transcription. B. A model for Isd-mediated heme-iron transport across the cell wall of S. aureus. IsdA, IsdB, and IsdH are anchored to the cell wall by sortase A and function as receptors for hemoprotein ligands, including haptoglobin (Hpt), hemoglobin (Hb), or heme. Upon binding to Isd receptors, heme is released from the hemoproteins by an as-yet-undefined mechanism and passaged through the cell wall in an IsdC-dependent manner. Treatment of staphylococcal cells with extracellular proteinase K completely degrades IsdB, only partially digests IsdA, and leaves IsdC intact, suggesting different degrees of surface exposure for each of these cell wall proteins. The heme molecule is then transported through the membrane transport system composed of IsdDEF into the cytoplasm. Upon entry into the cytoplasm, heme is degraded by IsdG and IsdI heme monooxygenases. This leads to the release of free iron for use by the bacterium as a nutrient source. (Adapted from reference 182 with permission from Elsevier.)
IsdC and Sortase B Contribute to Heme-Iron Transport
IsdC, a cell wall-anchored protein, is the only known substrate of sortase B. In contrast to IsdA, IsdB, and IsdH, each of which contains LPXTG-type sorting signals and is a substrate for sortase A, the IsdC sorting signal harbors an NPQTN motif. Cell wall anchoring of IsdC or Seb-IsdCCWS is abolished in a srtB mutant strain; however, srtA mutants attached both proteins to the envelope in a fashion similar to that of wild-type staphylococci (123). Deletion of the srtB gene did not interfere with the anchoring of 15 different surface proteins harboring LPXTG motif sorting signals. Thus, sortase B uniquely recognizes its IsdC substrate and tethers the polypeptide to the staphylococcal peptidoglycan. Unlike sortase A-anchored substrates that are displayed on the bacterial surface, cell wall-anchored IsdC remains buried within the cell wall envelope. Two lines of evidence support this conclusion. First, IsdC, but not IsdA or IsdB, is protected from digestion with extracellular protease unless the integrity of the cell wall envelope is perturbed by treatment with muralytic enzyme (121). Further, IsdA and IsdB are detectable by immunofluorescence microscopy, indicating that surface-displayed polypeptides bind to antibody. However, specific antibody added to intact staphylococci may not bind to cell wall-anchored IsdC (L. A. Marraffini and O. Schneewind, unpublished observation). Seb-SpaCWS, carrying a C-terminal fusion of enterotoxin B to the protein A sorting signal, is a substrate for sortase A (195). Cell wall-anchored Seb-SpaCWS is displayed on the bacterial surface and can be degraded by proteinase K digestion. In contrast, cell wall-anchored Seb-IsdCCWS is not displayed on the bacterial surface and can be degraded by extracellular protease only when the integrity of the cell wall envelope is perturbed by treatment with muralytic enzymes (115). Together these experiments demonstrate that sortase A and sortase B target their protein substrate to discrete locations within the cell wall envelope. Further, the information for the ultimate destination of a polypeptide in the staphylococcal envelope resides within its cell wall sorting signal. Proper targeting to discrete locations in the cell wall envelope requires polypeptide substrate interactions with cognate transpeptidases and specific yet distinct peptidoglycan substrates for each of the two sortases (115).
What is the purpose of anchoring IsdC at a discrete site within the cell wall envelope? A plausible explanation is that sortase A-anchored proteins, i.e., IsdA, IsdB, and IsdH, capture hemoproteins on the bacterial surface and dislodge heme from host polypeptides (Fig. 6B). Transfer of heme from sortase A-anchored polypeptides to sortase B-anchored IsdC in the cell wall envelope, followed by subsequent transfer of heme-iron to IsdD and IsdEF, is thought to provide for the passage of this essential nutrient across the 100-nm-thick cell wall envelope. Once transported across the plasma membrane, iron may be released from heme via IsdG- or IsdI-mediated tetrapyrrol cleavage (182).
The contribution of sortase B to heme-iron uptake was examined in srtB mutant staphylococci. Growth media were depleted of divalent cations and supplemented with heme-iron. While wild-type staphylococci were able to grow under these conditions, srtB mutant bacteria were not (121). Although S. aureus is capable of synthesizing heme-iron both in the presence and in the absence of srtB, only uptake of exogenous heme-iron was affected by deletion of the srtB gene. Measurement of the uptake of [55Fe]heme in wild-type and srtB mutant staphylococci with intact cell wall envelopes or in osmotically stabilized protoplasts showed that srtB is required for heme-iron uptake under both conditions (121). Thus, the lack of sortase B activity or the absence of anchored IsdC not only prevents the passage of heme-iron across the cell wall envelope but also prevents heme-iron transport across the plasma membrane into the bacterial cytoplasm.
Molecular Genetic Analysis of Sortase B (srtB) Function
Iron is an essential nutrient for most microbes, including staphylococci, and although iron is abundantly present in host tissues, its availability is severely restricted by sequestering iron via bound proteins and cellular compartments (17). To test whether sortase B is required for the pathogenesis of staphylococcal infections, virulence properties of srtB and srtA mutant staphylococci were compared with those of the wild-type parent strain S. aureus Newman. The calculated LD50 after intraperitoneal injection of a srtB mutant was not significantly different from that of wild-type S. aureus, indicating that sortase B is dispensable during acute early phases of infection (213). In the renal abscess model following intravenous infections of mice, isogenic S. aureus variants carrying a srtB deletion displayed a small defect in virulence, which became more pronounced during later stages of infection (123, 213). Using the rat infectious endocarditis model, no difference was observed in the number of wild-type or srtB mutant staphylococci multiplying on cardiac vegetations (213). Measurement of the arthritic index as well as the number of staphylococci present in the joints during the murine infectious arthritis model demonstrated that the srtB mutant is significantly less virulent than the wild-type strain (88, 213). The srtB defect is not as pronounced as that of srtA variants; however, in double mutant strains the srtA srtB deletions caused an additive defect in virulence compared to that of the single mutant strains (88, 213). Together these results revealed the contributions of sortase B to S. aureus pathogenesis, in particular to infections that require bacterial persistence in host tissues. The contribution of sortase B is additive to that of sortase A, indicating that the two enzymes perform nonredundant and complementary functions and that each promotes the establishment of staphylococcal disease.
Biochemistry of the Sortase B Reaction
Recombinant sortase B has been purified, and, as reported for sortase A, the N-terminal membrane anchor sequence was removed to obtain soluble enzyme. SrtBΔN cleaves NPQTN peptide substrate but not LPETG peptides (123). The enzyme is inhibited with MTSET and this inhibition can be relieved with DTT, indicating that sortase B also utilizes its sole cysteine residue (Cys223) for catalysis. In contrast to that of sortase A, sortase B activity is very low, which has thus far precluded a detailed biochemical analysis of its transpeptidation reaction (123). To reveal the site of cleavage at the IsdC substrate, i.e., the NPQTN motif, and to examine the peptidoglycan substrate for sortase B, the cell wall anchor structure of IsdC was determined. An engineered reporter protein, Seb-MHis6-IsdCCWS, is anchored to the cell wall envelope in a fashion similar to that for IsdC. After the cell wall envelope of S. aureus expressing the reporter was cleaved with lysostaphin (169), the polypeptide was purified and cleaved with cyanogen bromide, and C-terminal anchor peptides were purified by a second round of affinity chromatography. Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) analysis of anchor peptides revealed that sortase B cleaves NPQTN motif sorting signals between the threonine and the asparagine residues. The C-terminal threonine residue is amide linked to the amino groups of pentaglycine cross bridges within the staphylococcal cell wall (115). Thus, the chemical product of the sortase B reaction has striking similarity to that of the sortase A reaction. Nevertheless, detailed analysis of peptidoglycan treated with mutanolysin (21, 224) or Φ11 hydrolase (137) revealed only a very limited degree of cross-linking between IsdC anchor peptides compared to that between the anchor peptides generated by sortase A (115, 137). About 80 to 95% of all murein subunits of assembled peptidoglycan harbor cell wall tetrapeptides with cross-linked d-Ala at position four (57, 184, 188), and this can also be observed for sortase A-anchored surface proteins, which are embedded at any position in glycan chains with up to 11 MurNAc-GlcNAc disaccharide units and cross-linked to as many as 15 cell wall peptides (137). In contrast, sortase B-anchored product is attached to at most six or seven disaccharide subunits, and its wall peptides are either non-cross-linked (murein petapeptides) or linked to two or three peptidoglycan subunits (Fig. 7).
FIG. 7.
Cell wall anchor structure of staphylococcal IsdC. The C-terminal threonine of IsdC, generated by sortase B-mediated cleavage between the threonine and the asparagine of the NPQTN motif, is amide linked to the pentaglycine cross bridge of S. aureus cell wall peptidoglycan. Treatment of the staphylococcal peptidoglycan with lysostaphin (glycyl-glycine endopeptidase), mutanolysin [N-acetylmuramidase that cleaves the β(1-4) O-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine (GN)], amidase (N-acetylmuramoyl-l-Ala amidase), or Φ11 hydrolase (N-acetylmuramoyl-l-Ala amidase and d-Ala-Gly endopeptidase) releases surface protein with the predicted C-terminal cell wall anchor structures. In contrast to sortase A substrates, sortase B-anchored IsdC is attached to only six or seven disaccharide subunits and its wall peptides are either non-cross-linked (murein pentapeptides containing an extra C-terminal d-Ala) or linked to only two or three peptidoglycan subunits.
Sortase B Positions IsdC within the Cell Wall Envelope
The apparent lack of cross-linking of IsdC anchor peptides suggests that sortase B employs a unique cell wall substrate. Recent work proposed that the enzyme anchors IsdC to assembled (polymerized) peptidoglycan at a site with limited cross-linking (115). Considering that peptidoglycan strands may grow in a fashion that is perpendicular to the cell membrane (39) (as illustrated in Fig. 6B), attachment of protein to fully assembled peptidoglycan would enable a topology that encloses anchored protein in the cell wall envelope. It seems reasonable to presume that the enzyme properties in selecting peptidoglycan substrate are imprinted in the primary structures as well as the three-dimensional structures of sortase A and B. The polypeptide chain of sortase B is longer than that of sortase A. The two enzymes fold into very similar structures, i.e., β-barrels with short α helices connecting some of the β strands (225, 228) (Fig. 8). Two short α helices at the N terminus (α1 and α2) of sortase B, directly linked to the transmembrane segment of the enzyme, and a long α helix (α5) are absent in sortase A. It was proposed that α1 and α2 may project the enzyme active site towards the bacterial surface, whereas that of sortase A may face the plasma membrane (228). Is it really that simple? Turn the enzyme barrel 180° and, voila, the topology of anchored protein is changed? At this time, such a view is only speculation. Nevertheless, this hypothesis and others will guide future work aiming at the elucidation of the molecular mechanism whereby staphylococci position proteins at discrete locations in their cell wall envelope. As it is the case for sortase A, the two C-terminal β strands (β7 and β8) of sortase B form a groove where the active site resides. The structure in the presence of MTSET showed a disulfide bond between this inhibitor and the sulfhydryl group of Cys223 (228), supporting the notion that this residue is the equivalent to sortase A Cys184 (Fig. 8) (see above). The functional assignments of other residues within the sortase B active site are not at all clear. Zong et al. reported the presence of an arginine residue in the vicinity of Cys223, i.e., Arg233, the analog of Arg197 in sortase A, and proposed that sortase B, or in general all sortases, generate a cysteine-arginine dyad, in which the sulfhydryl group is activated by the guanidinium group for catalysis (228). Supporting this hypothesis, the crystal structure of sortase B bound to MTSET and triglycine showed that the free amino group of this substrate was in close proximity to Arg233 but far from His130 (the sortase A His120 analog) (228). Zhang et al. suggested that the active site of sortase B may contain a cysteine-histidine-asparagine triad (Fig. 8, Cys223-His130-Asn225) and proposed a catalytic mechanism that is similar to that of other cysteine proteases, in which the histidine imidazolium group assists the nucleophilic attack of the cysteine sulfhydryl group (225). Future advances towards understanding sortase B require improvements of the in vitro assay and experimental tests of structural predictions by analyzing single-residue substitutions.
FIG. 8.
Crystal structure of S. aureus sortase B. Sortase A and sortase B fold into very similar β-barrel structures; however, sortase B harbors three α helices that are absent in sortase A (here shown in orange) and that may contribute to the unique properties of sortase B substrate specificity and anchoring. Cys223, His130, and Arg233 are equivalent to sortase A Cys184, His120, and Arg197, respectively, and, along with Asn225, presumably constitute the active site of sortase B (the structure was generated from atomic coordinates deposited in Protein Data Bank, PDB ID 1QXA) (228).
SORTASE-CATALYZED POLYMERIZATION OF PILI
An additional and astonishing function of sortases is their involvement in the formation of pili in gram-positive bacteria. Pili, also known as fimbriae, represent proteinaceous filaments that protrude from microbial surfaces and typically function as a supramolecular structure, often with adhesive activity at their tip. The molecular mechanisms underlying the formation of pili in the outer membranes of gram-negative organisms are well understood (47). For gram-positive bacteria, electron microcopy experiments provided the first evidence for the presence of pili on the surfaces of Actinomyces spp., Corynebacterium spp., and Streptococcus spp. (27, 28, 102, 217, 218). As already reported for sortase-anchored surface proteins, the cell wall envelopes of gram-positive microbes appear to again serve as the assembly site for pili, which perform important functions during the pathogenesis of human or animal infections (203).
Actinomyces naeslundii
A. naeslundii is a human pathogen that can be isolated from the oral cavity and from supragingival dental plaque. Initial colonization of tooth or mucosal surfaces with Actinomyces spp. provides a biofilm substrate for the adherence of other plaque bacteria, including oral streptococci and several gram-negative bacterial species, which eventually leads to the formation of dental caries. Two types of fimbriae mediate adhesion of Actinomyces to tissue surfaces of the oral cavity. Type 1 fimbriae are required for binding tooth hydroxyapatite, whereas type 2 fimbriae mediate interaction with other bacteria and promote binding to the mucosal tissues of the host (203). Pioneering work carried out by Maria Yeung, John Cisar, and colleagues suggested an involvement of sortases in the formation of A. naeslundii fimbriae (222, 223). By generating Actinomyces plasmid expression libraries in E. coli and screening for plasmid clones that provided for immunoreactivity with antifimbrial serum, the genes encoding major pilin subunits of type I and type II fimbriae were isolated and named fimP and fimA, respectively (40, 219-221). DNA sequencing and analysis of genes surrounding fimP revealed its location within an operon containing seven open reading frames (ORFs): orf3-orf2-orf1-fimP-orf4-orf5-orf6 (223). Of these, orf1 and fimP encode a surface protein with LPXTG motif-type sorting signals, whereas the orf4 protein product displays homology to sortase. Insertional mutagenesis of orf4, as well as orf1, orf2, orf3, or fimP, abolished bacterial adherence, suggesting that these genes are indeed required for the assembly of pili. Consistent with this hypothesis, FimP and Orf1 subunits accumulate in the envelope the orf4 mutant strain, and fimbrial filaments are not assembled. Similar results were obtained for type 2 fimbriae after the analysis of the DNA sequences surrounding fimA (222), which is located in an operon containing three genes, orf977-fimA-orf365. While the first two genes encode surface proteins with LPXTG sorting signals, the product of orf365 specifies a sortase homolog. Interruption of orf365 abolished type II fimbrial assembly and led to the accumulation of FimA subunits with uncleaved sorting signals in the bacterial plasma membrane. These results suggest that A. naeslundii sortases are required for fimbrial assembly from precursor molecules that carry N-terminal signal peptides and C-terminal sorting signals.
Corynebacterium diphtheriae
C. diphtheriae is the causative agent of diphtheria, a deadly human disease that involves bacterial adherence to the upper respiratory tract and tissue damage via secretion of diphtheria toxin. The molecular mechanisms whereby C. diphtheriae binds to mucous membranes in the human pharynx and establishes a productive infection are still unknown. Following toxin-mediated tissue destruction and formation of pseudomembranes, associated inflammatory responses block human airways and precipitate respiratory failure (68). More than 30 years ago, Yanagawa and colleagues described pili on the surfaces of several different corynebacterial species, including C. diphtheriae (100, 217).
Bioinformatic analysis of the genome sequence of C. diphtheriae NCTC13129 identified six sortase-like genes (srtA to -F) (204). Five sortase genes are surrounded by ORFs encoding proteins with N-terminal signal peptides and C-terminal sorting signals, all clustered together in three separate loci on the bacterial chromosome. To analyze the expression and surface display of proteins, fragments of recombinant genes encoding signal peptides and sorting signals were expressed in E. coli and purified and antibody reagents were generated. Immunogold labeling of C. diphtheriae NCTC13129 followed by electron microscopy revealed that several of these antibodies stained pili on the bacterial surface. For example, antibodies raised against the SpaA (sortase-mediated pilin assembly A) protein stained filaments of 0.1 to 1 μm in length. SpaA protein is encoded by an operon comprised of four other open reading frames, spaA-srtA-spaB-spaC. Antibodies raised against purified SpaB also stained pili in immunogold labeling experiments, albeit that the gold particles were deposited at spaced intervals, whereas SpaA antibodies produced uniform staining. Antibodies raised against SpaC bound to the tip of the fiber and stained the same pili as SpaA- and SpaB-specific antibodies (Fig. 9).
FIG. 9.
Corynebacterium diphtheriae pili. A. Genetic organization of the spa locus of C. diphtheriae NCTC13129. Predicted promoters as well as the direction of transcription are shown with arrows. B to D. Corynebacterial pili stained with specific antiserum (anti-SpaA [B], anti-SpaB [C], or anti-SpaC [D]) and IgG-conjugated 12-nm gold particles. Samples were viewed by transmission electron microscopy. Bars indicate a distance of 0.2 μm. (Adapted from reference 204 with permission of Blackwell Publishing.)
Deletion of the spaA or the srtA gene completely abolished the assembly of SpaA pili as well as staining of pili with SpaB and SpaC antibodies. In contrast, deletion of spaC and spaB did not abrogate SpaA pilus formation. Transformation of pilin or srtA mutants with plasmid-carried wild-type genes restored the formation of pili and antibody staining (204). Pilus assembly can also be studied by immunoblotting. Pilin immune-reactive material must first be released from the bacterial envelope with muralytic enzyme and can then be loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). High-molecular-weight species that fail to separate on SDS-PAGE as well as monomeric pilin precursor molecules can be detected in this manner. When solubilized from extracts of srtA mutants, only SpaA pilin precursor, and not the assembled high-molecular-weight species, is detectable. These observations are consistent with the notion that SrtA catalyzes the assembly of SpaA pili and their deposition in the cell wall envelope. Polymerization of SpaA, SpaB, and SpaC into high-molecular-weight pili was examined in mutant strains lacking any one of the six different sortase genes. Only deletion of srtA prevented the incorporation of SpaA, SpaB, and SpaC into high-molecular-weight pili (204). SrtA alone appears to be sufficient for the polymerization of SpaA pili, as a variant with deletions of srtB to -F still produced high-molecular-weight immune-reactive material (199). As already observed by immunoelectron microscopy, deletion of spaA abrogated the incorporation of both SpaB and SpaC into pili when assayed by immunoblotting. In contrast, deletion of spaB or spaC did not abrogate the polymerization of SpaA (198, 204).
Taken together, these results suggest that C. diphtheriae pilus assembly is achieved by the activity of sortase A on pilin subunit precursors. As high-molecular-weight pili are resistant to boiling in SDS, it seems plausible that pilin subunits may be covalently cross-linked to one another. The requirement for sortase suggests that pilin subunit assembly involves not only the initiation of signal peptide-bearing precursors into the secretory pathway but also cleavage of sorting signals. SpaA, which represents the main pilin subunit, is uniformly present in the pilus shaft but apparently absent from the tip. Without SpaA, SpaB and SpaC cannot assemble into pili. SpaB, on the other hand, appears to decorate the shaft of SpaA pili, while SpaC may be positioned at the tip of this structure. Surprisingly, formation of this complex structure of SpaA pili requires only a single sortase gene, srtA. Whether this sortase can actually form transpeptidation products between proteins and, if so, what the nature of these linkages between pilus subunits is remain major research questions in this field. Interestingly, expression of the A. naeslundii pilin precursor FimA in C. diphtheriae led to formation of FimA pili (198). C. diphtheriae srtD, but not srtA, was required for the assembly of FimA pili, indicating that the mechanism of pilin polymerization is conserved among gram-positive bacteria.
How do sortases anchor some proteins to the cell wall envelope while assembling others into pili and still recognize precursor protein substrates at C-terminal sorting signals with strikingly similar properties? Pairwise comparison of the amino acid sequences of FimA, FimP, and SpaA identified four conserved sequence elements: (i) an N-terminal signal peptide, (ii) a C-terminal sorting signal, (iii) a central conserved domain with the amino acid sequence WxxxVxVYPK named the “pilin motif” (204), and (iv) a conserved domain with the amino acid sequence YxLxETxAPxGY, otherwise designated the “E box” (198). Alanine substitution experiments revealed that lysine 490 of the SpaA pilin motif is absolutely required for the polymerization of SpaA pili. As expected, mutations that perturb the LPLTG motif of the C-terminal sorting signal produced the same phenotype (204). Amino acid substitutions in the E box did not abrogate the polymerization of SpaA. Further, insertion of the pilin motif sequence into Seb flanked by an N-terminal signal peptide and a C-terminal sorting signal led to the polymerization of the reporter into high-molecular-weight polymers. Thus, the signal peptide, pilin motif, and sorting signal represent three topogenic elements that are necessary and sufficient for SpaA pilin polymerization by sortase.
The requirement for the single lysine residue within the pilin motif for assembly of pili is a compelling observation. Sortases require nucleophilic attack of amino groups at their acyl enzyme intermediates for product synthesis, i.e., a transpeptide bond between its two substrates. Therefore, it seems plausible that the ɛ-NH2 group of the conserved lysine residue may perform a nucleophilic attack at sortase acyl enzymes charged with pilin substrate, thereby cross-linking two adjacent SpaA pilin subunits via a transpeptide bond (Fig. 10). SpaC does not harbor a pilin motif. The C-terminal sorting signal of SpaC permits formation of sortase acyl enzyme but cannot provide a pilin motif amino group for its resolution. It would follow that SpaC would be the first subunit incorporated into pili during the sortase-mediated pilin assembly pathway. Therefore, SpaC can be located only at the tip of the fiber that is assembled by a sequence of transpeptidation reactions. Sortase-mediated pilus assembly must occur in the vicinity of the plasma membrane, and the acylated enzyme may on occasion accept lipid II as a nucleophile, a mechanism that could provide for termination of assembly and for the anchoring of pili in the cell wall envelope. The incorporation of SpaB into SpaA pilin requires the conserved glutamic acid residue of the SpaA E box. However, alanine substitution of the conserved glutamic acid also interferes with SpaC incorporation into SpaA pili. Thus, the mechanism whereby SpaB is incorporated into SpaA pili remains unknown. Nevertheless, the intellectual framework provided by these studies has established a fertile testing ground for several different hypotheses that predict the polymerization of pili in several different gram-positive bacteria and the universality of the assembly mechanisms.
FIG. 10.
Model for sortase-mediated pilus polymerization in C. diphtheriae. Sortase is thought to catalyze the polymerization of pili on the corynebacterial cell surface. Pilin subunits are typical sortase substrates, containing an N-terminal signal peptide (SP) that promotes secretion through the Sec system and a C-terminal cell wall sorting signal. SpaC is thought to be the first subunit to be incorporated into pili; if so, this might account for the detection of SpaC at the tip of the pilin fiber. The sortase-SpaC acyl intermediate may be attacked by the free amino group of a conserved lysine residue (K) present in the pilin motif of SpaA. The SpaA sorting signal would be in turn cleaved by sortase and linked to the lysine of a second SpaA pilin subunit. The remainder of the filament may then assemble by a sequence of similar transpeptidation reactions, and the polymerized pili may then be transferred to cell wall cross bridges for immobilization in the bacterial envelope. (Adapted from reference 203 with permission from Elsevier.)
Streptococcus agalactiae
Identification of four topogenic sequence elements involved in the polymerization of pili on the surface of C. diphtheriae permitted bioinformatic analysis of other gram-positive genomes for the presence of pilus assembly genes. In addition to A. naeslundii and C. diphtheriae, Bacillus cereus, Clostridium perfringens, Enterococcus faecalis, Streptococcus agalactiae, and Streptococcus pneumoniae all carry pilin genes and associated sortases. Experimental verification of the elaboration of pili on the surfaces of these microbes is rapidly being garnered (102). Thus, why were these pili not detected in earlier studies that examined the morphology of gram-positive bacteria by electron microscopy? The answer seems surprisingly simple. The diameter of gram-positive pili is significantly less than that of their gram-negative counterparts, and the structures were often mistaken for artifacts (102).
Focusing on the development of a new vaccine for the prevention of group B streptococcal meningitis, researchers identified almost 600 open reading frames in the genomes of eight different S. agalactiae human meningitis isolates that encode secreted proteins or surface-associated factors (111). By purifying 312 recombinant proteins that are conserved among all eight isolates and testing these polypeptides in a newborn mouse model of group B streptococcal meningitis for protective immune responses, vaccine efficacy was assigned to a cocktail of four proteins: one secreted factor and three proteins bearing both an N-terminal signal peptide and a C-terminal sorting signal. Antibodies raised against two of the purified recombinant surface proteins stained S. agalactiae pili in immunogold electron microscopy experiments (102). Although the contribution of pilus fibers to the pathogenesis of S. agalactiae meningitis has not yet been established, it seems likely that the identified pili play important roles in bacterial attachment to host cells or invasion of specific tissues.
SORTASE AND SURFACE PROTEIN FUNCTION IN SELECT GRAM-POSITIVE BACTERIA
Listeria monocytogenes
L. monocytogenes, a food-borne, facultatively intracellular human pathogen, causes listeriosis. During the pathogenesis of human disease, listeriae cross intestinal, placental, or blood-brain barriers and, following invasion across the plasma membrane and escape from phagocytic vacuoles, replicate within epithelial cells or macrophages in an effort to escape innate immune responses (43, 208). Essential for this invasion strategy are two surface proteins named internalin A (InlA) and internalin B (InlB), which allow Listeria to target host cells by binding to specific surface receptors (33). Bacterial attachment to host cells triggers a signaling cascade that culminates in cytoskeletal rearrangements and phagocytic uptake of bacteria. The best-studied surface protein of L. monocytogenes is internalin A, a surface protein that mediates bacterial entry into intestinal epithelial cells (56). Internalin A binds E-cadherin receptors located primarily in the adherens junctions of epithelial cells (126). The N-terminal signal peptide of internalin A is followed by 15 leucine-rich repeats, which together are responsible for binding to E-cadherin receptor (103, 175), and a C-terminal sorting signal bearing an LPXTG motif. The genome of Listeria monocytogenes carries 43 genes with predicted sorting signals, the largest number of surface proteins encountered so far in a microbial genome (19, 63). Nineteen genes display homology with internalin A, indicating that an entire family of bacterial ligands for host cell receptors may be responsible for mediating listerial invasion of different host species or different cell types. Two sortase homologs have been identified in the sequenced L. monocytogenes genomes (63).
The cell wall anchor structure of internalin A in L. monocytogenes was elucidated. A methionyl-six-histidyl affinity tag was inserted just upstream from the LPXTG motif, and recombinant internalin A was expressed in L. monocytogenes EGD and purified after solubilization of the cell wall with endolysin (38). Phage-encoded endolysin functions as an endopeptidase and cleaves the l-Ala-d-isoGlu amide bond of listerial cell wall peptides (109). Mass spectrometry analysis of C-terminal internalin A anchor peptides indicated that the LPTTG motif is cleaved between the threonine and the glycine residues and that the C-terminal threonine forms an amide bond with the amino group of m-diaminopimelic acid, the cell wall cross bridge of listerial peptidoglycan (38). Thus far, this is the only surface protein anchor structure that was solved for a nonstaphylococcal protein. Nevertheless, the data indicate that sortase-mediated anchoring is a universal process recognizing shared features of polypeptide and peptidoglycan substrates.
As already mentioned, the L. monocytogenes genome sequence encodes two sortases. The 222-residue sortase A is 28% identical to S. aureus sortase A (11, 58). As expected, deletion of the srtA gene abolishes anchoring of internalin A to the cell wall (11). Immunoelectron microscopy as well as immunofluorescence microscopy showed that InlA is not displayed on the surface of srtA mutant Listeria. Immunoblotting of bacterial cell fractions indicated that internalin A is missorted to the medium, cytoplasm, and membrane in srtA mutant strains (11). The internalin sorting defect could be complemented in trans by introducing a plasmid encoding sortase A in mutant bacteria (11). Tandem mass spectrometry of peptides solubilized from purified peptidoglycan with trypsin indicated the absence of at least 13 LPXTG-containing surface proteins from the surface of srtA Listeria, with 6 of them absent from the nonpathogenic species L. innocua (11, 157). Interestingly, L. monocytogenes sortase A was able to anchor a fusion protein between internalin B, a protein otherwise targeted to the envelope by binding to lipoteichoic acid, and the S. aureus protein A sorting signal, revealing conservation of functional elements of the cell wall sorting pathway between these two bacterial species (11).
The role of sortase A in the anchoring of InlA and other internalins, which are known virulence factors, prompted an investigation of the effects of a srtA deletion on the pathogenesis of L. monocytogenes. The invasion properties of a srtA mutant were assessed in vitro in a gentamicin survival assay. Caco-2 epithelial cells and HepG-2 hepatocytes were infected with wild-type or mutant bacteria, and gentamicin was added to kill all noninternalized bacteria. The results revealed a severe defect in the internalization of srtA listeriae (11, 58), with values similar to those obtained for an inlA mutant (11). Interestingly, complementation of the srtA deletion with a single-copy insertion elsewhere in the chromosome (58), but not with the gene introduced with a high-copy plasmid (11), allowed the recovery of listerial invasiveness. This suggests that sortase A overexpression causes a dominant negative effect on the invasion of L. monocytogenes, probably by anchoring an excess of surface proteins that mask other surface factors required for invasion. In addition, wild-type and srtA bacteria were equally able to multiply inside macrophages, showing that the defect is specific to epithelial cells and hepatocytes (10). The contribution of sortase A to L. monocytogenes virulence was also examined following oral and intravenous infections. After oral inoculation of mice, L. monocytogenes is able to cross the intestinal barrier and colonize different organs in a manner that does not require internalin A. Quantification of bacteria in the liver and the spleen at 3 days postinfection indicated that the srtA mutant displayed, in comparison with the wild type, a 1- to 2-log-unit decrease in bacterial replication (11). As inlA mutants do not display a phenotype in this assay, it follows that other surface proteins must be important to establish listeriosis in this model. Intravenous injection of mice with wild-type L. monocytogenes is lethal in animals infected with a dose of 104 to 105 CFU. A similar level of mortality could be achieved by injecting 106 to 107 CFU of srtA bacteria, indicating an important defect in virulence for this strain (58). For example, when infected with 106 CFU of wild-type L. monocytogenes, mice succumb to infection within 4 days, whereas animals infected with srtA mutant bacteria survive this challenge. Quantification of srtA L. monocytogenes in spleen, liver, brain, and blood over a period of 7 days showed an increase in the bacterial counts during the first 4 days of infection, followed by a sharp decrease and bacterial clearance (58). The importance of srtA for listerial pathogenesis was corroborated in the guinea pig model of oral infection (167). After oral administration of L. monocytogenes EGDe or the srtA or inlA isogenic deletion mutant, the abilities of these strains to cross the intestinal barrier and colonize different organs were assessed. Bacterial counts for the srtA strain decreased by 3 log units in the intestine and 2 log units in the mesenteric lymph nodes compared to those for the wild type and by 1 log unit in both organs compared with those for the inlA strain. These experiments establish sortase A as a virulence factor of L. monocytogenes and suggest that in addition to InlA, other sortase substrates contribute to the observed defects of srtA mutants in listerial pathogenesis.
In an effort to identify such sortase A substrates, LPXTG-containing surface proteins present only in pathogenic Listeria species were analyzed to determine their role during infection. These studies revealed two new virulence factors: Vip (20) and InlJ (167). Vip, an LPKAG motif surface protein, does not belong to the internalin family but contains a proline-rich region. Immunofluorescence detection of this protein indicated that it is present in the bacterial cell wall, and this localization is dependent on the presence of sortase A but not sortase B (20). Using an L. monocytogenes vip mutant isogenic strain, it was determined that Vip is required for bacterial entry into Caco-2 and L2071 cells in vitro. In vivo, the contribution of Vip to listeriosis was assessed after oral infection of mice with wild-type, vip mutant, and inlA mutant strains. Quantification of bacteria in the intestine, lymph nodes, liver, and spleen indicated a reduction of several log units in the number of vip mutant bacteria compared with the wild type in all organs analyzed. As already mentioned, srtA but not inlA mutants display a similar phenotype; it follows that the absence of cell wall-anchored Vip is responsible, at least in part, for the srtA mutant virulence defect in this model. In addition, vip was shown to be required for listerial pathogenesis in the mouse model of intravenous injection and in the guinea pig model of oral inoculation (20). The analysis of proteins from Caco-2 and L2071 cell extracts with the ability to interact with Vip allowed the identification of the Vip ligand as Gp96, a protein present in eukaryotic cells and involved in the modulation of innate and immune responses (20). It is then hypothesized that Vip binding to Gp96 may impair its physiological function, thereby subverting the host immune response in a manner that facilitates listerial infections.
InlJ is one of the 19 InlA homologs present in L. monocytogenes, containing 13 leucine-rich repeat sequences (rich in cysteine) (167) and an LPKTG motif sorting signal. The isogenic inlJ mutant strain was impaired in its ability to colonize the liver and spleen after intravenous injection of mice compared to wild-type L. monocytogenes (167). However, the inlJ mutant showed no defect in the invasion of epithelial or endothelial cells, hepatocytes, or macrophages, challenging the classification of InlJ as an internalin. These results indicate that, while InlJ function remains elusive, this surface protein constitutes a novel virulence factor that contributes to the virulence defects of L. monocytogenes srtA mutants.
L. monocytogenes sortase B, a protein of 246 amino acids, is encoded by the srtB gene (11), which resides in an operon with an organization similar to that reported for staphylococcal isd (see above) (10, 181). The first gene of the operon, lmo2186, specifies an IsdC homolog containing an NPKSS motif (157) and is followed by svpA, encoding a polypeptide with weak homology to S. aureus IsdA and a sorting signal with a putative NAKTN motif. lmo2184, lmo2183, and lmo2182 encode a putative lipoprotein, a membrane-anchored protein, and an ABC protein, similar to IsdD, IsdE, and IsdF of S. aureus, respectively. The srtB gene is positioned between isdCDEF and isdG, and the last gene of this locus encodes a protein with homology to the listerial phage protein Gp46. A Fur box is present in the promoter region of the listerial isd operon. The function of this element was assessed experimentally by placing gfp under the control of the srtB operon promoter (140). As expected, fluorescence of bacteria transformed with the gfp construct and grown in minimal medium was eliminated upon addition of FeSO4. The notion that the listerial isd locus is expressed under iron-restrictive conditions is also supported by the observation that expression of SvpA is dramatically increased in media lacking iron and inside Caco-2 and HepG-2 cells.
The role of sortase B in listerial surface protein anchoring was investigated in a srtB mutant. Using polyclonal serum raised against proteins present in purified peptidoglycan, cell wall proteome expression was compared between wild-type, srtA, and srtB strains. While sortase A anchors the great majority of surface proteins to the cell wall envelope, sortase B is responsible for the surface localization of only a few polypeptides, among them SvpA (10). Immunofluorescence analysis supported this observation, as SvpA could be detected only on the surfaces of wild-type or srtB-complemented bacteria and not on the surfaces of srtB mutants. Interestingly, SvpA was detected either laterally along the bacterial cylinder or at one pole of Listeria, an observation that suggests some specialized type of anchoring by sortase B. Lmo2186, the S. aureus IsdC homolog, is also anchored by SrtB, as tandem mass spectrometry of peptides solubilized from purified peptidoglycan with trypsin indicated the absence of SvpA and Lmo2186 from the surface of srtB Listeria (157). Moreover, fusions of SvpA or Lmo2186 sorting signals to InlB were absent from the peptidoglycan fraction of srtB Listeria, a result that unequivocally defines these surface proteins as SrtB substrates.
Streptococcus pyogenes
S. pyogenes (group A streptococcus [GAS]) is a gram-positive extracellular human pathogen and is responsible for a wide spectrum of disease, ranging from localized suppurative infections such as pharyngitis to pyoderma to severe systemic illnesses (for example, pneumonia or septicemia) and to serious postinfection autoimmune sequelae exemplified by acute rheumatic fever and glomerulonephritis (34). Surface proteins play a major role in streptococcal virulence and have been studied for more than 50 years (50). Several surface proteins of S. pyogenes can be classified as LPXTG motif sortase substrates; among them are the adhesins protein M (75) and protein F (a fibronectin binding protein (177), the C5a peptidase (ScpA) (25), and a protein G-related α2-macroglobulin-binding protein (159). Protein T, a trypsin-resistant surface protein, also harbors a C-terminal LPXTG motif sorting signal (172) and polymerizes into pilus structures that can be detected by immunoelectron microscopy (129). The antigenic properties of the M and T proteins are the basis for the serotype classification of GAS strains, and a combination of the recombinant versions of these proteins confer protection against mucosal challenge of mice with these pathogens (129).
A genetic screen was used to search for sortase genes, with the assumption that sortase-defective streptococci would fail to display protein F on the bacterial surface (7). Mutants generated by transposon mutagenesis were pooled and subjected to several rounds of immunoprecipitation with IgM, which precipitates bacteria that display protein F on their surface. Mutants unable to sediment upon incubation with IgM were mapped by DNA sequencing of insertion sites. One isolate harbored an insertion in the sortase A gene (srtA), and a deletion of this gene was generated by allelic replacement. The presence of several surface proteins in the bacterial envelopes of wild-type (M6 serotype) and srtA streptococci was detected by dot blotting using specific antibodies. Surprisingly, while the srtA mutant failed to display protein F, protein M, ScpA, and protein G-related α2-macroglobulin-binding protein on the bacterial surface, the anchoring and surface display of T protein were not affected by deletion of the srtA gene in spite of the presence of an LPXTG motif sorting signal in this polypeptide (7). However, deletion of a second sortase gene, named srtB, abolished the cell wall anchoring of T protein but had no effect on the anchoring and surface display of protein F, protein M, or ScpA. The srtB gene was found after bioinformatic searches of the S. pyogenes M1 genome for sortase homologs (7). The gene encodes a sortase with an N-terminal signal peptide and a C-terminal membrane anchor domain that is not structurally related to S. aureus SrtB. Additionally, in contrast to the case for S. aureus and L. monocytogenes, the S. pyogenes srtB gene is not associated with the isd locus but is present in an ∼11-kilobase pathogenicity island known as the fibronectin-binding, collagen-binding T antigen (FTC) region (9). While the genetic composition of the FTC island is highly variable, genes encoding T protein and SrtB homologs are always present in this region (129). As mentioned above, T proteins seem to be major subunits of S. pyogenes pili, whose assembly and surface display are dependent on sortase B (129). Together these results indicate that while S. pyogenes sortase A is able to anchor most LPXTG motif surface proteins, only sortase B can provide for the special linkage required for the polymerization of high-molecular-weight pili from T proteins.
S. pyogenes genomes harbor a variable number of four sortase genes (5, 48, 66, 132, 183, 189). Bioinformatic and Southern blot analyses of 12 different M serotypes showed that srtA is present in all strains, whereas srtB is present in only five of these isolates (7). Allele-specific PCR designed to detect all four sortases corroborated this finding (6). Analysis of 18 S. pyogenes isolates indicated that srtA is present in all strains examined, whereas srtB is present in fewer than half of all strains. Two other sortase genes (srtC1 and srtC2) are only sometimes found in GAS strains. Interestingly, srtC1 and srtC2 have not yet been found together in streptococcal isolates. The srtC1 and srtC2 genes are flanked by ORFs that likely encode their surface protein substrates. In S. pyogenes MGAS315 (M3 serotype), the operon contains five ORFs: cpa-sipA2-SPyM3_100-srtC2-SPyM3_102, where sipA2 encodes a putative signal peptidase and cpa, SPyM3_100, and SPyM3_102 encode cell surface proteins with VPPTGL, QVPTGV, and LPLAGE sorting signal motifs, all of which diverge from the canonical LPXTG sequence. This finding triggered the question of which, if any, of these proteins were sortase C2-specific substrates. DNA sequences encompassing sipA2-SPyM3_100 and slpA2-SPyM3_100-srtC2 were cloned in an S. pyogenes plasmid and introduced into strain JRS4, a serotype M6 strain that lacks the srtC2 locus (6). Detection of SPyM3_100 on the cell surface by dot blotting or in cell wall envelope fractions was shown to be dependent on the presence of srtC2. Replacement of the QVPTGV sequence with LPSTGE abrogated the anchoring of the mutant surface protein to the peptidoglycan. Thus, SrtC2 specifically recognizes and anchors proteins containing QVPTGV motif sorting signals. Interestingly, the amount of anchored product was significantly larger when srtC2 and its substrates were expressed in a srtA strain than when they were expressed in wild-type streptococci. This observation suggests that sortases A and C2 may compete for the same cell wall substrate of the sorting reaction, presumably lipid II (see above).
Oral Streptococci
The production of acid and the ability to form biofilms with other microbes are attributes of Streptococcus mutans that aid in the development of human tooth decay (caries). Several cell wall-anchored surface proteins are involved in the attachment of bacteria to tooth surfaces or to other streptococci and actinomycetes that are present in mixed biofilms and in dental plaque (127). Surface protein P1 (also known as antigen I/II or Pac) is an adhesin that promotes bacterial colonization of tooth surfaces by binding to salivary agglutinin, a glycoprotein that coats teeth (127). P1 is a surface protein with an N-terminal signal peptide and a C-terminal LPXTG motif sorting signal. Cell wall anchoring and surface display of P1 require the sortase A gene (srtA) of S. mutans (81, 105). In contrast to srtA deletions in S. aureus, deletion of srtA caused mutant streptococci to secrete P1 into the culture medium (81, 105). The secreted species reacted with a polyclonal antibody raised against the sorting signal of P1 (105) and migrated more slowly than the species present in wild-type bacterial pellets (81), indicating that the uncleaved precursor is released into the medium by srtA mutants. Similar experiments demonstrated that srtA is required for the cell wall anchoring and surface display of glucan binding protein C (GbpC) (82) and dextranase (80), surface proteins with LPXTG motif sorting signals. GbpC promotes bacterial aggregation through binding of many different microbes to the same substrate molecule. As glucan is present on tooth surfaces, GbpC-mediated aggregation may contribute to plaque formation (127). Dextranase, on the other hand, is an enzyme that hydrolyzes the α-1,6 bonds of certain glucan molecules. This results in the alteration of the solubility and adhesive properties of the glucan substrate and promotes S. mutans biofilm formation (80). Other S. mutans LPXTG surface proteins are presumably anchored by sortase and involved in formation of oral cavities by this microbe, and these include fructosidase (18) and WapA (49, 158). As may be expected from the lack of anchoring of several adhesins, S. mutans srtA variants displayed a remarkable reduction in their ability to form biofilms (107), in the adhesion to saliva-coated hydroxyapatite in vitro and the colonization of rat teeth in vivo (105), and in the ability to aggregate in the presence of dextrose (82).
Interestingly, two S. mutans clinical isolates contain deleterious mutations in the srtA gene. S. mutans Ingbritt contains an 11-base-pair deletion in the srtA ORF that generates a premature stop codon (79). As a result, bacteria secrete P1, GbpC, and dextranase. Similarly, S. mutans NG5 carries a missense mutation in the srtA gene that results in the production of truncated, nonactive enzyme (106). This strain also secretes P1 and is unable to adhere to hydroxyapatite and to aggregate in the presence of saliva. These mutant phenotypes were reversed to the wild-type phenotype when sortase A from strain NG8 was expressed in trans (105).
Streptococcus gordonii, a commensal of the human oral cavity, also displays proteins on its surface that are essential for adhesion and colonization of the oral cavity. A combination of degenerate PCR and BLAST searches on the partially sequenced genome of S. gordonii allowed the identification and inactivation of the srtA gene in this bacterium (13). Mutant bacteria were unable to bind fibronectin, consistent with the lack of anchoring of several LPXTG motif surface proteins. More importantly, the ability to colonize the oral cavities of mice was significantly reduced in the srtA mutant compared with the wild-type strain.
Streptococcus pneumoniae: Surface Proteins and Pili
The host specificity of S. pneumoniae is mainly restricted to humans, where the organism colonizes the nasopharynx and respiratory tract and is the causative agent of otitis media, pneumonia, bacteremia, and meningitis (73). Like for many other gram-positive pathogens, several surface proteins with LPXTG motif sorting signals are known virulence factors of S. pneumoniae, including hyaluronidase (hylA) and neuraminidase (nanA), hydrolytic enzymes that degrade polysaccharides in the extracellular matrix and other bacteria (127). The two S. pneumoniae strains sequenced to date, R6 and TIGR4, each harbor a sortase A gene homolog, srtA. The srtA gene has been deleted using the R6 strain as a parent (92), and β-galactosidase surface display and NanA cell wall anchoring were abolished in the srtA mutant strain. The srtA mutant displayed defects in the ability of S. pneumoniae to attach and invade pharyngeal cells in vitro (92). A virulence defect could be detected in vivo in competitive infection studies using murine models for pneumonia, bacteremia, and nasopharyngeal colonization (150) and in the chinchilla model of nasopharyngeal colonization (26). These results suggest that srtA contributes to pneumococcal disease.
Signature-tagged mutagenesis experiments identified other sortase genes as virulence factors of the S. pneumoniae TIGR4 (serotype 4) encapsulated clinical isolate (71). Pooled insertional mutants, a total of 6,149 strains, were examined for the ability to cause lung infections in mice, and 387 insertional variants displayed an attenuated phenotype. Two of these strains contained transposon insertions in rlrA (for RofA-like regulator) and srtD (encoding a sortase homolog). In S. pyogenes rofA encodes a transcription factor that regulates the expression of protein F, a sortase A-anchored virulence factor (see above). Interestingly, the rlrA and srtD genes are located on a pathogenicity island, flanked by IS1167 transposon elements (71) and present in a subset of clinical strains (150). rlrA is divergently transcribed from six other genes, three of which encode sortase homologs (srtB, srtC, and srtD) while the other three encode proteins with cell wall sorting signals. As expression of surface protein and sortase genes requires the RlrA transcription factor (72), the surface proteins were named RrgA, RrgB, and RrgC (RlrA-regulated gene). These proteins contain sorting signal motifs that diverge from the canonical LPXTG: YPRTG, IPQTG, and VPDTG, respectively. To examine the contribution of sortase and surface protein substrate genes to the pathogenesis of pneumococcal disease, all genes located in the pathogenicity island were mutated by in vitro transposition, and the virulence of the mutants obtained was assessed (71). Variants with transposon insertions in rlrA, rrgA, and srtD presented a virulence defect during murine lung infection. Moreover, when tested for colonization of the nasopharynx, rrgA and srtB variants were attenuated. Only the rlrA mutant displayed defects in the acute lethal disease following intraperitoneal injection of S. pneumoniae. It should be noted that the rrgB gene product encompasses not only an N-terminal signal peptide and C-terminal sorting signal but also a pilin motif and E-box sequence element. Although this has not yet been demonstrated experimentally, in accordance with the model for pilus assembly discussed above, the rlrA-regulated pathogenicity island of S. pneumoniae would be expected to provide for the expression of adhesive pili that aid in the pathogenesis of pneumococcal disease.
Streptococcus suis
S. suis, a chain-forming gram-positive pathogen, infects pigs and causes arthritis, meningitis, pneumonia, and endocarditis. S. suis is also known to cause meningitis in humans, and the microbe has been isolated from the respiratory and intestinal tracts of several ruminants. Two major S. suis virulence factors have been reported, a muramidase-released protein (Mrp) and an extracellular factor (EF). The physiological and biochemical properties of these virulence factors are, however, still unknown (185). Mrp, a surface protein with a C-terminal sorting signal, is thought to be covalently anchored to the S. suis cell wall envelope, and this has stimulated the study of sortases in this microbe. As S. suis genomic sequences are not yet available, PCR amplification with degenerate primers was used to identify five sortase gene homologs, srtA to -E, in the genome of S. suis serotype 2, the most common disease-associated serotype (146). The srtB to -D genes are clustered with two genes encoding putative surface protein substrates, named orf203 and orf204, specifying IPYTG and LPATG sorting signal motifs, respectively. srtA and srtE are located elsewhere on the chromosome. To characterize the role of these sortases in the anchoring of proteins to the cell wall of S. suis, three mutant strains lacking either srtA, srtBCD, or srtE were constructed. Purified S. suis cell wall sacculi from wild-type and mutant strains were treated with muramidase, and the solubilized proteins were separated by two-dimensional PAGE. Compared with the cell wall proteome of wild-type streptococci, the envelope of the srtA strain lacked several polypeptides. In contrast, no differences in cell wall proteome were observed between the wild-type, srtBCD mutant, or srtE mutant strains. Edman degradation was used to identify Mrp and three other surface proteins with LPXTG motif sorting signals whose presence in the cell wall envelope required the srtA gene, consistent with the notion that sortase A anchors surface proteins with LPXTG motif sorting signals to the cell wall envelope. S. suis strains can be classified into 35 serotypes according to their polysaccharide capsular antigens, and this classification provides predictions of virulence and disease phenotypes (185). Not all serotypes appear to harbor the srtA gene, as Southern blot analysis failed to detect srtA in serotypes 20, 22, and 26 (145). It is not yet known whether the presence of srtA coupled with the ability to anchor surface proteins with LPXTG motif sorting signals exerts a general impact on the virulence of S. suis isolates.
Bacillus anthracis
B. anthracis is the causative agent of anthrax, a gram-positive spore-forming bacterium that predominantly infects herbivores (96). Humans and many different animal species are susceptible to B. anthracis infection. B. anthracis spores represent the infectious form of the pathogen and enter the host in three different ways, i.e., through a minor skin lesion, via inhalation, or via ingestion. Each of these entry routes leads to a different disease spectrum, commonly referred to as cutaneous, pulmonary, or gastrointestinal anthrax, respectively (128). The main virulence factors of B. anthracis include two secreted toxins, lethal toxin and edema toxin, as well as the cell wall-anchored poly-γ-d-glutamic acid capsule (22), which confers antiphagocytic properties on the vegetative form of bacilli. The genes encoding anthrax toxins or factors required for capsule production are located on two virulence plasmids, pXO1 and pXO2 (128).
The genome of B. anthracis carries three sortase genes (160). The predicted product of one of these genes, designated srtA, displays a high degree of amino acid sequence homology with S. aureus sortase A. The second gene is located within the isd locus of B. anthracis, which is predicted to be regulated by Fur and encodes surface proteins, an ABC transporter, a heme oxygenase, and a sortase B homolog (srtB). A third sortase gene, srtC, displays homology with Bacillus sp., Streptomyces sp., and Clostridium sp. sortases, enzymes that are found only in sporulating bacteria (41). Several genes encoding predicted surface proteins with LPXTG motif-type sorting signals were identified through bioinformatic searches of B. anthracis genome sequences. These surface proteins include many polypeptides of unknown function, several collagen binding proteins (216), and the receptor for the γ phage, a bacteriolytic phage specific for B. anthracis strains (35). To determine whether these polypeptides are anchored to the cell wall by sortase A, the srtA gene was deleted from the genome of B. anthracis strain Sterne, a variant lacking the pXO2 virulence plasmid, which provides for capsule biosynthesis (59). The cell wall anchoring of BasC (Bacillus anthracis surface protein C), a surface protein that functions as a collagen adhesin (216), was analyzed in wild-type as well as in srtA bacilli (59). Cleavage of the peptidoglycan strands with muramidase released FLAG epitope- and His6-tagged BasC from the cell wall envelopes of bacilli. Recombinant BasC could be purified by affinity chromatography and detected by immunoblotting. BasC could not, however, be purified from the cell wall envelopes of srtA mutant bacilli, which harbored precursor polypeptide in membrane and cytoplasmic fractions. Cell wall anchoring of BasC in srtA mutant strains could be restored by expression of the srtA gene from a complementing plasmid. The contribution of srtA to virulence was assayed in the A/J mouse model of acute B. anthracis infection (59). A/J mice display a defect in the phagocytic killing of bacterial pathogens and, when infected with the attenuated B. anthracis strain Sterne, develop acute lethal disease symptoms. Mice were injected intravenously with wild-type and srtA spores. Time-to-death experiments indicated that the srtA mutant strain displayed no defect in the ability to cause an acute lethal infection in A/J mice. However, it was recently shown that B. anthracis srtA and srtB mutants are impaired in their ability to infect J774 macrophages (226), suggesting that, as is the case in the majority of gram-positive pathogens, B. anthracis sortases form part of the virulence repertoire of this microbe.
Recombinant B. anthracis sortase A was purified by affinity chromatography using His6 replacement of the N-terminal signal peptide/membrane anchor. B. anthracis SrtA displayed a level of activity that is comparable to that of S. aureus sortase A, providing for an analysis of substrate specificity with FRET peptides (59). B. anthracis SrtA cleaved LPETG and LPATG fluorescent substrates, but not LPNTA, LGATG, or NPKTG; the latter peptide motif sequences are present in sorting signals of other B. anthracis surface proteins and presumably represent substrates for sortases B and C. Mass spectrometric analysis of B. anthracis SrtA cleavage products revealed that the enzyme cleaves LPETG substrate between the threonine and the glycine residue. This activity can be abolished with MTSET, and treatment with DTT restores enzyme activity. Thus, SrtA specifically cleaves LPXTG motif sorting signals and anchors surface proteins to the cell wall envelope of B. anthracis, similar to the case for sortase A of S. aureus. The biological functions of SrtB and SrtC and their protein substrates remain unknown. Future work is also needed to unravel the contribution of the three sortases to the pathogenesis of anthrax infections by using the fully virulent B. anthracis isolates as parent strains for mutagenesis and complementation studies.
Hyphal Development in Streptomyces coelicolor
Streptomyces coelicolor, a gram-positive filamentous bacterium in the soil, produces many natural antibiotics (74). The life cycle of this microbe begins with the formation of a feeding (submerged) mycelium, which differentiates into aerial hyphae that septate into spore chains. The hyphal surface becomes hydrophobic, a property that promotes outgrowth into the air and facilitates the dispersion of spores, which give rise to new mycelia. Six streptomycete surface proteins, collectively called chaplins, are involved in aerial hypha formation (29, 45). All chaplins are synthesized during mycelium formation as precursors with an N-terminal signal sequence and a short hydrophobic domain (chaplin domain) of about 40 residues. Chaplins D to H are small polypeptides, whereas chaplins A to C are larger and carry a C-terminal sorting signal (LAXTG motif). Deletion of six chaplin genes (chpA to -H) hindered the formation of aerial hyphae, and the defect could be rescued by the addition of purified chaplin proteins (45). As mixtures of ChpD to -H isolated from cell walls of aerial hyphae are highly surface active (29), it is presumed that these proteins would lower the water surface tension and thus allow the emergence of aerial hyphae at the air-water interface. In this model, the large chaplins ChpA to -C would be anchored by sortase to the bacterial peptidoglycan and would serve as a scaffold for the assembly of the small chaplins ChpD to -H. In addition, the chaplin layer provides a hydrophobic surface for the formation of the RdlA-RdlB rodlet layer, the highly insoluble, outer layer of proteins that facilitate spore dispersion (30).
BIOINFORMATIC ANALYSIS OF SORTASES AND SUBSTRATES
Through a variety of bioinformatic searches, sortase and sortase-like genes, along with a plethora of substrate genes, have been found in almost all gram-positive bacterial genomes available to date (12, 31, 41, 148). Additionally, sortase enzymes have been identified in the gram-negative organisms Bradyrhizobium japonicum, Colwellia psychroerythraea, Microbulbifer degradans, Shewanella oneidensis, and Shewanella putrefasciens, as well as in Methanobacterium thermoautotrophicum, a thermophilic archaeon (31, 147). Interestingly, in the majority of genomes where sortase enzyme genes have been identified, usually multiple sortases are encoded. Based on homology, the sortases thus far identified can be grouped into four or five subgroups or classes (Table 2). Each subgroup, in addition to distinctions in sequence, can be distinguished from one another based on membrane topology, genome position, and preference for substrates with specific amino acids within the cell wall sorting signal pentapeptide motif (31, 41).
The prototypical sortase A, first identified in S. aureus, contains an N-terminal transmembrane domain and the sequence TLXTC at its active site, where C corresponds to the catalytic cysteine residue (Cys184 in S. aureus sortase A [see above]). Sortase A appears to anchor a large number and broad range of surface proteins, and unlike many other sortase genes, the sortase A gene is not found clustered with its substrates. It also appears that only a single sortase A homolog is encoded per bacterial genome (31). The sortase A subgroup of enzymes also seems to share a preference for the LPXTG cell wall sorting signal motif. The second subgroup of enzymes, sortase B, along with its substrate (IsdC in S. aureus), is encoded in an iron transport operon involved in heme-iron uptake (see above) (41, 121, 182). Enzymes belonging to the sortase B subgroup contain three amino acid segments not found in sortase A and recognize substrates containing an NPQTN motif rather than the canonical LPXTG (41). The third class, designated sortase C or subfamily 3, contains a C-terminal hydrophobic domain (31, 41). This group of sortase enzymes is often encoded in multiple copies per genome. Subfamily 3 enzymes also share a preference for substrates containing the LPXTG cell wall sorting signal motif, often followed by a second G residue. Unlike those for sortase A, the genes for subgroup 3 enzymes are predicted to anchor a much smaller set of substrates, which are typically clustered with the structural gene for the enzyme (31).
A fourth subgroup can be defined after alignment of sortase sequences. It has been noted as the sortase D subgroup (41) or subfamilies 4 and 5, as sortases in this subgroup can be distinguished based on the cell wall sorting signals of their associated substrates (31). Sortases belonging to subfamily 4 are predicted to anchor proteins bearing the unique LPXTA(ST) motif (31). An alanine residue in the last position of the substrate motif suggests that the subfamily 4 enzymes fulfill a nonredundant role within the cell (31). These sortases are typically found clustered with their substrates, which usually possess enzymatic function. Many high-G+C bacteria contain sortases belonging to subfamily 5, and, interestingly, most do not harbor sortase A homologs. This subgroup of sortase enzymes shares substrate specificity for proteins containing an LAXTG motif (31), and at least in Streptomyces coelicolor they are essential for mycelium and hypha development (29, 45).
Many sortase genes are found clustered with genes encoding their substrates. C. diphtheriae serves as an example where sortases and their substrates cooperate to assemble pili. Five sortase genes are found in three loci on the corynebacterial chromosome, along with the various surface protein substrates that assemble into three types of pili (204). Genomic analysis also suggests that sortases and substrates belonging to different subgroups can be merged to form distinct sorting pathways (31). The S. aureus isd locus, for example, encompasses sortase B, its NPQTN motif substrate IsdC, and two other surface protein genes which are anchored by sortase A (IsdA and IsdB) (121, 182). Thus, while parallel pathways for sorting surface proteins to the cell wall envelope do exist, intersecting pathways appear to increase structural and functional flexibility. Once again, bacteria have evolved to make the most of their genes.
CONCLUSIONS AND FUTURE DIRECTIONS
Sortases are transpeptidases that cleave protein substrates at defined sites once they have been translocated across the bacterial plasma membrane. The resulting acyl intermediate between substrate and sortase is resolved by nucleophilic attack of an amine, typically provided by cell wall cross bridges but presumably also occurring for amino groups of proteins. The end product is the formation of a single amide bond with a surface protein substrate. Obviously, this bond would not be there if it were not for the ability of sortase to catalyze these reactions, without the need for high-energy phosphates. Is there more? Can sortases link proteins not just to cell wall and polypeptides? We do not know the answer yet, but shouldn't we simply ask why not? Amino groups exist on carbohydrates and lipids, and hence they could also be substrates for sortases.
Many surface protein substrates, sorting signals, and sortases have been identified, and the three dimensional structure of sortase with and without peptide substrate has been unraveled. In spite of this progress, we do not truly appreciate how these enzymes bind their peptidoglycan or protein cosubstrates, and we do not fully understand how their active-site thiol is activated for catalysis. Perhaps the most pressing need for our research is not to fill in the remaining puzzle of sortase reactions and enzyme-substrate relationships or to study their role in pathogenesis. Instead, since we already know that sortases are immensely important, we should concentrate our efforts on the discovery of small-molecule inhibitors that specifically block the sortase-catalyzed transpeptidation reaction, which, after all, does not exist in humans or animals. With a little bit of luck, such a therapeutic inhibitor may become a reality in the foreseeable future.
Acknowledgments
We thank all members of our laboratory, past, present, and future, for unveiling the mysteries of sortases. You all made it fun, and it was again fun for us to write about it. We apologize to all those authors whose work we may not have adequately represented or may have even omitted. This did not occur with ill intent, but because we ran out of time. O.S. is indebted to Rockefeller University mentors Peter Model, Marjorie Russel, and Vincent Fischetti for their relentless interest and encouragement.
Work on sortases and surface proteins was made possible by funding from the National Institute of Allergy and Infectious Diseases, Infectious Diseases Branch (grants AI38897 and AI52474). O.S. acknowledges membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153).
REFERENCES
- 1.Abrahmsen, L., T. Moks, B. Nilsson, U. Hellman, and M. Uhlen. 1985. Analysis of signals for secretion in the staphylococcal protein A gene. EMBO J. 4:3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akabas, M. H., and A. Karlin. 1995. Identification of acetylcholine receptor channel-lining residues in the M1 segment of the alpha-subunit. Biochemistry 34:12496-12500. [DOI] [PubMed] [Google Scholar]
- 3.Albus, A., R. D. Arbeit, and J. C. Lee. 1991. Virulence studies of Staphylococcus aureus mutants altered in type 5 capsule production. Infect. Immun. 59:1008-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, T., and O. Schneewind. 2003. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185:2910-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190:727-738. [DOI] [PubMed] [Google Scholar]
- 6.Barnett, T. C., A. R. Patel, and J. R. Scott. 2004. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J. Bacteriol. 186:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayles, K. W., C. A. Wesson, L. E. Liou, L. K. Fox, G. A. Bohach, and W. R. Trumble. 1998. Intracellular Staphylococcus aureus escapes the endosome and induces apoptosis in epithelial cells. Infect. Immun. 66:336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bierne, H., C. Garandeau, M. G. Pucciarelli, C. Sabet, S. Newton, F. Garcia-del Portillo, P. Cossart, and A. Charbit. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, P. Dehoux, L. Jansch, F. G. Portillo, L. G., O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 12.Boekhorst, J., M. W. de Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 187:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolken, T. C., C. A. Franke, K. F. Jones, G. O. Zeller, C. H. Jones, E. K. Dutton, and D. E. Hruby. 2001. Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect. Immun. 69:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley, J. S. 2005. Newer antistaphylococcal agents. Curr. Opin. Pediatr. 17:71-77. [DOI] [PubMed] [Google Scholar]
- 15.Bremell, T., A. Adelnour, and A. Tarkowski. 1992. Histopathological and serological progression of experimental Staphylococcus aureus arthritis. Infect. Immun. 60:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bremell, T., S. Lange, A. Yacoub, C. Ryden, and A. Tarkowski. 1991. Experimental Staphylococcus aureus arthritis in mice. Infect. Immun. 59:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullen, J., E. Griffiths, H. Rogers, and G. Ward. 2000. Sepsis: the critical role of iron. Microbes Infect. 2:409-415. [DOI] [PubMed] [Google Scholar]
- 18.Burne, R. A., and J. E. Penders. 1992. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-beta-d-fructosidase. Infect. Immun. 60:4621-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabanes, D., P. Dehoux, O. Dussurget, L. Franeul, and P. Cossart. 2002. Surface proteins and pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 20.Cabanes, D., S. Sousa, A. Cebria, M. Lecuit, F. Garcia-del Portillo, and P. Cossart. 2005. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 24:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calandra, G. B., and R. Cole. 1980. Lysis and protoplast formation of group B streptococci by mutanolysin. Infect. Immun. 28:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candela, T., and A. Fouet. 2005. Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol. Microbiol. 57:717-726. [DOI] [PubMed] [Google Scholar]
- 23.Chang, T. K., D. Y. Jackson, J. P. Burnier, and J. A. Wells. 1994. Subtiligase: a tool for semisynthesis of proteins. Proc. Natl. Acad. Sci. USA 91:12544-12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee, A. N., and J. T. Park. 1964. Biosynthesis of cell wall mucopeptide by a particulate fraction from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 51:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, C. C., and P. P. Cleary. 1990. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J. Biol. Chem. 263:3161-3167. [PubMed] [Google Scholar]
- 26.Chen, S., G. K. Paterson, H. H. Tong, T. J. Mitchell, and T. F. Demaria. 2005. Sortase A contributes to pneumococcal nasopharyngeal colonization in the chinchilla model. FEMS Microbiol. Lett. 253:151-154. [DOI] [PubMed] [Google Scholar]
- 27.Cisar, J. O., and A. E. Vatter. 1979. Surface fibrils (fimbriae) of Actinomyces viscosus T14V. Infect. Immun. 24:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cisar, J. O., A. E. Vatter, W. B. Clark, S. H. Curl, S. Hurst-Calderone, and A. L. Sandberg. 1988. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect. Immun. 56:2984-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claessen, D., R. Rink, W. de Jong, J. Siebring, P. de Vreugd, F. G. Boersma, L. Dijkhuizen, and H. A. Wosten. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17:1714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claessen, D., I. Stokroos, H. J. Deelstra, N. A. Penninga, C. Bormann, J. A. Salas, L. Dijkhuizen, and H. A. Wosten. 2004. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53:433-443. [DOI] [PubMed] [Google Scholar]
- 31.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis idenitifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 72:2710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conolly, K. M., B. T. Smith, R. Pilpa, U. Ilangovan, M. E. Jung, and R. T. Clubb. 2003. Sortase from Staphylococcus aureus does not contain a thiolate-imidazolium ion pair in its active site. J. Biol. Chem. 278:34061-34065. [DOI] [PubMed] [Google Scholar]
- 33.Cossart, P., J. Pizarro-Cerda, and M. Lecuit. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell. Biol. 13:23-31. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davison, S., E. Couture-Tosi, T. Candela, M. Mock, and A. Fouet. 2005. Identification of the Bacillus anthracis (gamma) phage receptor. J. Bacteriol. 187:6742-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deisenhofer, J., T. A. Jones, R. Huber, J. Sjödahl, and J. Sjöquist. 1978. Crystallization, crystal structure analysis and atomic model of the complex formed by a human Fc fragment and fragment B of protein A from Staphylococcus aureus. Hoppe-Seyler's Z. Physiol. Chem. 359:975-985. [DOI] [PubMed] [Google Scholar]
- 37.Deivanayagam, C. C., E. R. Wann, W. Chen, M. Carson, K. R. Rajashankar, M. Hook, and S. V. Narayana. 2002. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 21:6660-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 39.Dmitriev, B. A., F. V. Toukach, O. Holst, E. T. Rietschel, and S. Ehlers. 2004. Tertiary structure of Staphylococcus aureus cell wall murein. J. Bacteriol. 186:7141-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donkersloot, J. A., J. O. Cisar, M. E. Wax, R. J. Harr, and B. M. Chassy. 1985. Expression of Actinomyces viscosus antigens in Escherichia coli: cloning of a structural gene (fimA) for type 2 fimbriae. J. Bacteriol. 162:1075-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156:289-297. [DOI] [PubMed] [Google Scholar]
- 42.Dryla, A., D. Gelbmann, A. von Gabain, and E. Nagy. 2003. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49:37-53. [DOI] [PubMed] [Google Scholar]
- 43.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 44.Dziewanowska, K., P. J. M., C. F. Deobald, B. K. W., W. R. Trumble, and G. A. Bohach. 1999. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect. Immun. 67:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elliot, M. A., N. Karoonuthaisiri, J. Huang, M. J. Bibb, S. N. Cohen, C. M. Kao, and M. J. Buttner. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17:1727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Escolar, L., J. Perez-Martin, and V. De Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez, L. A., and J. Berenguer. 2000. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol. Rev. 24:21-44. [DOI] [PubMed] [Google Scholar]
- 48.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferretti, J. J., R. R. B. Russel, and M. L. Dao. 1989. Sequence analysis of the wall-associated protein precursor of Streptococcus mutans antigen A. Mol. Microbiol. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 50.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 52.Flock, J. I., G. Fröman, K. J.önsson, B. Guss, C. Signäs, B. Nilsson, G. Raucci, M. Höök, T. Wadström, and M. Lindberg. 1987. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 6:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foster, T. J., and M. Höök. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 54.Foster, T. J., and D. McDevitt. 1994. Surface-associated proteins of Staphylococcus aureus: their possible roles on virulence. FEMS Microbiol. Lett. 118:199-206. [DOI] [PubMed] [Google Scholar]
- 55.Frankel, B. A., M. Bentley, R. G. Kruger, and D. G. McCafferty. 2004. Vinyl sulfones: inhibitors of SrtA, a transpeptidase required for cell wall protein anchoring and virulence in Staphylococcus aureus. J. Am. Chem. Soc. 126:3404-3405. [DOI] [PubMed] [Google Scholar]
- 56.Gaillard, J.-L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 57.Gally, D., and A. R. Archibald. 1993. Cell wall assembly in Staphylococcus aureus: proposed absence of secondary crosslinking reactions. J. Gen. Microbiol. 139:1907-1913. [DOI] [PubMed] [Google Scholar]
- 58.Garandeau, C., H. Reglier-Poupet, I. Dubail, J. L. Beretti, P. Berche, and A. Charbit. 2002. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 70:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaspar, A. H., L. A. Marraffini, E. M. Glass, K. L. Debord, H. Ton-That, and O. Schneewind. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 187:4646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghuysen, J.-M., and R. Hakenbeck (ed.). 1994. Bacterial cell wall. Elsevier Science B.S., Amsterdam, The Netherlands.
- 61.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis amd fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 64.Gomez, M. I., A. Lee, B. Reddy, A. Muir, G. Soong, A. Pitt, A. Cheung, and A. Prince. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842-848. [DOI] [PubMed] [Google Scholar]
- 65.Gouda, H., H. Torigoe, A. Saito, M. Sato, Y. rata, and I. Shimada. 1992. Three-dimensional solution structure of the B domain of staphylococcal protein A: a comparison of the solution and crystal structures. Biochemsitry 31:9665-9672. [DOI] [PubMed] [Google Scholar]
- 66.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. Lefebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192:760-770. [DOI] [PubMed] [Google Scholar]
- 67.Guss, B., M. Uhlén, B. Nilsson, M. Lindberg, J. Sjöquist, and J. Sjödahl. 1984. Region X, the-cell-wall-attachment part of staphylococcal protein A. Eur. J. Biochem. 138:413-420. [DOI] [PubMed] [Google Scholar]
- 68.Hadfield, T. L., P. McEvoy, Y. Polotsky, V. A. Tzinserling, and A. A. Yakovlev. 2000. The pathology of diphtheria. J. Infect. Dis. 181(Suppl. 1):S116-S120. [DOI] [PubMed] [Google Scholar]
- 69.Hartford, O., P. Francois, P. Vaudaux, and T. J. Foster. 1997. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 25:1065-1076. [DOI] [PubMed] [Google Scholar]
- 70.Hartleib, J., N. Kohler, R. Dickinson, G. Chhatwal, J. Sixma, O. Hartford, T. J. Foster, G. Peters, B. Kehrl, and M. Herrmann. 2000. Protein A is the von Willebrand factor binding protein of Staphylococcus aureus. Blood 96:2149-2156. [PubMed] [Google Scholar]
- 71.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 72.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hava, D. L., J. LeMieux, and A. Camilli. 2003. From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol. Microbiol. 50:1103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hodgson, D. A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47-238. [DOI] [PubMed] [Google Scholar]
- 75.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A streptococcus. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 76.Horsburgh, M., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang, X., A. Aulabaugh, W. Ding, B. Kapoor, L. Alksne, K. Tabei, and G. Ellestad. 2003. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry 42:11307-11315. [DOI] [PubMed] [Google Scholar]
- 78.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Igarashi, T. 2004. Deletion in sortase gene of Streptococcus mutans Ingbritt. Oral Microbiol. Immunol. 19:210-213. [DOI] [PubMed] [Google Scholar]
- 80.Igarashi, T., E. Asaga, and N. Goto. 2004. Roles of Streptococcus mutans dextranase anchored to the cell wall by sortase. Oral Microbiol. Immunol. 19:102-105. [DOI] [PubMed] [Google Scholar]
- 81.Igarashi, T., E. Asaga, and N. Goto. 2003. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol. Immunol. 18:266-269. [DOI] [PubMed] [Google Scholar]
- 82.Igarashi, T., E. Asaga, Y. Sato, and N. Goto. 2004. Inactivation of srtA gene of Streptococcus mutans inhibits dextran-dependent aggregation by glucan-binding protein C. Oral Microbiol. Immunol. 19:57-60. [DOI] [PubMed] [Google Scholar]
- 83.Ilangovan, U., J. Iwahara, H. Ton-That, O. Schneewind, and R. T. Clubb. 2001. Assignment of 1H, 13C and 15N signals of sortase. J. Biomol. NMR. 19:379-380. [DOI] [PubMed] [Google Scholar]
- 84.Ilangovan, U., H. Ton-That, J. Iwahara, O. Schneewind, and R. T. Clubb. 2001. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6056-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Izaki, K., M. Matsuhashi, and J. L. Strominger. 1966. Glycopeptide transpeptidase and d-alanine carboxypeptidase: penicillin sensitive enzymatic reactions. Proc. Natl. Acad. Sci. USA 55:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jensen, K. 1958. A normally occuring staphylococcus antibody in human serum. Acta Pathol. Microbiol. Scand. 44:421-428. [Google Scholar]
- 87.Jonsson, I. M., S. K. Mazamanian, O. Schneewind, M. Vendrengh, T. Bremell, and A. Tarkowski. 2002. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J. Infect. Dis. 185:1417-1424. [DOI] [PubMed] [Google Scholar]
- 88.Jonsson, I. M., S. K. Mazmanian, O. Schneewind, T. Bremell, and A. Tarkowski. 2003. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microb. Infect. 5:775-780. [DOI] [PubMed] [Google Scholar]
- 89.Jönsson, K., C. Signäs, H. P. Müller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 90.Josefsson, E., K. W. McCrea, D. Ní Eidhin, D. O'Connell, J. Cox, M. Höök, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387-3395. [DOI] [PubMed] [Google Scholar]
- 91.Josefsson, E., D. O'Connell, T. J. Foster, I. Durussel, and J. A. Cox. 1998. The binding of calcium to the B-repeat segment of SrdD, a cell surface protein of Staphylococcus aureus. J. Biol. Chem. 273:31145-31152. [DOI] [PubMed] [Google Scholar]
- 92.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 71:2758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim, S. H., D. S. Shin, M. N. Oh, S. C. Chung, J. S. Lee, I. M. Chang, and K. B. Oh. 2003. Inhibition of sortase, a bacterial surface protein anchoring transpeptidase, by beta-sitosterol-3-O-glucopyranoside from Fritillaria verticillata. Biosci. Biotechnol. Biochem. 67:2477-2479. [DOI] [PubMed] [Google Scholar]
- 94.Kim, S. H., D. S. Shin, M. N. Oh, S. C. Chung, J. S. Lee, and K. B. Oh. 2004. Inhibition of the bacterial surface protein anchoring transpeptidase sortase by sioquinoline alkaloids. Biosci. Biotechnol. Biochem. 68:421-424. [DOI] [PubMed] [Google Scholar]
- 95.Kim, S. W., I. M. Chang, and K. B. Oh. 2002. Inhibition of the bacterial surface protein anchoring transpeptidase sortase by medicinal plants. Biosci. Biotechnol. Biochem. 66:2751-2754. [DOI] [PubMed] [Google Scholar]
- 96.Koch, R. 1876. Die Aetiologie der Milzbrand-Krankheit, begruendet auf die Entwicklungsgeschichte des Bacillus Anthracis. Beitraege Biol. Pflanzen 2:277-310. [Google Scholar]
- 97.Kopp, U., M. Roos, J. Wecke, and H. Labischinski. 1996. Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb. Drug Resist. 2:29-41. [DOI] [PubMed] [Google Scholar]
- 98.Kruger, R. G., S. Barkallah, B. A. Frankel, and D. G. McCafferty. 2004. Inhibition of the Staphylococcus aureus sortase transpeptidase SrtA by phosphinic peptidomimetics. Bioorg. Med. Chem. 12:3723-3729. [DOI] [PubMed] [Google Scholar]
- 99.Kruger, R. G., B. Otvos, B. A. Frankel, M. Bentley, P. Dostal, and D. G. McCafferty. 2004. Analysis of the substrate specificity of the Staphylococcus aureus sortase transpeptidase SrtA. Biochemistry 43:1541-1551. [DOI] [PubMed] [Google Scholar]
- 100.Kuamazawa, N., and R. Yanagawa. 1972. Chemical properties of the pili of Corynebacterium renale. Infect. Immun. 5:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Labischinski, H., K. Ehlert, and B. Berger-Bachi. 1998. The targeting of factors necessary for expression of methicillin resistance in staphylococci. J. Antimicrob. Chemother. 41:581-584. [DOI] [PubMed] [Google Scholar]
- 102.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 103.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee, J. C., M. J. Betley, C. A. Hopkins, N. E. Perez, and G. B. Pier. 1987. Virulence studies, in mice, of transposon-induced mutants of Staphylococcus aureus differing in capsule size. J. Infect. Dis. 156:741-750. [DOI] [PubMed] [Google Scholar]
- 105.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee, S. F., and M. K. McGavin. 2004. Identification of a point mutation resulting in loss of cell wall anchoring activity of SrtA of Streptococcus mutans NG5. Infect. Immun. 72:4314-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Levesque, C. M., E. Voronejskaia, Y. C. Huang, R. W. Mair, R. P. Ellen, and D. G. Cvitkovitch. 2005. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 73:3773-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liew, C. K., B. T. Smith, R. Pilpa, N. Suree, U. Ilangovan, K. M. Connolly, M. E. Jung, and R. T. Clubb. 2004. Localization and mutagenesis of the sorting signal binding site on sortase A from Staphylococcus aureus. FEBS Lett. 571:221-226. [DOI] [PubMed] [Google Scholar]
- 109.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 110.Mack, J., C. Vermeiren, D. E. Heinrichs, and M. J. Stillman. 2004. In vivo heme scavenging by Staphylococcus aureus IsdC and IsdE proteins. Biochem. Biophys. Res. Commun. 320:781-788. [DOI] [PubMed] [Google Scholar]
- 111.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. Nardi Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maira-Litran, T., A. Kropec, D. A. Goldmann, and G. B. Pier. 2005. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl-beta-(1-6)-glucosamine. Infect. Immun. 73:6752-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mao, H. 2004. A self-cleavable sortase fusion for one-step purification of free recombinant proteins. Protein Expr. Purif. 37:253-263. [DOI] [PubMed] [Google Scholar]
- 114.Mao, H., S. A. Hart, A. Schink, and B. A. Pollok. 2004. Sortase-mediated protein ligation: a new method for protein engineering. J. Am. Chem. Soc. 126:2670-2671. [DOI] [PubMed] [Google Scholar]
- 115.Marraffini, L. A., and O. Schneewind. 2005. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J. Biol. Chem. 280:16263-16271. [DOI] [PubMed] [Google Scholar]
- 116.Marraffini, L. A., H. Ton-That, Y. Zong, S. V. Narayana, and O. Schneewind. 2004. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. A conserved arginine residue is required for efficient catalysis of sortase A. J. Biol. Chem. 279:37763-37770. [DOI] [PubMed] [Google Scholar]
- 117.Matsuhashi, M. 1994. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation, p. 55-72. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Biochemical Press, Amsterdam, The Netherlands.
- 118.Matsuhashi, M., C. P. Dietrich, and J. L. Strominger. 1967. Biosynthesis of the peptidoglycan of bacterial cell walls. III. The role of soluble ribonucleic acid and of lipid intermediates in glycine incorporation in Staphylococcus aureus. J. Biol. Chem. 242:3191-3206. [PubMed] [Google Scholar]
- 119.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 97:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 121.Mazmanian, S. K., E. P. Skaar, A. H. Gasper, M. Humayun, P. Gornicki, J. Jelenska, A. Joachimiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 122.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 123.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase enzyme anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 125.McDevitt, D., T. Nanavaty, K. House-Pompeo, E. Bell, N. Turner, L. McIntire, T. Foster, and M. Höök. 1997. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur. J. Biochem. 247:416-424. [DOI] [PubMed] [Google Scholar]
- 126.Mengaud, J., H. Ohayon, P. Gounon, M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of Listeria monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 127.Mitchell, T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1:219-230. [DOI] [PubMed] [Google Scholar]
- 128.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 129.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 102:15641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Murphy, T. M., J. M. Deitz, P. J. Petersen, S. M. Mikels, and W. J. Weiss. 2000. Therapeutic efficacy of GAR-936, a novel glycylcycline, in a rat model of experimental endocarditis. Antimicrob. Agents Chemother. 44:3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Naik, M. T., N. Suree, U. Ilangovan, C. K. Liew, W. Thieu, D. O. Campbell, J. J. Clemens, M. E. Jung, and R. T. Clubb. 2006. Staphylococcus aureus sortase A transpeptidase: Calcium promotes sorting signal binding by altering the mobility and structure of an active loop. J. Biol. Chem. 281:1817-1826. [DOI] [PubMed] [Google Scholar]
- 132.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nakagawa, J., S. Tamaki, S. Tomioka, and M. Matsuhashi. 1984. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. Penicillin-binding protein 1Bs of Escherichia coli with activities of transglycosylase and transpeptidase. J. Biol. Chem. 259:13937-13946. [PubMed] [Google Scholar]
- 134.Navarre, W. W., and O. Schneewind. 1996. Cell wall sorting of lipoproteins in Staphylococcus aureus. J. Bacteriol. 178:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 136.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and the mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. 1998. Anchor structure of staphylococcal surface proteins. II. COOH-terminal structure of muramidase and amidase-solubilized surface protein. J. Biol. Chem. 273:29135-29142. [DOI] [PubMed] [Google Scholar]
- 138.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. 1999. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage φ11. Identification of a d-alanyl-glycine endopeptidase activity. J. Biol. Chem. 274:15847-15856. [DOI] [PubMed] [Google Scholar]
- 139.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Newton, S. M., P. E. Klebba, C. Raynaud, Y. Shao, X. Jiang, I. Dubail, C. Archer, C. Frehel, and A. Charbit. 2005. The svpA-srtB locus of Listeria monocytogenes: Fur-mediated iron regulation and effect on virulence. Mol. Microbiol. 55:927-940. [DOI] [PubMed] [Google Scholar]
- 141.Ní Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Höök, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 142.Oh, K. B., S. H. Kim, J. Lee, W. J. Cho, T. Lee, and S. Kim. 2004. Discovery of diarylacrylonitriles as a novel series of small molecule sortase A inhibitors. J. Med. Chem. 47:2418-2421. [DOI] [PubMed] [Google Scholar]
- 143.Oh, K. B., M. N. Oh, J. G. Kim, D. S. Shin, and J. Shin. Inhibition of sortase-mediated Staphylococcus aureus adhesion to fibronectin via fibronectin-binding protein by sortase inhibitors. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 144.O'Riordan, K., and J. C. Lee. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2003. Allelic variation in srtAs of Streptococcus suis strains. FEMS Microbiol. Lett. 219:195-201. [DOI] [PubMed] [Google Scholar]
- 146.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2002. Characterization of Streptococcus suis genes encoding proteins homologous to sortase of gram-positive bacteria. J. Bacteriol. 184:971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 148.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases-a richness of substrates. Trends Microbiol. 9:97-101. [DOI] [PubMed] [Google Scholar]
- 149.Patel, A. H., P. Nowlan, E. D. Weavers, and T. Foster. 1987. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect. Immun. 55:3103-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Paterson, G. K., and T. J. Mitchell. 2005. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. In press. [DOI] [PubMed]
- 151.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:89-115. [DOI] [PubMed] [Google Scholar]
- 152.Patti, J. M., H. Jonsson, B. Guss, L. M. Switalski, K. Wiberg, M. Lindberg, and M. Höök. 1992. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267:4766-4772. [PubMed] [Google Scholar]
- 153.Perkins, S., E. J. Walsh, C. C. Deivanayagam, S. V. Narayana, T. J. Foster, and M. Hook. 2001. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J. Biol. Chem. 276:44721-44728. [DOI] [PubMed] [Google Scholar]
- 154.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241-16248. [DOI] [PubMed] [Google Scholar]
- 155.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ponnuraj, K., M. G. Bowden, S. Davis, S. Gurusiddappa, D. Moore, D. Choe, Y. Xu, M. Hook, and S. V. Narayana. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217-228. [DOI] [PubMed] [Google Scholar]
- 157.Pucciarelli, M. G., E. Calvo, C. Sabet, H. Bierne, P. Cossart, and F. Garcia-Del Portillo. 2005. Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomic analysis. Proteomics 5:4808-4817. [DOI] [PubMed] [Google Scholar]
- 158.Qian, H., and M. L. Dao. 1993. Inactivation of the Streptococcus mutans wall-associated protein A gene (wapA) results in a decrease in sucrose-dependent adherence and aggregation. Infect. Immun. 61:5021-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rasmussen, M., H.-P. Muller, and L. Bjorck. 1999. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding α2-macroglobulin. J. Biol. Chem. 274:15336-15344. [DOI] [PubMed] [Google Scholar]
- 160.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baille, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. T. Cline, C. Redmond, J. E. Thwaite, O. WHite, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 161.Rich, R. L., C. C. Deivanayagam, R. T. Owens, M. Carson, A. Hook, D. Moore, J. Symersky, V. W. Yang, S. V. Narayana, and M. Hook. 1999. Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus Cna MSCRAMM. J. Biol. Chem. 274:24906-24913. [DOI] [PubMed] [Google Scholar]
- 162.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 163.Rosch, J., and M. Caparon. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513-1515. [DOI] [PubMed] [Google Scholar]
- 164.Rosenstein, R., and F. Gotz. 2000. Staphylococcal lipases: biochemical and molecular characterization. Biochimie 82:1005-1014. [DOI] [PubMed] [Google Scholar]
- 165.Ruzin, A., A. Severin, F. Ritacco, K. Tabei, G. SIngh, P. A. Bradford, M. M. Siegel, S. J. Projan, and D. M. Shlaes. 2002. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J. Bacteriol. 184:2141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ryden, C., K. Rubin, P. Speziale, M. Höök, M. Lindberg, and T. Wadstrom. 1983. Fibronectin receptors from Staphylococcus aureus. J. Biol. Chem. 258:3396-3401. [PubMed] [Google Scholar]
- 167.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73:6912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sandermann, H. 1976. Moenomycin: an inhibitor of C55-isoprenoid-alcohol kinase from Staphylococcus aureus. Biochim. Biophys. Acta 444:783-788. [PubMed] [Google Scholar]
- 169.Schindler, C. A., and V. T. Schuhardt. 1964. Lysostaphin: a new bacteriolytic agent for the staphylococcus. Proc. Natl. Acad. Sci. USA 51:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 172.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characteristics of the trypsin-resistant T6 surface protein of group A streptococci. J. Bacteriol. 172:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 175.Schubert, W. D., C. Urbanke, T. Ziehm, V. Beier, M. P. Machner, E. Domann, J. Wehland, T. Chakraborty, and D. W. Heinz. 2002. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111:825-836. [DOI] [PubMed] [Google Scholar]
- 176.Scott, C. J., A. McDowell, S. L. Martin, J. F. Lynas, K. Vandenbroek, and B. Walker. 2002. Irreversible inhibition of the bacterial cysteine protease-transpeptidase sortase (SrtA) by substrate-derived affinity labels. Biochem. J. 366:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 178.Signas, C., G. Raucci, K. Jonsson, P.-E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sjödahl, J. 1977. Repetitive sequences in protein A from Staphylococcus aureus. Arrangement of five regions within the protein, four being highly homologous and Fc-binding. Eur. J. Biochem. 73:343-351. [DOI] [PubMed] [Google Scholar]
- 180.Sjöquist, J., B. Meloun, and H. Hjelm. 1972. Protein A isolated from Staphylococcus aureus after digestion with lysostaphin. Eur. J. Biochem. 29:572-578. [DOI] [PubMed] [Google Scholar]
- 181.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 182.Skaar, E. P., and O. Schneewind. 2004. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6:390-397. [DOI] [PubMed] [Google Scholar]
- 183.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Snowden, M. A., H. R. Perkins, A. W. Wyke, M. V. Hayes, and J. B. Ward. 1989. Cross-linking and O-acetylation of newly synthesized peptidoglycan in Staphylococcus aureus H. J. Gen. Microbiol. 135:3015-3022. [DOI] [PubMed] [Google Scholar]
- 185.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-407. [DOI] [PubMed] [Google Scholar]
- 186.Storer, A. C., and R. Menard. 1994. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 244:487-500. [DOI] [PubMed] [Google Scholar]
- 187.Strominger, J. L. 1968. Penicillin-sensitive enzymatic reactions in bacterial cell wall synthesis. Harvey Lectures 64:179-213. [PubMed] [Google Scholar]
- 188.Strominger, J. L., K. Izaki, M. Matsuhashi, and D. J. Tipper. 1967. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed. Proc. 26:9-18. [PubMed] [Google Scholar]
- 189.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 190.Switalski, L. M., J. M. Patti, W. Butcher, A. G. Gristina, P. Speziale, and M. Höök. 1993. A collagen receptor on Staphylococcus aureus strains isolated from patients with septic arthritis mediates adhesion to cartilage. Mol. Microbiol. 7:99-107. [DOI] [PubMed] [Google Scholar]
- 191.Takatsuki, A., K. Arima, and G. Tamura. 1971. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiotics. 24:215-223. [DOI] [PubMed] [Google Scholar]
- 192.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus penumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 193.Tipper, D. J., and J. L. Strominger. 1968. Biosynthesis of the peptidoglycan of bacterial cell walls. XII. Inhibition of cross-linking by penicillins and cephalosporins: studies in Staphylococcus aureus in vivo. J. Biol. Chem. 243:3169-3179. [PubMed] [Google Scholar]
- 194.Tipper, D. J., and J. L. Strominger. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-alanine. Proc. Natl. Acad. Sci. USA 54:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ton-That, H., K. F. Faull, and O. Schneewind. 1997. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272:22285-22292. [DOI] [PubMed] [Google Scholar]
- 196.Ton-That, H., H. Labischinski, B. Berger-Bächi, and O. Schneewind. 1998. Anchor structure of staphyococcal surface proteins. III. The role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J. Biol. Chem. 273:29143-29149. [DOI] [PubMed] [Google Scholar]
- 197.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ton-That, H., L. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53:251-261. [DOI] [PubMed] [Google Scholar]
- 199.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694:269-278. [DOI] [PubMed] [Google Scholar]
- 200.Ton-That, H., H. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. I. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J. Biol. Chem. 275:9876-9881. [DOI] [PubMed] [Google Scholar]
- 201.Ton-That, H., S. K. Mazmanian, L. Alksne, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. II. Cysteine 184 and histidine 120 of sortase A form a thiolate imidazolium ion pair for catalysis. J. Biol. Chem. 277:7447-7452. [DOI] [PubMed] [Google Scholar]
- 202.Ton-That, H., and O. Schneewind. 1999. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 274:24316-24320. [DOI] [PubMed] [Google Scholar]
- 203.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12:228-234. [DOI] [PubMed] [Google Scholar]
- 204.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of C. diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 205.Tung, H. S., B. Guss, U. Hellman, L. Persson, K. Rubin, and C. Ryden. 2000. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem. J. 345:611-619. [PMC free article] [PubMed] [Google Scholar]
- 206.Uhlén, M., B. Guss, B. Nilsson, S. Gatenbeck, L. Philipson, and M. Lindberg. 1984. Complete sequence of the staphylococcal gene encoding protein A. J. Biol. Chem. 259:1695-1702. (Correction, 259:13628.) [PubMed] [Google Scholar]
- 207.van Heijenoort, Y., M. Leduc, H. Singer, and J. van Heijenoort. 1987. Effects of moenomycin on Escherichia coli. J. Gen. Microbiol. 133:667-674. [DOI] [PubMed] [Google Scholar]
- 208.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Vernachio, J., A. S. Bayer, T. Le, Y. L. Chai, B. Prater, A. Schneider, B. Ames, P. Syribeys, J. Robbins, and J. M. Patti. 2003. Anti-clumping factor A immunoglobulin reduces the duration of methicillin-resistant Staphylococcus aureus bacteremia in an experimental model of infective endocarditis. Antimicrob. Agents Chemother. 47:3400-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Vernet, T., D. C. Tessier, J. Chatellier, C. Plouffe, T. S. Lee, D. Y. Thomas, A. C. Storer, and R. Menard. 1995. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 270:16645-16652. [DOI] [PubMed] [Google Scholar]
- 211.Walsh, C. T. 1993. Vancomycin resistance: decoding the molecular logic. Science 261:308-309. [DOI] [PubMed] [Google Scholar]
- 212.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 213.Weiss, W. J., E. Lenoy, T. Murphy, L. Tardio, P. Burgio, S. J. Projan, O. Schneewind, and L. Alksne. 2004. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal infection. J. Antimicrob. Chemother. 53:480-486. [DOI] [PubMed] [Google Scholar]
- 214.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 215.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterization of the ferric uptake regulator, fur, from Staphylococcus aureus. Microbiology 146:659-668. [DOI] [PubMed] [Google Scholar]
- 216.Xu, Y., X. Liang, Y. Chen, T. M. Koehler, and M. Hook. 2004. Identification and biochemical characterization of two novel collagen binding MSCRAMMs of Bacillus anthracis. J. Biol. Chem. 279:51760-51768. [DOI] [PubMed] [Google Scholar]
- 217.Yanagawa, R., and E. Honda. 1976. Presence of pili in species of human and animal parasites and pathogens of the genus Corynebacterium. Infect. Immun. 13:1293-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yanagawa, R., and K. Otsuki. 1970. Some properties of the pili of Corynebacterium renale. J. Bacteriol. 101:1063-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yeung, M. K., B. M. Chassy, and J. O. Cisar. 1987. Cloning and expression of a type 1 fimbrial subunit of Actinomyces viscosus T14V. J. Bacteriol. 169:1678-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yeung, M. K., and J. O. Cisar. 1988. Cloning and nucleotide sequence of a gene for Actinomyces naeslundii WVU45 type 2 fimbriae. J. Bacteriol. 170:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yeung, M. K., and J. O. Cisar. 1990. Sequence homology between the subunits of two immunologically and functionally distinct types of fimbriae of Actinomyces spp. J. Bacteriol. 172:2462-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yeung, M. K., J. A. Donkersloot, J. O. Cisar, and P. A. Ragsdale. 1998. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect. Immun. 66:1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yeung, M. K., and P. A. Ragsdale. 1997. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require expression of additional fimbria-associated genes. Infect. Immun. 65:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yokogawa, K., S. Kawata, S. Nishimura, Y. Ikeda, and Y. Yoshimura. 1974. Mutanolysin, bacteriolytic agent for cariogenic streptococci: partial purification and properties. Antimicrob. Agents Chemother. 6:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang, R.-G., G. Joachimiak, R.-Y. Wu, S. K. Mazmanian, D. M. Missiakas, O. Schneewind, and A. Joachimiak. 2004. Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site. Structure 12:1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zink, S. D., and D. L. Burns. 2005. Importance of srtA and srtB for growth of Bacillus anthracis in macrophages. Infect. Immun. 73:5222-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zong, Y., T. W. Bice, H. Ton-That, O. Schneewind, and S. V. Narayana. 2004. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J. Biol. Chem. 279:31383-31389. [DOI] [PubMed] [Google Scholar]
- 228.Zong, Y., S. K. Mazmanian, O. Schneewind, and S. V. Narayana. 2004. The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall. Structure 12:105-112. [DOI] [PubMed] [Google Scholar]