Abstract
All eukaryotic cells contain multiple acidic organelles, and V-ATPases are central players in organelle acidification. Not only is the structure of V-ATPases highly conserved among eukaryotes, but there are also many regulatory mechanisms that are similar between fungi and higher eukaryotes. These mechanisms allow cells both to regulate the pHs of different compartments and to respond to changing extracellular conditions. The Saccharomyces cerevisiae V-ATPase has emerged as an important model for V-ATPase structure and function in all eukaryotic cells. This review discusses current knowledge of the structure, function, and regulation of the V-ATPase in S. cerevisiae and also examines the relationship between biosynthesis and transport of V-ATPase and compartment-specific regulation of acidification.
INTRODUCTION
All eukaryotic cells contain multiple acidic organelles, including lysosomes, early and late endosomes, and the late Golgi apparatus (86). Organelle acidification is implicated in protein sorting in the biosynthetic and endocytic pathways, proteolytic activation of zymogen precursors, and transmembrane transport of viral contents and toxins, but may also impact many other aspects of cell physiology (86, 103, 139). In plants and fungi, the lysosome-like vacuole is adapted to additional functions, including storage of metabolic building blocks, calcium homeostasis, and osmotic control (72), and vacuolar acidification is critical for these functions as well. The central player in organelle acidification in all eukaryotes is the vacuolar proton-translocating ATPase (V-ATPase).
V-ATPases are evolutionary descendants of a family of archaeal proton pumps and ATP synthases that also gave rise to the F1F0-ATP synthases of mitochondria and chloroplasts (38, 43, 91). Although archaeal and bacterial V-ATPases show remarkable versatility, exhibiting the ability to transport Na+ or H+, and to synthesize or hydrolyze ATP in different contexts (60, 91), eukaryotic V-ATPases are dedicated proton pumps in vivo. In fungi and most other eukaryotic cells, their primary role is ATP-driven transport of protons from the cytosol into acidic organelles. Eukaryotic V-ATPases appear to have relinquished some of the versatility of their bacterial precursors, but they are still associated with an amazing range of cellular functions and regulated at many different levels.
This review focuses on the V-ATPase of Saccharomyces cerevisiae, with reference to work in other fungi in some cases. The yeast V-ATPase has become the major model system for the study of eukaryotic V-ATPases, and current knowledge of the structure, assembly, and regulation of this enzyme will be discussed, along with its functions in yeast.
STRUCTURE AND MECHANISM OF V-ATPases
Structure of the Yeast V-ATPase
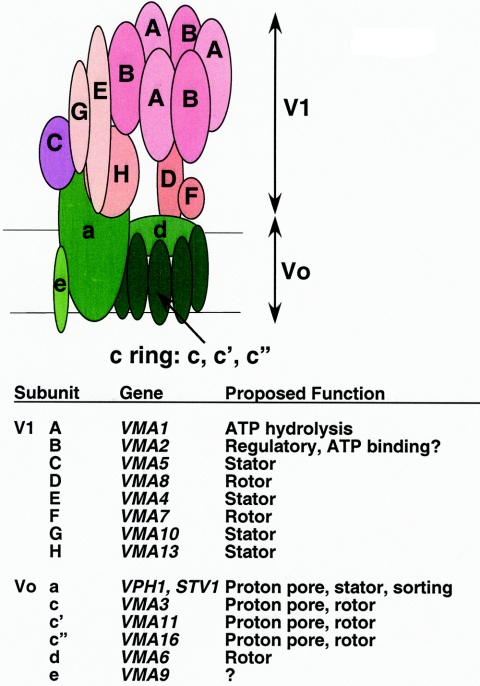
V-ATPases are multisubunit enzymes composed of a peripheral complex (called V1 by analogy to F1 of the F1F0-ATP synthase) attached to a membrane-bound complex called V0 (100, 103). A proposed structure and subunit composition for the yeast V-ATPase is shown in Fig. 1. The V1 sector contains three copies of the catalytic A subunit, which is responsible for ATP hydrolysis, that alternate with three copies of the B subunit, which also contains potential nucleotide binding sites, but are believed to play a regulatory role (42, 139, 159). These two subunits are homologous to the β and α subunits, respectively, of F1F0-ATP synthases, with amino acid identities ranging from 20 to 25%, with the highest conservation in the nucleotide binding regions (38, 102). In addition, the yeast V1 sector contains six other subunits, designated C, D, E, F, G, and H, that do not have readily identifiable homologs in F-type ATPases (103). These subunits appear to be arranged in two or more stalks that connect the catalytic headgroup, consisting of the A and B subunits, with the membrane domain (152, 157).
FIG. 1.
Subunit composition and structural model of the yeast V-ATPase. V1 subunits are shown in pink and purple and V0 subunits are shown in green. A single stator stalk containing subunits C, E, G, and H is shown, but recent electron microscopy evidence suggests that there may be two peripheral stalks in the closely related Neurospora crassa V-ATPase (152). If there are two stator stalks in the yeast enzyme, it is likely that each stalk will contain subunits E and G, but one stalk may contain subunit H, and the other subunit C (152).
The membrane-bound V0 domain consists of six subunits, designated a, c, c′, c", d, and e (126, 139). The c, c′, and c" subunits show homology to the c subunits of the F1F0-ATP synthases, but appear to have undergone gene duplication, resulting in a four-transmembrane domain protein rather than the two-transmembrane domains typically found in c subunits of F1F0 (48). (The c" subunit may actually have five transmembrane domains [32].) The discovery that the c, c′, and c" subunits are all required for V-ATPase activity in yeast and that all are present in a single complex was surprising (120). This mixing of different c-like subunits is not observed in any F1F0-ATP synthase and has important implications for enzyme mechanism and assembly.
The a subunit of V-ATPases shows limited sequence resemblance to any F1F0-ATPase subunit, but its placement in the V0 sector suggested that it might play a similar role to the a subunit of F0-ATPases in rotational catalysis, and further mutational analysis has supported this (69, 75). Subunit a is the only yeast V-ATPase subunit that is present as multiple isoforms (84). These isoforms, encoded by the VPH1 and STV1 genes, are targeted to the vacuole and Golgi apparatus/endosome, respectively, by the mechanisms discussed below (68, 84). The d and e subunits are unique to V-ATPases (5, 126). A bacterial d subunit has been crystallized, and based on the crystal structure, it is proposed to rest on a “nest” formed by multiple copies of the c subunits (57). The e subunit is part of the final V-ATPase complex, both in yeast and in other eukaryotes, and is essential for its assembly and function, but nothing is known about its placement or function in the V0 sector (79, 87, 126).
Catalytic Mechanism of V-ATPases
The core catalytic mechanism of the yeast V-ATPase parallels that of F1F0-ATP synthases operating in the ATP hydrolysis direction. Both enzymes use a rotational catalytic mechanism in which conformational changes accompanying ATP hydrolysis are communicated through rotation of a central stalk attached to the c subunits and rotation of the ring of c subunits results in proton transport (50, 54). The c subunits of F1F0 are present as a ring of 10 to 14 subunits (58, 85). In current models of rotational catalysis, the c ring sequentially binds protons to an intramembrane acidic amino acid present in each c subunit and then releases the protons to the opposite side of the membrane when the proton-bearing c subunits are brought into register with the a subunit through rotation (2, 31).
In the V-ATPase, there appears to be only one intramembrane glutamate responsible for proton binding in each c-like subunit, even though these subunits are twice as large as the F1F0 c subunits (48, 105). In the c and c′ subunits, this glutamate is in the fourth transmembrane domain, while in the c" subunit, it is in the third transmembrane domain in models predicting five transmembrane domains (48). (This would be the second transmembrane domain if the first predicted domain does not, in fact, enter the bilayer [104].)
Given the central role of the c ring in rotational catalysis, it would appear to be very important that the hetero-oligomeric c rings of V-ATPases be carefully assembled to properly position the catalytically essential glutamates in order to avoid interruptions in the catalytic cycle. Although there is evidence that the different c subunits are present in a defined stoichiometry (120), and there may be specific mechanisms for their incorporation during biosynthesis (41, 80), the incorporation of the c" subunit into the c ring, particularly into the rotational mechanism, is not well understood.
In F0, interaction of a critical arginine in the a subunit, arginine 210 in Escherichia coli, is directly involved in proton transfer from the rotating ring of c subunits. Mutagenesis of many charged and polar amino acids in predicted transmembrane domains of VPH1 suggested several candidate amino acids in the last four transmembrane domains that might assume this function in V-ATPases (75). Subsequent work demonstrated that arginine 735 of Vph1p is absolutely required from proton transport (69), and arginine 735 is now generally believed to play a catalytic role in the yeast V-ATPase similar to that of arginine 210 in the E. coli F-ATPase. There is also very weak homology between the final transmembrane domain of Vph1p and the a subunit of F0. Mutagenesis of this region of VPH1, particularly E789 and R799, suggested that these amino acids support the function of arginine 735, much as glutamate 219 and asparagine 214 of the E. coli a subunit support arginine 210 (69).
Another structural requirement of rotational catalysis is that part of the enzyme, including at minimum the A, B, and a subunits, must be held together in order for rotation of the central stalk and the c ring to drive proton translocation. In E. coli F1F0, one of the best-studied examples, this stable linkage of the a subunit and the catalytic head group is achieved through a single peripheral stalk, including a dimer of the highly elongated b subunit, a transmembrane protein associated with the a subunit in F0, in complex with the δ subunit, an F1 subunit that binds to the top of the catalytic headgroup (22, 23). In V-ATPases, this “stator” function appears to be accomplished by several of the V1 subunits, one or more of which interact with the large (∼45-kDa) cytoplasmic domain of the a subunit (55, 59, 152, 158). Consistent with a more complex subunit composition for the stator, the stalks of V-ATPases appear to be more complex than those of F1F0 when viewed by electron microscopy (21, 152, 159), and there is even evidence of multiple peripheral stalks in V-ATPases (152).
The G subunit was definitively placed in the stator by the original set of rotational catalysis experiments which demonstrated rotation of the c subunit relative to subunit G (50). The E, C, and H subunits are also localized to the peripheral face of the catalytic headgroup and thus are candidates for the stator as well (4, 55, 158). In addition, a small domain of the catalytic subunit, which has been called the nonhomologous region because it appears to represent an insertion into the V-ATPase catalytic subunits that is not present in any F1F0 catalytic subunit, also appears to function as part of the stator (133).
V-ATPase FUNCTION AND CELL PHYSIOLOGY
Phenotypes Arising from Loss of V-ATPase Function
The first V-ATPase subunit genes were cloned by reverse genetic approaches. The nucleotide binding (A and B) subunits of Neurospora crassa, carrot, and Arabidopsis thaliana were identified by probing cDNA libraries with oligonucleotides based on peptide sequences from subunits of the isolated enzyme or by probing expression libraries with subunit-specific antibodies (8, 11, 82, 163). The c subunit from the bovine chromaffin granule V-ATPase was cloned at the same time by similar methods (81). Because these subunits are very highly conserved among V-ATPases, identification of the homologous genes in other organisms was accelerated by publication of the first few subunit sequences, although some of the initial yeast V-ATPase subunit genes were also identified by reverse genetic approaches (96, 98).
The definition of the yeast V-ATPase subunit composition was greatly assisted by genetic approaches, however, after a set of phenotypes highly characteristic of loss of V-ATPase activity (Vma− phenotypes) was uncovered. Nelson and Nelson (99) first proposed that loss of V-ATPase function in yeast led to a pH-dependent conditional lethality in which the mutant strains failed to grow at pH 7 or higher, but were able to grow at pH 5 to 5.5. Ohya et al. (107) subsequently showed that mutants that were both sensitive to high concentrations of extracellular calcium and unable to grow on nonfermentable carbon sources were likely to contain defective V-ATPases, and thus identified a number of V-ATPase subunit genes from their set of cls (calcium-sensitive) mutants.
The combination of sensitivity to high pH and high extracellular calcium concentrations came to be the “trademark” Vma− phenotype. With the exception of the VPH1 and STV1 genes, which exhibit the full Vma− phenotype only when both genes are disrupted (84), all of the subunit genes listed in Fig. 1 result in a very similar pH-dependent, calcium-sensitive growth phenotype when disrupted. Deletion of any of these genes also leads to the loss of all organelle acidification, as judged by accumulation of the fluorescent, lysosomotropic amine quinacrine, and the loss of ATPase activity in isolated vacuolar membranes (160). Although a limited set of mutants with deletions in other genes mimic the Vma− growth phenotype, only subunits of the V-ATPase and a small set of dedicated assembly factors (see below) result in both the growth phenotype and a total loss of quinacrine accumulation and vacuolar ATPase activity when deleted (125).
The Vma− growth phenotype has proved to be extremely valuable as an experimental tool. It has been used to screen for new subunit genes, assembly factors, and V-ATPase regulators (51, 107, 110, 125), to test proteins identified biochemically as associated with the V-ATPase for effects on V-ATPase function (143), and to rapidly identify mutations in V-ATPase subunit genes that compromise function. As it became increasingly clear that the yeast V-ATPase was extremely similar to V-ATPases of higher organisms, identification of potential subunits in other organisms was sometimes confirmed by identifying the corresponding gene in yeast and examining a deletion for the Vma− phenotype (40, 97). In fact, the Vma− phenotype is largely the reason that yeast has emerged as the predominant model system for studies of V-ATPase function.
Other fungi, including Neurospora crassa, Candida albicans, and Schizosaccharomyces pombe, also show a pH-dependent growth phenotype upon disruption of V-ATPase subunits (10, 56, 119), but disruption of V-ATPase function in higher eukaryotes is lethal (15, 108, 142). The exception to this is mutations in tissue-specific isoforms of certain subunits in mammalian cells. V-ATPases containing these isoforms provide additional specific functions beyond the constitutive function in organelle acidification, and these V-ATPases are often found at the plasma membrane. Mammalian plasma membrane V-ATPases have been implicated in bone resorption, urinary acidification, and pH regulation (155). In these cases, disruption of the tissue-specific isoform gives a more restricted phenotype, arising from loss of V-ATPase activity only in specific locations and associated with the specialized function of V-ATPases containing that isoform (67, 136).
Although the combination of pH-dependent growth and calcium sensitivity is characteristic of loss of V-ATPase activity, it should be noted that these are far from being the only physiological defects in mutants lacking V-ATPase function. Even at pH 5, vma mutants grow significantly more slowly than wild-type cells (160). They are acutely sensitive to a wide variety of heavy metals (25, 26) and hypersensitive to many different drugs (114). They have been characterized as having severe defects in sporulation and germination (27), but the germination defect, in particular, can be suppressed at low extracellular pH.
The reason for the “petite” phenotype, which was documented very early, is still not fully understood; vma mutant cells have been reported to have relatively normal activities in isolated mitochondria (107). A number of functions not readily associated with organelle acidification, for example, cell wall structure and function, cytoskeletal organization, and phospholipid content and distribution, are also affected in vma mutants (16, 107, 162). This array of defects suggests that V-ATPase activity is connected to many other cellular functions, but also needs to be considered when V-ATPase mutants are used as experimental tools.
vma Mutants Reveal Cellular Functions of V-ATPases
Some of the phenotypes of the vma mutants were expected, and others were surprising. However, among the phenotypes that were expected to arise from loss of V-ATPase activity was loss of vacuolar protease activity, because the acidic pH of the vacuole was believed to be critical for proteinase A activation and initiation of the cascade of vacuolar protease activations. Surprisingly, vma mutants do have active vacuolar proteases (160), although the overall levels of these proteases and their rates of maturation are lower than in wild-type cells (94, 137). This slow activation may come both from reduced activity of the activating proteases themselves and from slower transport to the vacuole (73, 94, 161). The deficiency in protease activity at the vacuole in vma mutants is serious enough to compromise digestion of autophagic bodies in cells undergoing autophagy or to slow down the vacuolar proteolysis induced by starvation (94).
Vacuolar protein sorting was also expected to be severely compromised in the vma mutants, but vacuolar proteases were missorted and secreted at a much lower level than in many of the vacuolar protein sorting (vps) mutants (73, 160). Endocytosis also appears to be slowed but not prevented in the vma mutants. Uptake of lucifer yellow was reported to be inhibited and uptake of FM4-64 slowed in the vma mutants (115, 150). However, the vma mutations are synthetically lethal with a number of endocytosis (end) mutations, suggesting that endocytosis not only occurs in the vma mutants but is critical for their survival (92). Taken together, these results suggest that although organelle acidification is certainly a factor in proteolytic activation and protein sorting in yeast, it is not completely essential.
The V0 sector of the V-ATPase itself has been proposed to play a critical role in the terminal steps of vacuole-vacuole fusion in yeast (6, 116), and more recently, in the late steps of synaptic vesicle exocytosis in Drosophila melanogaster (44). vma mutants do exhibit some defects in vacuolar morphology that might be consistent with problems in vacuolar fusion (6, 130), but these phenotypes are generally not as severe as those of SNARE or rab deletion mutations, which often show severe vacuolar fragmentation and in some cases are lethal (130). The more modest vacuolar morphology phenotype of the vma mutants may argue against a general role for V0 in membrane fusion, or could result from compensation by other proteins capable of catalyzing fusion in the absence of V0 sectors. Further experiments are necessary to distinguish these possibilities.
The yeast V-ATPase does play a central role in pH and calcium homeostasis, however. The full physiological basis for the pH- and calcium-dependent conditional lethality of the vma mutants is still not fully understood. Because vma and end mutations exhibit synthetic lethality, it has been proposed that vma mutants grow at low extracellular pH because they are able to acidify the vacuole by uptake of acidic extracellular fluid under these conditions (92). However, such a compensatory mechanism would require a very high rate of endocytosis compared to the rate of passive proton leakage, and thus seems somewhat unlikely. It was later proposed that the pH of the vacuole was lowered by passive uptake (not endocytosis) of weak acids from the extracellular fluid (118). More recently, it has been pointed out that copper and iron become limiting nutrients at high extracellular pH, suggesting that the defects of the vma mutants in metal uptake and/or distribution might underlie their intrinsic pH sensitivity (132). Resolving the role of the V-ATPase in pH homeostasis may require careful comparison of the vacuolar and cytosolic pHs of vma mutants and wild-type cells at various extracellular pHs, and these experiments have not been done.
The calcium sensitivity of the vma mutants is somewhat better understood. The Ca2+/H+ antiporter in the vacuole, encoded by VCX1, appears to act as the high-capacity regulator of cytosolic Ca2+ concentrations in yeast (14, 88). Loss of V-ATPase activity leads to loss of Vcx1p activity (33, 88). Under these conditions, cells appear to maintain calcium homeostasis only by constitutive activation of calcineurin, which in turn leads to upregulation of Pmr1p and Pmc1p, two calcium pumps (36, 145). (It should be noted that the effects of the V-ATPase extend even beyond driving Vcx1p because the vcx1Δ mutant has a much less pronounced phenotype than the vma mutants [33].) Consistent with a critical role for the V-ATPase in calcium homeostasis, V-ATPase and calcineurin mutations are synthetically lethal, and vma mutants rapidly lose calcium homeostasis and viability in the presence of the calcineurin inhibitor cyclosporine A (33, 36, 145).
The phenotypes of the vma mutants also highlight the importance of the vacuole as a storage organelle involved in many aspects of cellular homeostasis (72). Although the vma mutants can mate, they are quite defective in sporulation, particularly germination. This may arise from their inability to store nutrients in the vacuole for use during germination and is a feature shared by other fungi lacking V-ATPase activity (10, 34). Yeast vma mutants are also defective in many aspects of metal ion homeostasis (26). Some of these defects may come directly from the inability to store and/or detoxify metals in the vacuole (25), but others are more complex. For example, an acidic environment in the late Golgi apparatus or endosome is essential for maturation of the high-affinity plasma membrane iron transporter Fet3p (76). Inability to mature Fet3p may be the source of sensitivity to multiple heavy metals, because it can result in upregulation of less-specific transporters (76).
There has not yet been a thorough dissection of which metal ions have a direct dependence on the V-ATPase for their transport (for example, through the activity of antiporters dependent on a pH gradient in the vacuole) and which have a more indirect dependence. However, a recent genomewide analysis of homeostasis of multiple metals and other trace elements made it clear that the V-ATPase is a central player in metal ion homeostasis (26).
There are also connections between cell morphology and V-ATPase in fungi that are not understood. This was shown dramatically in Neurospora crassa vma-1 mutants, which lack the V-ATPase catalytic subunit (10). The mutants do not extend hyphae, but instead form short, very highly branched hyphae and are unable to form conidiae. These properties are mimicked but are less severe in N. crassa grown in the presence of the specific V-ATPase inhibitor concanamycin A (9). Although the morphology of yeast vma mutants does not appear to be altered dramatically, a temperature-conditional vma mutation, vma4-1(Ts), exhibited dramatically altered morphologies (with elongated or multiple buds) after a shift to the nonpermissive temperature (162).
The more pronounced morphological defects in the vma4-1(Ts) mutant may arise from a lag in activating compensatory mechanisms, such as calcineurin activation, that blunt or prevent these defects in the vma deletion mutants, but a specific effect of the vma4-1(Ts) mutation on cell morphology cannot be eliminated. Actin organization was found to be aberrant in both the vma4-1(Ts) mutant and a vma4Δ mutant (as well as other vma mutants), and this could underlie or at least contribute to the morphological defects (162).
The defects in pH and/or calcium homeostasis could potentially account for both cytoskeletal defects and the morphological changes in fungi. However, it is intriguing that direct interactions between V-ATPase subunits and the actin cytoskeleton have been observed in other systems, opening the possibility that disruption of the V-ATPase could directly affect cytoskeletal architecture (53, 153, 154). In one case, a V-ATPase subunit (subunit C) was even shown to cross-link actin filaments in vitro (153). The X-ray crystal structure of yeast subunit C shows a striking structural similarity to profilin (20) at one end of the subunit, further supporting the possibility of a direct interaction between V-ATPases and the actin cytoskeleton.
ASSEMBLY AND TRANSPORT OF V-ATPases
Assembly of Partial Complexes in Mutant Strains
V-ATPases are multisubunit complexes that function in multiple organelles, so their biosynthesis, assembly, and transport pose a particular challenge to the cell. Proteins traveling from the endoplasmic reticulum to the vacuole through the biosynthetic pathway or from the cell surface to the vacuole through the endocytic pathway encounter compartments of progressively lower pH (86). The final pH of each compartment results from the combination of proton pumps, other transporters, channels, and buffers localized there, and the mechanisms available for regulating each of these (111). Among the transporters and channels implicated in determining the final pH of specific compartments in yeast are the ClC (mammalian chloride channel)-type chloride channel Gef1p in the Golgi apparatus and the sodium/proton antiporter Nhx1p in the late endosome (95, 129), and there are almost certainly more. Because the V-ATPase is the central player in organelle acidification in fungi, its distribution and regulation are major factors governing the final pH in different organelles. However, it is still unclear to what extent the final pH is these organelles is dictated directly by the distribution of the V-ATPase itself as opposed to regulation of V-ATPase activity. This section will discuss current knowledge about how the yeast V-ATPase is assembled and targeted, and the next section will focus on its regulation.
Experiments with mutants lacking individual V-ATPase subunits first indicated that there was not extensive coordinate regulation of subunit synthesis, because loss of one subunit did not generally lead to loss of others (63). Subsequent work has indicated that loss of any V1 subunit does not affect the levels of other V1 or V0 subunits (139). The exception to this is destabilization of the E subunit when the G subunit is deleted (147). Loss of any V0 subunit does not generally affect the levels of the V1 subunits, but steady-state levels of the remaining V0 subunits, particularly the a subunit, are often reduced (63).
Because at least some of the remaining subunits were still present in the deletion mutants, it was possible to determine the partial complexes that were present. Such experiments clearly indicated that the V1 and V0 sectors could be assembled independently; assembled V0 sectors could be isolated from mutants lacking any V1 subunit, and assembled V1 sectors could be isolated from mutants lacking any V0 subunit (19, 148). In addition, a “core V1 complex” as well as V0 complexes could be isolated in the absence of V1 subunit C (19, 51), and inactive V1V0 complexes could be isolated in the absence of V1 subunit H (52). Tomashek et al. went on to show that a number of other smaller complexes could be formed in the absence of one subunit and that these complexes could be incorporated into larger, membrane-bound complexes when lysates from different deletion mutants were combined (146, 148). These data have provided important information about subunit-subunit interactions, but do not necessarily define steps in biosynthesis of the V-ATPase complex because kinetically unfavorable pathways may become available in the absence of one of the V-ATPase subunits that are not used in the presence of a full complement of subunits.
Early Steps in Biosynthesis and Assembly of V-ATPases
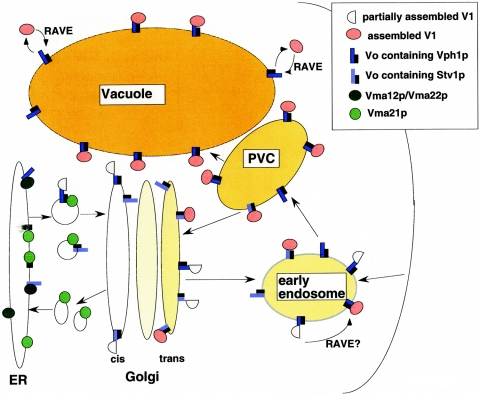
Figure 2 represents some of the steps in assembly and transport of the V-ATPase. It is clear that there are a number of requirements for assembly of V0 and these represent some of the earliest steps in V-ATPase assembly. Two genetic screens for vma mutants identified three gene products, Vma12p, Vma21p, and Vma22p, that were essential for any V-ATPase activity at the vacuole but were not part of the final, assembled V-ATPase complex (51, 107). When any one of these proteins is missing, Vph1p is destabilized, as it is in the absence of any of the other V0 subunits (46, 47, 49). This instability arises from endoplasmic reticulum-associated degradation of improperly assembled Vph1p (45). Vma12p and Vma21p proved to be integral membrane proteins of the endoplasmic reticulum, and Vma22p is a peripheral membrane protein that forms a complex with Vma12p (41, 46, 47).
FIG. 2.
Assembly and trafficking of the yeast V-ATPase. Possible steps in assembly and transport of Vph1p-containing and Stv1p-containing V-ATPases are shown here and described in more detail in the text. Shading of different organelles indicates the extent of acidification in that compartment; the vacuole is most intensely colored as the most acidic compartment in the yeast cell, and the first compartment in the secretory pathway showing any evidence of acidification in yeast is the Golgi apparatus. Vph1p-containing V-ATPases are known to travel to the vacuole via the prevacuolar compartment (PVC) (117) and are believed to reach the prevacuolar compartment via the early endosome, which is also likely to be somewhat acidic (134). Stv1p-containing V-ATPases appear to cycle between the prevacuolar compartment and the vacuole (68), and may travel through the early endosome as well. The RAVE complex is known to assist in reassembly of V1 and V0 complexes at the vacuole (131), and may also assist in assembly at the early endosome (134).
The functions of these three proteins, now believed to be dedicated assembly factors for the V-ATPase, have gradually become clearer. The Vma12p/Vma22p complex binds transiently and specifically to the newly synthesized Vph1p in the endoplasmic reticulum and promotes its stability (41). In a vma22Δ mutant, wild-type Vph1p assumes a wild-type membrane topology (45), and in a vma12Δ mutant, Vph1p has even been shown to associate with the other V0 subunits (80). However, in both of these mutants, Vph1p is rapidly degraded without transport to the Golgi apparatus. This suggests that the Vma12p/Vma22p somehow primes Vph1p for proper assembly with the other V0 subunits. The nature of this priming is unclear, but it protects Vph1p from degradation. Vma21p was more recently shown to bind directly to the c′ subunit, one of the three proteolipid subunits (80).
Coimmunoprecipitation experiments from mutants lacking individual V0 subunits suggested that Vma21p may bind to the ring of proteolipid subunits via its interaction with the c′ subunit, form a complex capable of recruiting subunit d (Vma6p), and then perhaps bind to Vph1p that has been prepared for assembly near the time of recruitment into vesicles. Using an in vitro assay for COPII vesicle formation, Malkus et al. demonstrated that Vma21p accompanies assembled V0 complexes into COPII vesicles and may accompany the V0 complex to the Golgi apparatus before dissociating and interacting with the endoplasmic reticulum retrieval machinery via its C-terminal dibasic retrieval sequence (80). Taken together, these results indicate that the initial steps of V0 assembly are very carefully orchestrated and coordinated with the initial stages of transport through the vacuolar network.
The stage at which V1 subunits associate with V0 subunits may be a critical factor in determining where in the secretory pathway acidification begins to occur. Malkus et al. (80) have shown that dissociation of Vma21p from assembled V0 sectors and association of the A subunit of the V1 sector with the V0 sector occur with similar kinetics. Based on this, Malkus and coworkers have argued that V1 associates with V0 after it is fully assembled. However, their data do not support assembly of a fully assembled V1 with V0, because the kinetics of association between V1 subunits A and B were shown to be slower than the kinetics of association between V1 subunit A and Vph1p (80).
Pulse-chase experiments examining the set of subunits coprecipitating with V1 subunits A and B (65) had also suggested that association of V1 and V0 subunits might occur before V1 assembly was completed. Based on these experiments, it was suggested that V-ATPase assembly in the presence of a full complement of subunits occurred via a concerted assembly pathway, with early associations between V1 and V0 subunits preceding full assembly of either complex, rather than by association of preassembled V1 and V0 sectors.
The primary disagreement between the two sets of experiments concerns whether full V0 assembly must precede any association with V1 subunits. Consistent with preassembly of V0, association of Vph1p with V1 subunits is decreased in a sec18 mutant, which blocks fusion of endoplasmic reticulum-derived vesicles with the Golgi apparatus and results in accumulation of V0 complexes associated with Vma21p (65, 80). With a sec18 block, however, coprecipitation of the proteolipid subunits with V1 subunits is completely prevented, suggesting that association of V1 with Vph1p can precede association with the proteolipid subunits (65). In addition, Vma21p clearly does not prevent V1 subunits from associating with V0, since a mutant form of Vma21p that lacks its endoplasmic reticulum retrieval sequence is transported to the vacuole in association with intact and active V-ATPase complexes (80).
All a subunits in eukaryotes consist of an N-terminal cytoplasmic domain of approximately 40 kDa that associates with a number of V1 subunits and other cellular proteins, and a C-terminal domain that contains multiple transmembrane segments (74, 103). The expressed soluble cytosolic domain of Vph1p is able to associate with V1 subunits in vivo, indicating that none of the endoplasmic reticulum-associated steps is absolutely required to set up this interaction (74). It may be that preassembly and transport of V0 to the Golgi apparatus followed by stepwise assembly of V1 after its initial attachment to V0 is the preferred pathway in wild-type cells, as shown in Fig. 2. However, as in the subunit deletion mutants that independently assemble V1 and V0 sectors, other pathways may become available when some aspect of this pathway is blocked.
A number of questions still surround the early steps of V-ATPase assembly. (i) Most early assembly studies have focused on defining when V1 subunits A and B become attached to the V0 subunits, because this attachment is necessary for catalytic activity of the enzyme (65, 80). It should be noted, however, that attachment of the A and B subunits to V0 is not sufficient for V-ATPase activity. In mutants lacking subunit H, for example, the rest of the V-ATPase is fully assembled but inactive (52). It is therefore important to understand when the stalk subunits, particularly subunit H, assemble with the V1V0 complexes in order to understand when and where the V-ATPase first becomes active. The incorporation of subunit H, or most of the other stalk subunits, has not been extensively studied.
(ii) The RAVE complex (described below) plays a role in both regulatory assembly and disassembly of the V-ATPase and, somewhat surprisingly, in biosynthetic assembly of the V-ATPase (131, 135). In the absence of the RAVE complex, V0 complexes are assembled and targeted to the vacuole, and there is some attachment of V1 to V0 (121), but the assembled complexes are unstable and there is very little ATPase activity in isolated vacuoles (135). Both the nature and the site of intervention by the RAVE complex in V-ATPase assembly are unknown.
(iii) Almost all of the V0 sector assembly experiments were performed prior to identification of the sixth V0 subunit, subunit e, encoded by the VMA9 gene (126). Sequence-based predictions suggest that subunit e has two transmembrane domains, and a C-terminal tag in the subunit appears to be vulnerable to vacuolar proteases. This suggests that the N and C termini of the protein are oriented toward the vacuolar lumen. Both the position of this subunit in the V-ATPase complex and the timing of its incorporation into the V-ATPase are completely unknown, but as an integral membrane V0 subunit, it almost certainly is initially inserted into the endoplasmic reticulum.
(iv) As described below, there are actually two isoforms of the a subunit, Vph1p and Stv1p (83, 84). Virtually all investigations of V0 and V-ATPase assembly have focused on Vph1p, which is present at a higher concentration than Stv1p (68, 84). Although it is assumed that the early assembly steps of the two a subunit isoforms are similar, this has yet to be proven. The best argument for a common early assembly pathway for the two a subunit isoforms is the strong phenotype of vma12Δ, vma21Δ, and vma22Δ mutants (51, 107), which would seem to indicate that all V-ATPase complexes require these assembly factors for their biosynthesis.
Role of the V0 a Subunit Isoforms in Targeting the V-ATPase
Although a number of V-ATPase subunits are present as multiple isoforms in plants and animals, there are isoforms of only the V0 a subunit in S. cerevisiae. The different phenotypes of the individual isoform deletion mutants, vph1Δ and stv1Δ, indicate that they fulfill different cellular functions (84). Consistent with this, the two isoforms are localized differently; Vph1p is localized to the vacuole and Stv1p cycles between the Golgi apparatus and prevacuolar endosome (68). Overexpression of STV1 can suppress the phenotypes of a vph1Δ strain, and under these conditions, Stv1p is present in the vacuole (84).
Comparison of the biochemical properties of vacuolar complexes containing Vph1p or Stv1p indicated that the a subunit isoforms impart significantly different properties on the enzyme complexes (70). Stv1p-containing complexes show less assembly with V1 subunits even when Stv1p and Vph1p are present in comparable amounts, and the assembled Stv1p-containing complexes couple ATP hydrolysis to proton transport much less efficiently (70). The cytoplasmic N-terminal domain of the two isoforms seems to dictate their localization. The N-terminal cytoplasmic domain of Stv1p is capable of directing a chimeric protein with a Vph1p C terminus to a steady-state Golgi apparatus localization, while a chimera containing the Vph1p N terminus and the Stv1p C-terminal domains is localized in the vacuole (68). It is likely that the Stv1p N-terminal domain contains a Golgi apparatus retrieval signal, but the exact nature of the targeting signal is still unknown (68). Vph1p, on the other hand, transits to the vacuole through the Golgi apparatus and prevacuolar compartment, i.e., via the carboxypeptidase Y vacuolar sorting pathway, and may not require a specific targeting signal (117).
Even though the two a subunit isoforms have different steady-state distributions, it is important to recognize that they do occupy the same compartments during their biosynthesis, starting presumably with the endoplasmic reticulum, then moving to the Golgi apparatus and prevacuolar endosome. This raises questions about how V1 is allocated to complexes containing the different a subunit isoforms, and how much each type of complex contributes to acidification of organelles in which both reside. The simplest model would have V1 subunits binding to both types of V0 sectors as they become available, and the contribution of Vph1p- and Stv1p-containing complexes to compartment acidification would be determined by their time of residence in different compartments. Thus, Vph1p-containing complexes might contribute to acidification of the late Golgi apparatus during transit, but their contribution might be less than that of Stv1p-containing complexes because they spend less time in the Golgi apparatus. Similarly, both Vph1p- and Stv1p-containing complexes might contribute to endosomal acidification during transit, but the relative rates at which Stv1p-containing complexes are retrieved to the Golgi apparatus and Vph1p-containing complexes are sent on to the vacuole, along with the relative ratios of the two complexes, would determine their contributions to endosomal acidification (115).
Under conditions where transport between compartments is blocked, the contributions of the two types of V-ATPase complex to the overall acidification might change. For example, in a vps27Δ mutant, in which both forward and reverse transport out of the late endosome/multivesicular body is inhibited, both Stv1p- and Vph1p-containing V-ATPase complexes accumulate in the enlarged endosome, which appears to become hyperacidified (68, 117, 121).
Compartment Identity and V-ATPase Assembly and Activity
The simplified model for assembly and function of V-ATPases containing different a subunits described above would suggest that acidification by Vph1p- and Stv1p-containing complexes depends only on the availability of a full complement of subunits and residence time in different compartments. However, there is evidence that V-ATPase activity can be altered by the environment in different cellular compartments. This suggests that compartments exert an effect on the V-ATPase that could, in turn, regulate the final level of acidification in those compartments. For example, although Stv1p-containing complexes in their normal location do not appear to dissociate in response to glucose deprivation, Stv1p-containing complexes at the vacuole do show some level of disassembly (70). Conversely, Vph1p-containing complexes, which normally show extensive disassembly when cells are deprived of glucose, lose some of this disassembly when they are trapped in earlier compartments by a vps21Δ or vps27Δ mutation (70).
There are many levels at which a compartment could exert a direct effect on V-ATPase activity or biochemical properties. If, as described below, the V-ATPase senses compartment pH (133), then some of the apparent differences between V-ATPase properties in different compartments could come from pH regulation of the V-ATPase, not a direct interaction with other components of the organelle. Work with other systems has emphasized the importance of other channels and transporters in determining the final pH of different compartments (17, 71, 144), and in this context, these proteins could be seen as indirectly regulating V-ATPase activity by contributing to the lumenal pH of the organelle.
It is also worth noting, however, that there is substantial evidence that V-ATPase activity is sensitive to the lipid environment, and this is certainly a potential mechanism by which V-ATPase activity could be regulated in different compartments. Deletion of either of two proteins involved in sphingolipid biosynthesis, Sur4p and Fen1p, results in a milder version of the Vma− growth phenotype and reduced quinacrine accumulation at the vacuole (12). Surprisingly, these mutations appear to affect the structure of the peripheral V1 sector rather than the integral membrane V0 sector, because membranes containing V0 sectors from the mutants can form functional complexes when provided with wild-type V1, but V1 sectors from the mutants are unable to reconstitute activity with vacuolar membranes containing wild-type V0 (12). These results seem to imply that either the maturation of the V-ATPase or its function relies on sphingolipids, but the biochemical connection is still not understood.
A number of experiments also connect inositol phospholipids and organelle acidification, but it is still not clear whether the effects on acidification arise from direct effects on the V-ATPase. A subset of the vacuolar protein sorting (vps) mutants are defective in quinacrine uptake into the vacuole, indicating a vacuolar acidification defect (121, 124). Among these are the vps34 and vps15 mutants, which are defective in phosphatidylinositol 3-kinase activity (128, 138). vps34Δ and vps15Δ mutants were also identified in a genomic screen for nonessential deletion mutants that exhibit sensitivity to high pH and extracellular calcium concentrations and poor growth on nonfermentable carbon sources characteristic of the vma mutants (125). Vacuoles isolated from the vps34Δ and vps15Δ have reduced V-ATPase activity, but also have reduced levels of several V-ATPase subunits (125). This may suggest that the vacuolar acidification defect arises from improper localization of the V-ATPase, but it is notable that other vps mutants with defects in V-ATPase targeting, such as the class E vps mutants, do not exhibit a Vma− growth phenotype.
In addition, there is very recent evidence that Vps34p and subunit F of the V-ATPase may directly interact in Candida albicans (24). Mutations in the phosphatidylinositol 3,5-kinase Fab1p and its regulators, Vac7p and Vac14p, also result in a loss of vacuolar acidification (7, 37). These mutants have not been reported to exhibit a Vma− phenotype, but the primary site of action of Fab1p is the vacuole (7, 37), and as described above, loss of V-ATPase function in the vacuole alone, for example, by inactivation of Vph1p but not Stv1p, does not generate the full Vma− phenotype (83, 84). In fab1 mutants, the V-ATPase appears to be properly localized to the vacuole (7), and experiments with a fab1 mutant that has very low activity have established that only low levels of phosphatidylinositol 3.5-kinase appear to be necessary for vacuolar acidification (37).
It is not clear whether the acidification defect arises from defective proton transport by the V-ATPase itself, or effects on other components of the vacuole that might change the ability to establish and maintain a proton gradient. There is much to be done to clarify the links between inositol phospholipids, the V-ATPase, and organelle acidification, but given the emerging importance of inositol phospholipids as compartment identifiers (106, 123), the possibility that they regulate ATPase activity at some level is extremely interesting.
In summary, many questions remain about how V-ATPases are localized and transported, and how these processes are linked to their overall activity at different cellular locations. Because the internal pH of an organelle is a fundamental determinant of the organelle's identity and function, it seems likely that there are factors that control V-ATPase transport and/or the activity of the V-ATPase during its transport. However, these factors have yet to be clearly defined.
REGULATION OF V-ATPase ACTIVITY
The final pH of different intracellular compartments depends not only on the localization of V-ATPases, but also on regulation of their activity. In addition, the requirement for V-ATPase of many basic homeostatic processes suggests that its activity may be adjusted under different extracellular conditions. Therefore, it is not surprising that the regulation of V-ATPases is rich and complex.
Reversible Disassembly of the Yeast V-ATPase
Reversible disassembly of V-ATPases was first observed independently in Manduca sexta and yeast (reviewed in references 62 and 156). In both cases, a drop in extracellular nutrients, initiated by a brief glucose deprivation in yeast and by a cessation of feeding during a molt or starvation in M. sexta, resulted in disassembly of fully assembled and active V-ATPase complexes into cytosolic V1 complexes and membrane bound V0 complexes (61, 141). Remarkably, this disassembly was reversible; restoration of extracellular glucose resulted in reassembly of V1 with V0 at the membrane (39, 61). These results suggested that even after its biosynthetic assembly, the level of V-ATPase activity might be adjusted by adjusting the level of assembly. In addition, the presence of a similar process in two systems as different as those of S. cerevisiae and M. sexta suggested that reversible disassembly might be a general property of V-ATPases. More recent evidence of controlled assembly of V-ATPase complexes in renal epithelial cells and maturing dendritic cells (127, 149) has suggested that V-ATPase activity is regulated at the level of assembly in mammalian cells as well.
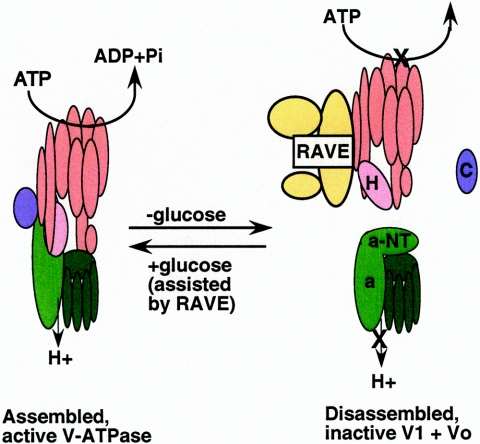
A model for reversible disassembly of the yeast V-ATPase is shown in Fig. 3. Biochemical characterization indicated that both disassembly and reassembly were very rapid and entirely posttranslational (61). In medium containing 2% glucose, the preferred carbon source, 55 to 70% of V0 sectors were assembled with V1, but after only 5 min of glucose deprivation, only 15 to 20% of V0 sectors had bound V1, and 5 min after glucose restoration, the level of assembly was again 55 to 70%. Both disassembly and reassembly occurred efficiently in the presence of 100 μg/ml cycloheximide. Immunofluorescence microscopy using a monoclonal antibody against a cryptic epitope in Vph1p, exposed only when V1 is not bound, revealed that the epitope was exposed at the vacuole with glucose deprivation and lost again with glucose readdition; these results make it unlikely that there is any trafficking of the complexes out of the vacuole required for reassembly (61).
FIG. 3.
Overview of reversible disassembly in yeast cells. Intact, active V-ATPase complexes are rapidly disassembled into free V1 and V0 complexes in response to glucose deprivation (61). Glucose readdition results in reassembly of the V-ATPase, and is slow and incomplete in the absence of the RAVE complex (131). RAVE binding to V1 is compromised in mutants lacking subunit E or G, suggesting that these two subunits are involved in the interaction of V1 and RAVE (135). Subunits that are believed to undergo major conformational changes during disassembly are identified: the H subunit shifts from being an activator of V1V0 complexes to being an inhibitor of free V1 complexes (113), the cytoplasmic N-terminal domain of the a subunit appears to fold down onto the rest of V0 (157), and the C subunit is released from both the V1 and the V0 sectors during disassembly (61).
Reversible disassembly proved to be more of a means of “tuning” V-ATPase assembly to an appropriate level than an all-or-none process. After overnight growth in a less favorable carbon source, such as raffinose, intermediate levels of V-ATPase assembly were observed (approximately 45% of V0 complexes bound to V1 in raffinose, for example). Remarkably, the free V1 sectors remained both stable in the cytosol and competent for rapid reassembly with V0 upon addition of glucose to the cells (61). Taken together, these results suggest that the V-ATPase assembly state is actively adjusted in response to the extracellular carbon source. Unlike F1, V1 loses all Mg2+-dependent ATP hydrolysis when it is removed from the membrane, and V0 sectors do not appear to be open proton pores (113, 157). Thus, disassembly of V-ATPases under less favorable conditions might conserve ATP when energy sources are limited, while rapid reassembly could help to prevent cytosolic acidification when rapid metabolism resumes.
The rapid and reversible disassembly of V-ATPase complexes at the vacuole suggests that there must be some means of signaling extracellular glucose availability to V-ATPases at the vacuolar membrane, but the nature of this signal is still unclear. Many glucose-dependent processes in yeast “sense” the level of glucose 6-phosphate, but further metabolism of glucose is required to sustain assembly of the V-ATPase (112). In addition, mutations in a number of signaling pathways implicated in glucose signaling, including the Snf1, cyclic AMP-dependent protein kinase, and protein kinase C pathways, affected neither disassembly nor reassembly of the V-ATPase (112). These results suggest that the signal for V-ATPase assembly is somewhat different from those previously characterized pathways, but it is certainly possible that some variation on these pathways could be involved.
One intriguing possibility is that some metabolite of glycolysis itself might bind to the V-ATPase and affect its assembly state. Although ATP is an attractive possibility, the levels of ATP during glucose deprivation and readdition do not appear to correlate to V-ATPase assembly state, and various levels of ATP and/or ADP in vitro do not trigger diassembly of V-ATPase complexes (112). However, Lu et al. have found that the glycolytic enzyme aldolase binds directly to the B and E subunits of the yeast V1 sectors and the a subunit of the V0 sector (77, 78). Intriguingly, mutants lacking aldolase grow better at pH 5 than at pH 7.5, possibly suggesting a connection to V-ATPase activity, and the level of free V1 sectors is increased in the mutants (77, 78).
None of these experiments has yet dissociated the effects of aldolase loss on glycolytic flux from direct effects of aldolase loss on V-ATPase structure, however. This is an important point, because loss of glucose metabolism in the aldolase mutant might mimic a glucose deprivation and thus induce V-ATPase disassembly indirectly. However, other glycolytic enzymes, in addition to aldolase, have been detected in association with V-ATPases in other systems (140). Whether the association of these enzymes with the V-ATPase proves to control V-ATPase assembly state or provide a localized source of ATP (as suggested for the kidney cell surface V-ATPase shown to associate with phosphofructokinase) (140), direct connections between glycolysis and V-ATPase activity are likely to be important in regulating V-ATPase activity.
Early experiments suggested that the V-ATPase itself had to be active for disassembly to occur, because both inactivating mutations in the enzyme and the specific inhibitor concanamycin A prevented release of V1 upon glucose deprivation (112). Because both concanamycin and the inactivating mutations also prevent vacuolar acidification, these experiments did not determine whether it was some aspect of catalysis that made the ATPase structure competent for disassembly or whether vacuolar acidification was important. Shao and Forgac recently showed that inhibiting vacuolar acidification using chloroquine could inhibit disassembly of the V-ATPase, even though the ATPase was fully active (133). This result suggests that the V-ATPase might itself contain a “vacuolar pH sensor” that can prevent inactivation of the enzyme by dissociation under conditions where the pH gradient across the vacuolar membrane is lost. Even under normal conditions, V-ATPase disassembly is never complete, and this result would suggest that a population of active enzymes is retained at the vacuole by stopping disassembly before the pH gradient is completely lost.
Many questions still remain about reversible disassembly of V-ATPases, ranging from the physiological implications of this process to its structural underpinnings. (i) Although it is hypothesized that the physiological purpose of reversible disassembly is conservation of cytoplasmic ATP, this has not been demonstrated directly, because there is still no mutant that has full V-ATPase activity but fails to disassemble. Such a mutant could reveal whether there is any physiological disadvantage to sustained V-ATPase activity in the presence of poor carbon sources, and possibly whether there is any additional role for disassembled V1 and V0 sectors.
(ii) Although there are intriguing clues to the molecular requirements for disassembly and reassembly of the V-ATPase, such as the aldolase association described above, the signals connecting extracellular carbon source availability and V-ATPase assembly state are still not clear, and this is an area of major interest.
(iii) The C subunit is lost from both the V1 and V0 sectors during disassembly, but it is still not clear whether the V1 and C subunits float independently into the cytosol or are retained in closer proximity to each other and the vacuolar membrane. In addition, the RAVE complex, discussed below, binds V1 after its release from the membrane and improves the efficiency of reassembly, but its mechanism is not understood.
(iv) The possibility of ongoing cycles of disassembly and reassembly places some important constraints on the structure of the V-ATPase itself, particularly on the stalk structures. While the stator and rotor stalks of the F1F0-ATPases appear to be dedicated to maintaining the structural connections between subunits that are essential for sustaining rotational catalysis, the V-ATPase stalks must balance the stability needed for rotational catalysis with the regulated instability implied by reversible disassembly. The V-ATPase stalk subunits show little resemblance to the stalk subunits of F-ATPases, and the need to balance stability and instability may account, at least in part, for this divergence.
The RAVE Complex
The RAVE complex was first discovered in a proteomic approach to identifying new binding partners of the highly conserved protein Skp1p, which is best known as a component of the SCF (Skp1-cullin-F-box) class of E3 ubiquitin ligases (131). Among the proteins bound to a Skp1p affinity column from a yeast lysate were two uncharacterized proteins encoded by yeast genes YJR033c and YDR202c, which were later named RAV1 and RAV2. Further characterization revealed that Skp1p, Rav1p, and Rav2p formed a complex that did not contain other components of SCF ubiquitin ligases and was not sensitive to mutations in these other components for its formation (131). This result suggested that this complex, named RAVE (regulator of H+-ATPase of vacuolar and endosomal membranes) after subsequent functional characterization, was not an SCF ubiquitin ligase even though it contained Skp1p.
Affinity chromatography under less-stringent conditions revealed that a number of V1 subunits copurified with the RAVE complex, and the functional importance of this interaction was supported by a partial (temperature-dependent) Vma− phenotype for the rav1Δ and rav2Δ mutants (131). Both rav1Δ and rav2Δ mutants proved to be unable to grow at elevated pH and calcium concentrations at 37°C, although they were able to grow under these conditions at 30°C, and consistent with the growth phenotype, there was a substantial decrease in quinacrine staining of the vacuole at low temperatures and a loss of staining at higher temperatures (131). These results indicated that RAVE was somehow regulating V-ATPase activity. Consistent with this, reassembly of V-ATPase complexes after glucose readdition to glucose-deprived cells was much slower in a rav1Δ mutant (121).
How does the RAVE complex affect reassembly of the V-ATPase? RAVE can bind to cytosolic V1, and among other possibilities, this association might transmit the elusive glucose signal, discussed above, to the V-ATPase or provide a chaperone-like protection of V1 that keeps it competent for reassembly. RAVE was shown to bind to V1 released from the vacuolar membrane by glucose deprivation, and to release V1 upon glucose readdition (135). The binding of RAVE to V1 is not intrinsically glucose sensitive, however, because RAVE also bound to the constitutively cytosolic V1 in a mutant with destabilized V0, and this binding was not altered by changing extracellular glucose. This result argues against RAVE as a glucose sensor, and might instead support a more general role in preserving V1 structure or stimulating its assembly.
Consistent with a more general role for RAVE in assembly, the V-ATPase in vacuoles isolated from rav1Δ and rav2Δ mutants proved to be very unstable, even in the presence of glucose (135). Although all of the subunits of the enzyme appear to be present at near-normal levels and there is enough activity to suppress the Vma− phenotype at lower temperatures, vacuoles isolated from the mutants have very low ATPase activity and low levels of V1 subunits. Immunoprecipitations from rav1Δ and rav2Δ mutants indicate that the V1 and V0 sectors are assembled, but there is little association of V1 with V0 (135).
As described above, the final level of assembly of the V-ATPase is established by both biosynthetic assembly of the enzyme and reversible disassembly of the assembled complexes. Because rav1Δ had been shown to affect reassembly of the V-ATPase after glucose readdition (131), it was hypothesized that the rav mutations were not affecting V-ATPase biosynthesis, but instead were perturbing the balance between disassembly and reassembly. An E145L mutation in the V-ATPase c′ subunit (vma11-E145L) abolishes all V-ATPase activity, results in high levels of assembled V-ATPase in the vacuole, and prevents disassembly in response to glucose deprivation (48, 112). If disassembly of the V-ATPase is necessary for the rav1Δ-dependent assembly defects to be observed, then the vma11-E145L mutation should be epistatic to the rav1Δ mutation and should restore V-ATPase assembly. Instead, it was found that there was still poor assembly of V1 with V0 in the vma11-E145L rav1Δ double mutant (135). Surprisingly, this result indicates that RAVE plays a role in both biosynthetic assembly and reassembly of the V-ATPase, two processes that were previously believed to be independent.
Further analysis of the RAVE-V1 interaction has provided some molecular clues to the potential functions of RAVE, although its activity is still not fully understood. The RAVE- V1 interaction is specifically disrupted by mutations in two subunits of the peripheral stalk, subunits E and G (135). This suggests the RAVE may be important for docking the peripheral stalk onto V0 sectors in the membrane. There are similarities between the assembly phenotypes of the ravΔ mutants and a vma5Δ mutant, which lacks the V1 C subunit: in both cases, a “core” V1 complex and V0 complexes are assembled, but V1 and V0 do not associate tightly (51, 135). The C subunit is associated with the E and G subunits in the peripheral stalk (55) and is lost from both V1 and V0 sectors during disassembly (61). The stage at which the C subunit becomes associated with V1 and/or V1V0 complexes during biosynthetic assembly is not known, but given the above data, it is tempting to speculate that RAVE plays a role in assembly of the C subunit both during reassembly and during biosynthetic assembly. Experiments are under way to address this possibility.
New clues to RAVE function were provided when the SOI3 gene, isolated via a mutation that suppresses the loss of the trans-Golgi network localization signal in Kex2p, was shown to be identical to RAV1 (1, 122, 134). These experiments demonstrated a substantial membrane-bound fraction of Rav1p by differential centrifugation, in addition to the cytosolic fraction that had been studied previously (134). Sucrose gradient fractionation of the membrane-bound Rav1p suggested that the membrane-bound portion of the RAVE complex might be localized to early endosomes, and a Rav1p-green fluorescent protein fusion localized in a pattern consistent with early endosomes (134). Furthermore, the Soi− phenotype and several other defects in the rav1 mutant are consistent with a role for the RAVE complex in the early endosome, and the authors hypothesized that RAVE may be critical to ensure acidification at this site (134).
One intriguing possibility is that the fraction of RAVE that is in the early endosome is responsible for the role of RAVE in V-ATPase biosynthesis and the fraction in the cytosol is involved in reassembly of the V-ATPase after it has been transported to the vacuole and disassembled. The biochemical function of RAVE could potentially be the same at both locations. Further experiments are necessary to sort out these possibilities.
To date, the RAVE complex has been studied only in yeast. It is not yet clear how general its role in V-ATPase assembly is. It is equally unclear how specific to V-ATPases its function is. Because the Vma− phenotype is so pronounced in yeast, defects in other complexes arising as a result of loss of RAVE function might be difficult to recognize. Most fungi have identifiable homologues of RAV1 and RAV2, but there has been no phenotypic characterization of mutants lacking these genes. Potential homologues of RAV1 are found in most eukaryotes (64), but these proteins are predicted to be much larger than yeast Rav1p, and they have not been connected biochemically to the V-ATPase. Interestingly, a potential mammalian homologue of yeast Rav1p, rabconnectin 3, was isolated as binding to both the Rab3 GTPase-activating protein and Rab3 GTPase exchange factor and was shown to be enriched in synaptic vesicles (93). These results suggested that rabconnectin was a scaffold protein, but provided no connection to V-ATPase function.
The involvement of Skp1p in the V-ATPase complex also remains rather mysterious. Skp1p appears both to be an essential component of the RAVE complex and to associate stably with Rav1p and Rav2p, even under conditions where RAVE cannot bind V1 (131, 135). In the SCF-type E3 ubiquitin ligases, Skp1p acts as an adaptor protein, bridging the cullin backbone of the ubiquitin ligase and a number of substrate recruitment proteins that are recognized by a sequence motif called the F-box (18). Although Skp1p is generally envisioned as a central component of SCF ubiquitin ligases, RAVE is not the first nonproteolytic complex to contain Skp1p. Skp1p is a structural component of the kinetochore complex CBF3 (66) and also interacts with Rcy1p to regulate localization of the plasma membrane v-SNARE Snc1p during endocytosis and recycling (35).
However, in both of these contexts, there are parallels with the functions of Skp1p in SCF complexes. In both, Skp1p binds to an F-box protein (Ctf13p in the CBF3 complex and Rcy1p in the Rcy1p-Skp1p complex) and may help to regulate the stability of one or more components involved in the respective functions of the complexes. However, no F-box protein has been identified in the RAVE complex or the V-ATPase, and there is no obvious link to protein stability. Skp1p may simply be an abundant protein that is “moonlighting” as part of a V-ATPase assembly complex, but it is still possible that Skp1p somehow provides a functionally important link between V-ATPase assembly and the many other cellular processes that involve Skp1p-containing complexes.
V-ATPase Regulation by pH and ATP—a Regulated “Slip” in Proton Transport?
Early biochemical characterization of V-ATPases from several sources suggested that the enzyme exhibited various degrees of coupling between ATP hydrolysis and proton transport, specifically, a loss of coupling efficiency at high ATP concentrations (3, 89, 151). This intrinsic uncoupling, or “slip,” between ATP hydrolysis and proton transport may represent an important mode of regulation for the V-ATPase. Arai et al. (3) demonstrated that the bovine clathrin-coated vesicle V-ATPase exhibited optimal rates of proton pumping at ATP concentrations of approximately 0.3 mM, with a decrease in pumping at higher ATP concentrations, even after purification and reconstitution of the enzyme (3, 90) These results suggested that variations in coupling efficiency with ATP concentration were a property of the V-ATPase itself. Muller et al. proposed that variations in coupling efficiency were in part responsible for differences in acidification in lemon fruit, where a steep pH gradient is established and maintained across the vacuolar membrane, and other parts of the plant, where the vacuolar pH is more typical of other eukaryotic cells (90).
More recently, Shao and Forgac provided further support for an intrinsic slip in ATP-dependent transport in the yeast V-ATPase by demonstrating that mutations in the nonhomologous domain of the catalytic subunit could change coupling between ATP hydrolysis and proton pumping (133). Significantly, not only were mutations with decreased coupling efficiency obtained, but at least one mutation (vma1-P217V) resulted in an increased coupling ratio, particularly at low ATP concentrations.
Decreased coupling efficiency might be accounted for by a variety of structural defects in the enzyme, but increased efficiency suggests that the wild-type enzyme is not optimally coupling ATP hydrolysis and proton transport. In a more global sense, these results suggest that at the millimolar concentrations of ATP commonly found in the cytosol of yeast with abundant carbon sources, a certain percentage of ATP hydrolysis by the V-ATPase is not productive in driving proton transport. This seems uncharacteristically wasteful, and might be seen as a flaw in V-ATPase architecture, perhaps arising from structural demands on the stator structure to balance coupling and the potential for disassembly. However, Nelson et al. have recently pointed out that intrinsic slips in transport systems may be beneficial to cells under certain circumstances, and in the case of V-ATPases may provide a means of controlling organelle acidification and providing adaptability, even at the expense of excess ATP hydrolysis (101).
This intrinsic uncoupling of the ATPase is also a potential site of intervention for other mechanisms for regulating V-ATPase activity in response to changing conditions or providing compartment-specific control. For example, Crider and Xie have recently shown that phospholipids can regulate the coupling efficiency of mammalian V-ATPases in vitro (13). Thus, this is another mechanism by which the phospholipid content of different membranes could help to account for the final internal pHs of different organelles.
Other Mechanisms of V-ATPase Regulation
Feng and Forgac first demonstrated that disulfide bond formation with a cysteine near the catalytic site in the A subunits inhibited the bovine clathrin-coated vesicle V-ATPase activity (28, 30). They went on to show that this inhibition is reversible by disulfide interchange among three conserved cysteines in the A subunit (29). In yeast, a cys4Δ mutation mimics the phenotype of the vma mutants, in part because it appears to reduce cellular glutathione levels (109). Mutation of the conserved catalytic site cysteine in the yeast A subunit partially suppresses the effects of the cys4Δ mutation (109). This suggests that there may be some control of V-ATPase activity in yeast by oxidation and reduction, but it is still unclear when this type of regulation might occur in wild-type cells.
There are likely to be other mechanisms of V-ATPase regulation as well. Work to date has made it clear that V-ATPases are entwined in many aspects of cell physiology and that regulation of V-ATPase activity is multilayered and complex. In coming years, detailed biochemical analysis of the V-ATPase itself and genomic analysis of its place in the cell are likely to lead to both greater understanding and new questions. Given the high degree of conservation of both the V-ATPase itself and the known mechanisms for its regulation, work on fungal V-ATPases will continue to provide important insights into the interplay between organelle acidification, pH homeostasis, and other aspects in cell physiology that are likely to be applicable to all eukaryotic cells.
Acknowledgments
The work from my laboratory described in this review was supported by NIH grants R01 GM50322 and R01 GM63742.
Many thanks to Anne Smardon and Maria Sambade for comments on the manuscript and to my colleagues in the V-ATPase field for years of collegiality and exciting science. I apologize for the omission of any work I was unable to cite here.
REFERENCES
- 1.Abazeed, M. E., J. M. Blanchette, and R. S. Fuller. 2004. Cell-free transport from the TGN to late endosome requires factors involved in formation and consumption of clathrin-coated vesicles. J. Biol. Chem. 280:4442-4450. [DOI] [PubMed] [Google Scholar]
- 2.Aksimentiev, A., I. A. Balabin, R. H. Fillingame, and K. Schulten. 2004. Insights into the molecular mechanism of rotation in the F0 sector of ATP synthase. Biophys. J. 86:1332-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai, H., S. Pink, and M. Forgac. 1989. Interaction of anions and ATP with the coated vesicle proton pump. Biochemistry 28:3075-3082. [DOI] [PubMed] [Google Scholar]
- 4.Arata, Y., J. D. Baleja, and M. Forgac. 2002. Localization of subunits D, E, and G in the yeast V-ATPase complex using cysteine-mediated cross-linking to subunit B. Biochemistry 41:11301-11307. [DOI] [PubMed] [Google Scholar]
- 5.Bauerle, C., M. N. Ho, M. A. Lindorfer, and T. H. Stevens. 1993. The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H+-ATPase membrane sector. J. Biol. Chem. 268:12749-12757. [PubMed] [Google Scholar]
- 6.Bayer, M. J., C. Reese, S. Buhler, C. Peters, and A. Mayer. 2003. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J. Cell Biol. 162:211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonangelino, C. J., N. L. Catlett, and L. S. Weisman. 1997. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 7:6847-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman, B. J., R. Allen, M. A. Wechser, and E. J. Bowman. 1988. Isolation of genes encoding the Neurospora vacuolar ATPase. Analysis of vma-2 encoding the 57-kDa polypeptide and comparison to vma-1. J. Biol. Chem. 263:14002-14007. [PubMed] [Google Scholar]
- 9.Bowman, E. J., and B. J. Bowman. 2000. Cellular role of the V-ATPase in Neurospora crassa: analysis of mutants resistant to concanamycin or lacking the catalytic subunit A. J. Exp. Biol. 203:97-106. [DOI] [PubMed] [Google Scholar]
- 10.Bowman, E. J., R. Kendle, and B. J. Bowman. 2000. Disruption of vma-1, the gene encoding the catalytic subunit of the vacuolar H+-ATPase, causes severe morphological changes in Neurospora crassa. J. Biol. Chem. 275:167-176. [DOI] [PubMed] [Google Scholar]
- 11.Bowman, E. J., K. Tenney, and B. J. Bowman. 1988. Isolation of genes encoding the Neurospora vacuolar ATPase. Analysis of vma-1 encoding the 67-kDa subunit reveals homology to other ATPases. J. Biol. Chem. 263:13994-14001. [PubMed] [Google Scholar]
- 12.Chung, J. H., R. L. Lester, and R. C. Dickson. 2003. Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J. Biol. Chem. 278:28872-28881. [DOI] [PubMed] [Google Scholar]
- 13.Crider, B. P., and X. S. Xie. 2003. Characterization of the functional coupling of bovine brain vacuolar-type H+-translocating ATPase. Effect of divalent cations, phospholipids, and subunit H (SFD). J. Biol. Chem. 278:44281-44288. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, K. W., and G. R. Fink. 1996. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2226-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, S. A., S. F. Goodwin, D. C. Kelly, Z. Wang, M. A. Sozen, K. Kaiser, and J. A. Dow. 1996. Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J. Biol. Chem. 271:30677-30684. [DOI] [PubMed] [Google Scholar]
- 16.Davis-Kaplan, S. R., D. M. Ward, S. L. Shiflett, and J. Kaplan. 2004. Genome-wide analysis of iron-dependent growth reveals a novel yeast gene required for vacuolar acidification. J. Biol. Chem. 279:4322-4329. [DOI] [PubMed] [Google Scholar]
- 17.Demaurex, N., W. Furuya, S. D'Souza, J. S. Bonifacino, and S. Grinstein. 1998. Mechanism of acidification of the trans-Golgi network (TGN). In situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J. Biol. Chem. 273:2044-2051. [DOI] [PubMed] [Google Scholar]
- 18.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 19.Doherty, R. D., and P. M. Kane. 1993. Partial assembly of the yeast vacuolar H+-ATPase in mutants lacking one subunit of the enzyme. J. Biol. Chem. 268:16845-16851. [PubMed] [Google Scholar]
- 20.Drory, O., F. Frolow, and N. Nelson. 2004. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 5:1148-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dschida, W. J., and B. J. Bowman. 1992. Structure of the vacuolar ATPase from Neurospora crassa as determined by electron microscopy. J. Biol. Chem. 267:18783-18789. [PubMed] [Google Scholar]
- 22.Dunn, S. D., and J. Chandler. 1998. Characterization of a b2delta complex from Escherichia coli ATP synthase. J. Biol. Chem. 273:8646-8651. [DOI] [PubMed] [Google Scholar]
- 23.Dunn, S. D., D. T. McLachlin, and M. Revington. 2000. The second stalk of Escherichia coli ATP synthase. Biochim. Biophys. Acta 1458:356-363. [DOI] [PubMed] [Google Scholar]
- 24.Eck, R., M. Nguyen, J. Gunther, W. Kunkel, and P. F. Zipfel. 2005. The phosphatidylinositol 3-kinase Vps34p of the human pathogenic yeast Candida albicans is a multifunctional protein that interacts with the putative vacuolar H+-ATPase subunit Vma7p. Int. J. Med. Microbiol. 295:57-66. [DOI] [PubMed] [Google Scholar]
- 25.Eide, D. J., J. T. Bridgham, Z. Zhao, and J. R. Mattoon. 1993. The vacuolar H+-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol. Gen. Genet. 241:447-456. [DOI] [PubMed] [Google Scholar]
- 26.Eide, D. J., S. Clark, T. M. Nair, M. Gehl, M. Gribskov, M. L. Guerinot, and J. F. Harper. 2005. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 6:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enyenihi, A. H., and W. S. Saunders. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng, Y., and M. Forgac. 1992. Cysteine 254 of the 73-kDa A subunit is responsible for inhibition of the coated vesicle (H+)-ATPase upon modification by sulfhydryl reagents. J. Biol. Chem. 267:5817-5822. [PubMed] [Google Scholar]
- 29.Feng, Y., and M. Forgac. 1994. Inhibition of vacuolar H+-ATPase by disulfide bond formation between cysteine 254 and cysteine 532 in subunit A. J. Biol. Chem. 269:13224-13230. [PubMed] [Google Scholar]
- 30.Feng, Y., and M. Forgac. 1992. A novel mechanism for regulation of vacuolar acidification. J. Biol. Chem. 267:19769-19772. [PubMed] [Google Scholar]
- 31.Fillingame, R. H., C. M. Angevine, and O. Y. Dmitriev. 2003. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 555:29-34. [DOI] [PubMed] [Google Scholar]
- 32.Flannery, A. R., L. A. Graham, and T. H. Stevens. 2004. Topological characterization of the c, c′, and c" subunits of the vacuolar ATPase from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 279:39856-39862. [DOI] [PubMed] [Google Scholar]
- 33.Forster, C., and P. M. Kane. 2000. Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 275:38245-38253. [DOI] [PubMed] [Google Scholar]
- 34.Forster, C., M. A. Santos, S. Ruffert, R. Kramer, and J. L. Revuelta. 1999. Physiological consequence of disruption of the VMA1 gene in the riboflavin overproducer Ashbya gossypii. J. Biol. Chem. 274:9442-9448. [DOI] [PubMed] [Google Scholar]
- 35.Galan, J. M., A. Wiederkehr, J. H. Seol, R. Haguenauer-Tsapis, R. J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 11:3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrett-Engele, P., B. Moilanen, and M. S. Cyert. 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+-ATPase. Mol. Cell. Biol. 5:4103-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gary, J. D., A. E. Wurmser, C. J. Bonangelino, L. S. Weisman, and S. D. Emr. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143:65-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gogarten, J. P., H. Kibak, P. Dittrich, L. Taiz, E. J. Bowman, B. J. Bowman, M. F. Manolson, R. J. Poole, T. Date, T. Oshima, et al. 1989. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc. Natl. Acad. Sci. USA 86:6661-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graf, R., W. R. Harvey, and H. Wieczorek. 1996. Purification and properties of a cytosolic V1-ATPase. J. Biol. Chem. 271:20908-20913. [DOI] [PubMed] [Google Scholar]
- 40.Graf, R., A. Lepier, W. R. Harvey, and H. Wieczorek. 1994. A novel 14-kDa V-ATPase subunit in the tobacco hornworm midgut. J. Biol. Chem. 269:3767-3774. [PubMed] [Google Scholar]
- 41.Graham, L. A., K. J. Hill, and T. H. Stevens. 1998. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 142:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber, G., M. Radermacher, T. Ruiz, J. Godovac-Zimmermann, B. Canas, D. Kleine-Kohlbrecher, M. Huss, W. R. Harvey, and H. Wieczorek. 2000. Three-dimensional structure and subunit topology of the V1 ATPase from Manduca sexta midgut. Biochemistry 39:8609-8616. [DOI] [PubMed] [Google Scholar]
- 43.Gruber, G., H. Wieczorek, W. R. Harvey, and V. Muller. 2001. Structure-function relationships of A-, F- and V-ATPases. J. Exp. Biol. 204:2597-2605. [DOI] [PubMed] [Google Scholar]
- 44.Hiesinger, P. R., A. Fayyazuddin, S. Q. Mehta, T. Rosenmund, K. L. Schulze, R. G. Zhai, P. Verstreken, Y. Cao, Y. Zhou, J. Kunz, and H. J. Bellen. 2005. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell 121:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill, K., and A. A. Cooper. 2000. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 19:550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill, K. J., and T. H. Stevens. 1994. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H+-ATPase complex. Mol. Biol. Cell 5:1039-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill, K. J., and T. H. Stevens. 1995. Vma22p is a novel endoplasmic reticulum-associated protein required for assembly of the yeast vacuolar H+-ATPase complex. J. Biol. Chem. 270:22329-22336. [DOI] [PubMed] [Google Scholar]
- 48.Hirata, R., L. A. Graham, A. Takatsuki, T. H. Stevens, and Y. Anraku. 1997. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J. Biol. Chem. 272:4795-4803. [DOI] [PubMed] [Google Scholar]
- 49.Hirata, R., N. Umemoto, M. N. Ho, Y. Ohya, T. H. Stevens, and Y. Anraku. 1993. VMA12 is essential for assembly of the vacuolar H+-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J. Biol. Chem. 268:961-967. [PubMed] [Google Scholar]
- 50.Hirata, T., A. Iwamoto-Kihara, G. H. Sun-Wada, T. Okajima, Y. Wada, and M. Futai. 2003. Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J. Biol. Chem. 278:23714-23719. [DOI] [PubMed] [Google Scholar]
- 51.Ho, M. N., K. J. Hill, M. A. Lindorfer, and T. H. Stevens. 1993. Isolation of vacuolar membrane H+-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H+-ATPase. J. Biol. Chem. 268:221-227. [PubMed] [Google Scholar]
- 52.Ho, M. N., R. Hirata, N. Umemoto, Y. Ohya, A. Takatsuki, T. H. Stevens, and Y. Anraku. 1993. VMA13 encodes a 54-kDa vacuolar H+-ATPase subunit required for activity but not assembly of the enzyme complex in Saccharomyces cerevisiae. J. Biol. Chem. 268:18286-18292. [PubMed] [Google Scholar]
- 53.Holliday, L. S., M. Lu, B. S. Lee, R. D. Nelson, S. Solivan, L. Zhang, and S. L. Gluck. 2000. The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J. Biol. Chem. 275:32331-32337. [DOI] [PubMed] [Google Scholar]
- 54.Imamura, H., M. Nakano, H. Noji, E. Muneyuki, S. Ohkuma, M. Yoshida, and K. Yokoyama. 2003. Evidence for rotation of V1-ATPase. Proc. Natl. Acad. Sci. USA 100:2312-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue, T., and M. Forgac. 2005. Cysteine-mediated cross-linking indicates that subunit C of the V-ATPase is in close proximity to subunits E and G of the V1 domain and subunit a of the V0 domain. J. Biol. Chem. 280:27896-27903. [DOI] [PubMed] [Google Scholar]
- 56.Iwaki, T., T. Goa, N. Tanaka, and K. Takegawa. 2004. Characterization of Schizosaccharomyces pombe mutants defective in vacuolar acidification and protein sorting. Mol. Genet. Genomics 271:197-207. [DOI] [PubMed] [Google Scholar]
- 57.Iwata, M., H. Imamura, E. Stambouli, C. Ikeda, M. Tamakoshi, K. Nagata, H. Makyio, B. Hankamer, J. Barber, M. Yoshida, K. Yokoyama, and S. Iwata. 2004. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc. Natl. Acad. Sci. USA 101:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones, P. C., and R. H. Fillingame. 1998. Genetic fusions of subunit c in the F0 sector of H+-transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking analysis. J. Biol. Chem. 273:29701-29705. [DOI] [PubMed] [Google Scholar]
- 59.Jones, R. P., L. J. Durose, J. B. Findlay, and M. A. Harrison. 2005. Defined sites of interaction between subunits E (Vma4p), C (Vma5p), and G (Vma10p) within the stator structure of the vacuolar H+-ATPase. Biochemistry 44:11924. [DOI] [PubMed] [Google Scholar]
- 60.Kakinuma, Y., I. Yamato, and T. Murata. 1999. Structure and function of vacuolar Na+-translocating ATPase in Enterococcus hirae. J. Bioenerg Biomembr. 31:7-14. [DOI] [PubMed] [Google Scholar]
- 61.Kane, P. M. 1995. Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem. 270:17025-17032. [PubMed] [Google Scholar]
- 62.Kane, P. M. 2000. Regulation of V-ATPases by reversible disassembly. FEBS Lett. 469:137-141. [DOI] [PubMed] [Google Scholar]
- 63.Kane, P. M., M. C. Kuehn, I. Howald-Stevenson, and T. H. Stevens. 1992. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H+-ATPase. J. Biol. Chem. 267:447-454. [PubMed] [Google Scholar]
- 64.Kane, P. M., and A. M. Smardon. 2003. Assembly and regulation of the yeast vacuolar H+-ATPase. J. Bioenerg Biomembr. 35:313-321. [DOI] [PubMed] [Google Scholar]
- 65.Kane, P. M., M. Tarsio, and J. Liu. 1999. Early steps in assembly of the yeast vacuolar H+-ATPase. J. Biol. Chem. 274:17275-17283. [DOI] [PubMed] [Google Scholar]
- 66.Kaplan, K. B., A. A. Hyman, and P. K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91:491-500. [DOI] [PubMed] [Google Scholar]
- 67.Karet, F. E., K. E. Finberg, R. D. Nelson, A. Nayir, H. Mocan, S. A. Sanjad, J. Rodriguez-Soriano, F. Santos, C. W. Cremers, A. Di Pietro, B. I. Hoffbrand, J. Winiarski, A. Bakkaloglu, S. Ozen, R. Dusunsel, P. Goodyer, S. A. Hulton, D. K. Wu, A. B. Skvorak, C. C. Morton, M. J. Cunningham, V. Jha, and R. P. Lifton. 1999. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat. Genet. 21:84-90. [DOI] [PubMed] [Google Scholar]
- 68.Kawasaki-Nishi, S., K. Bowers, T. Nishi, M. Forgac, and T. H. Stevens. 2001. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276:47411-47420. [DOI] [PubMed] [Google Scholar]
- 69.Kawasaki-Nishi, S., T. Nishi, and M. Forgac. 2001. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc. Natl. Acad. Sci. USA 98:12397-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawasaki-Nishi, S., T. Nishi, and M. Forgac. 2001. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276:17941-17948. [DOI] [PubMed] [Google Scholar]
- 71.Kim, J. H., C. A. Lingwood, D. B. Williams, W. Furuya, M. F. Manolson, and S. Grinstein. 1996. Dynamic measurement of the pH of the Golgi complex in living cells using retrograde transport of the verotoxin receptor. J. Cell Biol. 134:1387-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klionsky, D. J., H. Nelson, and N. Nelson. 1992. Compartment acidification is required for efficient sorting of proteins to the vacuole in Saccharomyces cerevisiae. J. Biol. Chem. 267:3416-3422. [PubMed] [Google Scholar]
- 74.Landolt-Marticorena, C., K. M. Williams, J. Correa, W. Chen, and M. F. Manolson. 2000. Evidence that the NH2 terminus of vph1p, an integral subunit of the V0 sector of the yeast V-ATPase, interacts directly with the Vma1p and Vma13p subunits of the V1 sector. J. Biol. Chem. 275:15449-15457. [DOI] [PubMed] [Google Scholar]
- 75.Leng, X. H., M. F. Manolson, Q. Liu, and M. Forgac. 1996. Site-directed mutagenesis of the 100-kDa subunit (Vph1p) of the yeast vacuolar (H+)-ATPase. J. Biol. Chem. 271:22487-22493. [DOI] [PubMed] [Google Scholar]
- 76.Li, L., and J. Kaplan. 1998. Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J. Biol. Chem. 273:22181-22187. [DOI] [PubMed] [Google Scholar]
- 77.Lu, M., L. S. Holliday, L. Zhang, W. A. Dunn, Jr., and S. L. Gluck. 2001. Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276:30407-30413. [DOI] [PubMed] [Google Scholar]
- 78.Lu, M., Y. Y. Sautin, L. S. Holliday, and S. L. Gluck. 2004. The glycolytic enzyme aldolase mediates assembly, expression and activity of V-ATPase. J. Biol. Chem. 279:8732-8739. [DOI] [PubMed] [Google Scholar]
- 79.Ludwig, J., S. Kerscher, U. Brandt, K. Pfeiffer, F. Getlawi, D. K. Apps, and H. Schagger. 1998. Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules. J. Biol. Chem. 273:10939-10947. [DOI] [PubMed] [Google Scholar]
- 80.Malkus, P., L. A. Graham, T. H. Stevens, and R. Schekman. 2004. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell 15:5075-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mandel, M., Y. Moriyama, J. D. Hulmes, Y. C. Pan, H. Nelson, and N. Nelson. 1988. cDNA sequence encoding the 16-kDa proteolipid of chromaffin granules implies gene duplication in the evolution of H+-ATPases. Proc. Natl. Acad. Sci. USA 85:5521-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manolson, M. F., B. F. Ouellette, M. Filion, and R. J. Poole. 1988. cDNA sequence and homologies of the “57-kDa” nucleotide-binding subunit of the vacuolar ATPase from Arabidopsis. J. Biol. Chem. 263:17987-17994. [PubMed] [Google Scholar]
- 83.Manolson, M. F., D. Proteau, R. A. Preston, A. Stenbit, B. T. Roberts, M. A. Hoyt, D. Preuss, J. Mulholland, D. Botstein, and E. W. Jones. 1992. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H+-ATPase. J. Biol. Chem. 267:14294-14303. [PubMed] [Google Scholar]
- 84.Manolson, M. F., B. Wu, D. Proteau, B. E. Taillon, B. T. Roberts, M. A. Hoyt, and E. W. Jones. 1994. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H+-ATPase subunit Vph1p. J. Biol. Chem. 269:14064-14074. [PubMed] [Google Scholar]
- 85.Meier, T., P. Polzer, K. Diederichs, W. Welte, and P. Dimroth. 2005. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308:659-662. [DOI] [PubMed] [Google Scholar]
- 86.Mellman, I., R. Fuchs, and A. Helenius. 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55:663-700. [DOI] [PubMed] [Google Scholar]
- 87.Merzendorfer, H., M. Huss, R. Schmid, W. R. Harvey, and H. Wieczorek. 1999. A novel insect V-ATPase subunit M9.7 is glycosylated extensively. J. Biol. Chem. 274:17372-17378. [DOI] [PubMed] [Google Scholar]
- 88.Miseta, A., R. Kellermayer, D. P. Aiello, L. Fu, and D. M. Bedwell. 1999. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 451:132-136. [DOI] [PubMed] [Google Scholar]
- 89.Moriyama, Y., and N. Nelson. 1988. The vacuolar H+-ATPase, a proton pump controlled by a slip. Prog. Clin. Biol. Res. 273:387-394. [PubMed] [Google Scholar]
- 90.Muller, M. L., M. Jensen, and L. Taiz. 1999. The vacuolar H+-ATPase of lemon fruits is regulated by variable H+/ATP coupling and slip. J. Biol. Chem. 274:10706-10716. [DOI] [PubMed] [Google Scholar]
- 91.Muller, V., and G. Gruber. 2003. ATP synthases: structure, function and evolution of unique energy converters. Cell. Mol. Life Sci. 60:474-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Munn, A. L., and H. Riezman. 1994. Endocytosis is required for the growth of vacuolar H+-ATPase-defective yeast: identification of six new END genes. J. Cell Biol. 127:373-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nagano, F., H. Kawabe, H. Nakanishi, M. Shinohara, M. Deguchi-Tawarada, M. Takeuchi, T. Sasaki, and Y. Takai. 2002. Rabconnectin-3, a novel protein that binds both GDP/GTP exchange protein and GTPase-activating protein for Rab3 small G protein family. J. Biol. Chem. 277:9629-9632. [DOI] [PubMed] [Google Scholar]
- 94.Nakamura, N., A. Matsuura, Y. Wada, and Y. Ohsumi. 1997. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. (Tokyo) 121:338-344. [DOI] [PubMed] [Google Scholar]
- 95.Nass, R., and R. Rao. 1998. Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast. Implications for vacuole biogenesis. J. Biol. Chem. 273:21054-21060. [DOI] [PubMed] [Google Scholar]
- 96.Nelson, H., S. Mandiyan, and N. Nelson. 1989. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J. Biol. Chem. 264:1775-1778. (Erratum, 264:5313.) [PubMed] [Google Scholar]
- 97.Nelson, H., S. Mandiyan, and N. Nelson. 1994. The Saccharomyces cerevisiae VMA7 gene encodes a 14-kDa subunit of the vacuolar H+-ATPase catalytic sector. J. Biol. Chem. 269:24150-24155. [PubMed] [Google Scholar]
- 98.Nelson, H., S. Mandiyan, T. Noumi, Y. Moriyama, M. C. Miedel, and N. Nelson. 1990. Molecular cloning of cDNA encoding the C subunit of H+-ATPase from bovine chromaffin granules. J. Biol. Chem. 265:20390-20393. [PubMed] [Google Scholar]
- 99.Nelson, H., and N. Nelson. 1990. Disruption of genes encoding subunits of yeast vacuolar H+-ATPase causes conditional lethality. Proc. Natl. Acad. Sci. USA 87:3503-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nelson, N., and W. R. Harvey. 1999. Vacuolar and plasma membrane proton-adenosinetriphosphatases. Physiol. Rev. 79:361-385. [DOI] [PubMed] [Google Scholar]
- 101.Nelson, N., A. Sacher, and H. Nelson. 2002. The significance of molecular slips in transport systems. Nat. Rev. Mol. Cell. Biol. 3:876-881. [DOI] [PubMed] [Google Scholar]
- 102.Nelson, N., and L. Taiz. 1989. The evolution of H+-ATPases. Trends Biochem. Sci. 14:113-116. [DOI] [PubMed] [Google Scholar]
- 103.Nishi, T., and M. Forgac. 2002. The vacuolar (h+)-ATPases—nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3:94-103. [DOI] [PubMed] [Google Scholar]
- 104.Nishi, T., S. Kawasaki-Nishi, and M. Forgac. 2003. The first putative transmembrane segment of subunit c" (Vma16p) of the yeast V-ATPase is not necessary for function. J. Biol. Chem. 278:5821-5827. [DOI] [PubMed] [Google Scholar]
- 105.Noumi, T., C. Beltran, H. Nelson, and N. Nelson. 1991. Mutational analysis of yeast vacuolar H+-ATPase. Proc. Natl. Acad. Sci. USA 88:1938-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Odorizzi, G., M. Babst, and S. D. Emr. 2000. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25:229-235. [DOI] [PubMed] [Google Scholar]
- 107.Ohya, Y., N. Umemoto, I. Tanida, A. Ohta, H. Iida, and Y. Anraku. 1991. Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet- phenotype are ascribable to defects of vacuolar membrane H+-ATPase activity. J. Biol. Chem. 266:13971-13977. [PubMed] [Google Scholar]
- 108.Oka, T., and M. Futai. 2000. Requirement of V-ATPase for ovulation and embryogenesis in Caenorhabditis elegans. J. Biol. Chem. 275:29556-29561. [DOI] [PubMed] [Google Scholar]
- 109.Oluwatosin, Y. E., and P. M. Kane. 1997. Mutations in the CYS4 gene provide evidence for regulation of the yeast vacuolar H+-ATPase by oxidation and reduction in vivo. J. Biol. Chem. 272:28149-28157. [DOI] [PubMed] [Google Scholar]
- 110.Oluwatosin, Y. E., and P. M. Kane. 1998. Mutations in the yeast KEX2 gene cause a Vma−-like phenotype: a possible role for the Kex2 endoprotease in vacuolar acidification. Mol. Cell. Biol. 8:1534-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paroutis, P., N. Touret, and S. Grinstein. 2004. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda) 19:207-215. [DOI] [PubMed] [Google Scholar]
- 112.Parra, K. J., and P. M. Kane. 1998. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 8:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parra, K. J., K. L. Keenan, and P. M. Kane. 2000. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275:21761-21767. [DOI] [PubMed] [Google Scholar]
- 114.Parsons, A. B., R. L. Brost, H. Ding, Z. Li, C. Zhang, B. Sheikh, G. W. Brown, P. M. Kane, T. R. Hughes, and C. Boone. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22:62-69. [DOI] [PubMed] [Google Scholar]
- 115.Perzov, N., V. Padler-Karavani, H. Nelson, and N. Nelson. 2002. Characterization of yeast V-ATPase mutants lacking Vph1p or Stv1p and the effect on endocytosis. J. Exp. Biol. 205:1209-1219. [DOI] [PubMed] [Google Scholar]
- 116.Peters, C., M. J. Bayer, S. Buhler, J. S. Andersen, M. Mann, and A. Mayer. 2001. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409:581-588. [DOI] [PubMed] [Google Scholar]
- 117.Piper, R. C., N. J. Bryant, and T. H. Stevens. 1997. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J. Cell Biol. 138:531-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plant, P. J., M. F. Manolson, S. Grinstein, and N. Demaurex. 1999. Alternative mechanisms of vacuolar acidification in H+-ATPase-deficient yeast. J. Biol. Chem. 274:37270-37279. [DOI] [PubMed] [Google Scholar]
- 119.Poltermann, S., M. Nguyen, J. Gunther, J. Wendland, A. Hartl, W. Kunkel, P. F. Zipfel, and R. Eck. 2005. The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology 151:1645-1655. [DOI] [PubMed] [Google Scholar]
- 120.Powell, B., L. A. Graham, and T. H. Stevens. 2000. Molecular characterization of the yeast vacuolar H+-ATPase proton pore. J. Biol. Chem. 275:23654-23660. [DOI] [PubMed] [Google Scholar]
- 121.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Redding, K., J. H. Brickner, L. G. Marschall, J. W. Nichols, and R. S. Fuller. 1996. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol. Cell. Biol. 6:6208-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roth, M. G. 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84:699-730. [DOI] [PubMed] [Google Scholar]
- 124.Rothman, J. H., C. T. Yamashiro, C. K. Raymond, P. M. Kane, and T. H. Stevens. 1989. Acidification of the lysosome-like vacuole and the vacuolar H+-ATPase are deficient in two yeast mutants that fail to sort vacuolar proteins. J. Cell Biol. 109:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sambade, M., M. Alba, A. M. Smardon, R. W. West, and P. M. Kane. 2005. A genomic screen for yeast vacuolar membrane ATPase mutants. Genetics 170:1539-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sambade, M., and P. M. Kane. 2004. The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and bovine e subunits that is essential for function. J. Biol. Chem. 279:17361-17365. [DOI] [PubMed] [Google Scholar]
- 127.Sautin, Y. Y., M. Lu, A. Gaugler, L. Zhang, and S. L. Gluck. 2005. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+- ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol. Cell. Biol. 5:575-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schu, P. V., K. Takegawa, M. J. Fry, J. H. Stack, M. D. Waterfield, and S. D. Emr. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260:88-91. [DOI] [PubMed] [Google Scholar]
- 129.Schwappach, B., S. Stobrawa, M. Hechenberger, K. Steinmeyer, and T. J. Jentsch. 1998. Golgi localization and functionally important domains in the NH2 and COOH terminus of the yeast CLC putative chloride channel Gef1p. J. Biol. Chem. 273:15110-15118. [DOI] [PubMed] [Google Scholar]
- 130.Seeley, E. S., M. Kato, N. Margolis, W. Wickner, and G. Eitzen. 2002. Genomic analysis of homotypic vacuole fusion. Mol. Biol. Cell 13:782-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Seol, J. H., A. Shevchenko, and R. J. Deshaies. 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3:384-391. [DOI] [PubMed] [Google Scholar]
- 132.Serrano, R., D. Bernal, E. Simon, and J. Arino. 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279:19698-19704. [DOI] [PubMed] [Google Scholar]
- 133.Shao, E., and M. Forgac. 2004. Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J. Biol. Chem. 279:48663-48670. [DOI] [PubMed] [Google Scholar]
- 134.Sipos, G., J. H. Brickner, E. J. Brace, L. Chen, A. Rambourg, F. Kepes, and R. S. Fuller. 2004. Soi3p/Rav1p functions at the early endosome to regulate endocytic trafficking to the vacuole and localization of trans-Golgi network transmembrane proteins. Mol. Biol. Cell 15:3196-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smardon, A. M., M. Tarsio, and P. M. Kane. 2002. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 13:13. [DOI] [PubMed] [Google Scholar]
- 136.Smith, A. N., J. Skaug, K. A. Choate, A. Nayir, A. Bakkaloglu, S. Ozen, S. A. Hulton, S. A. Sanjad, E. A. Al-Sabban, R. P. Lifton, S. W. Scherer, and F. E. Karet. 2000. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat. Genet. 26:71-75. [DOI] [PubMed] [Google Scholar]
- 137.Sorensen, S. O., H. B. van den Hazel, M. C. Kielland-Brandt, and J. R. Winther. 1994. pH-dependent processing of yeast procarboxypeptidase Y by proteinase A in vivo and in vitro. Eur. J. Biochem. 220:19-27. [DOI] [PubMed] [Google Scholar]
- 138.Stack, J. H., D. B. DeWald, K. Takegawa, and S. D. Emr. 1995. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stevens, T. H., and M. Forgac. 1997. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu. Rev. Cell Dev. Biol. 13:779-808. [DOI] [PubMed] [Google Scholar]
- 140.Su, Y., A. Zhou, R. S. Al-Lamki, and F. E. Karet. 2003. The a-subunit of the V-type H+-ATPase interacts with phosphofructokinase-1 in humans. J. Biol. Chem. 278:20013-20018. [DOI] [PubMed] [Google Scholar]
- 141.Sumner, J. P., J. A. Dow, F. G. Earley, U. Klein, D. Jager, and H. Wieczorek. 1995. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270:5649-5653. [DOI] [PubMed] [Google Scholar]
- 142.Sun-Wada, G., Y. Murata, A. Yamamoto, H. Kanazawa, Y. Wada, and M. Futai. 2000. Acidic endomembrane organelles are required for mouse postimplantation development. Dev. Biol. 228:315-325. [DOI] [PubMed] [Google Scholar]
- 143.Supekova, L., F. Supek, and N. Nelson. 1995. The Saccharomyces cerevisiae VMA10 is an intron-containing gene encoding a novel 13-kDa subunit of vacuolar H+-ATPase. J. Biol. Chem. 270:13726-13732. [DOI] [PubMed] [Google Scholar]
- 144.Swallow, C. J., S. Grinstein, R. A. Sudsbury, and O. D. Rotstein. 1993. Relative roles of Na+/H+ exchange and vacuolar-type H+ ATPases in regulating cytoplasmic pH and function in murine peritoneal macrophages. J. Cell Physiol. 157:453-460. [DOI] [PubMed] [Google Scholar]
- 145.Tanida, I., A. Hasegawa, H. Iida, Y. Ohya, and Y. Anraku. 1995. Cooperation of calcineurin and vacuolar H+-ATPase in intracellular Ca2+ homeostasis of yeast cells. J. Biol. Chem. 270:10113-10119. [DOI] [PubMed] [Google Scholar]
- 146.Tomashek, J. J., B. S. Garrison, and D. J. Klionsky. 1997. Reconstitution in vitro of the V1 complex from the yeast vacuolar proton-translocating ATPase. Assembly recapitulates mechanism. J. Biol. Chem. 272:16618-16623. [DOI] [PubMed] [Google Scholar]
- 147.Tomashek, J. J., L. A. Graham, M. U. Hutchins, T. H. Stevens, and D. J. Klionsky. 1997. V1-situated stalk subunits of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 272:26787-26793. [DOI] [PubMed] [Google Scholar]
- 148.Tomashek, J. J., J. L. Sonnenburg, J. M. Artimovich, and D. J. Klionsky. 1996. Resolution of subunit interactions and cytoplasmic subcomplexes of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 271:10397-10404. [DOI] [PubMed] [Google Scholar]
- 149.Trombetta, E. S., M. Ebersold, W. Garrett, M. Pypaert, and I. Mellman. 2003. Activation of lysosomal function during dendritic cell maturation. Science 299:1400-1403. [DOI] [PubMed] [Google Scholar]
- 150.Umemoto, N., T. Yoshihisa, R. Hirata, and Y. Anraku. 1990. Roles of the VMA3 gene product, subunit c of the vacuolar membrane H+-ATPase on vacuolar acidification and protein transport. A study with VMA3-disrupted mutants of Saccharomyces cerevisiae. J. Biol. Chem. 265:18447-18453. [PubMed] [Google Scholar]
- 151.Van Dyke, R. W., B. F. Scharschmidt, and C. J. Steer. 1985. ATP-dependent proton transport by isolated brain clathrin-coated vesicles. Role of clathrin and other determinants of acidification. Biochim. Biophys. Acta 812:423-436. [DOI] [PubMed] [Google Scholar]
- 152.Venzke, D., I. Domgall, T. Kocher, J. Fethiere, S. Fischer, and B. Bottcher. 2005. Elucidation of the stator organization in the V-ATPase of Neurospora crassa. J. Mol. Biol. 349:659-669. [DOI] [PubMed] [Google Scholar]
- 153.Vitavska, O., H. Merzendorfer, and H. Wieczorek. 2005. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J. Biol. Chem. 280:1070-1076. [DOI] [PubMed] [Google Scholar]
- 154.Vitavska, O., H. Wieczorek, and H. Merzendorfer. 2003. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J. Biol. Chem. 278:18499-18505. [DOI] [PubMed] [Google Scholar]
- 155.Wieczorek, H., D. Brown, S. Grinstein, J. Ehrenfeld, and W. R. Harvey. 1999. Animal plasma membrane energization by proton-motive V-ATPases. Bioessays 21:637-648. [DOI] [PubMed] [Google Scholar]
- 156.Wieczorek, H., G. Grber, W. R. Harvey, M. Huss, H. Merzendorfer, and W. Zeiske. 2000. Structure and regulation of insect plasma membrane H+V-ATPase. J. Exp. Biol. 203:127-135. [DOI] [PubMed] [Google Scholar]
- 157.Wilkens, S., and M. Forgac. 2001. Three-dimensional structure of the vacuolar ATPase proton channel by electron microscopy. J. Biol. Chem. 276:44064-44068. [DOI] [PubMed] [Google Scholar]
- 158.Wilkens, S., T. Inoue, and M. Forgac. 2004. Three-dimensional structure of the vacuolar ATPase. Localization of subunit H by difference imaging and chemical cross-linking. J. Biol. Chem. 279:41942-41949. [DOI] [PubMed] [Google Scholar]
- 159.Wilkens, S., E. Vasilyeva, and M. Forgac. 1999. Structure of the vacuolar ATPase by electron microscopy. J. Biol. Chem. 274:31804-31810. [DOI] [PubMed] [Google Scholar]
- 160.Yamashiro, C. T., P. M. Kane, D. F. Wolczyk, R. A. Preston, and T. H. Stevens. 1990. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol. Cell. Biol. 10:3737-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yaver, D. S., H. Nelson, N. Nelson, and D. J. Klionsky. 1993. Vacuolar ATPase mutants accumulate precursor proteins in a prevacuolar compartment. J. Biol. Chem. 268:10564-10572. [PubMed] [Google Scholar]
- 162.Zhang, J. W., K. J. Parra, J. Liu, and P. M. Kane. 1998. Characterization of a temperature-sensitive yeast vacuolar ATPase mutant with defects in actin distribution and bud morphology. J. Biol. Chem. 273:18470-18480. [DOI] [PubMed] [Google Scholar]
- 163.Zimniak, L., P. Dittrich, J. P. Gogarten, H. Kibak, and L. Taiz. 1988. The cDNA sequence of the 69-kDa subunit of the carrot vacuolar H+-ATPase. Homology to the beta-chain of F0F1-ATPases. J. Biol. Chem. 263:9102-9112. [PubMed] [Google Scholar]