Abstract
Coat proteins of non-enveloped, icosahedral viruses must perform a variety of functions during their life cycle such as assembly of the coat protein subunits into a closed shell, specific encapsidation of the viral nucleic acid, maturation of the capsid, interaction with host receptors and disassembly to deliver the genetic information into the newly-infected cell. A thorough understanding of the multiple capsid properties at the molecular level is required in order to identify potential targets for antiviral therapy and the prevention of viral disease. The system we have chosen for study are the astroviruses, a family of icosahedral, single-stranded RNA viruses that cause disease in mammals and birds. Very little is known about what regions of the coat protein contribute to the diverse capsid functions. This review will present novel structural predictions for the coat protein sequence of different astrovirus family members. Based on these predictions, we hypothesize that the assembly and RNA packaging functions of the astrovirus coat protein constitutes an individual domain distinct from the determinants required for receptor binding and internalization. Information derived from these structural predictions will serve as an important tool in designing experiments to understand astrovirus biology.
INTRODUCTION
Astroviruses are small, icosahedral RNA viruses that infect both mammals and birds. Human astroviruses (HAstV), along with rotavirus and calicivirus, are a major cause of viral gastroenteritis and a significant public health concern (8,61). While research into the molecular epidemiology of HAstVs has advanced over the years, relatively little is known concerning the molecular biology or structural aspects of the astrovirus capsid. Because the viral capsid is the primary determinant of cell tropism, stimulates the host’s protective immune response and may play a part in the pathogenesis of the virus, our ignorance of the role of the astrovirus coat protein in the virus life cycle has delayed progress in understanding astrovirus virulence and transmission. This review will summarizes our present knowledge of astrovirus molecular biology as well as present structural predictions we have generated for the coat protein of different astrovirus family members. We anticipate that these novel structural calculations, in addition to providing insight into regions of the coat protein involved in particle assembly, nucleic acid packaging, maturation and tropism, will also inform us of unique and shared properties of the astrovirus coat proteins.
ASTROVIRUSES
The astrovirus family
Astroviruses are non-enveloped, icosahedral viruses with a single-stranded, messenger-sense RNA genome. The Astroviridae are split into two genera according to the International Committee on Taxonomy of Viruses: the Mamastrovirus, which infects animals and the Avastrovirus, which infects birds (39). Astroviruses were first detected by electron microscopy in stools of infants hospitalized with diarrhea (34). The particles are approximately 33 nm in diameter and share a unique 5- or 6- pointed surface star which is discernable on the surface of approximately 10% of the virion population and give the virus its name (astron, Greek for “star”). Since the initial observation of astroviruses in children, viral particles of similar size and morphology have been demonstrated to infect piglets (54), calves (68), lambs (56), red deer (59), mink kits (45), puppies (67), kittens (20), mice (29), ducklings (16), turkeys (40) and chickens (21). In addition to causing acute infantile gastroenteritis in humans, the Mamastroviruses are the etiological agents for various syndromes in other animal species such as mild diarrhea in lambs (55) and outbreaks of diarrhea in pre-weaning mink kits (10). Avastroviruses cause severe disease in birds from poult enteritis and mortality syndrome in turkeys (30,69) to acute nephritis in chickens (21) and a fatal hepatitis in ducks (16).
Human astroviruses
Eight astrovirus serotypes have been described among human astroviruses (HAstV 1-8), with serotype 1 being the most prevalent worldwide (37). All eight antigenic types have been adapted to grow in tissue culture, in which trypsin is used as a supplement in the medium. A cDNA of HAstV-1 (Oxford strain) has been cloned and, when capped message generated from the cDNA is transfected into CaCo-2 cells, infectious virus is produced (12). The small size of the astroviral genome, along with the experimental advantages of growth in tissue culture and availability of a cDNA clone have made HAstV-1 a tractable system in which to study virus assembly.
Genomic organization of HAstVs
Since HAstV-1 (Oxford strain) is the best studied member of the Astroviridae, it will be used as the prototype to describe the molecular biology of astroviruses. HAstVs genomes are single-stranded, messenger-sense RNA molecules of approximately 6.8 kb in length, excluding the poly(A) tail at the 3′ end (Fig. 1A). The genome encodes three open reading frames (ORFs): ORFs 1a, 1b and 2. ORFs 1a and 1b at the 5′ end of the genome have protein sequence motifs indicative of non-structural proteins: a 3C-like serine protease with a nuclear localization signal in ORF1a and a putative RNA-dependent RNA polymerase in ORF1b (23,31,64). An interesting feature of ORF1a and 1b in the HAstV genome is that while the protease and viral replicase are encoded by separate ORFs, both proteins are thought to be translated initially as a polyprotein (37). A 70 nucleotide region between ORF 1a and 1b is highly conserved among HAstV serotypes and contains a signal and downstream stem-loop structure indicative of ribosomal frameshifting (23,32,33,35) as seen in other virus families (5,22). The product of ORF1a has been demonstrated to proteolytically activate and cleave itself both in vitro (27) and in vivo (11). For HAstV-8, in addition to cleaving itself, the protease has been demonstrated to cleave ORF1b as well (43). There is no data to suggest that ORF2 is a substrate for this protease. The product(s) of ORF1a has also been shown to accumulate in the nucleus of infected cells (63) and it has recently been reported that the nonstructural protein p38, which is derived from ORF1a, leads to apoptosis of the host cell resulting in efficient virus replication (17) and particle release (42) (see below).
FIG. 1.
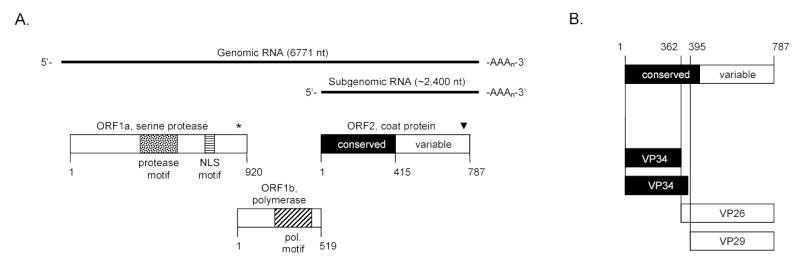
Genomic organization and morphogenesis of HAstV-1 prototype (Oxford strain). (A) Genomic and subgenomic RNA are indicated by the solid black line with their length indicated in nucleotides (nt). The open reading frames (ORF) along with their protein products are shown as rectangles. ORF1a encodes a 3C-like serine protease motif (stippled box) along with a nuclear localization (NLS) motif (horizontally striped box). The asterisk (*) denotes the ribosomal frameshifting signal. ORF1b encodes the viral polymerase and the polymerase motif is indicated (box with diagonal lines). ORF2 encodes the coat protein. The first half of the coat protein sequence (amino acids 1 to 415, heavily shaded) is highly conserved among all known members of the Astroviridae whereas the last half of the coat protein (amino acids 416–787, no shading) is highly variable. The arrowhead (▾) indicates the approximate location of putative caspase cleavage site(s) conserved among astrovirus capsid proteins (42). The numbers below the rectangles refer to length of the protein products in amino acids. (B) Cleavage cascade of the coat protein precursor as described in the text. Trypsin treatment cleaves the particle into its mature products of VP34 (of which two species are predicted), VP26 and VP29.
ORF2 is at the 3′ end of the genome and encodes an approximately 87–90 kDa coat protein precursor (2,31,41,46,51). In infected cells, both the full-length genomic RNA and an ORF2-specific subgenomic RNA of 2.4 kb are detected in large amounts (46). Translation of ORF2 most likely occurs from the subgenomic RNA and results in high levels of coat protein synthesis to assemble viral particles efficiently and encapsidate the viral genome. Translation of structural proteins from a subgenomic RNA is a strategy utilized by many positive-strand RNA viruses (48,52).
Precursor coat protein domain organization
As illustrated in Figure 1A, based upon the amino acid sequence identity of different astrovirus family members, the coat protein precursor is predicted to have two domains (24,25,44,45,62,65,66). Residues 1-415 compose the first domain and are highly conserved among the Astroviridae. In contrast, residues 416 to the end are highly divergent among all known serotypes and it has been reported that neutralizing monoclonal antibodies map to this second domain (3,51). Thus it is reasonable to assume that this hypervariable region is located on the surface of the viral particle and constitutes a region of the capsid that contributes to the strain-specific tropism of the virus. A novel role for the conserved and variable domains of the astrovirus coat protein in particle assembly will be discussed below.
Precursor coat protein cleavage
The coat protein precursor has previously been demonstrated to assemble readily into authentic virus (13) and more recently as virus-like particles using a recombinant vaccinia virus expression system (9) and a recombinant baculovirus expression system (6). After the particle is assembled, it is cleaved by trypsin to produce multiple smaller proteins; cleavage renders the particle infectious (2,41) (Fig. 1B). Attempts to identify the number and size of the mature structural proteins of the human astroviruses have yielded discrepancies, with proteins from 20 to 90 kDa being observed (2,4,9,41,46,51). In one report, HAstV-2 particles were cleaved from an 87 kDa precursor to three proteins, of 32, 29 and 26 kDa (9). Bass and Qiu (2) reported that the HAstV-1 structural protein is initially synthesized as an 87 kDa precursor and is then cleaved intracellularly at Arg70 to produce a 79 kDa protein. However, the existence of this smaller cleavage product (residues 1-70) was not observed in HAstV-1 infected cells in a separate study (13) and it has been recently demonstrated that the first 70 residues are dispensable for virus-like particle formation of HAstV-1 coat protein (6). The HAstV-1 87 kDa coat protein precursor assembles into particles that are cleaved by trypsin into three products of 34, 29 and 26 kDa. The 29 and 26 kDa products are found consistently in trypsin-digested particles (2,4,9,46,51) and result from cleavage of HAstV-1 and 2 coat protein at Arg361 and Arg395, respectively, with the C terminus of both being identical (2,51).
For HAstV-8, Méndez and colleagues (41) observed that the 90 kDa coat protein is first cleaved intracellularly at the C terminus, yielding a 70 kDa species. The 70 kDa protein then undergoes a complicated cascade of cleavages to generate VP34, VP27 and VP25, the predominant proteins found within fully cleaved particles and homologs to the three protein species found in mature HAstV-1 and 2 virions. It has been recently demonstrated that 90 kDa coat protein of HAstV-8 assembles into precursor particles within the cell and that the C-terminal cleavage of the coat protein is mediated by cellular caspases and is necessary to facilitate processing of the capsid precursor and release of the viral particles (42). The authors go on to demonstrate that putative caspase cleavage motifs found in cellular proteins (e.g., DXXD and XEXD) are present in the C terminus of the HAstV-8 capsid precursor and that acidic amino acid residues are highly conserved for other astrovirus family members despite the fact that this region of the coat protein is variable among the Astroviridae. These observations suggest that caspase-mediated cleavage of the coat protein may be a general mechanism for astrovirus particle release.
As outlined above, considerable progress has been made in understanding the basics of the astrovirus life cycle, however there are significant gaps in our knowledge concerning the fundamentals of particle assembly, RNA encapsidation, capsid maturation, tropism and disassembly. In the following sections, we present computer-generated predictions for the coat protein that will begin to elucidate the structural determinants of the astrovirus capsid.
STRATEGY FOR MAKING STRUCTURAL PREDICTIONS FOR THE ASTROVIRUS COAT PROTEIN
Very little is known about the astrovirus coat protein precursor and how it assembles into particles. In an attempt to understand what regions of this protein might contribute to particle assembly and receptor binding, we have utilized a structural prediction program to analyze the coat protein amino acid sequence. This analysis has led to predictions about what domains of the coat protein are necessary for protein assembly, as well as for host cell tropism. Except for within the Astroviridae, the astrovirus coat protein exhibits no significant sequence identity with any known viral capsid protein. As illustrated in Figure 1 above, the coat protein of both the Mamastrovirus and Avastrovirus has been divided into conserved (residues 1-415) and variable (residues 416-end) regions. Because of the lack of sequence identity seen in the variable region of the astrovirus coat protein (65) and based on the fact that neutralizing antibodies map to this area, (3,51) it has been proposed that the variable region is exposed on the surface of the particle and is a determinant for cellular tropism. In light of the conserved nature of residues 1-415, we reasoned that this region of the coat protein may represent a core “assembly domain”, a building block for capsid assembly. In support of this, a threonine residue at amino acid 227 of the HAstV-1 coat protein was demonstrated to be required for proper particle assembly (38). Mutation of this residue resulted in the absence of visible particles within the cytoplasm of CaCo-2 cells.
We used three-dimensional position-specific scoring matrix (3D-PSSM, version 2.6.0) to investigate whether conserved and variable regions of the coat protein assume a recognizable protein-folding motif (26). 3D-PSSM combines knowledge of three-dimensional structures with secondary-structure matching and solvation potentials to recognize protein folds when sequence homologies are remote. A number of parameters go into assessing confidence of the predictions of this program: Expectation Value (or Confidence Value), alignment quality and 3D molecule quality. Confidence Value scores are confirmed by checking the sequence alignment. A lack of significant gaps in the alignment and a match of the secondary structural elements are signs of good alignment. For model quality, a lack of significant deletions in the template (model) structure is important. We considered these factors to make predictions about structural information contained in the astrovirus capsid gene sequence. Previously, we applied this program to determine the domain organization of another icosahedral RNA virus that was verified by cryo-electron microscopy (58).
CONSERVED DOMAIN
Assembly domain functions
Table 1 outlines the astrovirus family members with fully sequenced capsid genes that were analyzed in this study. The amino acid sequence of the conserved domain of HAstV-1 (residues 1-415) was entered into the web server of 3D-PSSM (http://www.sbg.bio.ic.ac.uk/~3dpssm/) to search for folds in the database of known structures homologous to those predicted from astrovirus capsid gene structures. We initially investigated this sequence in September, 2002. However, all of the sequences examined in this study were again entered into 3D-PSSM in October, 2004 to take advantage of any new structures that had been added to the fold library during this time. In analyzing all of our predictions, a Confidence Value of 50% and greater was considered significant. However, it should be noted that top ranking non-confident hits (<50%) are often correct and should therefore be examined. Four structures were identified as significantly comparable, with residues 1-415 having a Confidence Value ranging from 57% to >93% (Table 2). All four structures were from viruses with icosahedral symmetry and that have canonical, uninterrupted β-sandwich folds. The comparisons predicted structural homology in overlapping regions of the HAstV-1 coat protein sequence: 20-287 (Theiler’s murine encephalomyelitis virus, DA strain [TMEV]), 72-273 (bean pod mottle virus [BPMV]); 80-349 (carnation mottle virus [CtMV]) and 79-413 (tomato bushy stunt virus [TBSV]). It is striking that these structures were identified as being comparable when they share sequence identity of 20% or less.
Table 1.
Astrovirus Family Members with Fully Sequenced Capsid Genes
| Host | Serotype | Virusa | Accession Number |
|---|---|---|---|
| Human | 1b | HAstV-1 | L23513 |
| 2 | HAstV-2 | L06802 | |
| 3 | HAstV-3 | AF117209 | |
| 4 | HAstV-4 | Z33883 | |
| 5 | HAstV-5 | AB037274 | |
| 6 | HAstV-6 | Z46658 | |
| 7 | HAstV-7 | Y08632 | |
| 8 | HAstV-8 | AF260508 | |
| Cat | – | FAstV | AF056197 |
| Pig | – | PAstV | Y15938 |
| Sheep | – | OAstV | Y15937 |
| Mink | – | MiAstV | AY179509 |
| Turkey | 1 | TAstV-1 | Y15936 |
| 2 | TAstV-2 | AF206663 | |
| Chicken | – | ANV | AB033998 |
HAstV, human astrovirus; FAstV, feline astrovirus; PAstV, porcine astrovirus; OAstV, ovine astrovirus; MiAstV, mink astrovirus; TAstV, turkey astrovirus; ANV, avian nephritis virus.
Two prevalent strains of HAstV-1 are described in the literature, Oxford and Newcastle. The Oxford strain analyzed here is identical to Newcastle in ORF2 (12).
Table 2.
The Top Four Protein Folds Comparable to that of HAstV-1 Coat Protein Residues 1-415 by 3D-PSSM Threading
| Templatea | Template length (no. of residues) | Sequence Identity (%) | Confidenceb(%) |
|---|---|---|---|
| TMEV | 267 | 13 | >93 |
| BPMV | 185 | 18 | >76 |
| CMtV | 267 | 20 | >72 |
| TBSV | 286 | 13 | >57 |
TMEV, Theiler’s murine encephalomyelitis virus DA strain; BPMV, bean pod mottle virus; CMtV, carnation mottle virus; TBSV, tomato bushy stunt virus.
Confidence is defined as 100 X probability of a match being correct (26)
Next, we tested whether the structural prediction for residues 1-415 would also hold true for VP34, which has been shown to be the mature protein product derived from the conserved region of the coat protein for HAstV-1,-2, and -8 (2,9,41,51). For HAstV-1, Bass and Qiu (2) reported two species of VP34, which are cleaved at residues 361 and 394 (Fig. 1B). When these sequences were entered into 3D-PSSM both forms of VP34 matched with the same top three hits for residues 1-415 (data not shown). Both forms of VP34 also aligned identically to TMEV, BPMV and CMtV as seen with residues 1-415. In harmony with these results, Caballero and colleagues have recently used 3D-PSSM to analyze VP34 of HAstV-1 and found the same homologies matches (6; see Discussion). To evaluate the consistency of these predictions across the known genomic diversity of HAstVs, we ran the conserved region of the coat protein of all the known human astrovirus serotypes through 3D-PSSM, as well as that of other animal and avian astrovirus capsid sequences (Table 3). The comparisons yielded the same protein template, identified for HAstV-1 (Table 2) for each of the other astrovirus capsid sequences analyzed: TMEV, BPMV, CMtV and TBSV. As with the HAstV-1 comparisons, homologous structural predictions occurred despite limited (12–19%) sequence identity between the templates recognized and the astrovirus sequences. These results suggest that residues 1-415 of all astrovirus coat proteins fold in a manner similar to that of well-studied icosahedral virus coat proteins.
Table 3.
The Top Protein Folds Comparable to that of the Astrovirus Coat Protein Residues 1-415 by 3D-PSSM Threading
| Astrovirus | Templatea | Template length (no. of residues) | Sequence identity (%) | Confidence |
|---|---|---|---|---|
| HAstV-1 | TMEV | 267 | 13 | >93 |
| HAstV-2 | TMEV | 267 | 12 | >96 |
| HAstV-3 | TMEV | 267 | 14 | >93 |
| HAstV-4 | BPMV | 185 | 19 | >64 |
| HAstV-5 | BPMV | 185 | 17 | >69 |
| HAstV-6 | TMEV | 267 | 15 | >91 |
| HAstV-7 | BPMV | 185 | 18 | >82 |
| HAstV-8 | TMEV | 267 | 13 | >89 |
| FAstV-1 | BPMV | 185 | 18 | >80 |
| PAstV-1 | CMtV | 267 | 19 | >83 |
| OAstV-1 | CMtV | 267 | 18 | >94 |
| MiAstV | TBSV | 321 | 16 | >90 |
| TAstV-1 | CMtV | 267 | 16 | >90 |
| TAstV-2 | CMtV | 267 | 17 | >84 |
| ANV | TBSV | 321 | 14 | >78 |
Multiple templates that had a significant comparability (>50%) were found for each astrovirus sequence; for clarity, only the top protein template is illustrated.
Inferences from known structural properties of viruses with homologies predicted by 3D-PSSM for the conserved domain
So what do the coat proteins of TMEV, BPMV, CMtV and TBSV tell us about the predicted capsid structure of the astroviruses? All four of these viruses have icosahedral virions containing an RNA genome. TMEV is a member of the Picornaviridae and its shell has 180 β-barrel domains formed from three proteins, VP1, VP2, and VP3, each approximately 30 kDa, to form a pseudo T=3 capsid. BPMV is in the Comoviridae family and its shell has a pseudo T=3 capsid composed of 180 β-barrel domains formed from two different protein types, L of 42 kDa and S of 24 kDa. In these structural predictions, astrovirus sequences always aligned with VP2 of TMEV and the S domain of BPMV. The tertiary and quaternary structures of comoviruses and animal picornaviruses have been demonstrated to be remarkably similar (7) and would explain why they were consistently present in the comparisons with the conserved domain of the astrovirus coat protein. CMtV and TBSV are in the Tombusviridae family and have a T=3 virion with 180 identical subunits (of approximately 38–40 kDa) in the capsid. The protein fold of both CMtV and TBSV subunits is very similar in tertiary and quaternary structure to each other and to that of other icosahedral plant viruses (47).
Detailed ultrastructure studies of trypsin-cleaved HAstV-2 virions have revealed icosahedral particles with an array of spikes protruding from the surface of the virion (50). A low-resolution cryoelectron microscopy image for trypsin-cleaved HAstV-1 particles has also been reported (37,38). These images show a solid, stippled icosahedral shell with a 33 nm diameter and 30 dimeric spikes that extend from the surface. Assuming that 180 copies of the VP34 protein are required to give a particle of 33 nm with T=3 symmetry and that the spikes are formed by VP26 and VP29 would be consistent with our structural predictions suggesting that VP34 is a structural homolog to known icosahedral coat proteins and that the variable domain (VP26/VP29) is involved in receptor binding. It is unclear, however, how only 30 dimeric spikes (60 subunits) of VP26/VP29 would be displayed on a 180-subunit shell.
Putative RNA binding domain
All astrovirus coat proteins for which the amino acid sequence has been determined contain a high concentration of basic amino acids in the N terminus. For HAstV-1, 22 lysine and arginine residues reside within the first 70 amino acids. The high level of basic amino acids suggests that this region of the coat protein may be involved in packaging the viral RNA molecule. Highly basic N termini of other icosahedral RNA virus coat proteins have been well-documented and shown to be involved in the specific encapsidation of the viral genomic RNA (1,14,36,49,53). It has recently been demonstrated that the first 70 amino acid residues of the HAstV-1 coat protein are not required for virus-like particle formation in a recombinant baculovirus expression system (6). In support of this, most of the sequence predictions that we performed for VP34 or residues 1-415 of the coat protein aligned downstream of residue 70 suggesting that these residues do not contribute to particle formation (see text above). The same conclusion was arrived at for VP34 of HAstV-1 by Caballero et al. (6). While not much is known about encapsidation of the viral RNA in the astrovirus system, the presence of numerous positively charged residues at the N terminus of the coat protein, the location of the β-sandwich folds C-terminal to this domain and inferences from coat protein molecules of other icosahedral, messenger-sense RNA viruses suggest that the N terminus of the coat protein may be involved in viral RNA packaging.
VARIABLE DOMAIN
Receptor-interaction functions
Although the sequences of residues 1-415 of the astrovirus coat protein are highly conserved, residues 416-end vary significantly and attempts to align capsid genes from different human serotypes and other animal astroviruses are suboptimal (24). To evaluate protein folding predictions of these diverse sequences, all 15 astrovirus coat protein sequences from residues 416-end were run in 3D-PSSM (Table 4). Six hits with significant confidence levels occurred for the fifteen sequences analyzed. Three of these were virus-related proteins and receptors, whereas the other three were of non-viral receptor-ligand interactions. The sequences for HAstV-2 had homology to the poliovirus receptor (PVR). PVR is also known as CD155 and is long and slender with a transmembrane domain and three Ig-like folds (18). It is expressed in the human intestinal epithelium and Peyer’s patches. Although its binding to poliovirus is known, its function otherwise is unknown. It is not immediately clear why HAstV-2 would encode this type of domain on the surface of their capsids, however, poliovirus, like astrovirus, is an enteric pathogen. The most striking match (>98% confidence) was that of HAstV-3 with Sindbis virus spike glycoprotein, E1. The crystal structure of E1 shows a long, slender molecule that interacts with Sindbis E2 to form a heterohexamer on the surface of this icosahedral particle (70). E2 is thought to have a role in the pH-dependent fusion process of cell entry. Homology of E1 with the HAstV-3 variable region is consistent with the notion that this portion of the coat protein is exposed on the surface of the capsid and has a role in viral entry.
Table 4.
The Top Protein Folds Comparable to that of the Astrovirus Coat Protein Residues 416-End by 3D-PSSM Threading
| Astrovirus | Template | Template length (no. of residues) | Sequence identity (%) | Confidence |
|---|---|---|---|---|
| HAstV-2 | Poliovirus receptor | 301 | 15 | >62 |
| HAstV-3 | Sindbis virus spike glycoprotein E1 | 369 | 15 | >98 |
| HAstV-4 | Interleukin 6 receptor, Alpha chain | 299 | 18 | >65 |
| FAstV-1 | BAFF-BAFF-R complex | 288 | 10 | >91 |
| ANVa | Neutrophil gelatinase-associated lipocalin | 174 | 12 | >84 |
| ANV | Adenovirus fiber protein head domain (knob domain) | 191 | 19 | >51 |
Two templates that had a significant comparability (>50%) were found for ANV.
Along with a non-viral receptor-ligand hit (see below), ANV demonstrated homology with the head domain of the adenovirus fiber binding protein. Adenovirus is an icosahedral, double-stranded DNA virus that causes disease in humans (19). Recently, the head domain of the adenovirus fiber has been demonstrated to be structurally similar to another virus attachment protein, reovirus s1 and the distinct cellular receptors that bind both of these attachment proteins also have significant structural resemblance (57). While adenovirus and reovirus belong to different virus families and bind different cellular receptors, the structural similarities between the attachment proteins and receptors point to an evolutionary conservation of function among diverse, icosahedral viruses and are consistent with the structural homologies defined for the variable domain of the astrovirus coat protein.
The other three hits with significant homology were with non-viral receptor-ligand interactions: the interleukin-6 receptor alpha chain that binds IL-6 (60), neutrophil gelatinase associated lipocalin (NGAL) which is expressed in immature neutrophil precursors and in epithelial cells (15) and with B-cell activating factor (BAFF) binding its receptor (BAFF-R). BAFF forms a virus-like cage that binds BAFF-R in a 1:1 molar ratio (28). Although only five out of fifteen astroviruses predicted significant homology to known structures, all of these were receptor-ligand interactions that lend support to the premise that the variable region encodes receptor-binding domains. These results also suggest that the findings for astrovirus capsid studies may be distinct and surprising.
CONCLUSIONS
Figure 2 illustrates the functional domains of the astrovirus coat protein. Based on the structural predictions generated here, biological data culled from the literature and what has been published on capsid assembly of other icosahedral, RNA viruses, we propose that the N-terminal half of the astrovirus coat protein constitutes the particle assembly domain (AD) needed to drive formation of a closed capsid and encapsidation of the viral genome. The C-terminal half of the coat protein forms the receptor-interaction domain (RID) which dictates host cell tropism. The consistency of these predictions is evident across the known genomic diversity of the Astroviridae and demonstrates a remarkable conservation of coat protein function in the absence of sequence identity.
FIG. 2.
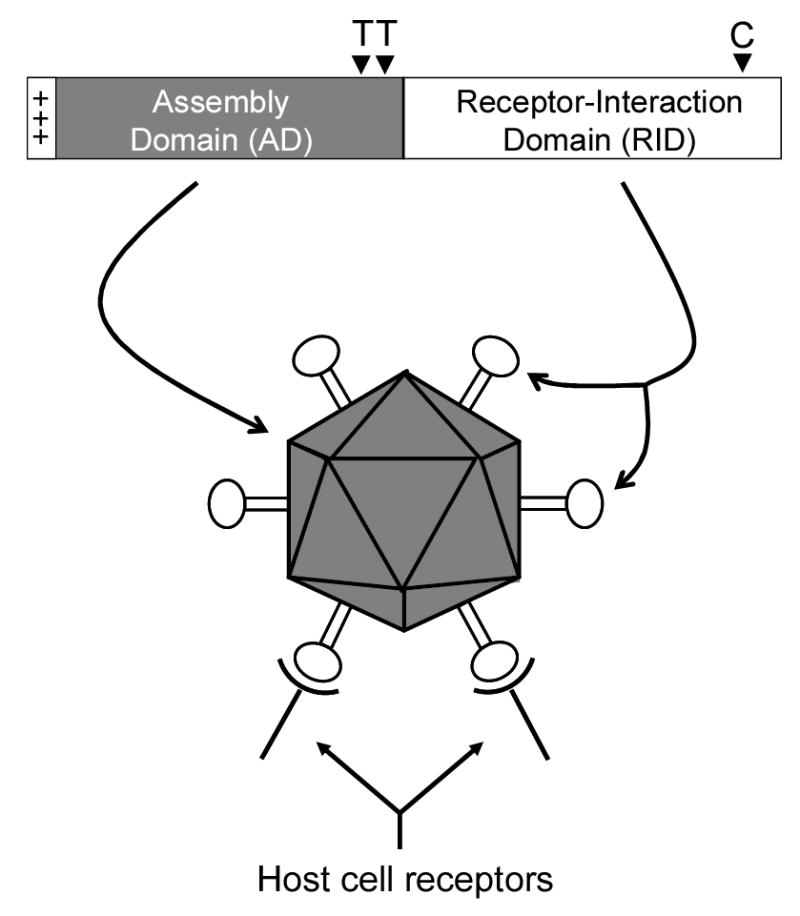
Proposed functional domains of the astrovirus coat protein. The coat protein is illustrated with the conserved region in the first half (heavily shaded) and the variable region in the last half (no shading) of the molecule. The positively charged N terminus of the coat protein proposed to be involved in viral RNA packaging is indicated (+++) along with the approximate location of trypsin cleavage sites (T▾) and putative caspase cleavage site(s) (C▾). Based on the structural predictions outlined in the text, we propose that the conserved region of the coat protein encodes the assembly domain (AD) required to form the viral capsid and encapsidate the viral RNA whereas the variable region encodes the receptor-interaction domain (RID) that binds to specific receptors on the surface of the host cell.
The next step will be to test whether functional predictions of regions of the coat protein required for virion assembly, particle maturation, and RNA encapsidation are exhibited in biological experiments, by characterizing the phenotypes of a series of coat protein mutations and deletions. It will also be of interest to test the modularity of the coat protein “assembly domain” by constructing chimeric coat protein molecules between different serotypes and species of the Astroviridae. The rationale for these studies is that insight into the molecular mechanisms of astrovirus assembly, nucleic acid packaging, maturation and tropism will inform us of unique and shared properties of astrovirus coat proteins. Experiments to address these questions are currently underway in the laboratory.
Acknowledgments
I thank Karin Bok, Lauren L. Keim and Dawn Marshall for review of the manuscript. The author acknowledges support from NIH grant AI 608734.
References
- 1.Baer ML, Houser F, Loesch-Fries LS, Gehrke L. Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 1994;13:727–735. doi: 10.1002/j.1460-2075.1994.tb06312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass DM, Qiu S. Proteolytic processing of the astrovirus capsid. J Virol. 2000;74:1810–1814. doi: 10.1128/jvi.74.4.1810-1814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass DM, Upadhyayula U. Characterization of human serotype 1 astrovirus-neutralizing epitopes. J Virol. 1997;71:8666–8671. doi: 10.1128/jvi.71.11.8666-8671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belloit G, Laveran H, Monroe SS. Capsid protein composition of reference strains and wild isolates of human astroviruses. Virus Res. 1997;49:49–57. doi: 10.1016/s0168-1702(97)01457-3. [DOI] [PubMed] [Google Scholar]
- 5.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal framshifting signal: requirement for a RNA psudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caballero S, Guix S, Ribes E, Bosch A, Pintó RM. Structural requirements of astrovirus virus-like particles assembled in insect cells. J Virol. 2004;78:13285–13292. doi: 10.1128/JVI.78.23.13285-13292.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Stauffacher C, Li Y, Schmidt T, Bomu W, Kamer G, Shanks M, Lomonossoff G, Johnson JE. Protein-RNA interactions in an icosahedral virus at 3.0 angstroms resolution. Science. 1989;245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 8.Clark B, McKendrick M. A review of viral gastroenteritis. Curr Opin Infect Dis. 2004;17:461–469. doi: 10.1097/00001432-200410000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Dalton RM, Pastrana EP, Sánchez-Fauquier A. Vaccinia virus recombinant expressing an 87-kilodalton polyprotein that is sufficient to form astrovirus-like particles. J Virol. 2003;77:9094–9098. doi: 10.1128/JVI.77.16.9094-9098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Englund L, Chriél M, Dietz HH, Hedlund KO. Astrovirus epidemiologically linked to pre-weaning diarrheoa in mink. Vet Microbiol. 2002;85:1–11. doi: 10.1016/s0378-1135(01)00472-2. [DOI] [PubMed] [Google Scholar]
- 11.Geigenmüller U, Chew T, Ginzton N, Matsui SM. Processing of nonstructural protein 1a of human astrovirus. J Virol. 2002;76:2003–2008. doi: 10.1128/JVI.76.4.2003-2008.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geigenmüller U, Ginzton NH, Matsui SM. Construction of a genome-length cDNA cone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J Virol. 1997;72:1713–1717. doi: 10.1128/jvi.71.2.1713-1717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geigenmüller U, Ginzton NH, Matsui SM. Studies on intracellular processing of the capsid protein of human astrovirus serotype 1 in infected cells. J Gen Virol. 2002;83:1691–1695. doi: 10.1099/0022-1317-83-7-1691. [DOI] [PubMed] [Google Scholar]
- 14.Geigenmüller-Gnirke U, Nitschko H, Schlesinger S. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J Virol. 1993;67:1620–1626. doi: 10.1128/jvi.67.3.1620-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz DH, Willie ST, Armen RS, Bratt T, Borregaard N, Strong RK. Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin. Biochemistry. 2000;39:1935–1941. doi: 10.1021/bi992215v. [DOI] [PubMed] [Google Scholar]
- 16.Gough RE, Collins MS, Borland E, Keymer LF. Astrovirus-like particles associated with hepatitis in ducklings. Vet Res. 1984;114:279. doi: 10.1136/vr.114.11.279-a. [DOI] [PubMed] [Google Scholar]
- 17.Guix S, Bosch A, Ribes E, Martínez LD, Pintó RM. Apoptosis in astrovirus-infected CaCo-2 cells. Virology. 319:249–261. doi: 10.1016/j.virol.2003.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, V, Bowman D, Mueller S, Bator CM, Bella J, Peng X, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci (USA) 2000;97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwitz, M.S. 2001. Adenoviruses, pp. 2301–2326. In: D.M. Knipe and P.M. Howley (eds.), Fields Virology, 4th ed. Lippincott-Ravan, Philadelphia.
- 20.Hoshino Y, Zimmer JF, Moise NS, Scott FW. Detection of astroviruses in feces of a cat with diarrhea. Arch Virol. 1981;70:373–376. doi: 10.1007/BF01320252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imada T, Yamaguchi S, Mase M, Tsukamoto K, Kubo M, Morooka A. Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA. J Virol. 2000;74:8487–8493. doi: 10.1128/jvi.74.18.8487-8493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang B, Monroe SS, Koonin EV, Steine SE, Glass RI. RNA sequence of astrovirus: Distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci (USA) 1993;90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonassen CM, Jonassen TO, Saif YM, Snodgrass DR, Ushijima H, Shimizu M, Grinde B. Comparison of capsid sequences from human and animal astroviruses. J Gen Virol. 2001;82:1061–1067. doi: 10.1099/0022-1317-82-5-1061. [DOI] [PubMed] [Google Scholar]
- 25.Kakizawa J, Ushijima H, Wen L, Ikeda Y, Oseto M. Genetic analysis of the capsid region of human astrovirus serotype 3 isolated in Japan. Microbiol Immunol. 1997;41:637–40. doi: 10.1111/j.1348-0421.1997.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 26.Kelly LA, MacCallum RM, Sternberg MJE. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 27.Kiang D, Matsui SM. Proteolytic processing of a human astrovirus nonstructural protein. J Gen Virol. 2002;83:25–34. doi: 10.1099/0022-1317-83-1-25. [DOI] [PubMed] [Google Scholar]
- 28.Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, Yoo OJ, Lee H, Lee JO. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat Struct Biol. 2003;10:342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- 29.Kjeldsberg E, Hern A. Detection of astroviruses in gut contents of nude and normal mice. Arch Virol. 1985;84:135–140. doi: 10.1007/BF01310560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koci MD, Seal BS, Schultz-Cherry S. Molecular characterization of an avian astrovirus. J Virol. 2000;74:6173–6177. doi: 10.1128/jvi.74.13.6173-6177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis TL, Greenberg HB, Hermann JE, Smith LS, Matsui SM. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J Virol. 1994;68:77–83. doi: 10.1128/jvi.68.1.77-83.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis TL, Matsui SM. An astrovirus frameshift signal induces ribosomal frameshifting in vitro. Arch Virol. 1995;140:1127–1135. doi: 10.1007/BF01315421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis TL, Matsui SM. Astrovirus frameshifting in an infection-transfection transient expression system. J Virol. 1996;70:2869–2875. doi: 10.1128/jvi.70.5.2869-2875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madeley CR, Cosgrove BP. Viruses in infantile gastroenteritis. Lancet. 1975;2:124. doi: 10.1016/s0140-6736(75)90020-3. [DOI] [PubMed] [Google Scholar]
- 35.Marczinke B, Bloys AJ, Brown TDK, Willcocks MM, Carter MJ, Brierley I. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J Virol. 1994;68:5588–5595. doi: 10.1128/jvi.68.9.5588-5595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall D, Schneemann A. Specific packaging of nodaviral RNA2 requires the N terminus of the capsid protein. Virology. 2001;285:165–175. doi: 10.1006/viro.2001.0951. [DOI] [PubMed] [Google Scholar]
- 37.Matsui, S.M. and H.B. Greenberg. 2001. Astroviruses. pp. 875–893. In: D.M. Knipe and P.M. Howley (eds.), Fields Virology, 4th ed. Lippincott-Ravan, Philadelphia.
- 38.Matsui SM, Kiang D, Ginzton N, Chew T, Geigenmüller-Gnirke U. Molecular biology of astroviruses: selected highlights. Novartis Found Symp. 2001;238:219–236. doi: 10.1002/0470846534.ch13. [DOI] [PubMed] [Google Scholar]
- 39.Mayo MA. Virology division news: ICTV at the Paris ICV: results of the plenary session and the binomial ballot. Arch Virol. 2002;147:2254–2260. [Google Scholar]
- 40.McNulty MS, Curran WL, McFerran JB. Detection of astrovirus in turkey faeces by direct electron microscopy. Vet Res. 1980;106:561. doi: 10.1136/vr.106.26.561. [DOI] [PubMed] [Google Scholar]
- 41.Méndez E, Fernández-Luna T, López S, Méndez-Toss M, Arias CF. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J Virol. 2002;76:7996–8002. doi: 10.1128/JVI.76.16.7996-8002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Méndez E, Salas-Ocampo E, Arias CF. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J Virol. 2004;78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Méndez E, Salas-Ocampo MPE, Munguía ME, Arias CF. Protein products of the open reading frames encoding nonstructural proteins of human astrovirus serotype 8. J Virol. 2003;77:11378–11384. doi: 10.1128/JVI.77.21.11378-11384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Méndez-Toss M, Romero-Guido P, Munguía ME, Méndez E, Arias CF. Molecular analysis of a serotype 8 human astrovirus genome. J Gen Virol. 2000;81:289–2897. doi: 10.1099/0022-1317-81-12-2891. [DOI] [PubMed] [Google Scholar]
- 45.Mittelholzer C, Hedlund KO, Englund L, Dietz HH, Svensson L. Molecular characterization of a novel astrovirus associated with disease in mink. J Gen Virol. 2003;84:3087–3094. doi: 10.1099/vir.0.19267-0. [DOI] [PubMed] [Google Scholar]
- 46.Monroe SS, Steine SE, Gorelkin L, Herrmann JE, Blacknow NR, Glass RI. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J Virol. 1991;65:641–648. doi: 10.1128/jvi.65.2.641-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgunova EYu, Dauter Z, Fry E, Stuart DI, Stel’mashchuk VYa, Mikhailov AM, Wilson KS, Vainshtein BK. The atomic structure of carnation mottle virus capsid protein. FEBS Lett. 1994;338:267–271. doi: 10.1016/0014-5793(94)80281-5. [DOI] [PubMed] [Google Scholar]
- 48.Neill JD, Mengeling WL. Further characterization of the virus-specific RNAs in feline calicivirus infected cells. Virus Res. 1988;11:59–72. doi: 10.1016/0168-1702(88)90067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao AL, Grantham GL. Molecular studies of bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement and pathology. Virology. 1996;201:312–320. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 50.Risco C, Carrascosa JL, Pedregosa AM, Humphrey CD, Sánchez-Fauquier A. Ultrastructure of human astrovirus serotype 2. J Gen Virol. 1995;76:2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 51.Sánchez-Fauquier, Carrascosa AL, Carrascosa JL, Otero A, Glass RI, Lopez JA, San Martin C, Melero JA. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology. 1994;201:312–320. doi: 10.1006/viro.1994.1296. [DOI] [PubMed] [Google Scholar]
- 52.Schlesinger, S. and M.J. Schlesinger. 1990. Replication of togaviridae and flaviviridae, pp. 697–711. In: B.M. Fields, D.M. Knipe, et al. (eds.), Virology. Raven Press, New York.
- 53.Schmitz I, Rao AL. Deletions in the conserved amino-terminal basic arm of cucumber mosaic virus coat protein disrupt virion assembly but do not abolish infectivity and cell-to-cell movement. Virology. 1998;248:323–331. doi: 10.1006/viro.1998.9257. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu M, Shirai J, Narita M, Yamane T. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J Clin Microbiol. 1990;28:201–206. doi: 10.1128/jcm.28.2.201-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snodgrass DR, Angus KW, Gray EW, Menzies JD, Paul G. Pathogenesis of diarrhoea caused by astrovirus infectious in lambs. Arch Virol. 1979;60:217–226. doi: 10.1007/BF01317493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snodgrass DR, Gray EW. Detection and transmission of 30 nm virus particles (astroviruses) in faeces of lambs with diarrheoea. Arch Virol. 1977;55:287–291. doi: 10.1007/BF01315050. [DOI] [PubMed] [Google Scholar]
- 57.Stehle T, Dermody TS. Structural similarities in the cellular receptors used by adenovirus and reovirus. Viral Immunol. 2004;17:129–143. doi: 10.1089/0882824041310621. [DOI] [PubMed] [Google Scholar]
- 58.Tang L, Lin CS, Krishna NK, Yeager M, Schneemann A, Johnson JE. Virus-like particles of a fish nodavirus display a capsid subunit domain organization different from that of insect nodaviruses. J Virol. 2002;76:6370–6375. doi: 10.1128/JVI.76.12.6370-6375.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzipori S, Menzies JD, Gray EW. Detection of astroviruses in the faeces of red deer. Vet Rec. 1981;108:286. doi: 10.1136/vr.108.13.286. [DOI] [PubMed] [Google Scholar]
- 60.Varghese JN, Moritz RL, Lou MZ, van Donkelaar A, Ji H, Ivancic N, Branson KM, Hall NE, Simpson RJ. Structure of the extracellular domains of the human interleukin-6 receptor α-chain. Proc Natl Acad Sci (USA) 2002;99:15959–15964. doi: 10.1073/pnas.232432399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter JE, Mitchell DK. Astrovirus infection in children. Curr Opin Infect Dis. 2003;16:247–253. doi: 10.1097/00001432-200306000-00011. [DOI] [PubMed] [Google Scholar]
- 62.Wang QH, Kakizawa J, Wen LY, Shimizu M, Nishio O, Fang ZY, Ushijima H. Genetic analysis of the capsid region of astroviruses. J Med Virol. 2001;64:245–255. doi: 10.1002/jmv.1043. [DOI] [PubMed] [Google Scholar]
- 63.Willcocks MM, Boxall AS, Carter MJ. Processing and intracellular location of human astrovirus non-structural proteins. J Gen Virol. 1999;80:2607–2611. doi: 10.1099/0022-1317-80-10-2607. [DOI] [PubMed] [Google Scholar]
- 64.Willcocks MM, Brown TDK, Madeley CR, Carter MJ. The complete sequence of a human astrovirus. J Gen Virol. 1994;75:1785–1788. doi: 10.1099/0022-1317-75-7-1785. [DOI] [PubMed] [Google Scholar]
- 65.Willcocks MM, Carter MJ. Identification and sequence determination of the capsid protein gene of human astrovirus serotype 1. FEMS Microbiol Lett. 1993;114:1–8. doi: 10.1111/j.1574-6968.1993.tb06542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willcocks MM, Kurtz JB, Lee TW, Carter MJ. Prevalence of human astrovirus 4: capsid protein sequence and comparison with other strains. Epidemiol Infect. 1995;114:385–391. doi: 10.1017/s0950268800058015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams FP., Jr Astrovirus-like, coronavirus-like, and parvovirus-like particles detected in the diarrheal stools of beagle pups. Arch Virol. 1980;66:215–226. doi: 10.1007/BF01314735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woode GN, Bridger JC. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J Med Microbiol. 1978;11:441–452. doi: 10.1099/00222615-11-4-441. [DOI] [PubMed] [Google Scholar]
- 69.Yu M, Tang Y, Guo M, Zhang Q, Saif YM. Characterization of a small round virus associated with the poult enteritis and mortality syndrome. Avian Dis. 2000;44:600–610. [PubMed] [Google Scholar]
- 70.Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


