Abstract
Systematic genome-wide scans to date have shown that genes of major effect are not common causes of schizophrenia, but independent linkage studies looking for schizophrenia susceptibility genes are converging on a number of key chromosomal locations. Microarray expression analysis may identify new candidate genes and pathways, and a number of intriguing preliminary findings have already been reported.
Schizophrenia is a severe and debilitating mental disorder characterized by profound disturbances of cognition, emotion and social functioning. The risk of developing it in a lifetime (the lifetime morbid risk) is surprisingly uniform, at slightly less than 1%, across different populations and different cultures. Family, twin and adoption studies have revealed a substantial genetic contribution to schizophrenia; estimates of heritability are all greater than 60%, and siblings of schizophrenic people are ten times more likely than others to be schizophrenic (that is, the sibling recurrence risk ratio, λ s, is approximately 10). At present, however, the number of susceptibility loci, the disease risk conferred by each locus and the degree of interaction between them all remain unknown (reviewed in [1]). Estimates based on the recurrence risks in the relatives of individuals with schizophrenia are incompatible with the existence of a single locus of λ s = 3 (that is, a locus that increases the risk in siblings threefold), but more likely to involve a number of loci of λ s < 2 [2]. Despite decades of research, our understanding of the pathophysiology of schizophrenia is little more than rudimentary, and the evidence for genetic risk factors remains our strongest clue to its cause. There is therefore a compelling case for studies aimed at identifying disease susceptibility genes.
Systematic genome-wide linkage scans
To date, the majority of systematic genome-wide scans for schizophrenia-linked genes have focused on large pedigrees containing multiple affected individuals. This approach has been successful in identifying highly penetrant disease alleles for common disorders such as breast cancer, non-insulin-dependent diabetes and Alzheimer's disease [3,4,5]. Unfortunately it seems that families that clearly have segregating schizophrenia alleles with large effects are, at best, extremely rare and quite possibly nonexistent. But several loci showing some linkage have been reported (Figure 1). These include chromosome 22q11-12, 6p24-22, 8p22-21 and 6q [6,7,8]. There are also a number of other promising areas of putative linkage. These include 13q14.1-q32 [9,10,11,12], 5q21-q33 [12,13,14,15], 10p15-p11 [13,16,17] and 15q14 [18,19,20,21]. One other region that is notable for the strength of evidence (maximum heterogeneity LOD score of 6.5) from a single study, rather than the more important convergence of evidence from different sources is 1q21-q22 [11]. The finding on 1q21-q22 more than meets standard criteria for claiming 'significant' linkage. Early experience in psychiatric genetics has, however, impressed the need for replication firmly upon the minds of most researchers, and although there have been other reports of linkage to markers on chromosome 1 [15,22,23], these are distal to 1q21-q22 and it is not yet clear that these findings are consistent with the existence of a single susceptibility locus on 1q. Moreover, a recent report at the 2001 World Congress on Psychiatric Genetics in St Louis described the analysis of 16 microsatellite markers spanning 107 centiMorgans (cM) of 1q21-22, which were genotyped through a combined sample of 779 schizophrenia families from an international linkage consortium [24]. No linkage to schizophrenia was identified in any of the eight different independent samples forming the consortium, suggesting that while any schizophrenia susceptibility genes present in this region may indeed carry a large genetic contribution in the reported linked families [11,15,22,23], they are likely to have only a small genetic effect in the overall population.
Figure 1.
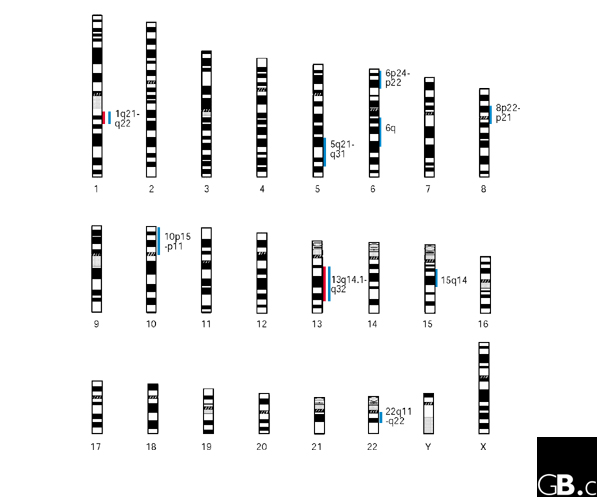
Human chromosomes showing the major chromosomal regions that have been implicated in schizophrenia by linkage studies. Blue lines indicate regions for which evidence of linkage has been found in more than one data set; red lines indicate regions for which evidence of linkage has been found with statistical significance.
As an alternative to linkage based upon extended families with several affected individuals, some groups have advocated the use of smaller families with two or more affected siblings. Such families may be more suited for the analysis of complex traits because their analysis uses information from only affected family members; also, being much more common, they may also be more representative of the occurrence of the disease in the general population. We and colleagues [25] have recently reported the largest systematic search for linkage yet published, using a sample of 196 affected sibling pairs. This study was designed to have a power of over 0.95 to detect a susceptibility locus of λ s = 3 (the maximum effect size estimated by Risch [2]). None of the findings approached statistical significance, however. We were able to exclude susceptibility genes with an effect of λ s = 3 and λ s = 2 from 82.8% and 48.7% of the genome, respectively, but genes with an effect size of λ s = 1.5 could be excluded from only 9.3% of the genome.
At present, therefore, the linkage literature supports predictions from genetic epidemiological studies that there are unlikely to be commonly occurring schizophrenia-susceptibility loci of effect size λ s > 3. There is suggestive evidence implicating a number of regions, however, which is consistent with the existence of some susceptibility alleles of moderate effect (λ s 1.5-3) and possibly also of uncommon loci of larger effect that can be identified in specific large families with multiple affected members.
To some, the results of linkage studies have seemed to be disappointing, as individual studies suggesting linkage have often not provided compelling evidence and failures to replicate proposed linkages are common. Similar difficulties are being faced for other common diseases, however [26,27,28]. Also, we should expect true positive linkages to be difficult to replicate when predisposition to a disorder involves the combined action of several genes of moderate effect [29]. Indeed, excluding Mendelian subtypes of disorders, the only successful examples of positional cloning of a susceptibility gene for a complex disorder are the recent identification of NOD2 and Calpain 10 as susceptibility loci for Crohn's disease and type II diabetes, respectively [30,31,32].
The difficulties of detecting and replicating linkage for complex disorders leave us with a major problem: how can results be generated that are sufficiently robust to justify the expense of large-scale efforts needed for identifying disease alleles? One way of resolving this issue will be to study much larger samples, say of 600-800 nuclear families, which should be sufficient to detect susceptibility genes of moderate effect size, and studies of this sort should now become a priority. Of course, the proof that a positive linkage is correct comes when the disease gene is identified, and a number of groups are already attempting to identify disease genes in candidate regions. These efforts should be encouraged by findings on Crohn's disease and type II diabetes [30,31,32] as well as by converging evidence for linkage to several regions.
Selection of candidate genes
If genetic susceptibility to schizophrenia is entirely due to the operation of many genes of small effect, then even large-scale linkage studies will have difficulty identifying such loci. As an alternative, candidate-gene association studies offer a potentially powerful approach to identifying susceptibility loci of small effect. A gene is typically defined as a candidate if it is located within a chromosomal region previously implicated by positional cloning and/or it is of functional relevance to a neurobiological hypothesis for schizophrenia. As there is moderate but inconclusive evidence for many different chromosomal regions, each of which tends to be large (typically over 50 cM) and poorly demarcated, the number of potential candidate genes is extremely large.
The situation with functionally defined candidate genes is equally difficult. There are many neurobiological theories of schizophrenia, dominated for many years by neuropharmacology, which has implicated monoamine neurotransmission, in particular the dopaminergic and serotonergic systems. It is unclear, however, to what extent any neurochemical findings reflect primary rather than secondary pathology, compensatory mechanisms or environmental influences, and there are alternative neurochemical and neuropharmacological hypotheses. Consequently, it is difficult to use the neuropharmacological literature to define with any degree of conviction a group of candidate genes that it is both highly likely to include a true susceptibility gene and also small enough to be comprehensively analyzed using current laboratory techniques. Other hypotheses of schizophrenia meet the same numerical obstacle: for example, the view that schizophrenia is a disorder of neurodevelopment (reviewed by Weinberger [33]), which is supported by some neuropathological studies [34]. The regulation of the development and maturation of the brain is extremely complex and poorly understood, but it certainly involves the dynamic interplay of a large number of genetic, environmental and stochastic factors. Thus, despite some broad clues, the precise biological mechanisms underlying putative neurodevelopmental disturbances remain largely speculative, which makes it difficult to limit rational selection of candidate genes and pathways using the neurodevelopmental hypothesis.
Microarray expression analysis
High-throughput microarray platforms are capable of simultaneously measuring the expression of several thousands of genes; this gives us the potential to identify genes that are consistently dysregulated in tissues from subjects with any given disease. Although such methods have huge potential, their limitations are most apparent when they are applied to analysis of the human brain. These limitations include variation in expression as a result of polymorphisms in genes that are not related to the disease, variation in the environment, variation as a consequence of the phenotype (for example, self-neglect, drugs or nutrition), variation in mRNA quality (as a result, for example, of post mortem delay or the condition of the patient immediately before death), variation in cause of death, and variation in tissue preparation. Nevertheless, highly parallel expression studies do offer a complementary approach to linkage studies in that they are genome-wide, have the potential to reveal novel molecular mechanisms that contribute to disease, and can provide an alternative source of candidate genes derived without regard to our preconceptions of disease mechanisms.
To date, two independent studies have used microarray mRNA expression profiling to assess expression levels in schizophrenic people relative to controls. Both focused on the dorsolateral prefrontal cortex (DLPC), an area of the brain that has been implicated in cognitive disturbances in schizophrenia. Hakak et al. [35] assayed the expression levels of over 6,000 genes in the DPLC in 12 schizophrenics and matched controls. Hakak et al. [35] also applied a method of hierarchical clustering to the expression data in order to identify groups of genes that have similar expression profiles and found 89 genes that were differentially expressed; these are involved in a wide range of biological functions, including synaptic plasticity, neuronal development, neurotransmission and signal transduction. The most striking finding was the transcriptional downregulation in schizophrenic patients of six different genes (those encoding T-cell differentiation protein, 2',3'-cyclic nucleotide 3' phosphodiesterase, myelin-associated glycoprotein, transferrin, gelsolin and the epidermal growth factor receptor ErbB-3), all of which have been reported to be strongly expressed in oligodendrocytes and implicated in the formation of myelin sheaths. Oligodendrocytes are known to increase the rate of conduction through neurons by sheathing them with myelin and they also provide extrinsic trophic factors that promote neuronal maturation and survival of axons. The downregulation of these six genes in schizophrenia led the authors [35] to propose that some of the brain abnormalities in schizophrenia that have been reported by neuropathology and neuro-imaging studies may be a consequence of abnormal myelination and/or oligodendrocyte function.
The study by Mirnics et al. [36] examined the expression of over 7,800 genes in the DLPC in six schizophrenic patients and six controls. They reported two main findings. First, all patients studied had alterations in the expression of a number of genes related to presynaptic secretory function. The combination of such genes with reduced expression was different in each schizophrenic person studied, implying that the gene expression pattern is more complex than the more uniform reduced expression of the myelin-related genes reported by Hakak et al. [35]. Second, Mirnics et al. [36] identified reduced expression of the gene encoding the regulator of G-protein signaling 4 (RGS4) [37], one of a group of proteins that modulate the duration of response of G-protein coupled receptors after binding to their specific ligands. There was no overall change in the expression of the other genes (over 200) studied from the G-protein-signaling group, however.
At first glance, it may appear the two microarray studies are inconsistent. Like Mirnics et al. [36], however, Hakak et al. [35] also identified changes in the expression levels of some genes involved in presynaptic secretory function. Also, the two studies used different types of microarray, with a very imperfect overlap between the genes that were represented. For example, the array used by Mirnics et al. [36,37] contained probes for only one of the six myelination genes identified by Hakak et al. [35]. Conversely, the majority of the most significantly affected presynaptic function genes identified by Mirnics et al. [36,37] were not represented on the microarray platform used by the other group [35]. We will therefore know how much weight we can place on these findings only when they are replicated in independent samples using the same microarray platforms. Although this does not prove the myelination/oligodendrocyte hypothesis, it is at least very encouraging that the data of Hakak's group can be replicated.
In conclusion, positive signals from independent linkage studies are beginning to converge on similar chromosomal locations, so we can be optimistic concerning that some genes of moderate effect will be found in some of these regions. These regions are large, however, and contain large numbers of candidate genes. It is possible that new genes and pathways might be implicated by emerging transcriptomic approaches. Some of the gene expression changes identified in this way may be a result of environmental factors, chance, or other confounding variables associated with post mortem studies of brains. Nevertheless, combining the two hypothesis-independent approaches - linkage and gene expression profiling - may simplify the process of moving from putative linkage to gene identification. For example, RGS4, which was implicated in schizophrenia by expression studies [36], maps to 1q21-22, a region that has been implicated in schizophrenia by linkage [11]; it is therefore an outstanding candidate gene for schizophrenia. But just as chance dictates that every large linkage region contains at least one apparently 'good' candidate gene on the basis of function that in fact turns out to be a false positive, so, given the number of putative linkages for schizophrenia and their combined physical size, similar coincidences will no doubt apply to candidate genes found on the basis of their altered expression.
References
- McGuffin P, Owen MJ, O'Donovan MC, Thapar A, Gottesman II. Seminars in Psychiatric Genetics London: Gaskell; 1994.
- Risch N. Linkage strategies for genetically complex traits. 2. The power of affected relative pairs. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- Hall J, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell Gi, Zouali H, Lesage S, Velho G, Iris F, Passa P, et al. Nonsense mutation in the glucokinase gene causes early-onset non-insulin-dependent diabetes-mellitus. Nature. 1992;356:721–722. doi: 10.1038/356721a0. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimers-disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6, and 8 Additional support for schizophrenia linkage on chromosomes 6 and 8: a multicenter study. Am J Med Genet. 1996;67:580–594. doi: 10.1002/(SICI)1096-8628(19961122)67:6<580::AID-AJMG11>3.3.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Gill M, Vallada H, Collier D, Sham P, Holmans P, Murray R, McGuffin P, Nanko S, Owen M, Antonarakis S, et al. A combined analysis of D22s278 marker alleles in affected sib-pairs - support for a susceptibility locus for schizophrenia at chromosome 22q12. Am J Med Genet. 1996;67:40–45. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.3.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Holmans P, Straub RE, Owen MJ, Wildenauer DB, Gejman PV, Pulver AE, Laurent C, Kendler KS, Walsh D, et al. Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am J Hum Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MW, Curtis D, Williams N, Arranz M, Nanko S, Collier D, McGuffin P, Murray R, Owen M, Gill M, Powell J. Suggestive evidence for linkage of schizophrenia to markers on chromosome 13q14.1-q32. Psychiatr Genet. 1995;5:117–126. doi: 10.1097/00041444-199505030-00004. [DOI] [PubMed] [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lassetter VK, Wolniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kascg L, et al. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EWC, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver DL, Holcomb J, Mapua FM, Wilson R, Barnes B. Schizophrenia spectrum disorders: an autosomal-wide scan in multiplex pedigrees. Schizophr Res. 2001;52:145–160. doi: 10.1016/s0920-9964(01)00157-8. [DOI] [PubMed] [Google Scholar]
- Schwab SG, Hallmayer J, Albus M, Lerer B, Eckstein GN, Borrmann M, Segman RH, Hanses C, Freymann J, Yakir A, et al. A genome-wide autosomal screen for schizophrenia susceptibility loci in 71 families with affected siblings: support for loci on chromosome 10p and 6. Mol Psychiatry. 2000;5:638–649. doi: 10.1038/sj/mp/4000791. [DOI] [PubMed] [Google Scholar]
- Straub RE, Maclean CJ, Oneill FA, Walsh D, Kendler KS. Support for a possible schizophrenia vulnerability locus in region 5q22-31 in Irish families. Mol Psychiatry. 1997;2:148–155. doi: 10.1038/sj/mp/4000258. [DOI] [PubMed] [Google Scholar]
- Gurling HMD, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, et al. Genome-wide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, Zambuto CT, Schmitt K, Meyer J, et al. Genome scan of European-American schizophrenia pedigrees: results of the NIMH genetics initiative and millennium consortium. Am J Med Genet. 1998;81:290–295. doi: 10.1002/(SICI)1096-8628(19980710)81:4<290::AID-AJMG3>3.3.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Straub RE, Maclean CJ, Martin RB, Ma YL, Myakishev MV, Harris-Kerr C, Webb BT, O'Neill FA, Walsh D, Kendler KS. A schizophrenia locus may be located in region 10p15-p11. Am J Med Genet. 1998;81:296–301. doi: 10.1002/(SICI)1096-8628(19980710)81:4<296::AID-AJMG4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Leonard S, Gault J, Moore T, Hopkins J, Robinson M, Olincy A, Adler LE, Cloninger CR, Kaufmann CA, Tsuang MT, et al. Further investigation of a chromosome 15 locus in schizophrenia: Analysis of affected sibpairs from the NIMH Genetics Initiative. Am J Med Genet. 1998;81:308–312. doi: 10.1002/(SICI)1096-8628(19980710)81:4<308::AID-AJMG6>3.3.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, Zambuto CT, Schmitt K, Matise TC, et al. NIMH Genetics Initiative Millennium Schizophrenia Consortium: Linkage analysis of African-American pedigrees. Am J Med Genet. 1998;81:282–289. doi: 10.1002/(SICI)1096-8628(19980710)81:4<282::AID-AJMG2>3.3.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Riley BP, Makoff A, Mogudi-Carter M, Jenkins T, Williamson R, Collier D, Murray R. Haplotype transmission disequilibrium and evidence for linkage of the CHRNA7 gene region to schizophrenia in Southern African Bantu families. Am J Med Genet. 2000;96:196–201. doi: 10.1002/(SICI)1096-8628(20000403)96:2<196::AID-AJMG15>3.3.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Liu CM, Hwu HG, Lin MW, Ou-Yang WC, Lee SFC, Fann CSJ, Wong SH, Hsieh SH. Suggestive evidence for linkage of schizophrenia to markers at chromosome 15q13-14 in Taiwanese families. Am J Med Genet. 2001;105:658–661. doi: 10.1002/ajmg.1547. [DOI] [PubMed] [Google Scholar]
- Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, Juvonen H, Varilo T, Arajärvi R, Kokko-Sahin ML, et al. Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet. 2000;9:1049–1057. doi: 10.1093/hmg/9.7.1049. [DOI] [PubMed] [Google Scholar]
- Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajärvi R, Juvonen H, Kokko-Sahin MJ, Väisänen L, Mannila H, et al. Genome-wide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet. 1999;65:1114–1125. doi: 10.1086/302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Holmans PA. Multicenter linkage study of schizophrenia candidate regions on chromosomes 1q, 15q, 18p and 22q. Am J Med Genet. 2001;105:562. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Rees MI, Holmans P, Norton N, Cardno AG, Jones LA, Murphy KC, Sanders RD, McCarthy G, Gray MY, et al. A two-stage genome scan for schizophrenia susceptibility genes in 196 affected sibling pairs. Hum Mol Genet. 1999;8:1729–1739. doi: 10.1093/hmg/8.9.1729. [DOI] [PubMed] [Google Scholar]
- Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O'Rahilly S, Frayling TM, Bell JI, Lathrop GM, et al. A genome-wide scan for loci predisposing to type 2 diabetes in a U.K. population (the diabetes UK Warren 2 repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet. 2001;69:553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake DS, O'Connor DT, Parmer RJ. Molecular genetics of essential hypertension: recent results and emerging strategies. Curr Opin Nephrol Hypertens. 2001;10:71–79. doi: 10.1097/00041552-200101000-00012. [DOI] [PubMed] [Google Scholar]
- Cookson WO, Moffatt MF. Genetics of asthma and allergic disease. Hum Mol Genet. 2000;9:2359–2364. doi: 10.1093/hmg/9.16.2359. [DOI] [PubMed] [Google Scholar]
- Suarez BK, Hampe CL, Van Eerdewegh P. Problems of replicating linkage claims in psychiatry. In Genetic approaches to mental disorders Edited by Gershon ES, Cloninger CR Washington, DC: American Psychiatric; 1994. pp. 23–46.
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li XQ, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PEH, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. From neuropathology to neurodevelopment. Lancet. 1995;346:552–557. doi: 10.1016/s0140-6736(95)91386-6. [DOI] [PubMed] [Google Scholar]
- Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterisation of schizophrenia viewed by microarray analysis of the prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj/mp/4000866. [DOI] [PubMed] [Google Scholar]


