The relevance of a particular signaling pathway or a protein family correlates with the number of functions associated with it and their physiological and pathophysiological implications. In the case of the nitric oxide (NO) signaling pathway and the NO synthases (NOS1–3), their relevance became also apparent in recent years from the complexity of their regulation. Not only are NOS isozymes “cofactor-accumulating” enzymes but also are associated with a jungle of more than 20 interacting proteins (Table 1). These affect both the activity and spatial organization of NO synthesis within the cell, and some of these interactions may even have functions beyond NO formation.
Table 1.
NOS-interacting proteins
| Protein
|
NOS isoform
|
Binding site on | Other complex members
|
Function
|
Refs.
|
|
|---|---|---|---|---|---|---|
| Binding protein | NOS | |||||
| CaM | 1–3 | C- and N-terminal domains | CaM-binding domain | Activation | 1 | |
| Caveolin-1 | 3 | aa 61–101; 135–178 | aa 350–358 | Inhibition | 5, 37 | |
| Caveolin-3 | 1–3 | aa 65–84; 135–178 | N terminus | Inhibition | 4, 38 | |
| Bradykinin B2 and angiotensin AT1 receptor | 3 | ID 4 | Inhibition | 6 | ||
| CAT1 | 3 | Substrate supply | 8 | |||
| Dynamin-2 | 3 | Activation | 22 | |||
| Porin | 3 | Activation | 7 | |||
| Protein kinase B/Akt | 3 | ? | ? | Hsp90 | Activation | 9, 10 |
| HSP90 | 1,3 | aa 442–600 | N terminus (aa 300–400) | sGCβ1 | Allosteric; scaffold, coupling for Akt | 16, 17; R. J. Venema and J. D. Catravas, personal communication |
| NOSTRIN | 3 | SH3 | aa 98–366 | i.c. trafficking | 2 | |
| NOSIP | 1,3 | aa 366–486 | Inhibition | 3 | ||
| PDZ domain proteins | ||||||
| α1-Syntrophin | 1 | PDZ | N-terminal PDZ | 23, 25 | ||
| CAPON | 1 | C terminus | N-terminal PDZ | Dexras | Competition with PSD | 26, 27 |
| PSD-95 | PDZ | N-terminal PDZ | SAP90; sGCα2 | Coupling to NMDA receptor | 23, 30, 39 | |
| PSD-93 | 1 | PDZ | N-terminal PDZ | Coupling to NMDA receptor | 40 | |
| Phosphofructokinase M (PFK-M) | 1 | PDZ | 41 | |||
| EBP50 | 2 | PDZ | C terminus | 33 | ||
| PIN | 1 | β-strands of dimer | aa 228–244 | Inhibition | 28, 42–45 | |
| Rac2 | 2 | Oxygenase domain | Activation | 34 | ||
| Kalirin | 2 | aa 570–753 | N-terminal 70 aa | Prevents dimer formation | 36 | |
| NAP110 | 2 | N-terminal 70 aa | Prevents dimer formation | 35 | ||
Although the mechanisms regulating overall NOS activity by increases in Ca2+ or phosphorylation or by expression are quite well understood, subcellular regulation of NO formation by binding proteins with respect to time and place is much more complex. The first NOS-binding protein discovered was calmodulin (CaM) (1). It is required for electron flux and thus NO production by all three NOS isoforms. Other aspects of NOS regulation such as membrane attachment and activity modulation are also mediated by (often isoform-specific) protein-interaction networks. Moreover, trafficking of NOS within the cell is an important issue, because these interactions are not static. Zimmermann et al. (2) report in this issue of PNAS the identification of a previously uncharacterized player in this important aspect of NOS transport, NOSTRIN (NOS3 traffic inducer), which regulates the relocation of NOS3 in different cell types. Together with the recently identified other NOS3-binding protein, NOS3 interacting protein (NOSIP) (3), NOSTRIN is a “hot” candidate for regulating the intracellular trafficking of NOS3 and perhaps other proteins in the plasma membrane.
NOS3 Activation
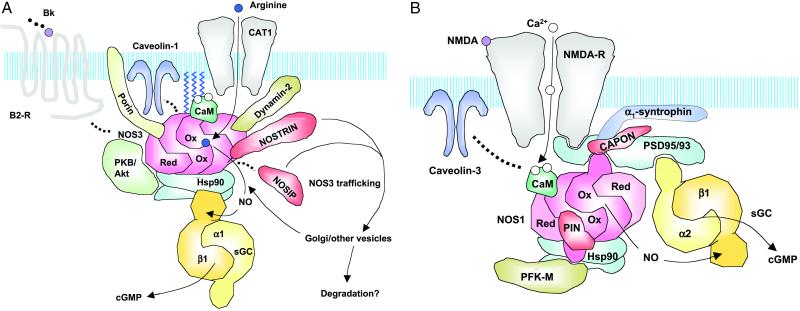
Protein–protein interactions are crucial and best studied for the regulation of NOS3 activity. Docking of NOS3 to the plasma membrane in complex with caveolin-1 [caveolin-3 in cardiomyocytes (4)] keeps NOS3 in an inactive yet activatable state (5). Similarly, direct interaction with the intracellular domain 4 of the bradykinin B2 receptor inhibits NOS3 (6). After activation of endothelial cells with bradykinin or Ca2+ ionophore, NOS3 dissociates from its complexes and becomes active (see Fig. 1A). Stimulation of endothelial cells with bradykinin or Ca2+ ionophore also promotes the interaction between NOS3 and the voltage-dependent anion/cation channel (Porin) to further augment NO production (7). Activation of NOS3 after stimulation with vascular endothelial growth factor also results in a dynamic reorganization of the protein complexes between NOS3 and its binding proteins. The “early” Ca2+-dependent activation of NOS3 is followed by a “late” phosphorylation-dependent activation by protein kinase B/Akt (9, 10) and protein kinase A (11). Heat-shock protein 90 (Hsp90), which binds both NOS3 (12, 13) and the neuronal isoform NOS1 (14), facilitates CaM-dependent disruption of NOS3 binding with caveolin (15) and mediates the interaction between NOS3 and protein kinase B/Akt (16, 17). Efficient supply with substrate during all this is ensured by localization of the arginine transporter cationic amino acid transporter (CAT1) in caveolae and its direct interaction with NOS3 (8).
Fig 1.
NOS-associated proteins forming signaling clusters and regulating NOS trafficking. The presentation illustrates possible interactions of NOS and NOS-binding proteins and does not represent the composition of actual complexes. (A) NOS3 and NOS3-binding proteins including NOSTRIN. (B) NOS1 and NOS1-binding proteins. For details, see text.
NOS3 Traffic
To ensure that all these events can occur, NOS3 needs to be in the right place at the right time. In confluent endothelial cells, NOS3 localizes to the Golgi region and plasmalemmal caveolae where it is activated (18). Further co- and posttranslational fatty acid modifications are required (19–21). The role of protein–protein interactions in this process is more elusive. A great number of NOS3-interacting proteins within caveolae can be important factors determining the localization of NOS3 in this compartment in addition to regulating activity. Other proteins such as dynamin-2 (22) or NOSIP (3) may be involved in the localization of the enzyme to the Golgi apparatus and other intracellular compartments (Fig. 1A).
Redistribution of NOS3 is one of the events following its activation. However, demonstrations that NO production is a more rapid process than the enzyme relocation suggest that intracellular NOS3 traffic plays a role in terminating rather than initiating NO release. Zimmermann et al. (2) demonstrate now that overexpression of NOSTRIN in Chinese hamster ovary cells results in a profound relocation of NOS3 from plasma and Golgi membranes to vesicle-like structures spread over the cytosol. Inhibition of NO production in NOSTRIN-overexpressing cells may be the consequence of this redistribution of NOS3. It is now of great interest whether NOSTRIN, which is expressed in intact endothelial cells, participates in NOS3 traffic in these cells.
NOS1
The neuronal isoform of NOS, NOS1, is also under tight control by binding proteins with several parallels to NOS3. Here, PDZ domain-containing proteins play a central role in forming multiprotein signaling complexes. In neuronal cells, the N terminus of NOS1 binds via PDZ–PDZ interaction to postsynaptic density proteins, PSD95 or PSD93, which further couple NOS1 to N-methyl-D-aspartate (NMDA) receptors (23, 24). In case of skeletal muscle cells, NOS1 is attached to sarcolemma via the dystrophin-binding and PDZ protein, syntrophin (23, 25). Although in the latter case, the function of the membrane attachment of NOS1 remains to be clarified, proximity of NOS1 to the calcium channel of NMDA receptors in neuronal cells may increase the efficacy of NOS1 stimulation in response to glutamate-induced influx of Ca2+ (Fig. 1B). Moreover, a postsynaptic localization of NOS1 simplifies the participation of the putative retrograde messenger, NO, in the regulation of synaptic plasticity. Proteins competing with PSD proteins for the PDZ domain of NOS1 are supposed to be negative regulators of NO production in NOS1-expressing cells. One such protein is C-terminal PDZ ligand of NOS1 (CAPON) (26), which is expressed in neuronal cells, interacts with the NOS1 PDZ domain, and disrupts PSD-95–NOS1 complexes when overexpressed. An alternative role of CAPON may be the coupling of NOS1 to NO targets, as has been demonstrated for Dexras-1, an NO-activated member of the Ras family (27).
In addition to activity and localization, also the stability of NOS1 homodimers can be affected by protein–protein interactions. A recently described protein inhibitor of neuronal NOS1, PIN, binds to the N terminus of NOS1 and destabilizes the NOS1 homodimers (28); a similar mechanism has been described for NOS2 (see below).
Soluble Guanylyl Cyclase (sGC) Coupling
Besides the aspect of regulating NOS activity and its localization and trafficking within the cell, NOS–protein interactions have gained an additional interesting aspect recently: the spatial coupling of NO synthesis to its ubiquitous receptor enzyme, sGC. Despite its name attribute “soluble,” sGC may attach to some extent to cellular membranes involving proteins that also bind to NOS (29). One such complex seems to involve NOS1 and specifically the sGCα2/β1 heterodimer. The C terminus of the α2 subunit binds to the third PDZ domain of PSD-95 (30), the same protein that integrates with its second and first PDZ domain, neuronal NOS1 and the NMDA receptor, respectively. This may allow for tight colocalization of sGCα2/β1 and NOS1 in postsynaptic densities (see Fig. 1B).
The second case involves endothelial cells, where both NOS3 and sGC were found in caveolae (29). However, the mechanism of this colocalization remains to be established. Hsp90 may play an essential role in this scenario, because it has been found to interact with both NOS3 and sGC (R. J. Venema and J. D. Catravas, personal communication). Hsp90, just like PSD-95 for NOS1 and sGCα2/β1, serves as a linker between NOS3 and sGCβ1 (see Fig. 1A). In both cases, NO would not need to travel through a cell to reach its cytosolic receptor sGC, but NO–cGMP signaling would be confined to a protein complex, presumably at or near the cell membrane. Besides such intracellular NO effects, the paracrine mode of signaling between NO generator and effector cells is well established for blood vessels and retrograde synapses. In the latter case, sGC localizes close to the presynaptic membrane (31). These findings may modify our understanding of NO signaling, from NO clouds and diffusion gradients penetrating several cell layers to a much more closely organized and spatially confined puff of NO at or between two membranes but leaving the rest of the cell untouched, which is quite similar to submembrane Ca2+ spikes versus large cellular Ca2+ waves.
NOS2
Much less is known about proteins interacting with NOS2. NOS2 serves not so much for signaling purposes but generates large amounts of NO to kill bacteria and tumor cells. It also differs from the other isoforms in that it binds CaM at free Ca2+ levels of resting cells. As early as 1995 Nathan and coworkers (32) observed a membrane localization for NOS2, apparently in specialized vesicles not corresponding to peroxisomes or lysosomes. In polarized epithelial cells, NOS2 has been demonstrated to bind the PDZ protein ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (33), which participates in localization of NOS2 to the apical cell compartment. Recently, interaction between NOS2 and a Rho GTPase member, Rac2, has been demonstrated (34). Overexpression of Rac2 in macrophages results in augmentation of lipopolysaccharide-stimulated NO production, suggesting that Rac2 serves as a positive allosteric regulator of NOS2. Similar to PIN-destabilizing NOS1 homodimers, two proteins, NOS-associated protein of 110 kDa (NAP110) and kalirin, affect NOS2 by preventing dimerization (35, 36).
In conclusion, protein–protein interactions play an essential role in our understanding of NOS regulation. Reorganization of local NOS protein complexes regulates NOS activation. Moreover, the active redistribution of NOS, bringing it to or away from NO targets, is now clearly controlled by NOS-interacting proteins. We are curious whether the newly discovered NOSTRIN in particular will become the protein connecting separated NOS intracellular pools to a unified dynamic and multiply regulated system and along that line also extend our knowledge about traffic of other membrane proteins in general.
See companion article on page 17167.
References
- 1.Bredt D. S. & Snyder, S. H. (1990) Proc. Natl. Acad. Sci. USA 87, 682-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmermann K., Opitz, N., Dedio, J., Renné, C., Müller-Esterl, W. & Oess, S. (2002) Proc. Natl. Acad. Sci. USA 99, 17167-17172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dedio J., Konig, P., Wohlfart, P., Schroeder, C., Kummer, W. & Muller-Esterl, W. (2001) FASEB J. 15, 79-89. [DOI] [PubMed] [Google Scholar]
- 4.Feron O., Belhassen, L., Kobzik, L., Smith, T. W., Kelly, R. A. & Michel, T. (1996) J. Biol. Chem. 271, 22810-22814. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Cardena G., Martasek, P., Masters, B. S., Skidd, P. M., Couet, J., Li, S., Lisanti, M. P. & Sessa, W. C. (1997) J. Biol. Chem. 272, 25437-25440. [DOI] [PubMed] [Google Scholar]
- 6.Ju H., Venema, V. J., Marrero, M. B. & Venema, R. C. (1998) J. Biol. Chem. 273, 24025-24029. [DOI] [PubMed] [Google Scholar]
- 7.Sun J. & Liao, J. K. (2002) Proc. Natl. Acad. Sci. USA 99, 13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald K. K., Zharikov, S., Block, E. R. & Kilberg, M. S. (1997) J. Biol. Chem. 272, 31213-31216. [DOI] [PubMed] [Google Scholar]
- 9.Dimmeler S., Fleming, I., Fisslthaler, B., Hermann, C., Busse, R. & Zeiher, A. M. (1999) Nature 399, 601-605. [DOI] [PubMed] [Google Scholar]
- 10.Fulton D., Gratton, J. P., McCabe, T. J., Fontana, J., Fujio, Y., Walsh, K., Franke, T. F., Papapetropoulos, A. & Sessa, W. C. (1999) Nature 399, 597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butt E., Bernhardt, M., Smolenski, A., Kotsonis, P., Frohlich, L. G., Sickmann, A., Meyer, H. E., Lohmann, S. M. & Schmidt, H. H. H. W. (2000) J. Biol. Chem. 275, 5179-5187. [DOI] [PubMed] [Google Scholar]
- 12.Venema V. J., Marrero, M. B. & Venema, R. C. (1996) Biochem. Biophys. Res. Commun. 226, 703-710. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Cardena G., Fan, R., Shah, V., Sorrentino, R., Cirino, G., Papapetropoulos, A. & Sessa, W. C. (1998) Nature 392, 821-824. [DOI] [PubMed] [Google Scholar]
- 14.Bender A. T., Silverstein, A. M., Demady, D. R., Kanelakis, K. C., Noguchi, S., Pratt, W. B. & Osawa, Y. (1999) J. Biol. Chem. 274, 1472-1478. [DOI] [PubMed] [Google Scholar]
- 15.Gratton J. P., Fontana, J., O'Connor, D. S., Garcia-Cardena, G., McCabe, T. J. & Sessa, W. C. (2000) J. Biol. Chem. 275, 22268-22272. [DOI] [PubMed] [Google Scholar]
- 16.Fontana J., Fulton, D., Chen, Y., Fairchild, T. A., McCabe, T. J., Fujita, N., Tsuruo, T. & Sessa, W. C. (2002) Circ. Res. 90, 866-873. [DOI] [PubMed] [Google Scholar]
- 17.Brouet A., Sonveaux, P., Dessy, C., Balligand, J. L. & Feron, O. (2001) J. Biol. Chem. 276, 32663-32669. [DOI] [PubMed] [Google Scholar]
- 18.Fulton D., Fontana, J., Sowa, G., Gratton, J. P., Lin, M., Li, K. X., Michell, B., Kemp, B. E., Rodman, D. & Sessa, W. C. (2002) J. Biol. Chem. 277, 4277-4284. [DOI] [PubMed] [Google Scholar]
- 19.Busconi L. & Michel, T. (1993) J. Biol. Chem. 268, 8410-8413. [PubMed] [Google Scholar]
- 20.Sessa W. C., Garcia-Cardena, G., Liu, J., Keh, A., Pollock, J. S., Bradley, J., Thiru, S., Braverman, I. M. & Desai, K. M. (1995) J. Biol. Chem. 270, 17641-17644. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Cardena G., Oh, P., Liu, J., Schnitzer, J. E. & Sessa, W. C. (1996) Proc. Natl. Acad. Sci. USA 93, 6448-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao S., Yao, J., McCabe, T. J., Yao, Q., Katusic, Z. S., Sessa, W. C. & Shah, V. (2001) J. Biol. Chem. 276, 14249-14256. [DOI] [PubMed] [Google Scholar]
- 23.Brenman J. E., Chao, D. S., Gee, S. H., McGee, A. W., Craven, S. E., Santillano, D. R., Wu, Z., Huang, F., Xia, H., Peters, M. F., et al. (1996) Cell 84, 757-767. [DOI] [PubMed] [Google Scholar]
- 24.Christopherson K. S., Hillier, B. J., Lim, W. A. & Bredt, D. S. (1999) J. Biol. Chem. 274, 27467-27473. [DOI] [PubMed] [Google Scholar]
- 25.Chao D. S., Gorospe, J. R., Brenman, J. E., Rafael, J. A., Peters, M. F., Froehner, S. C., Hoffman, E. P., Chamberlain, J. S. & Bredt, D. S. (1996) J. Exp. Med. 184, 609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffrey S. R., Snowman, A. M., Eliasson, M. J., Cohen, N. A. & Snyder, S. H. (1998) Neuron 20, 115-124. [DOI] [PubMed] [Google Scholar]
- 27.Fang M., Jaffrey, S. R., Sawa, A., Ye, K., Luo, X. & Snyder, S. H. (2000) Neuron 28, 183-193. [DOI] [PubMed] [Google Scholar]
- 28.Jaffrey S. R. & Snyder, S. H. (1996) Science 274, 774-777. [DOI] [PubMed] [Google Scholar]
- 29.Zabel U., Kleinschnitz, C., Oh, P., Nedvetsky, P., Smolenski, A., Muller, H., Kronich, P., Kugler, P., Walter, U., Schnitzer, J. E. & Schmidt, H. H. H. W. (2002) Nat. Cell Biol. 4, 307-311. [DOI] [PubMed] [Google Scholar]
- 30.Russwurm M., Wittau, N. & Koesling, D. (2001) J. Biol. Chem. 276, 44647-44652. [DOI] [PubMed] [Google Scholar]
- 31.Burette A., Zabel, U., Weinberg, R. J., Schmidt, H. H. H. W. & Valtschanoff, J. G. (2002) J. Neurosci. 22, 8961-8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vodovotz Y., Russell, D., Xie, Q. W., Bogdan, C. & Nathan, C. (1995) J. Immunol. 154, 2914-2925. [PubMed] [Google Scholar]
- 33.Glynne P. A., Darling, K. E., Picot, J. & Evans, T. J. (2002) J. Biol. Chem. 277, 33132-33138. [DOI] [PubMed] [Google Scholar]
- 34.Kuncewicz T., Balakrishnan, P., Snuggs, M. B. & Kone, B. C. (2001) Am. J. Physiol. 281, F326-F336. [DOI] [PubMed] [Google Scholar]
- 35.Ratovitski E. A., Bao, C., Quick, R. A., McMillan, A., Kozlovsky, C. & Lowenstein, C. J. (1999) J. Biol. Chem. 274, 30250-30257. [DOI] [PubMed] [Google Scholar]
- 36.Ratovitski E. A., Alam, M. R., Quick, R. A., McMillan, A., Bao, C., Kozlovsky, C., Hand, T. A., Johnson, R. C., Mains, R. E., Eipper, B. A. & Lowenstein, C. J. (1999) J. Biol. Chem. 274, 993-999. [DOI] [PubMed] [Google Scholar]
- 37.Ju H., Zou, R., Venema, V. J. & Venema, R. C. (1997) J. Biol. Chem. 272, 18522-18525. [DOI] [PubMed] [Google Scholar]
- 38.Venema V. J., Ju, H., Zou, R. & Venema, R. C. (1997) J. Biol. Chem. 272, 28187-28190. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi M., Hata, Y., Hirao, K., Toyoda, A., Irie, M. & Takai, Y. (1997) J. Biol. Chem. 272, 11943-11951. [DOI] [PubMed] [Google Scholar]
- 40.Brenman J. E., Christopherson, K. S., Craven, S. E., McGee, A. W. & Bredt, D. S. (1996) J. Neurosci. 16, 7407-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Firestein B. L. & Bredt, D. S. (1999) J. Biol. Chem. 274, 10545-10550. [DOI] [PubMed] [Google Scholar]
- 42.Fan J. S., Zhang, Q., Li, M., Tochio, H., Yamazaki, T., Shimizu, M. & Zhang, M. (1998) J. Biol. Chem. 273, 33472-33481. [DOI] [PubMed] [Google Scholar]
- 43.Liang J., Jaffrey, S. R., Guo, W., Snyder, S. H. & Clardy, J. (1999) Nat. Struct. Biol. 6, 735-740. [DOI] [PubMed] [Google Scholar]
- 44.Haraguchi K., Satoh, K., Yanai, H., Hamada, F., Kawabuchi, M. & Akiyama, T. (2000) Genes Cells 5, 905-911. [DOI] [PubMed] [Google Scholar]
- 45.Tochio H., Ohki, S., Zhang, Q., Li, M. & Zhang, M. (1998) Nat. Struct. Biol 5, 965-969. [DOI] [PubMed] [Google Scholar]