Abstract
The rice KNOX protein OSH15 consists of four conserved domains: the MEINOX domain, which can be divided into two subdomains (KNOX1 and KNOX2); the GSE domain; the ELK domain; and the homeodomain (HD). To investigate the function of each domain, we generated 10 truncated proteins with deletions in the conserved domains and four proteins with mutations in the conserved amino acids in the HD. Transgenic analysis suggested that KNOX2 and HD are essential for inducing the abnormal phenotype and that the KNOX1 and ELK domains affect phenotype severity. We also found that both KNOX2 and HD are necessary for homodimerization and that only HD is needed for binding of OSH15 to its target sequence. Transactivation studies suggested that both the KNOX1 and ELK domains play a role in suppressing target gene expression. On the basis of these findings, we propose that overproduced OSH15 probably acts as a dimer and may ectopically suppress the expression of target genes that induce abnormal morphology in transgenic plants.
INTRODUCTION
Homeobox genes were first characterized as transcriptional regulatory genes that control morphogenesis in Drosophila species (Gehring, 1987). Products of these genes share a unique domain known as a homeodomain (HD). The HD consists of a highly conserved 60–amino acid stretch containing three α-helices that form a helix-turn-helix–type DNA binding motif (Desplan et al., 1988; Otting et al., 1990). This motif recognizes and binds to specific DNA sequences; thus, HD proteins are believed to regulate the expression of batteries of target genes by acting as transcriptional factors (Affolter et al., 1990; Hayashi and Scott, 1990; Kissinger et al., 1990; Laughon, 1991).
The first plant homeobox gene, knotted1 (kn1), was identified by cloning of the gene affected in the maize Knotted1 mutant (Vollbrecht et al., 1991). Afterward, knotted1-like homeobox (knox) genes encoding KNOX proteins were isolated from various plants, including rice, barley, Arabidopsis, soybean, tomato, and tobacco (Matsuoka et al., 1993; Lincoln et al., 1994; Ma et al., 1994; Müller et al., 1995; Harven et al., 1996; Tamaoki et al., 1997). The loss-of-function mutants of kn1 in maize and shootmeristemless (stm) in Arabidopsis show defects in shoot apical meristem (SAM) development or maintenance (Long et al., 1996; Kerstetter et al., 1997). The opposite phenotype, namely, ectopic meristem formation in leaves, has been reported in transformants that ectopically express knox genes (Matsuoka et al., 1993; Sinha et al., 1993; Chuck et al., 1996; Nishimura et al., 2000; Sentoku et al., 2000). On the basis of this evidence, many KNOX proteins are considered to play a critical role in the maintenance of the indeterminate properties of cells in the SAM (Reiser et al., 2000), although their direct function remains unresolved.
All KNOX proteins have a highly conserved atypical HD that consists of a 63–amino acid stretch, whereas typical HDs consist of 60 amino acids (Figure 1). The HDs of KNOX proteins have three extra amino acids situated between the first and second helices. These invariant extra residues also occur in the loop between the first and second helices of several HD proteins from other organisms, although they are not found in the typical HD proteins such as Antennapedia (Bertolino et al., 1995). Based on this unique feature, these proteins have been designated TALE (three–amino acid loop extension) HD proteins, and all KNOX proteins are members of this superclass (Bürglin, 1997).
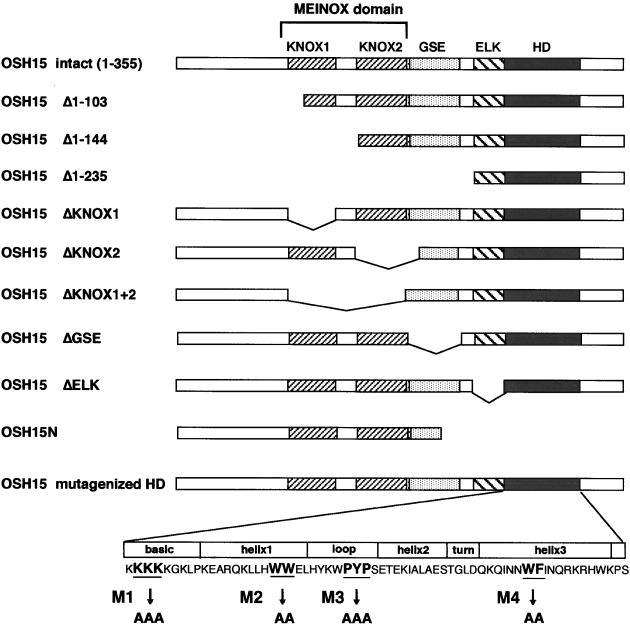
Figure 1.
Scheme of the Mutagenized OSH15 Proteins.
The full-length OSH15 (amino acids 1 to 355) contains four putative functional domains that are conserved in plant KNOX proteins: the MEINOX domain (which can divided into two subdomains, KNOX1 and KNOX2); the GSE domain; the ELK domain; and the HD. Deletions from the N- or C-terminal ends and internal deletions of each domain are indicated. In the HD, four sites of highly conserved amino acid residues in the basic, helix1, loop, and helix3 regions were replaced with alanine residues (M1, M2, M3, and M4, respectively).
The sequence immediately upstream of the HD, termed the ELK domain (Vollbrecht et al., 1991; Kerstetter et al., 1994), also is conserved (Figure 1). The ELK domain has been speculated to form a novel amphipathic helix (Kerstetter et al., 1994) and could function as a nuclear localization signal (Meisel and Lam, 1996). The ELK domain also is considered to act as a protein–protein interaction domain (Vollbrecht et al., 1991), but the precise role of this domain has not been determined. In addition to the conserved ELK and TALE HDs, a stretch of ∼100 amino acids located at the N terminus of almost all KNOX proteins also is conserved (Figure 1). This conserved region, known as the MEINOX domain, may function in protein–protein interactions (Bürglin, 1997). A relatively smaller and less well-conserved amino acid motif is located between the MEINOX and ELK domains. The function of this conserved region, termed the GSE domain, has not yet been determined.
To understand the function of KNOX proteins in plant development, it is necessary to characterize the biochemical properties of these proteins; however, few studies of plant KNOX proteins to date have taken this approach. In this article, we characterize the biochemical properties of the conserved domains of a rice KNOX protein, OSH15, and report that these domains can mediate DNA binding, dimerization, and target gene regulation.
RESULTS
Overexpression of Mutagenized OSH15 Proteins in Transgenic Rice Plants
It is well known that the overexpression of class I–type knox genes often results in abnormal plant morphologies. To characterize the domains of OSH15 that were required for the abnormal plant morphologies induced in the overexpression studies, we overexpressed mutant OSH15 proteins with deletion or point mutations, as summarized in Figure 1, in transgenic rice plants under the control of the rice actin promoter (ACT1), which is a strong constitutive promoter in rice organs (Zhang et al., 1991). If the OSH15 derivatives are functional, in terms of directing abnormal plant morphogenesis, in transgenic rice plants, the plants should show altered morphologies, as seen in plants that overproduce intact OSH15 proteins (Sentoku et al., 2000).
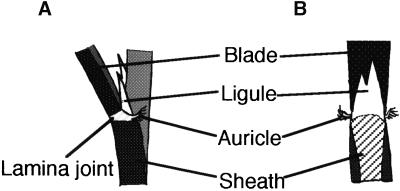
Some of the OSH15 derivatives induced altered leaf morphology in transformants, whereas others did not induce an abnormal phenotype. We categorized the phenotypes of the transgenic plants into six groups based on their leaf morphologies. In wild-type rice leaves, the ligule and a pair of auricles are located in the region between the leaf blade and the leaf sheath. The rice ligule is a thin, white, tongue-like organ of triangular shape that rises from the adaxial surface at the proximal part of the sheath. A pair of auricles projects as horn-like tissues from both sides of the lamina joint (Figure 2) (Hoshikawa, 1989). Plants categorized as having the wild-type phenotype (category I) did not display any abnormalities in the leaves or alterations in gross morphology (Figures 3A and 3G). The second category (II) includes plants showing an asymmetry phenotype that has a perturbed boundary between the leaf sheath and blade (Figure 3B), which caused a ligule to split (Figure 3B, arrows) and also caused asymmetric positioning of the paired auricles (Figure 3H, arrowheads). Plants exhibiting a knot phenotype (category III), which was similar to that of the dominant Kn1 mutant in maize (Smith and Hake, 1994), showed a disruption in the parallel nature of the lateral veins and the formation of a knot-like structure in the leaf blades with a perturbation of the boundary between the sheath and the blade (Figures 3C and 3I).
Figure 2.
Scheme of the Lamina Joint Region of Rice Leaf in the Wild- Type Plant.
(A) Abaxial view.
(B) Adaxial view.
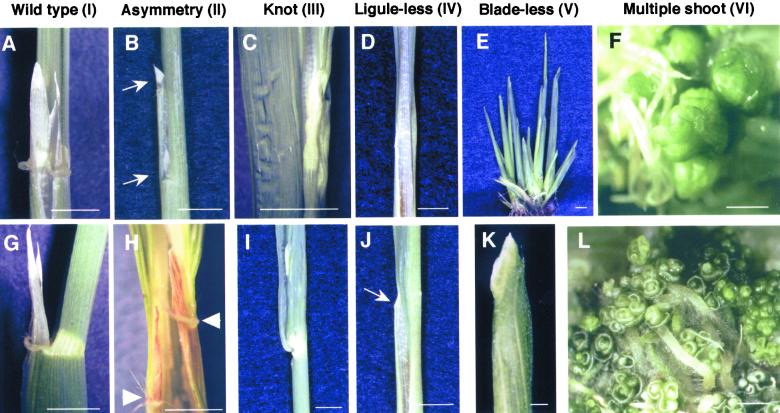
Figure 3.
Typical Phenotypes of Rice Leaves in Transgenic Plants That Overproduce the OSH15 Derivatives.
(A) and (G) Wild-type phenotype (I): adaxial (A) and abaxial (G) views show the lamina joint region of a developed leaf.
(B) and (H) Asymmetry phenotype (II): abaxial (B) and adaxial (H) views around the lamina joint region. The arrows in (B) indicate the split ligule, and the arrowheads in (H) indicate asymmetrical formation of auricles.
(C) and (I) Knot phenotype (III): close-up (C) and abaxial (I) views of a leaf blade with a knot.
(D) and (J) Ligule-less phenotype (IV): adaxial (D) and abaxial (J) views around the putative lamina joint region. There was no typical lamina joint, only the development of malformed auricles indicated by the arrow in (J).
(E) and (K) Blade-less phenotype (V): whole plant (E) and abaxial (K) views of a blade-less leaf.
(F) and (L) Multiple shoot phenotype (VI).
Bars in (A) to (E) and (G) to (J) = 5 mm; bars in (F), (K), and (L) = 1 mm.
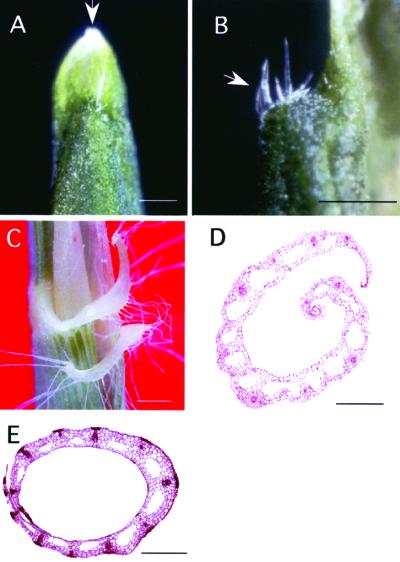
Transformants in category IV showed a ligule-less phenotype involving the disappearance of the ligule and auricles (Figure 3D). Ectopic formation of small segments of ligule or auricle-like organs often was seen in the leaf blade region (Figure 3J). The fifth category (V) showed a blade-less phenotype (Figure 3E). The leaves of plants in this category consisted of only the leaf sheath region and did not form any parts of a typical leaf blade structure (Figure 3K). When the top of a blade-less leaf was viewed at higher magnification, the formation of a tongue-like pale green organ with a triangular shape was evident, which appeared similar to a ligule (Figure 4A, arrow). There were also auricle-like organs, with well-developed hairs similar to those of the wild-type plants, present at the boundary between the tongue-like organ and the leaf sheath (Figures 4B and 4C). Although the leaf blade failed to develop, the formation of the leaf sheath appeared to be normal and its internal structure was similar to that of nontransgenic rice plants (cf. Figures 4D and 4E). Transformants categorized as multiple shoot (category VI) showed the most severe phenotype. Leaves of these plants did not develop normally, and neither blade-sheath differentiation nor the formation of a ligule and auricles was evident (Figures 3F and 3L).
Figure 4.
Morphology of Blade-Less Leaves.
(A) Close-up view of the top of a blade-less leaf. The arrow indicates a tongue-like pale green organ.
(B) Boundary region between the tongue-like organ and the leaf sheath of a blade-less leaf. The arrow indicates the auricle-like organ.
(C) Close-up view of the auricle of a wild-type plant.
(D) Transverse section of the leaf from a blade-less plant.
(E) Transverse section of the leaf sheath from a wild-type plant.
Bars = 1 mm.
Using these six phenotypic categories, we assessed the phenotypes of transgenic rice plants that overproduced the various OSH15 derivatives. As shown in Table 1, the majority (∼90%) of plants transformed with the intact OSH15 construct exhibited the multiple shoot phenotype (VI). A similar distribution of the severe phenotype was seen in the transgenic plants carrying M1. This observation suggests that the lysine stretch in the basic region of the HD is not necessary for the induction of the abnormal phenotype of leaves in transgenic rice plants. Plants transformed with ΔKNOX1, ΔKNOX2, ΔKNOX1+2, M2, M3, and M4 primarily showed the wild-type phenotype, whereas plants carrying ΔKNOX1 also exhibited a less severe phenotype, asymmetry. This suggests that these domains are important for the induction of altered leaf morphology. Plants carrying the ΔELK construct showed a high frequency (∼87%) of the blade-less phenotype. Interestingly, this unusual phenotype was unique to the ΔELK protein and was not observed in transformants overproducing any other OSH15 derivatives, with the exception of one plant carrying the intact OSH15 construct. The fact that this phenotype is specific to the ΔELK protein leads us to speculate that the ELK domain may have a specific function(s). Plants with the ΔGSE protein exhibited only the severe phenotype: multiple shoots. It is noteworthy that the phenotypic severity of the ΔGSE plants was stronger than that observed in plants carrying the intact OSH15 construct. Whereas all of the ΔGSE plants showed the multiple shoot phenotype without exception, 10% of the OSH15 plants displayed a less severe phenotype. It is possible that the GSE domain may have a role in suppressing the induction of altered leaf morphology (see below).
Table 1.
Distribution of Each Phenotype in Transgenic Rice Plants Overproducing the OSH15 Derivatives
| Phenotype
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Construct | I Wild type |
II Asymmetry |
III Knot |
IV Ligule-less |
V Blade-less |
VI Multiple shoot |
Total | ||||||
| Intact OSH15 | 1a | 1.3b | 0 | 0.0 | 4 | 5.3 | 2 | 2.7 | 1 | 1.3 | 67 | 89.3 | 75 |
| ΔKNOX1 | 20 | 55.6 | 16 | 44.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 36 |
| ΔKNOX2 | 72 | 100.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 72 |
| ΔKNOX1+2 | 53 | 100.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 53 |
| ΔGSE | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 92 | 100.0 | 92 |
| ΔELK | 5 | 13.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 32 | 86.5 | 0 | 0.0 | 37 |
| M1 (KKK) | 11 | 9.5 | 0 | 0.0 | 1 | 0.9 | 6 | 5.1 | 0 | 0.0 | 98 | 84.5 | 116 |
| M2 (WW) | 25 | 100.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 25 |
| M3 (PYP) | 15 | 100.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 15 |
| M4 (WF) | 22 | 100.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 22 |
Number of transformants categorized into the phenotype.
Percentage.
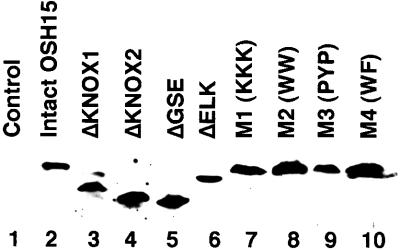
We wondered whether the difference in the phenotypes was caused by differences in the level of the OSH15 derivative proteins in transgenic plants. To investigate this possibility, we directly tested the level of protein expression by protein gel blot analysis using an anti-OSH15 antiserum (Figure 5). Each of the nine different transgenes examined induced a similar level of expression of the OSH15 derivative protein in the transformants. This result suggests that the phenotypic differences are not attributable to a significant difference in the level of expression of the OSH15 derivative proteins in the transgenic plants carrying the different transgenes.
Figure 5.
Immunodetection of OSH15 Derivative Proteins.
Total protein was extracted from 10 hygromycin-resistant calli transformed with the intact OSH15 transformants (lane 2), ΔKNOX1 (lane 3), ΔKNOX2 (lane 4), ΔGSE (lane 5), ΔELK (lane 6), M1 (lane 7), M2 (lane 8), M3 (lane 9), M4 (lane 10), or the pBI empty vector (lane 1) as a negative control. Twenty micrograms of total protein was subjected to SDS-PAGE, electroblotted onto nitrocellulose membrane, and probed with antiserum against OSH15.
Nuclear Localization of OSH15
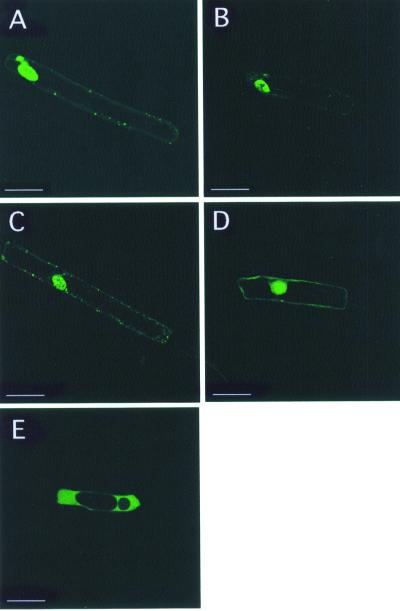
Because OSH15 is believed to act as a transacting factor, nuclear localization should be necessary for the function of OSH15. Part of the reason for the failure of induction in the abnormal phenotypes may be a failure in nuclear localization of the OSH15 derivatives. To test this possibility, we analyzed the subcellular localization of the OSH15 mutant proteins with the aid of the green fluorescent protein (GFP). The fusion proteins were expressed transiently in onion epidermal cells. As shown Figure 6A, the intact OSH15-GFP was localized primarily to the nucleus. ΔKNOX1+2-GFP, ΔELK-GFP, and OSH15N-GFP showed localization patterns that were almost the same as that of the intact OSH15 (Figures 6B, 6C, and 6D, respectively). In contrast, the maize cytoplasm-localized protein PEPC-GFP was observed only in the cytoplasm (Figure 6E). These results demonstrate that the MEINOX domain, the ELK domain, or the C-terminal region containing ELK-HD is not essential for the nuclear localization of OSH15.
Figure 6.
Subcellular Localization of OSH15 Derivatives in Onion Cells in a Transient Assay.
(A) Intact OSH15-GFP.
(B) ΔKNOX1+2-GFP.
(C) ΔELK-GFP.
(D) OSH15N-GFP.
(E) PEPC-GFP.
Each panel shows a confocal image. Bars = 50 μm.
DNA Binding Property of OSH15 Proteins
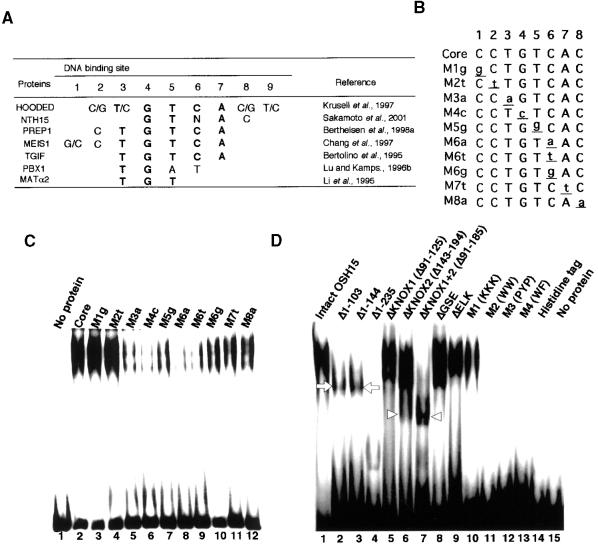
OSH15 belongs to the TALE superclass of HD proteins, which includes the animal PBC class HD, vertebrate MEIS1, human PREP1, and yeast MAT α2 proteins. The target sequences of some of the TALE HD proteins have been identified previously, and these sequences are similar, with a core motif of TGTCA (Figure 7A) (Bertolino et al., 1995; Chang et al., 1997; Krusell et al., 1997; Berthelsen et al., 1998a; Sakamoto et al., 2001). We predicted that OSH15 also may interact with the TGTCA motif or a similar sequence. To examine this possibility, we performed an electrophoretic mobility shift assay (EMSA) using a series of oligonucleotides relating in sequence to the TGTCA motif (Figure 7B). As expected, recombinant OSH15 protein expressed in Escherichia coli cells interacted with the sequences with the TGTCA motif and also with some related sequences, but it bound most strongly to the sequences with the TGTCAC motif (Figure 7C). Consequently, we used the oligonuleotide with the TGTCAC motif for further studies.
Figure 7.
DNA Binding Property of OSH15.
(A) Target sequences of TALE HD proteins that have been reported. Most TALE HD proteins share the same target sequence (i.e., the TGTCAC motif).
(B) The TGTCAC motif and its derivative sequences. Oligonucleotides containing these sequences were used in this study.
(C) Recombinant histidine-tagged OSH15 was subjected to EMSA using different DNA probes as indicated above each lane. Probe sequences are shown in (B).
(D) EMSA was performed using histidine-tagged OSH15 derivatives, as indicated above the gel lanes. Both the histidine tag and no protein were used as negative controls. The arrowheads indicate the positions of the faster mobility bands (lanes 6 and 7), in contrast to the shifted bands with slower mobility, which are indicated by arrows (lanes 2 and 3).
We tested the DNA binding activity of the OSH15 derivatives (Figure 7D). DNA binding activity was lost completely in proteins mutagenized in the HD region, such as M2 (lane 11), M3 (lane 12), and M4 (lane 13). In contrast, proteins mutagenized at the basic region of HD (M1, lane 10), deleted in the MEINOX domain (lanes 5, 6, and 7), the GSE domain (lane 8), or the ELK domain (lane 9), or missing all of the N-terminal region (lanes 2 and 3) retained their DNA binding activity. Furthermore, even a peptide with only the ELK HD (lane 4) still interacted with the probe. These observations demonstrate that the amino acid residues in the HD, such as WW in helix1, PYP in the loop, and WF in helix3, are essential for the interaction between OSH15 and its target sequence. In contrast to the HD, other domains or regions located at the N-terminal end of the HD, such as the N-terminal region itself, the MEINOX domain, the GSE domain, and the ELK domain, are not necessary for the DNA binding. The higher mobility of the sifted bands of some truncated proteins, such as Δ1–103 and Δ1–144 (lanes 2 and 3, arrows), may reflect the smaller molecular weights of the truncated proteins. However, in the case of ΔKNOX1+2 (lane 7, arrowheads) and the lower shifted band of ΔKNOX2 (lane 6, arrowheads), the higher mobility of these bands cannot be attributed solely to the smaller molecular weights of the proteins because the molecular weights of ΔKNOX2 and ΔKNOX1+2 are larger than those of Δ1–103 and Δ1–144, whereas the mobilities of shifted bands of ΔKNOX2 and ΔKNOX1+2 (arrowheads) were faster than those of Δ1–103 and Δ1–144 (arrows). This may be caused by another factor, probably resulting from a failure of dimer formation (see below).
Dimer Formation of OSH15
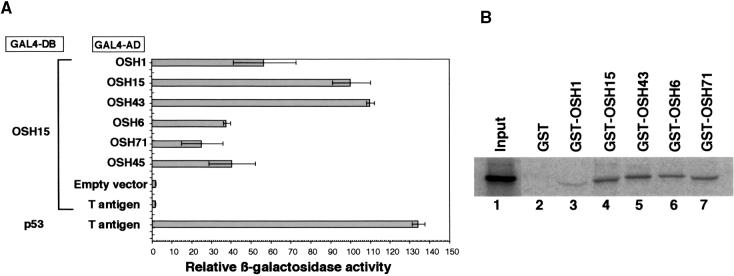
The shifted bands with higher mobility that were caused by some of the truncated proteins in the EMSA suggest that OSH15 may form a homodimer in vitro. This possibility also is supported by previous observations that animal TALE HD proteins often interact with other HD proteins to regulate their function (Chan et al., 1994; Rieckhof et al., 1997; Berthelsen et al., 1998b). To examine this possibility, we investigated the protein–protein interactions that occur between OSH15 itself and other rice KNOX (OSH) proteins using a quantitative yeast two-hybrid assay. As shown in Figure 8A, OSH15 interacted with all of the OSH proteins. Particularly strong LacZ activity was observed in the case of homodimer formation by OSH15 and heterodimer formation between OSH15 and OSH43. The fact that OSH15 did not interact with an unrelated protein (T antigen) indicates that the interaction between OSH15 and OSH proteins is specific.
Figure 8.
Interaction between OSH15 and Rice KNOX Proteins.
(A) Quantitative yeast two-hybrid assay with OSH15 and rice KNOX (OSH) proteins. For the bait construct, OSH15 was fused to the GAL4 DNA binding domain (GAL4-DB). For the prey constructs, OSH proteins were fused to the GAL4 activation domain (GAL4-AD). The pairwise combination of the bait and prey constructs for each row is shown at left. Relative β-galactosidase (LacZ) activity was calculated by standardizing each LacZ activity with that of intact OSH15/OSH15. We set the activity of OSH15/OSH15 at 100. Error bars represent the standard deviations calculated from three independent yeast clones. The combinations of OSH15–empty vector and OSH15–T antigen are negative controls of interaction, whereas the combination of p53–T antigen is a positive control.
(B) In vitro pull-down assay with 35S-methionine–labeled OSH15. The fusion proteins consisted of OSH proteins and GST: GST-OSH1 (lane 3); GST-OSH15 (lane 4); GST-OSH43 (lane 5); GST-OSH6 (lane 6); and GST-OSH71 (lane 7). The input protein, 35S-methionine–labeled OSH15, was loaded onto the gel alone as a control (lane 1). GST alone was used as negative control for interaction (lane 2).
To confirm the interaction between OSH15 and OSH proteins, we tested directly the interaction between 35S-methionine–labeled OSH15 and fusion proteins consisting of glutathione S-transferase (GST) and OSH proteins (GST-OSH1, GST-OSH15, GST-OSH43, GST-OSH6, GST-OSH71, or GST alone) in an in vitro pull-down experiment. As shown in Figure 8B, the labeled OSH15 was precipitated with the GST-OSH proteins (lanes 3 to 7) but not with GST alone (lane 2). The amount of OSH15 precipitated by GST-OSH1 was much less than that precipitated by the other GST-OSH proteins (lane 3) because almost all of the GST-OSH1 fusion protein was accumulated in the inclusion body of the recombinant E. coli and not extracted in the soluble fraction (data not shown). These results confirm that OSH15 can interact both with itself and with other OSH proteins without binding to its target DNA sequence.
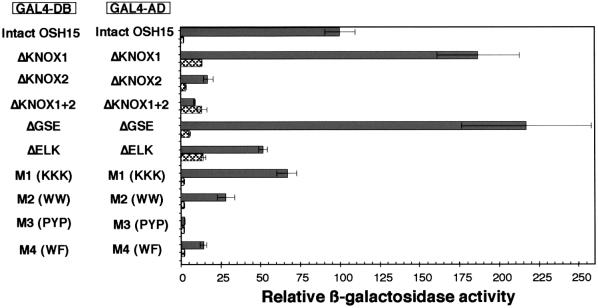
Considering the fact that OSH15 can form a homodimer in vitro and in yeast, it is possible that OSH15 also forms homodimers in transgenic rice plants that overproduce OSH15 and that the dimerization of OSH15 may be required for the induction of abnormal leaf morphology. To test the relationship between homodimer formation and induction of the abnormal leaf morphology in transgenic rice plants, we examined the formation of homodimers by various mutagenized constructs of OSH15 in a yeast two-hybrid assay (Figure 9). Overall, the results of the yeast two-hybrid assay correlated well with the results of overexpression studies. For example, ΔGSE, ΔELK, and M1, all of which can induce the abnormal phenotypes in transgenic plants, showed strong or moderate LacZ activities, whereas ΔKNOX2, ΔKNOX1+2, M3, and M4, which do not induce any morphological alterations, did not show significant LacZ activities. These results suggest that the dimerization of OSH15 may be required for the induction of abnormal morphologies. However, a truncated protein with strong or moderate LacZ activity does not always induce an abnormal phenotype in transgenic plants. Indeed, ΔKNOX1 showed higher LacZ activity than the intact OSH15 but could induce only the asymmetry phenotype with a low frequency. Likewise, M2 showed some LacZ activity but could not induce the abnormal phenotype. Considering the fact that M2 could not bind the putative target sequence (Figure 7D), this result may be reasonable. Thus, the induction of abnormal morphologies seems to require several properties of OSH15, such as DNA binding, transcriptional regulation (see below), and dimerization.
Figure 9.
Homodimer Formation of OSH15 in a Quantitative Yeast Two-Hybrid Assay.
Homodimer formation of OSH15 derivatives in yeast. Shown at left is the pairwise combination of OSH15 derivatives. The shaded bars show the relative LacZ activity of each pairwise combination of the OSH15 derivatives. The cross-hatched bars show the relative LacZ activity of GAL4-DB fused with each OSH15 derivative protein alone used as negative control. Error bars represent standard deviations. Relative LacZ activity was calculated by standardizing each LacZ activity with that of intact OSH15/OSH15. We set the activity of OSH15/OSH15 at 100.
Transcriptional Activity of OSH15
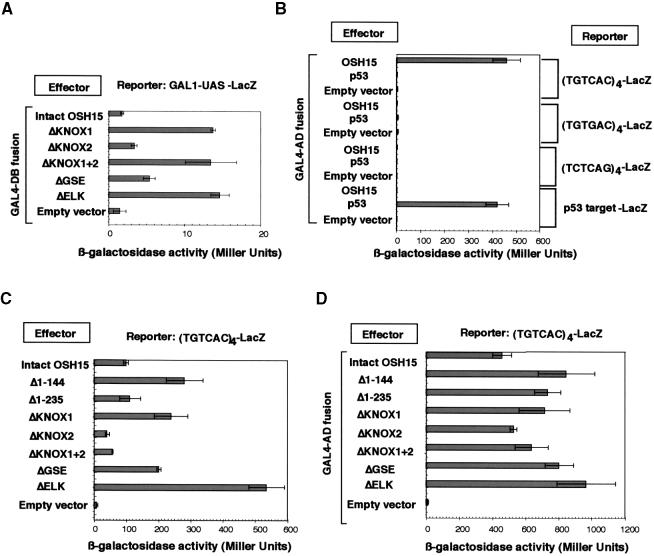
In the yeast two-hybrid analysis of OSH15, we observed low but reproducible background activity when some of the truncated OSH15 domains, such as ΔKNOX1, ΔKNOX1+2, and ΔELK, were fused with the GAL4 DNA binding domain (GAL4-DB; amino acids 1 to 147), whereas this background activity was negligible for fusion proteins with the intact OSH15 or GAL4-DB alone (Figure 10A). This finding suggests that a specific region(s) of OSH15 may function as a transactivation domain and may transactivate the expression of a reporter gene without the interaction of GAL4 activation domain (GAL4-AD; amino acids 768 to 881) fused proteins.
Figure 10.
Transcriptional Activity of OSH15 Derivatives.
(A) Transcriptional activity only of GAL4-DB–fused OSH15 derivatives in yeast. GAL1-UAS stands for the enhancer-like sequence controlled by GAL4-DB.
(B) DNA binding activity of GAL4-AD–fused OSH15 in yeast. Each reporter gene ([TGTCAC]4-LacZ; [TGTGAC]4-LacZ; [TCTCAG]4-LacZ; or p53 target–LacZ) was introduced into yeast strain YM4271. GAL4-AD–fused OSH15 was introduced into each yeast strain sequentially. The empty vector or GAL4-AD–fused p53 served as a negative control, whereas the combination of GAL4-AD–fused p53 with the p53 target–LacZ reporter was used as a positive control.
(C) Transcriptional activity of OSH15 derivatives depending on their DNA binding context in yeast. OSH15 derivatives were introduced into the (TGTCAC)4-LacZ reporter strain. The empty vector served as a negative control.
(D) DNA binding activity of GAL4-AD–fused OSH15 derivatives in yeast. GAL4-AD–fused OSH15 derivatives were introduced into the (TGTCAC)4-LacZ reporter strain. The empty vector served as a negative control.
The bars show the LacZ activity, presented as the measured value in Miller units, of each of the effector proteins. Error bars represent standard deviations.
We performed further analyses to examine the transactivation activity of OSH15 using a quantitative yeast one-hybrid analysis. First, we tested whether the TGTCAC motif functions as a cis-acting target of OSH15 in yeast. We transformed yeast with three different reporter constructs consisting of four repeated cis-acting motifs of TGTCAC, TGTGAC, or TCTCAG at the front of the core promoter (yeast iso-1-cytochrome C) for reporter gene expression. As shown in Figure 10B, the fusion protein GAL4-AD-OSH15, which consisted of the GAL4-AD and the intact OSH15 (labeled as OSH15 in Figure 10B), was able to transactivate the expression of the (TGTCAC)4-LacZ reporter to a level ∼50-fold higher than that of the negative control (empty vector) in yeast. In contrast, GAL4-AD-OSH15 did not activate either the (TGTGAC)4-LacZ or the (TCTCAG)4-LacZ reporter. These results indicate that GAL4-AD-OSH15 specifically recognizes the (TGTCAC)4 motif in the reporter and that this recognition can activate the expression of LacZ mainly via the transactivation activity of the GAL4-AD. OSH15 binding with the cis-acting motifs was much more specific in the yeast cells than in vitro. In fact, GAL4-AD-OSH15 could not transactivate the reporter genes carrying the (TGTGAC)4 or (TCTCAG)4 motifs, even though OSH15 bound to these sequences with moderate affinity in the EMSA (Figure 7C). No LacZ activity was induced by the GAL4-AD-p53 fusion protein containing the human DNA binding protein, p53 (Figure 10B), or by the combination of the empty vector and the (TGTCAC)4-LacZ reporter, indicating that the LacZ activity was induced specifically by GAL4-AD-OSH15.
Using the yeast carrying the (TGTCAC)4-LacZ reporter, we tested the transactivating activity of the OSH15 derivatives. As shown in Figure 10C, the intact OSH15 led to an ∼10-fold activation relative to the basal activity. This finding indicates that OSH15 itself has a transactivation function for expression of the reporter gene. Interestingly, deletion of the 144 amino acid residues of the N-terminal region (Δ1–144, which lacks the KNOX1 domain) caused a threefold increase in the transactivation activity of OSH15. However, further deletions to 235 amino acid residues (Δ1–235, lacking both the MEINOX and GSE domains) abolished the enhanced transactivation activity of Δ1–144 but still resulted in a level of activity that was almost equivalent to that of the intact OSH15. Similarly, deletion of KNOX1 (ΔKNOX1) enhanced the activity to a level approximately threefold higher than that of the intact OSH15, and this enhancement also was canceled completely by entire deletion of the MEINOX domain (ΔKNOX1+2). This suppressive effect on the enhanced transactivating activity of OSH15 may be abolished by a failure of dimer formation through the KNOX2 region. In addition, deletion of the ELK domain also increased the transactivation activity, suggesting that this domain functions as a suppressor of OSH15 activation. As a control for these experiments, we constructed fusion genes encoding the chimeric protein with the GAL4-AD and each mutagenized OSH15 protein and tested the transactivation activity of these proteins (Figure 10D). All of these proteins induced a high level of LacZ activity, indicating that the deletion of each region does not affect the interaction between the truncated derivatives and the cis-acting motif of the reporter gene.
DISCUSSION
The Function of the HD
We have demonstrated that the HD is essential for the interaction between OSH15 and its target sequence in vitro. In particular, the conserved amino acids WW in helix1, PYP in the loop, and WF in helix3 are important for DNA binding, but the amino acid residues (KKK) in the basic region of the HD are not (Figure 7C). Analysis of the crystal structure of an animal TALE HD has shown that PYP and WF, which are conserved between plant and animal TALE families, interact directly with the target DNA (Passner et al., 1999). On this basis, PYP in the loop and WF in helix3 of OSH15 HD are believed to interact directly with the OSH15 target sequence. In contrast to the loop and helix3, helix1 does not interact directly with DNA, but nuclear magnetic resonance analyses of Antennapedia have revealed that some hydrophobic amino acids in helix1 form a hydrophobic core that can stabilize the HD structure and the HD–DNA interaction (Qian et al., 1989). It is possible then that WW in helix1 of OSH15 may stabilize the HD conformation as part of a hydrophobic core.
Interestingly, the HD of OSH15 also is necessary for homodimer formation. The importance of the TALE HD for dimer formation has been reported in the case of interactions between Drosophila Ultrabithorax (Ubx; typical HD protein) and Extradenticle (Exd; TALE HD). Crystal structural analysis has revealed that the Ubx-Exd complex is formed through the interaction between the Exd HD surface structure, known as a hydrophobic pocket, which is formed by hydrophobic amino acids of both the loop and helix3, and the YPWM amino acid motif located at the N-terminal end of Ubx (Passner et al., 1999). We found that exchanges of PYP in the loop and WF in helix3 with alanine residues severely impede homodimer formation; therefore, it may be possible that the OSH15 HD also forms a hydrophobic pocket for interaction. In animal HDs, it has been proposed that the basic region located at the N-terminal end of the HD is necessary for recognition of the target sequences (Qian et al., 1989; Kissinger et al., 1990; Passner et al., 1999) and that the basic amino acid cluster in this region functions as a nuclear localization signal (Abu-Shaar et al., 1999). OSH15 also has the basic region at the front of the HD, although the OSH15 protein mutagenized at this basic region (M1) was no different from the intact OSH15 in all of the biochemical properties we tested, including DNA binding and nuclear localization. Moreover, transgenic rice plants that overproduce M1 showed a distribution of abnormal phenotypes similar to that seen in transgenic plants carrying the intact OSH15. These observations indicate that, in contrast to animal HDs, the lysine cluster in the basic region does not appear to have any function in terms of DNA interaction in vitro, nuclear localization, or induction of abnormal morphology.
The Function of the MEINOX Domain
The MEINOX domain was first characterized as a highly conserved domain outside of the HD in plant KNOX proteins (Bürglin, 1997). This domain also shows apparent similarity to the MEIS and PBC domains, both of which are found in animal TALE HD proteins (Bürglin, 1998). Recent biochemical analyses of the MEIS/PBC domain have revealed that these domains have several functions, such as transrepression, heterodimer formation, and nuclear export signaling (Lu and Kamps, 1996a, 1996b; Rieckhof et al., 1997; Berthelsen et al., 1998b, 1999; Abu-Shaar et al., 1999). The precise alignment of the KNOX and MEIS/PBC proteins has suggested that the MEINOX domain can be divided into two subdomains (KNOX1 and KNOX2 in the case of the KNOX proteins) that are joined by a flexible linker (Bürglin, 1997). In this study, we have proposed that the MEINOX domain also can be divided functionally into two domains and that these domains are important (KNOX1) or essential (KNOX2) for the induction of altered leaf morphology in transgenic plants.
According to the nuclear localization experiments and EMSA analysis, these subdomains are not essential for nuclear localization or DNA binding. The transactivation experiments using a reporter gene under the control of the cis-acting motif of OSH15 revealed that KNOX1 has a suppressive function against the transcription of the reporter gene in yeast, whereas KNOX2 does not have this function. A previous study of another rice KNOX protein, OSH45, demonstrated that the N-terminal region of OSH45 (amino acids 1 to 170, including KNOX1) works as a suppressive region against the transcription of a reporter gene in rice protoplasts (Tamaoki et al., 1995). Furthermore, recent studies have demonstrated that the abnormal leaf morphology of transgenic tobacco plants that overproduce a tobacco KNOX protein, NTH15, is caused by the ectopic suppression of a gibberellin biosynthetic gene, GA C-20 oxidase (Tanaka-Ueguchi et al., 1998). Biochemical analyses indicate that ectopically produced NTH15 interacts with the target sequence in the GA C-20 oxidase gene and suppresses its transcription (Sakamoto et al., 2001). These results support the possibility that KNOX1 has a suppressive function against the expression of a target gene(s) of OSH15.
Experiments with transgenic tobacco plants that overproduce KNOX proteins have demonstrated that KNOX2 is a key factor in determining phenotypic severity in plants (Sakamoto et al., 1999). Our results show that KNOX2 is required for dimer formation, which leads us to speculate that homodimer formation is a prerequisite for the induction of the abnormal phenotype.
The Function of the GSE Domain
Deletion of the GSE domain caused an increase in the severity of abnormalities in the transformants, and all plants showed a multiple shoot phenotype. This enhancement of phenotypic severity may be attributable to a unique feature of the GSE domain. The GSE domain is enriched with the amino acids proline (P), serine (S), and glutamate (E) at its C-terminal end (amino acids 205 to 231; KSEGVGSSEDDMSPSGRENEPPEIDPR). Peptide regions enriched with P, E, S, and threonine (T) (PEST sequence) have been known to act as signals for promoting protein degradation (Rogers et al., 1986). Deletion of this domain may prolong the half-life of OSH15, resulting in the induction of severe phenotypes of the transformants.
The Function of the ELK Domain
Based on its predicted secondary structure, it has been suggested that the ELK domain may be involved in protein–protein interactions (Mushegian and Koonin, 1996; Sakamoto et al., 1999). It also has been proposed that this domain may function as a nuclear localization signal (Meisel and Lam, 1996). In this study, we have suggested that the ELK domain is not essential for nuclear targeting, DNA binding, or homodimer formation. The apparent effect of the deletion of the ELK domain was the enhancement of the transactivation activity against a reporter gene under the control of the cis-acting motif of OSH15. As was the case for the deletion of KNOX1, the ELK domain may have a suppressive function against the expression of a target gene(s). However, in contrast to ΔKNOX1, ΔELK induced a unique phenotype (blade-less) in which leaves lacked leaf blade formation but showed normal formation of the sheath. This finding leads us to speculate that this domain not only has the suppressive function but also may have another function (e.g., interaction with a specific protein). Further studies are necessary to reveal the precise function of the ELK domain.
How Does OSH15 Function in Vivo?
Based on the yeast one-hybrid experiment using a reporter gene controlled by the OSH15 target cis motif, we conclude that the transrepressive function of OSH15 is one of the important factors for the induction of the abnormal phenotypes of transformants. However, this does not mean that OSH15 itself does not have a transactivating function against its target gene in vivo. Indeed, our studies revealed that the intact OSH15 caused an ∼10-fold increase in LacZ activity relative to the basal activity. A dual function of this type also has been reported for the HOX (typical HD)-PBX (TALE HD) complex. The HOX-PBX complex can be converted from a repressor to an activator through differential interactions with other coregulators, depending on the cellular context (Saleh et al., 2000). By analogy, we suggest that OSH15 may regulate the expression of its target genes as both a transactivator and a transrepressor, depending on the nature of the target genes and/or the cellular context, by the formation of multimeric complexes in vivo.
According to the expression analyses of OSH genes, OSH15 can interact with other OSH proteins because the spatial expression pattern of OSH15 sometimes overlaps with that of other OSH genes. During the late globular embryo stage, for example, OSH15 expression is observed in the area of subsequent shoot development and overlaps with that of OSH1, OSH43, OSH6, and OSH71. After the formation of the SAM, the expression of OSH15 is downregulated in the SAM and in turn is localized at the boundaries of the shoot lateral organs, overlapping the expression of OSH6 and OSH71 (Sato et al., 1998; Sentoku et al., 1999). Although it is possible that other TALE HD proteins or other transcriptional regulators may interact with OSH15, OSH15 can change its interaction with other OSH proteins through different developmental stages. This speculation is attractive because the state of the SAM at various stages, including before its formation, could be determined by the various combinations of OSH proteins present. The combination of different kinds of OSH proteins may be involved in the determination of the state of the SAM.
METHODS
Construction of OSH15 Derivatives
The intact OSH15 (amino acids 1 to 355) was constructed by amplifying the OSH15 coding region (in pBluescript SK−) by polymerase chain reaction (PCR) using a 5′ forward primer including the EcoRI site and a 3′ reverse primer encompassing the SalI site. The PCR products were inserted at the EcoRI–SalI site of pBluescript SK (Stratagene). To construct the OSH15 Δ1–103, OSH15 Δ1–144, and OSH15 Δ1–235 derivatives, the intact OSH15 (described above) coding region was amplified by PCR in the same way. The OSH15 ΔKNOX1 (Δ91–125), ΔKNOX2 (Δ143–194), ΔKNOX1+2 (Δ91–185), ΔGSE (Δ184–230), ΔELK (Δ235–256), M1, M2, M3, and M4 derivatives were constructed by PCR-mediated mutagenesis using the TaKaRa in vitro mutagenesis kit (TaKaRa, Siga, Japan). All OSH15 derivatives were verified by sequence analysis.
Plant Materials and Transformation Procedure
Rice plants (Oryza sativa cv Nipponbare) were used for the analyses. Transgenic rice plants were grown in a growth chamber maintained at 30°C (day) and 24°C (night). Procedures for rice tissue culture and transformation with Agrobacterium tumefaciens were as described previously (Hiei et al., 1994). For the transformation, the hygromycin-resistant binary vector containing the ACT1 promoter and the NOS terminator was used as reported previously (Sentoku et al., 2000).
Histological Analysis
Plant materials were fixed overnight at 4°C in 4% paraformaldehyde and 0.25% glutaraldehyde in a 0.1 M sodium phosphate buffer pH 7.4, dehydrated through a graded ethanol series followed by a t-butanol series (Sass, 1958), and finally embedded in Paraplast Plus (Sherwood Medical, St. Louis, MO). Microtome sections of 7 to 10 μm thick were applied to silan-coated glass slides (Matsunami Glass, Osaka, Japan). The sections were deparaffinized in xylene, rehydrated through a graded ethanol series, and dried overnight before staining with hematoxylin.
Protein Gel Blot Analysis
Protein gel blot analysis with anti-OSH15 antiserum was performed as described previously (Sentoku et al., 2000).
Transient Expression Assay
For the construction of green fluorescent protein (GFP) chimeric proteins of the OSH15 derivatives, the OSH15 derivative cDNAs were amplified to replace the stop codon with the XbaI site by PCR. These PCR products were digested with appropriate restriction enzymes, and the fragments were ligated into the cassette vector (Chiu et al., 1996) containing the 35S promoter of cauliflower mosaic virus (35S promoter)-GFP-NOS terminator between the 35S promoter and GFP. These clones were sequenced to confirm that no nucleotide substitution occurred during amplification. OSH15N-GFP was constructed by ligating the EcoRI–AvaII fragment of the intact OSH15 between the 35S promoter and GFP. GFP-fused proteins were expressed in onion bulb epidermal cells using particle bombardment (Bio-Rad).
Electrophoretic Mobility Shift Assay (EMSA)
To produce proteins of the OSH15 derivatives, the OSH15 derivative cDNAs were inserted into the pET-32a expression vector (Novagen, Madison, WI) in the sense orientation and expressed in BL21 (DE3) Escherichia coli cells (Stratagene). DNA probes containing the TGTCA motif or its derived sequences were excised with restriction endonucleases, purified on 8% polyacrylamide gels, labeled with 32P-dCTP using Klenow fragment, and purified on Sephadex G-50 columns. DNA binding reactions were performed at 4°C for 30 min in 10 mM Tris, pH 7.5, 75 mM NaCl, 1 mM EDTA, 500 μM DTT, 10% glycerol, 0.05% Nonidet P-40, and 50 mg L−1 poly(dI-dC)·poly(dI-dC) (Amersham Pharmacia Biotech) and subjected to EMSA using 5 to 7% polyacrylamide gels in 0.25 × Tris-borate-EDTA buffer.
Yeast Two-Hybrid Analysis
The MATCHMAKER yeast two-hybrid system (Clontech, Palo Alto, CA) was used. The OSH15 derivatives were inserted into the yeast expression vectors pACT2 and pGBT9. To determine the interaction affinity, a yeast strain, Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met−, gal80Δ, URA3::GAL1UAS-GAL1TATA-LacZ), was used. The β-galactosidase liquid assay was performed according to the Clontech manual.
In Vitro Pull-Down Assay
Fusion proteins between glutathione S-transferase (GST) and OSH proteins were generated in the pGEX-4T-1 vector (Amersham Pharmacia Biotech). GST-OSH proteins or GST were expressed in E. coli according to the manufacturer's instructions. 35S-Methionine–labeled OSH15 protein was synthesized using the TNT T7 coupled wheat germ extract system (Promega) with linearized pGADT7 (Clontech)-OSH15 plasmid according to the manufacturer's instructions. The production of labeled protein was confirmed by SDS-PAGE. Three micrograms of purified GST or GST-OSH fusion protein was preincubated with glutathione–Sepharose 4B (Amersham Pharmacia Biotech) in 500 μL of binding buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol, 1% Nonidet P-40, and 1% BSA) for 60 min at 4°C. After the addition of 35S-methionine–labeled OSH15 protein, the samples were incubated for another 60 min at 4°C. The Sepharose beads were washed three times in binding buffer and another two times in binding buffer without BSA. Bound proteins were eluted by boiling in 2 × sample buffer for 5 min before loading onto 12.5% SDS-polyacrylamide gels.
Transactivation Experiment in Yeast
The MATCHMAKER yeast one-hybrid system (Clontech) was used. The OSH15 derivatives were inserted into the yeast expression vector pGADT7 (EcoRI–SalI site) to produce the GAL4 activation domain chimera proteins or into pGADT7 (KpnI–BamHI site), which contains the simian virus 40 nuclear localization signal but not the GAL4 activation domain. To produce the reporter gene construct, the fragments containing four repeats of the TGTCAC sequence or its derivatives (5′-GAATT[CCTGTCACCA]4GTCGAC-3′, 5′-GAATT[CCTG-TGACCA]4GTCGAC-3′, 5′-GAATT[CCTCTCAGCA]4GTCGAC-3′) were inserted into pLacZi at the EcoRI–SalI site. In this experiment, the yeast strain YM4271 (MATa, ura3-52, his3-200, ade2-101, lys2-801, leu2-3, 112, trp1-901, tyr1-501, gal4-Δ512, gal80-Δ538, ade5::hisG) was used.
Acknowledgments
We are grateful to Dr. Yasuo Niwa (Shizuoka Prefectural University, Japan) for providing the 35S-GFP-NOS cassette vector. This study was partially supported by a Grant-in-Aid for Scientific Research on Priority Areas (Molecular Mechanisms Controlling Multicellular Organization of Plants) from the Ministry of Education, Science, Sports, and Culture of Japan.
References
- Abu-Shaar, M., Ryoo, H.D., and Mann, R.S. (1999). Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter, M., Schier, A., and Gehring, W.J. (1990). Homeodomain proteins and regulation of gene expression. Curr. Opin. Cell Biol. 2 485–495. [DOI] [PubMed] [Google Scholar]
- Berthelsen, J., Zappavigna, V., Ferretti, E., Mavilio, F., and Blasi, F. (1998. a). The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 17 1434–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen, J., Zappavigna, V., Mavilio, F., and Blasi, F. (1998. b). Prep1, a novel functional partner of Pbx proteins. EMBO J. 17 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen, J., Kilstrup-Nielsen, C., Blasi, C., Mavilio, F., and Zappavigna, V. (1999). The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino, E., Reimund, B., Wildt-Perinic, D., and Clerc, R.G. (1995). A novel homeobox protein which recognize a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270 31178–31188. [DOI] [PubMed] [Google Scholar]
- Bürglin, T.R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin, T.R. (1998). The PBC domain contains a MEINOX domain: Coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 208 113–116. [DOI] [PubMed] [Google Scholar]
- Chan, S.K., Jaffe, L., Capovilla, M., Botas, J., and Mann, S.R. (1994). The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with Extradenticle, another homeoprotein. Cell 78 603–615. [DOI] [PubMed] [Google Scholar]
- Chang, C.P., Jacobs, Y., Nakamura, T., Jenkins, N.A., Copeland, N.G., and Cleary, M.L. (1997). Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol. 17 5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W.-L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan, C., Theis, J., and O'Farrell, P.H. (1988). The sequence specificity of homeodomain-DNA interaction. Cell 54 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, W.J. (1987). Homeoboxes in the study of development. Science 236 1245–1252. [DOI] [PubMed] [Google Scholar]
- Harven, D., Gutfinger, T., Parnis, A., Eshed, Y., and Lofschitz, E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84 735–744. [DOI] [PubMed] [Google Scholar]
- Hayashi, S., and Scott, M.P. (1990). What determines the specificity of action of Drosophila homeodomain proteins? Cell 63 883–894. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hoshikawa, K. (1989). Leaf. In The Growing Rice Plant. (Tokyo: Nobunkyo), pp. 86–121.
- Kerstetter, R., Vollbrecht, E., Lowe, B., Veit, B., Yamaguchi, J., and Hake, S. (1994). Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter, R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124 3045–3054. [DOI] [PubMed] [Google Scholar]
- Kissinger, S.R., Liu, B., Martin-Blanco, E., Kornberg, T.B., and Pabo, C.O. (1990). Crystal structure of an engrailed homeodomain-DNA complex at 2.8Å resolution: A framework for understanding homeodomain-DNA interactions. Cell 63 579–590. [DOI] [PubMed] [Google Scholar]
- Krusell, L., Rasmussen, I., and Gausing, K. (1997). DNA binding sites recognised in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare). FEBS Lett. 408 25–29. [DOI] [PubMed] [Google Scholar]
- Laughon, A. (1991). DNA binding specificity of homeodomains. Biochemistry 30 11357–11367. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Lu, Q., and Kamps, M.P. (1996. a). Selective repression of transcriptional activators by pbx1 does not require the homeodomain. Proc. Natl. Acad. Sci. USA 93 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., and Kamps, M.P. (1996. b). Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: Proposal for a model of a Pbx1-Hox-DNA complex. Mol. Cell. Biol. 16 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H., McMuller, M.D., and Finer, J.J. (1994). Identification of a homeobox containing gene with enhanced expression during soybean (Glycine max L.) somatic embryo development. Plant Mol. Biol. 24 465–473. [DOI] [PubMed] [Google Scholar]
- Matsuoka, M., Ichikawa, H., Saito, A., Tada, Y., Fujimura, T., and Kano-Murakami, Y. (1993). Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell 5 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel, L., and Lam, E. (1996). The conserved ELK-homeodomain of KNOTTED-1 contains two regions that signal nuclear localization. Plant. Mol. Biol. 30 1–14. [DOI] [PubMed] [Google Scholar]
- Müller, K., Romano, N., Gerstner, O., Garcia-Maroto, F., Pozzi, C., Salamini, F., and Rohde, W. (1995). The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374 727–730. [DOI] [PubMed] [Google Scholar]
- Mushegian, A., and Koonin, E.V. (1996). Sequence analysis of eukaryotic developmental proteins: Ancient and novel domains. Genetics 144 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A., Tamaoki, M., Sakamoto, T., and Matsuoka, M. (2000). Over-expression of tobacco knotted1-type class 1 homeobox genes alters various leaf morphology. Plant Cell Physiol. 41 583–590. [DOI] [PubMed] [Google Scholar]
- Otting, G., Qian, Y.Q., Billeter, M., Muller, M., Affolter, M., Gehring, W.J., and Wuthrich, K. (1990). Protein-DNA contacts in the structure of a homeodomain-DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 9 3085–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passner, J.M., Ryoo, H.D., Shen, L., Mann, R.S., and Aggarwal, A.K. (1999). Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397 714–719. [DOI] [PubMed] [Google Scholar]
- Qian, Y.Q., Billeter, M., Otting, G., Müller, M., Gehring, W.J., and Wüthrich, K. (1989). The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: Comparison with prokaryotic repressors. Cell 59 573–580. [DOI] [PubMed] [Google Scholar]
- Reiser, L., Sanchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42 151–166. [PubMed] [Google Scholar]
- Rieckhof, G.E., Casares, F., Ryoo, H.D., Abu-Shaar, M., and Mann, R.S. (1997). Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeoprotein. Cell 91 171–183. [DOI] [PubMed] [Google Scholar]
- Rogers, S., Wells, R., and Rechsteiner, M. (1986). Amino acid sequence common to rapidly degraded proteins: The PEST hypothesis. Science 234 364–369. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., Nishimura, A., Tamaoki, M., Kuba, M., Tanaka, H., Iwahori, S., and Matsuoka, M. (1999). The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell 11 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, M., Rambaldi, I., Yang, X., and Featherstone, M.S. (2000). Cell signalling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferase. Mol. Cell. Biol. 20 8623–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass, A.E. (1958). Botanical Micro Technique, 3rd ed. (Ames, IA: Iowa State University Press).
- Sato, Y., Sentoku, N., Nagato, Y., and Matsuoka, M. (1998). Two separable functions of a rice homeobox gene, OSH15, in plant development. Plant Mol. Biol. 38 983–998. [DOI] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., Kurata, N., Ito, Y., Kitano, H., and Matsuoka, M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11 1651–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., and Matsuoka, M. (2000). Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220 358–364. [DOI] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeobox gene, Knotted-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7 787–795. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., and Hake, S. (1994). Molecular genetic approaches to leaf development: Knotted and beyond. Can. J. Bot. 72 617–625. [Google Scholar]
- Tamaoki, M., Tsugawa, H., Minami, E., Kayano, T., Yamamoto, N., Kano-Murakami, Y., and Matsuoka, M. (1995). Alternative RNA products from a rice homeobox gene. Plant J. 7 927–938. [DOI] [PubMed] [Google Scholar]
- Tamaoki, M., Kusaba, S., Kano-Murakami, Y., and Matsuoka, M. (1997). Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol. 38 917–927. [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi, M., Itoh, H., Oyama, N., Koshioka, M., and Matsuoka, M. (1998). Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J. 15 391–400. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Veit, B., Sinha, N., and Hake, S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350 241–243. [DOI] [PubMed] [Google Scholar]
- Zhang, W., McElroy, D., and Wu, R. (1991). Analysis of rice Act1 5′ region activity in transgenic rice plants. Plant Cell 3 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]