Abstract
The COP9 signalosome is a highly conserved protein complex initially identified as a repressor of photomorphogenesis. Here, we report that subunit 6 of the Arabidopsis COP9 signalosome is encoded by a family of two genes (CSN6A and CSN6B) located on chromosomes V and IV, respectively. The CSN6A and CSN6B proteins share 87% amino acid identity and contain a MPR1p and PAD1p N-terminal (MPN) domain at the N-terminal region. The CSN6 proteins share homology with CSN5 and belong to the Mov34 superfamily of proteins. CSN6 proteins present only in the complex form and coimmunoprecipitate with other known subunits of the COP9 signalosome. Partial loss-of-function strains of the COP9 signalosome created by antisense and cosuppression with CSN6A exhibit diverse developmental defects, including homeotic organ transformation, symmetric body organization, and organ boundary definition. Protein blot analysis revealed that the defective plants accumulate significant amounts of ubiquitinated proteins, supporting the conclusion that the COP9 signalosome regulates multifaceted developmental processes through its involvement in ubiquitin/proteasome–mediated protein degradation.
INTRODUCTION
The COP9 signalosome is a nucleus-enriched multisubunit protein complex conserved among diverged organisms, including fission yeast, Drosophila melanogaster, Arabidopsis, human, and probably Anacystis nidulans (Chamovitz et al., 1996; Seeger et al., 1998; Freilich et al., 1999; Karniol et al., 1999; Mundt et al., 1999). Biochemical and molecular characterizations in Arabidopsis and mammals have demonstrated that the COP9 signalosome is composed of eight distinct subunits designated CSN1 to CSN8 (Seeger et al., 1998; Mundt et al., 1999; Wei and Deng, 1999; Deng et al., 2000). Interestingly, the COP9 signalosome shares remarkable subunit-to-subunit similarity with the lid subcomplex of the 26S proteasome. In addition, the in vivo presence of the free lid-like complex (PR500) of the 26S proteasome depends on the integrity of the COP9 signalosome in Arabidopsis (Peng et al., 2001a).
The COP9 signalosome was initially discovered during genetic analysis of light-regulated Arabidopsis seedling development (Wei and Deng, 1992; Wei et al., 1994a; Chamovitz et al., 1996). Arabidopsis seedlings follow two contrasting developmental pathways: photomorphogenesis in the light and skotomorphogenesis or etiolation in darkness. Genetic screens for constitutive photomorphogenic seedling development in darkness resulted in the identification of 11 Arabidopsis loci, collectively known as the pleiotropic COP/DET/FUS genes (Castle and Meinke, 1994; Miséra et al., 1994; Wei et al., 1994b; Wei and Deng, 1996, 1999; Chory, 1997). The recessive nature of the cop/det/fus mutations suggests that their encoded proteins are negative regulators of photomorphogenesis. Among these 11 mutated loci, eight are required for the biogenesis of the COP9 signalosome (Kwok et al., 1998). All isolated alleles in those eight loci result in severely retarded growth at the early seedling stage and lethality after the seedling stage. Molecular and genetic studies have shown that CSN8, CSN7, CSN4, CSN1, and CSN3 are encoded by COP9, FUS5, COP8/FUS4, COP11, and FUS11, respectively (Wei et al., 1994a; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999; Peng et al., 2001b). On the other hand, no corresponding mutant has been identified for CSN5 because it is encoded by two redundant genes, CSN5A and CSN5B. Two-dimensional protein gel analyses have demonstrated that both CSN5A and CSN5B are components of the COP9 signalosome complex (Kwok et al., 1998). Although CSN5A/CSN5B and CSN7 can be presented in both the complex and the monomeric forms, CSN1, CSN3, CSN4, and CSN8 exist exclusively in the complex.
The molecular and biochemical nature of the involvement of the COP9 signalosome in cell signaling and development has just begun to be revealed. In mammals, transient overexpression of CSN5 (JAB1) instigated the proteasome-mediated degradation of the cyclin-dependent kinase inhibitor p27 (Tomoda et al., 1999) and the precursor of the lutropin/choriogonadotropin receptor (Li et al., 2000). Human CSN6 was identified as an interactive partner of the human immunodeficiency virus 1 accessory protein Vpr (also called hVIP for Vpr interactive protein) and found to be involved in the G2/M phase transition of the cell cycle (Mahalingam et al., 1998). Although CSN1 (GPS1) was shown to modulate the JNK/MAPK signaling pathway, CSN5 (JAB1) appears to play a role in c-Jun–dependent transcriptional regulation (Claret et al., 1996; Kallunki et al., 1996; Spain et al., 1996). It has also been reported that in mammals the COP9 signalosome targets p53 for degradation in the ubiquitin/proteasome pathway by a specific phosphorylation (Bech-Otschir et al., 2001). In plants, genetic and biochemical results show that the COP9 signalosome is required for proteasome-mediated degradation of HY5, whereas it is dispensable for proteasome-mediated degradation of phytochrome A (Osterlund et al., 2000; Peng et al., 2001b). Most recently, it has been demonstrated that the COP9 signalosome interacts with the SCF E3 ligase complex and promotes deconjugation of RUB1 from Cullin (Lyapina et al., 2001; Schwechheimer et al., 2001). It was hypothesized that the COP9 signalosome may regulate multiple cellular and developmental processes by modulating specific E3 ligases and thus proteasome-mediated protein degradation (Schwechheimer et al., 2001). A general regulatory role in the ubiquitin/proteasome pathway could explain all of the reported roles of the COP9 signalosome in diverse cellular pathways.
In this article, we report the molecular cloning and characterization of the Arabidopsis COP9 signalosome subunit 6 genes CSN6A and CSN6B. Our studies demonstrate that CSN6 is present exclusively in the complex form. By creating reduction-of-function transgenic plants of the COP9 signalosome, we generated viable partial loss-of-function plants for the COP9 signalosome and revealed many new roles of the COP9 signalosome in plant development.
RESULTS
Arabidopsis CSN6 Is Encoded by a Family of Two Genes
Although the COP9 signalosome has been shown to contain eight subunits (Wei et al., 1998; Wei and Deng, 1999), the molecular characterization of three Arabidopsis subunits remains to be completed. On the basis of the peptide sequences obtained from the purified cauliflower COP9 signalosome (Serino et al., 1999), the corresponding Arabidopsis expressed sequence tag clones were identified through a database search. The full-length cDNA sequences for two distinct Arabidopsis genes encoding the CSN6 subunit were characterized. While one gene is located on chromosome V (referred to as CSN6A), the other gene is located on chromosome IV (referred to as CSN6B). Genomic DNA gel blot analysis and further search for the completed genome sequence revealed that CSN6A and CSN6B are the only two closely related genes. The CSN6A and CSN6B genes encode proteins of 317 and 318 amino acids, respectively (Figure 1A). Both proteins contain a 140–amino acid MPR1p and PAD1p N-terminal (MPN) domain at the N-terminal region and share ∼40% overall amino acid identity with the human counterpart (Figure 1A) (Asano et al., 1997). Quantitative reverse transcriptase–mediated polymerase chain reaction analysis indicated that the expression of CSN6A was at least 10-fold greater than that of CSN6B at the seedling stage (data not shown). Among the five cauliflower peptides, peptides 2, 4, and 5 share greater identity with CSN6A, whereas peptides 1 and 3 are more closely related to CSN6B (Figure 1B), indicating that both proteins are present in the cauliflower COP9 signalosome. Although CSN6A and CSN6B are only 87% identical in their overall amino acid sequences, the two corresponding genes share complete nucleic acid sequence identity in eight blocks of 20 nucleotides or longer. Because 21- to 23-nucleotide fragments are ideal for causing RNA interference (Zamore et al., 2000), these identical DNA sequences in multiple regions suggest the possibility that these two genes can be affected simultaneously by cosuppression or antisense approaches.
Figure 1.
Sequence Comparison of CSN6 Proteins.
(A) Sequence comparison between the Arabidopsis COP9 signalosome subunit 6 (CSN6A and CSN6B) and its human homolog hCSN6. The MPN domain is indicated by the line below the sequence. Identical amino acids are shown in blackened type.
(B) Sequence comparison among the cauliflower peptides and the corresponding regions of CSN6A and CSN6B.
CSN6 Shares Homology with CSN5 and Belongs to the Mov34 Protein Family
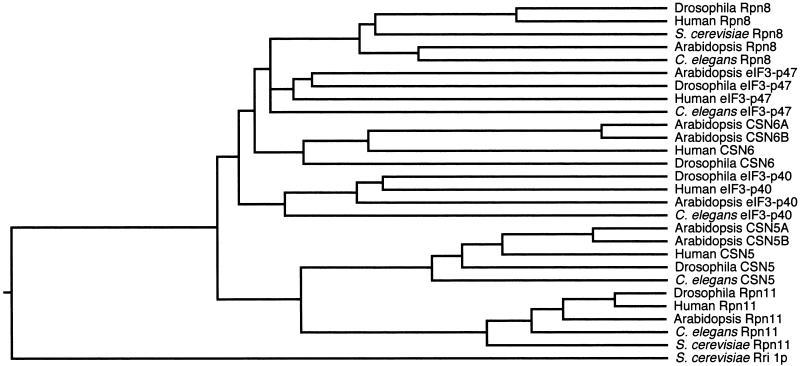
Like CSN6, CSN5 of the Arabidopsis COP9 signalosome also is encoded by two redundant genes, and two-dimensional protein gel analyses revealed that both gene products are present in the COP9 signalosome (Kwok et al., 1998). Interestingly, sequence analysis revealed that CSN6 and CSN5 share significant homology with each other and that both belong to the Mov34 protein family (Figure 2), a name taken from the first protein in the family, murine Mov34, a subunit of the 26S proteasome. Most members of the Mov34 protein family are components of the COP9 signalosome, translation initiation factor 3 (eIF3) complex, and the 26S proteasome (Asano et al., 1997). Figure 2 illustrates the phylogenetic relationship of 28 proteins from five diverse organisms. This genetic tree divides into two major cladistic branches. One contains CSN5 and Rpn11, a subunit of the 26-proteasome lid subcomplex. The other contains CSN6, Rpn8, eIF3-p47, and eIF3-p40, the latter three include one subunit of the 26S proteasome (Rpn8) and two subunits of the elF3 complex (elF3-p47 and elF3-p40). This result suggests that the lid subcomplex of the 26S proteasome and the COP9 signalosome probably are evolved from a common ancestor protein complex that contained two subunits of the Mov34 protein family. Although CSN5 and Rpn11 were evolved from one of these two ancestor proteins, CSN6, Rpn8, eIF3-p40, and eIF3-p47 were evolved from the other one. Rri 1p is a budding yeast protein with 30% identity to Arabidopsis CSN5 that does not appear to be a component of the three protein complexes.
Figure 2.
Phylogenetic Relationship of the Mov34 Family Proteins.
This phylogenetic tree was generated using the Jotun Hein algorithm in the Megalign program (gap penalty, 11; gap length, 3) (DNAstar, Madison, WI). The Mov34 group of proteins was selected from the following five organisms: human, Arabidopsis, Drosophila, C. elegans, and Saccharomyces cerevisiae. From top to bottom (from Drosophila Rpn8 to S. cerevisiae Rri 1p), the GenBank accession numbers for the proteins listed are AAF47199, S65491, T41067, O24412, T33096, U54561, AAF52101, NP_003745, T20338, AY048692, AF434762, NP_006824, AAF56022, AAF52210, NP_003747, AAG53614, T28786, AAC36344, AAC36343, XP_011713, AAF55321, T29320, AAF08394, NP_005796, BAA97246, T33344, NP_011194, and NP_010065.
CNS6 Cofractionates with Other Known Subunits of the COP9 Signalosome
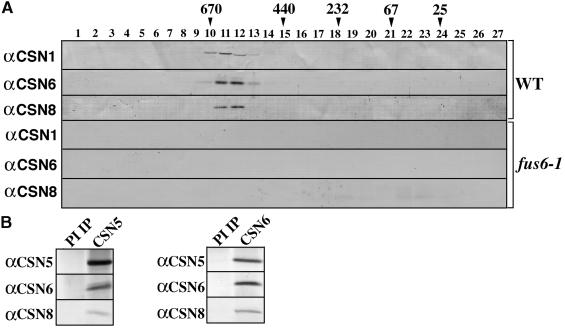
To confirm that CSN6 is a subunit of the COP9 signalosome, we performed cofractionation and coimmunoprecipitation analyses with CSN6 and other previously defined COP9 signalosome subunits. Because of the high similarity between CSN6A and CSN6B, the polyclonal antibodies raised against CSN6A could not differentiate CSN6A from CSN6B in protein blot studies (see Methods). Thus, we refer to the protein band detected in protein blots with antibodies against CSN6A as CSN6, which likely is composed of both CSN6A and CSN6B. As shown in Figure 3A, CSN6 cofractionated with other known COP9 signalosome subunits in a Superose 6 gel filtration column and was found exclusively in the complex form. In the mutant of the COP9 signalosome (fus6-1), however, the CSN6, CSN8, and CSN1 proteins were all absent, indicating that the cellular level of CSN6 depends on the integrity of the COP9 signalosome. To further substantiate this observation, we performed coimmunoprecipitation experiments with antibodies against CSN5 and CSN6. As shown in Figure 3B, CSN6 coimmunoprecipitated with CSN1 and CSN5 from total protein extracts with antibodies against both CSN1 and CSN5, demonstrating that CSN6 was in fact an integral component of the COP9 signalosome.
Figure 3.
CSN6 Is a Subunit of the COP9 Signalosome.
(A) Cofractionation of the putative COP9 signalosome subunit 6 (CSN6) with CSN1 and CSN8 on a Superose 6 H/R gel filtration column. The proteins were extracted from 6-day-old light-grown Arabidopsis seedlings (ecotype Columbia). Protein size markers are indicated at the top in kilodaltons. Lanes 1 to 27 indicate the corresponding fractions eluted from the gel filtration column (started at 7.5 mL, 0.5 mL per fraction). Rabbit polyclonal antibodies (α) against CSN6A, CSN1, and CSN8 were used for protein blots. WT, wild type.
(B) Coimmunoprecipitation between CSN6 and two other COP9 signalosome subunits (CSN5 and CSN8). The antibodies used for the immunoprecipitation are indicated at the top. The antibodies (α) used for protein blots are indicated at left. PI IP, preimmune serum immunoprecipitation.
CSN6 Interacts with Multiple Subunits of the COP9 Signalosome in Yeast Two-Hybrid Assays
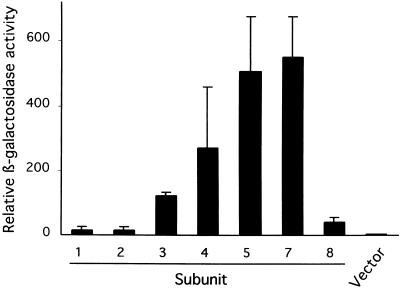
Several recent reports have described some subunit–subunit interaction within the COP9 signalosome (Serino et al., 1999; Kapelari et al., 2000; Tsuge et al., 2001). In Arabidopsis, it has been reported that CSN4 interacts with CSN1, CSN5, CSN7, and CSN8 in yeast (Serino et al., 1999). In addition, yeast two-hybrid assays have revealed that CSN1 interacts with itself, CSN5, and CSN7 and that CSN7 interacts with CSN8 (Kwok et al., 1998; Karniol et al., 1999). To further elucidate the overall subunit interaction of the COP9 signalosome, we examined the interactions of CSN6 with all other published subunits of the COP9 signalosome. As shown in Figure 4, CSN6 interacted strongly with CSN5 and CSN7 and interacted weakly with CSN3 and CSN4 in a yeast two-hybrid assay.
Figure 4.
CSN6A Interacts with Multiple Subunits of the COP9 Signalosome.
The interactions between CSN6A and the subunits of the COP9 signalosome were analyzed in a yeast two-hybrid assay. Full-length CSN6A was fused to the activation domain and assayed for its interaction with other COP9 signalosome subunits in a LexA-DNA binding domain fusion. The relative β-galactosidase (LacZ) activity shown is the average of at least six independent transformants. The LexA-CSN6A fusion caused lethality in yeast and thus was not available for examination of the CSN6–CSN6 interaction. Error bars represent the standard deviation from the mean of the independent transformants tested. Error bars represent the standard deviation from the mean of the independent transformants tested.
Reduction of the Cellular Level of the COP9 Signalosome Results in Multifaceted Developmental Defects
Because all available COP9 signalosome mutants exhibit severely retarded growth at the early seedling stage and are lethal after seedling development, we had been prevented from examining the possible role of the COP9 signalosome in other developmental stages. To circumvent this problem, reduction-of-function strains for the COP9 signalosome were obtained by introducing 35S promoter–driven sense and antisense CSN6A cDNA into wild-type Arabidopsis. We selected 16 of 45 sense and 19 of 53 antisense lines that exhibited early seedling phenotypes in the T2 generation for detailed studies in both T2 and subsequent generations. In all of the lines examined, the overall phenotypes in the selfed progeny were similar between the sense and antisense lines (Table 1). Protein blot analyses revealed that all of the 16 sense lines and 19 antisense lines had dramatically reduced amounts of COP9 signalosome compared with those of wild-type plants, indicating that the sense lines resulted in cosuppression of CSN6 genes (data not shown).
Table 1.
Summary of Plant Phenotypes in 10 Representative CSN6A Transgenic Linesa
| Lines | Total Plant | Purple Dwarf |
Apical Dominance |
Flower Defect |
Symmetry | Fusion | Missing Root/Leaf |
Lethal | Others | Normal |
|---|---|---|---|---|---|---|---|---|---|---|
| T2 | ||||||||||
| A-C-3 | 219 | 29 | 27 | 25 | 36 | 24 | 32 | 37 | 59 | 55 |
| A-C-5 | 167 | 11 | 24 | 8 | 21 | 12 | 14 | 32 | 43 | 48 |
| A-C-21 | 117 | 7 | 21 | 5 | 19 | 6 | 20 | 21 | 38 | 31 |
| A-C-24 | 112 | 0 | 1 | 0 | 27 | 0 | 87 | 89 | 19 | 1 |
| A-C-10 | 79 | 3 | 5 | 1 | 9 | 3 | 0 | 5 | 26 | 42 |
| S-C-18 | 302 | 18 | 51 | 21 | 55 | 27 | 13 | 31 | 79 | 105 |
| S-C-20 | 144 | 7 | 10 | 4 | 21 | 8 | 27 | 48 | 40 | 41 |
| S-C-3 | 104 | 4 | 15 | 7 | 27 | 10 | 5 | 12 | 26 | 36 |
| S-C-29 | 81 | 5 | 4 | 0 | 5 | 1 | 6 | 17 | 33 | 37 |
| S-C-1 | 67 | 11 | 4 | 1 | 14 | 3 | 0 | 12 | 16 | 21 |
| T3 | ||||||||||
| A-C-3-1 | 138 | 5 | 18 | 7 | 32 | 12 | 12 | 57 | 41 | 6 |
| A-C-3-2 | 177 | 12 | 23 | 9 | 31 | 6 | 14 | 37 | 48 | 71 |
| A-C-3-3 | 173 | 8 | 12 | 19 | 28 | 14 | 29 | 39 | 42 | 68 |
| S-C-18-1 | 146 | 4 | 12 | 4 | 19 | 14 | 21 | 29 | 29 | 58 |
| S-C-18-2 | 112 | 6 | 14 | 3 | 15 | 7 | 19 | 31 | 19 | 49 |
a The transgenic lines starting with the letter A or S are CSN6A antisense or sense cosuppression transgenic plants, respectively. The 35S promoter was used to drive both sense and antisense CSN6A cDNA. For plants in the T2 generation, the total number of plants represents the total number of gentamycin-resistant plants. For the T3 plants, the total number of plants includes gentamycin-sensitive plants (without gentamycin selection). Because many plants had more than one phenotype, the combined number of plants in all phenotype groups is larger than the total number of plants examined. Plant phenotypes are classified into general groups (see text for more details). Plants with single and triple cotyledons were considered to exhibit loss of symmetry. “Others” includes plants with many phenotypes not described here, such as bladeless and twisted leaves, inflorescence phenotypes, silique phenotypes, etc. “Normal” includes those plants that appeared to be normal morphologically.
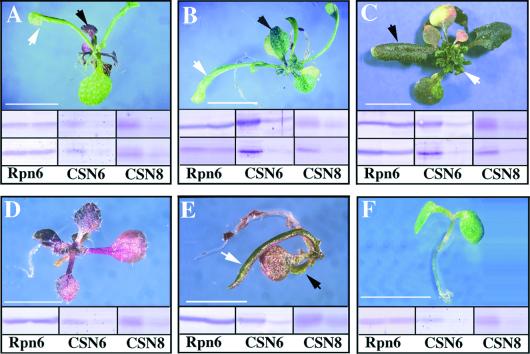
As shown in Table 1, a striking feature of these transgenic lines is that each independent line segregates a variety of different phenotypes regardless of the specific phenotype of the parental plant. Although the phenotype of a given progeny varies, the overall spectra of phenotypes within a given line are quite consistent. In fact, the phenotype spectra have been inherited through at least four tested generations so far. It was also common for an individual transgenic plant to exhibit more than one phenotype. Several typical examples of the phenotypes from the antisense line progeny are shown in Figure 5. Protein blots with antibodies to two representative COP9 signalosome subunits (CSN6 and CSN8) clearly indicated that the COP9 signalosome levels in these transgenic plants were reduced significantly.
Figure 5.
Phenotype Diversity of CSN6A Antisense Transgenic Plants.
All plants shown ([A] to [F]) are from two representative antisense lines in the T2 generation. Total proteins were extracted from rosette leaves or cotyledons if no true leaves were produced. For the protein gel blots shown below each plant, the two lanes represent the wild-type control on the left and the transgenic plant on the right. The proteasome subunit Rpn6 was used as an internal loading control. Antibodies against CSN6 and CSN8 were used to detect the levels of the COP9 signalosome. For the protein gel blots shown in (A) to (C), those in the top row were proteins extracted from the leaves indicated by black arrows, and those in the bottom row were proteins extracted from the leaves indicated by white arrows. Bars = 5 mm.
One frequent phenotype among the progeny was a purple, short, and small plant (<2 cm in diameter, 8 cm in height, without lateral shoots) (Table 1, Figures 5A and 5D). Interestingly, ∼10 to 40% (depending on the line) of these purple plants switched to produce green leaves at a specific development stage, generating plants carrying two sets of leaves with distinct colors (Figure 5A). Protein blot analyses of these two leaf types indicated that they both had significantly reduced levels of CSN6 and CSN8 (Figure 5A, bottom). These results were consistent with the fact that the structures of the green leaves also were aberrant in other respects (Figure 5A).
Phenotype transitions were common among the transgenic plants. Two additional examples are shown in Figures 5B and 5C. In these two plants, the leaf shape switched from one type (black arrow) to another (white arrow) during development. Other common developmental defects, including plants with bladeless leaves (Figure 5E) and seedlings lacking a shoot apical meristem function (Figure 5F), and their phenotypes are described below.
Flower Development
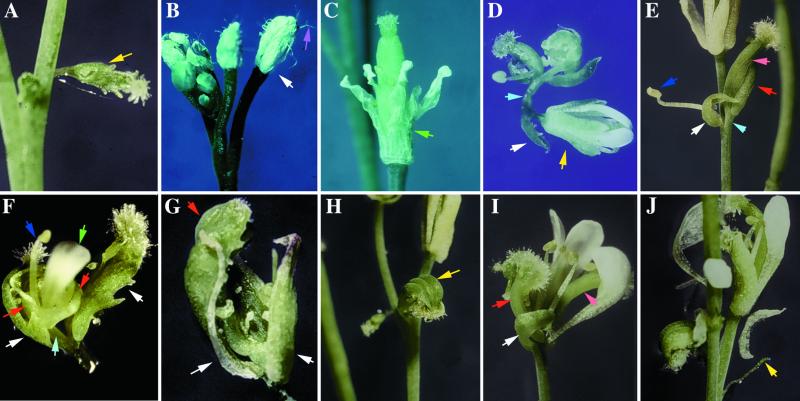
Although the COP9 signalosome is present at its highest level in floral tissue (Wei et al., 1994a; Chamovitz et al., 1996), little is known about its role in flower development. A close examination of the adult CSN6A cosuppression and antisense plants revealed a great variety of flower developmental defects (Table 1, Figure 6). The flower phenotype varies, even in the same plant, ranging from normal flowers to severely defective flowers. In many cases, defective flowers deviated so severely from the regular structure and whorl pattern that it became impossible to define whorl identities. Figure 6 presents some selected examples from both sense and antisense lines. The most common flower development defects were homeotic organ transformation and absence of floral organs. Examples include a cauline leaf transformed into a carpeloid structure with stigmatic papillae on the tip (Figure 6A), flowers having petal-like sepals with typical stellated sepal trichomes on the surface (Figure 6B), a flower with extra, fused petals while missing sepals (Figure 6C), and flowers with sepals and petals transformed into carpeloid structures and having ovules at the margins (Figures 6F and 6I). There also were defects in floral patterning, such as flowers formed at the axils of the first whorl organs (Figure 6D), flowers initiating floral organs spirally (Figures 6E and 6F), and flowers having internode elongation (Figures 6D to 6F). Other flower phenotypes observed were flowers with sepal/petal mosaic organs but completely missing the third and fourth whorl organs (Figure 6G), flowers replaced by a carpeloid structure generated by fusing three carpels (Figure 6H), and a flower replaced by a filamentous structure (Figure 6J). Some of these flower development phenotypes were similar to those of the various ufo mutants (Ingram et al., 1995; Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995; Lee et al., 1997). The defects include flowers with carpeloid structures replacing the first whorl organs, spiral initiation of floral organs, missing petals and stamens, “empty” flowers (Figure 6G), a filament structure replacing a flower, stellated trichromes on the petal-like first whorl organ, and organ fusion. However, the flower phenotypes observed in the CSN6A transgenic plants appeared to be more diverse than those seen in the ufo mutants. For example, the initiation of new flowers from the axils of first whorl organs (Figure 6D) resembles that of AP1 mutants (Irish and Sussex, 1990). Because similar flower phenotypes also have been observed in the transgenic plants of other COP9 signalosome subunits (Peng et al., 2001b), the flower phenotypes were clearly related to COP9 signalosome function, supporting a role of the COP9 signalosome in flower development.
Figure 6.
Representative Flower Phenotypes from CSN6 Antisense and Sense Cosuppression Transgenic Plants.
(A) Cauline leaf converted to a carpeloid structure with stigmatic tissue at the tip.
(B) Flowers with petal-like sepals (white arrow) containing stellated trichromes (purple arrow).
(C) Flower missing sepals while having extra petals; some petals were fused at the base.
(D) Normal flower (yellow arrow) produced at the axil of the first whorl organ in a flower with severe defects.
(E) Flower with spiral initiation of organs, except a sepal and a stamen; the other two organs are transformed into carpeloid structures. The flower does not possess petals and shows reductions in a number of other organs.
(F) Flower missing a gynoecium and with abnormal elongation of the internode between whorl 1 and whorl 2. The first whorl of the flower is converted to a carpeloid structure with ovules at the margins. The carpeloid structures at right face away from the floral axil. Once the internode elongated, two more carpeloid structures were produced, and a petal and a stamen were produced at the center.
(G) “Empty” flower with a sepal, a sepal/petal mosaic organ, and a carpeloid structure containing ovule-like structures. The center of the flower is empty.
(H) Flower with only three fused carpeloid structures (yellow arrow).
(I) Flower with sepals and petals converted to carpeloid structures. This flower also has a reduction in the number of sepals and stamens.
(J) Filament structure produced in place of a flower.
(A) to (E) show flowers from sense cosuppression line S-C-18, and (F) to (J) show flowers from antisense transgenic line A-C-3. All flowers were from plants in the T2 generation. The arrows point to organs or flowers of interest. The color coding is as follows: yellow for modified or misplaced flowers; white for first whorl organs or modified first whorl organs; purple for trichomes; green for petals or modified petals; sky blue for elongated flower axils; blue for stamens; red for modified second whorl organs; and pink for modified third or fourth whorl organs.
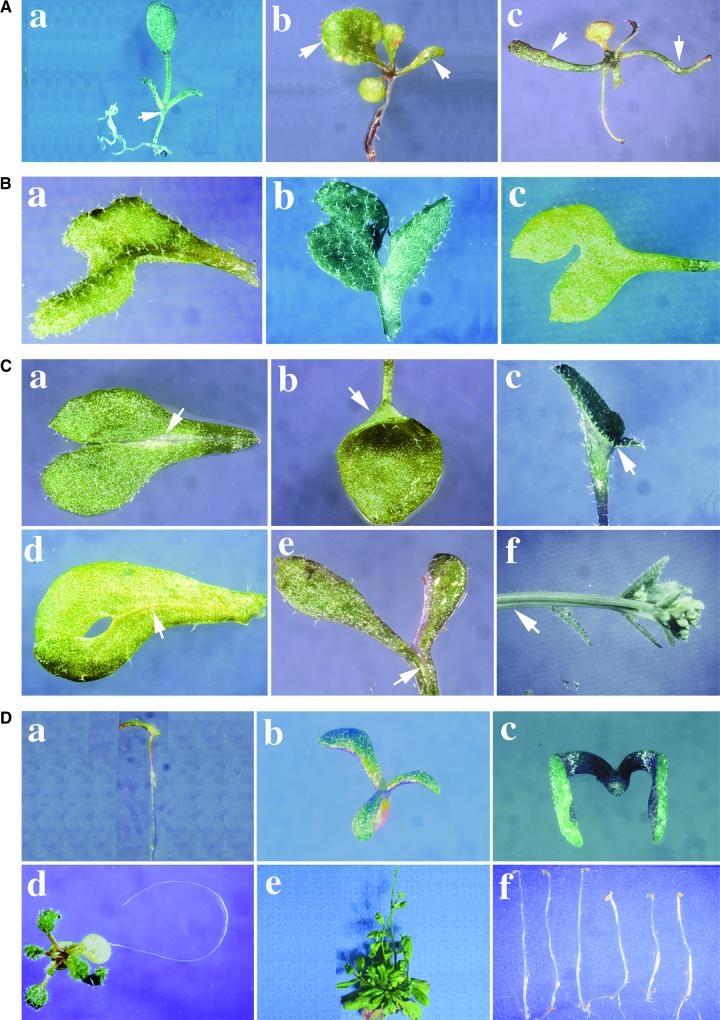
(A) Effects on phyllotaxis of the transgenic plants. The arrows point to the first two leaves or the positions of the first two leaves.
(B) Effects on body symmetry of the transgenic plants.
(C) Effects on organ boundary definition. In each panel, organ fusion positions are marked by arrows.
(D) Phenotypes with similarities to those of the auxin response mutants. The plants in (f) were in the T3 generation. The three seedlings at left were wild-type controls; the three seedlings at right were transgenic plants.
All plants were from antisense line A-C-3 in the T2 generation unless indicated otherwise.
Phyllotaxis and Body Symmetry
In Arabidopsis, the first two leaves are initiated at opposite positions simultaneously and are similar in size (Telfer and Poethig, 1994). After the juvenile stage, the leaves start to initiate in a spiral pattern. As shown in Figure 7A, panel a, this transgenic plant was missing one of the first pair of true leaves, suggesting a change in phyllotaxis. In addition, plants in panels b and c had the first two leaves in different sizes and shapes, indicating a defect in the timing of organ initiation or in organ size control. Body symmetric organization also was disrupted in some of the transgenic plants. The most striking symmetric defects were observed in leaves. As shown in Figure 7B, the three leaves in panels a, b, and c lost bilateral symmetry. Some of those leaf defects were similar to the leaf polarity defects described previously (McConnell and Barton, 1998). As summarized in Table 1, body symmetry defects are among the most commonly observed phenotype groups in CSN6A transgenic plants. In fact, many of the flowers shown in Figure 6 also lost typical radial symmetry. Therefore, it is evident that the COP9 signalosome plays an essential role in maintaining symmetric body development at multiple developmental stages.
Figure 7.
Effects of Antisense CSN6A on Multifaceted Plant Developmental Processes.
Organ Fusion
The definition of boundaries among developing organs is an essential process of pattern formation in plant development. Defects in the ability to define a boundary result in organ fusion. On the other hand, postgenital organ fusion is required for the development of a gynoecium in angiosperms. Abnormal petal and carpeloid fusions and failure of postgenital carpel fusion were observed frequently in the defective flowers of the CSN6A antisense and sense transgenic plants shown in Figure 6. In addition, improper organ fusions were observed between leaf midveins (Figure 7C, panel a), between leaf lobe margins within a leaf or between two leaves (Figure 7C, panels b to d), between petioles (Figure 7C, panel e), and between stems (Figure 7C, panel f). The overall frequency of improper organ fusion in the selected representative lines is summarized in Table 1. Currently, dozens of loci regulating organ fusion have been identified in Arabidopsis (Levin et al., 1998; Lolle et al., 1998). It will be interesting to determine if the COP9 signalosome affects the stability of some key regulators encoded by these genes.
Auxin-Regulated Development
Phenotype examination of the CSN6A transgenic lines found that some of the plants had phenotypes resembling those of the auxin response mutants, similar to the recently reported CSN5 partial loss-of-function strains (Schwechheimer et al., 2001). At the seedling stage, progeny of both antisense and cosuppression transgenic lines produced seedlings with a single cotyledon, triple cotyledons, or without roots at frequencies of <5% (Figure 7D, panels a to c, Table 1). Some transgenic plants were defective in producing lateral roots (Figure 7D, panel d); this 3-week-old transgenic plant is still missing lateral roots, whereas control plants produced 18 lateral roots on average (data not shown). In addition, many transgenic plants were short and had lost apical dominance (Table 1). Instead of producing a main inflorescence first, multiple secondary branches bolted simultaneously (Figure 7D, panel e). The seedlings shown in Figure 7D, panel f (right half), developed short hypocotyls and open cotyledons in the darkness, in contrast to the completely etiolated wild-type plants at left. Approximately 5 to 15% of the antisense transgenic plants in line A-C-3 possessed this partial photomorphogenic developmental phenotype. All of the phenotypes shown in Figure 7D were similar to the phenotypes reported for the auxin pathway mutants, such as PIN-FORMED and PINOID for triple and single cotyledons (Goto et al., 1987; Bennett et al., 1995). The tir1-1 mutant exhibited a reduction of lateral roots (Ruegger et al., 1998), and axr1-1 and axr1-12 exhibited loss of apical dominance, dwarfing, and the dark partial photomorphogenic phenotype (Estelle and Somerville, 1987; Gray and Estelle, 2000). It has been demonstrated that ubiquitin/proteasome–mediated protein degradation plays a key role in mediating auxin responses (Gray and Estelle, 2000; Worley et al., 2000). The phenotype similarity between both the CSN6A (this work) and CSN5 (Schwechheimer et al., 2001) transgenic plants and the auxin response pathway mutants further substantiates that the COP9 signalosome plays a role in regulating proteasome-mediated protein degradation in response to auxin.
Developmental Defects of CSN6A Transgenic Plants Are Likely Caused by Malfunction of the Ubiquitin/Proteasome Pathway Regulated by the COP9 Signalosome
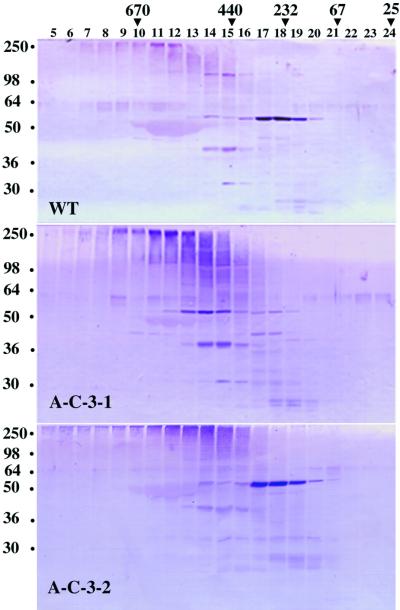
To determine whether the transgenic plants with reduced levels of the COP9 signalosome show altered degradation of cellular ubiquitinated proteins, we examined the profiles of ubiquitinated proteins in representative transgenic plants. The transgenic plant A-C-3-1 exhibited a strong and early phenotype similar to that shown in Figure 5D, and the transgenic plant A-C-3-2 exhibited a phenotype similar to that shown in Figure 7D, panel e. These two types of plants accumulated more polyubiquitinated proteins (high molecular mass smear in fractions 9 to 16) compared with those accumulated by the wild-type control, as shown in Figure 8. A-C-3-1 had a stronger phenotype than did A-C-3-2. Correspondingly, more polyubiquitinated proteins accumulated in plant A-C-3-1 (Figure 8). Comparison of the profiles of ubiquitinated proteins indicated that there was a general correlation between the phenotypic strength and the degrees of accumulation of the polyubiquitinated proteins among the transgenic plants examined (data not shown).
Figure 8.
Comparison of the Ubiquitinated Protein Profiles of Wild-Type and Selected CSN6A Antisense Transgenic Plants.
The proteins were extracted from 1-month-old rosette leaves. For line A-C-3-1, the protein extract was from CSN6A antisense transgenic plants, as shown in Figure 4D. For line A-C-3-2, the protein extract was from antisense transgenic plants, as shown in Figure 7D, panel e. For wild-type control (WT), the protein extract was from 1-month-old rosette leaves of Columbia wild type. The total protein extracts were size fractionated on a Superose 6 HR gel filtration column with TB buffer (see Methods). The collected fractions were concentrated with StrataClean beads and further separated on 10% SDS–polyacrylamide gels. Antibodies against ubiquitin were used in the protein gel blots to detect the ubiquitinated proteins. Lane numbers indicate gel filtration fraction numbers, and molecular mass markers (kilodaltons) are indicated at the top. Molecular mass markers (kilodaltons) for the SDS-PAGE are indicated at left. The polyubiquitinated proteins appear as high molecular mass smears in fractions 9 to 16.
DISCUSSION
In this work, we report the molecular cloning and characterization of two genes encoding subunit 6 of the COP9 signalosome in Arabidopsis. Our functional studies revealed that the COP9 signalosome plays a critical role in multifaceted developmental processes.
Arabidopsis CSN6 Is a Component of the COP9 Signalosome and Is Encoded by Two Redundant Genes
Several lines of evidence support the notion that Arabidopsis CSN6 is a component of the COP9 signalosome and is encoded by a family of two redundant genes. First, gel filtration and protein blot analyses show that CSN6 is present only in a complex of the same molecular size as the COP9 signalosome and has the same elution profile of other known subunits of the COP9 signalosome. In the fus6-1 mutant in which the COP9 signalosome is disrupted, CSN6 is absent in the complex fractions and in the total cell extracts, indicating that CSN6 is present exclusively in the COP9 signalosome and that its accumulation depends on the integrity of the COP9 signalosome. Second, CSN6 can be coimmunoprecipitated by antibodies against CSN5. Furthermore, CSN5 and CSN8 also can be coimmunoprecipitated by antibodies against CSN6, suggesting that these proteins are associated physically. Third, the peptide sequences obtained from purified cauliflower COP9 signalosome match the predicted amino acid sequences of CSN6A and CSN6B. Although three peptides more closely resemble CSN6A, the other two peptides match better with CSN6B. This finding indicates that both cauliflower counterparts of CSN6A and CSN6B are components of the COP9 signalosome. Because cauliflower and Arabidopsis are closely related species, it is likely that both Arabidopsis CSN6A and CSN6B are present in the Arabidopsis COP9 signalosome. Fourth, the fact that no corresponding mutants of CSN6A and CSN6B have been identified is consistent with our finding that they are two redundant genes encoding the same subunit. This is because the mutant screen has been near saturation (Miséra et al., 1994; Wei and Deng, 1999).
Because CSN6 and CSN5 both are encoded by two redundant genes, it is possible that the COP9 signalosome is a heterogeneous population in the cell with distinct forms of subunits CSN6 and CSN5. It is unlikely that the two forms of either CSN5 or CSN6 are present simultaneously in the same complex, because the abundance of the two versions of each subunit is drastically different. Furthermore, these two subunits seem to be encoded by a single gene in fission yeast, Caenorhabditis elegans, and Drosophila, which is consistent with the notion that the two forms of subunits 5 and 6 are functionally equivalent in the Arabidopsis COP9 signalosome.
Phenotypes of the COP9 Signalosome Reduction-of-Function Transgenic Lines and Their Functional Implications
Recently, it has been shown that the COP9 signalosome interacts with the core subunits of the SCF E3 ligase complex and regulates the levels of RUB modification of the Cullin 1 protein, a core subunit of the SCF E3 ligase (Lyapina et al., 2001; Schwechheimer et al., 2001). Furthermore, it was demonstrated using CSN5 partial loss-of-function lines that the COP9 signalosome plays an essential role in auxin response through direct interaction with SCFTIR1 (Schwechheimer et al., 2001). The CSN6A antisense and cosuppression plants also displayed phenotypes similar to those of the auxin response mutants. The auxin-like phenotypes in the CSN6 transgenic lines further substantiate the role of the COP9 signalosome in SCFTIR1-mediated auxin response.
Our CSN6A antisense and sense cosuppression transgenic plants displayed a great diversity of phenotypes, consistent with the hypothesis that the COP9 signalosome regulates multifaceted developmental processes by regulating distinct SCF E3 ligases. Our transgenic plant phenotypes covered almost every organ of the plant, including root, stem, cotyledon, rosette leaf, cauline leaf, trichome, inflorescence, flower organs, and siliques. These results are in agreement with the fact that the COP9 signalosome is expressed in all of these organs (Chamovitz et al., 1996; Wei and Deng, 1999). Some of the flower development defects in our transgenic lines resemble those of the ufo mutants, which clearly supports the hypothesis described above, because UFO is an F-box protein and a component of SCFUFO, a specific E3 ligase that regulates flower development (Samach et al., 1999).
A major characteristic feature of these transgenic plants is the stochastic nature of the phenotypic defects. Individual progeny of the same transgenic line often exhibited different phenotypes. Furthermore, it is common that different parts of the same plant have different phenotypes (Figure 5). For example, as shown in Figure 6D, the flower produced at the axil of the first whorl organ of a highly disturbed flower was actually a normal flower. Although the phenotypes among individual progeny vary greatly, several constant features are evident as a result of our studies. First, the spectra of the phenotypes for the reduction-of-function lines are quite similar when the population used was large enough (e.g., 100 or more). Second, for a given transgenic line, the spectrum of phenotypes is relatively constant from generation to generation and independent of the specific phenotype of the parent plant. Third, the phenotype defects developed at late developmental stages were in general milder, whereas the early phenotypes were more severe and often detrimental to plant development. Fourth, there seemed to be a trend in which the plants with the most severe phenotype had more drastic reductions in the abundance of the COP9 signalosome. However, it should be noted that the exact abundance of the COP9 signalosome in the proper cell types at the onset of the phenotype is the most relevant information, but it is not feasible to define this quantity with the available tools.
Why do individual progeny of the same transgenic line have different phenotypes and the different parts of the same transgenic plant have different phenotypes? It is possible that during plant development, the abundance of numerous regulatory proteins must be controlled in a precise temporal and spatial manner according to developmental and environmental cues. The COP9 signalosome plays a key role in controlling the levels of these proteins by regulating their degradation. The reduction of the COP9 signalosome may result in activities of these key regulators above or below their critical thresholds, leading to developmental defects. The stochastic nature of the phenotypes could be caused by the variation of regulatory protein abundance in different cells, tissues, and plants resulting from the reduction of COP9 signalosome levels. Although the onset of distinct developmental defects among different plants as well as in different organs of the same plant appeared to be quite stochastic, as a whole population these plants cover the most common phenotypes in all derivative lines examined as long as the population examined is sufficiently large. The stochastic nature of phenotype defects caused by aberrant protein degradation would be consistent with the observation that in a given ufo mutant plant, individual flowers also exhibit variation in their phenotypes (Ingram et al., 1995; Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995).
Both CSN6 and CSN5 Are Members of the Mov34 Superfamily and Evolved before the Divergence of the Proteasome “Lid” Subcomplex and the COP9 Signalosome
Among the eight subunits of the COP9 signalosome, the similarity between CSN5 and CSN6 is worth noting. Both CSN5 and CSN6 contain a MPN domain at the N-terminal region and are encoded by two redundant genes. More interestingly, CSN5 and CSN6 strongly interact with each other in the yeast two-hybrid assay. In addition, both CSN5 and CSN6 belong to the Mov34 superfamily of proteins. The members of this protein family are mainly components of the COP9 signalosome, the eIF3 complex, and the 26S proteasome. The PCI domain, a motif also shared by components in these three protein complexes, has been shown to be involved in incorporating CSN1 into the COP9 signalosome in mammals (Tsuge et al., 2001). Whether the MPN domain plays a role in protein–protein interaction and the assembly of these complexes remains to be determined.
Phylogenic relationship analysis with 28 proteins of the Mov34 family from five selected organisms revealed that CSN5 and CSN6 were separated earlier than were the COP9 signalosome and the lid subcomplex of the 26S proteasome. This finding implies the presence of an ancestor complex containing two subunits of the Mov 34 family. Although Rpn8, eIF3-p40, eIF3-p47, and CSN6 appear to be evolved from the same ancestor protein, CSN5 and Rpn11 appear to be evolved from another ancestor protein (Figure 2).
METHODS
Plant Materials and Arabidopsis Strains
The fus6-1 and other cop/fus mutants of Arabidopsis thaliana have been described previously (Castle and Meinke, 1994; Miséra et al., 1994; Wei et al., 1994b; Kwok et al., 1996). The seed were planted on agar plates containing growth medium and 1% sucrose (Wei et al., 1994a) and were cold treated at 4°C for 5 to 12 days before being transferred to specific growth chambers. The white light intensity used was 156 μmol·m−2·sec−1.
Identification of CSN6 cDNAs from Arabidopsis and Sequence Analyses
The peptide sequences of cauliflower CSN6 (Serino et al., 1999) were used to search the Arabidopsis expressed sequence tag (EST) database. Several EST clones representing two distinct but highly similar genes were identified. One of the EST clones is a full-length cDNA that was designated CSN6A and obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The other full-length EST was designated CSN6B. Comparison of the cDNA sequences with the Arabidopsis genomic sequences revealed the intron-exon structure of both genes and their respective genomic locations. The CSN6A gene is located on chromosome V, whereas CSN6B is located on chromosome IV.
Protein Extraction and Gel Filtration Chromatography
Arabidopsis seedlings were homogenized in Tris buffer (TB) containing 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 5 mM DTT, and 10% glycerol, with freshly added protease inhibitor phenylmethylsulfonyl fluoride at a final concentration of 2 mM and protease inhibitor cocktail (Boehringer Mannheim) as instructed by the manufacturer. The homogenate was microcentrifuged for 15 min, and the supernatant was filtered through a 0.2-μm filter (Gelman Sciences, Ann Arbor, MI) before loading onto a Superose 6 (HR 10/30) gel filtration column (Pharmacia). The column was equilibrated with a buffer containing 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 2 mM DTT, and 10% glycerol, and the proteins were eluted in the same buffer at a flow rate of 0.2 mL/min. All manipulations were performed at 4°C. Fractions of 0.5 mL were collected starting from the onset of the column void volume (7.5 mL) and were concentrated using StrataClean Resin (Stratagene) as described by Kwok et al. (1998). Equal volumes of each fraction were loaded onto a SDS–polyacrylamide gel, followed by immunoblot analysis. The molecular mass standards used for gel filtration size estimation were thyroglobulin (669 kD), apoferritin (443 kD), catalase (232 kD), aldolase (158 kD), and chymotrypsinogen (25 kD).
For protein extraction from transgenic plants, the tissues were frozen in liquid nitrogen and homogenized in microcentrifuge tubes with TB buffer. Total proteins were used for protein blot and gel filtration.
Immunoblot Analysis and Immunoprecipitation
All protein blot and immunoprecipitation analyses were performed as described previously (Staub et al., 1996) with minor modifications. The antibodies coupled with protein A beads were mixed with the protein extracts for 3 hr instead of 6 hr, and the beads were washed with PBS buffer supplemented with 0.4 M NaCl and 0.2% SDS. The immunoprecipitated proteins were released from the protein A beads and analyzed by SDS-PAGE. To detect the ubiquitinated proteins, the filter was autoclaved for 10 min to completely denature ubiquitin after the proteins were transferred to the filter and before protein blot analysis. The antibodies for ubiquitin were purchased from Sigma.
Plant Transformation and Analyses of the Transgenic Plants
The full-length cDNA of Arabidopsis CNS6A (ecotype Columbia) was cloned into the binary vector pPZPY122 in either the sense or antisense orientation under the control of the 35S promoter (Yamamoto et al., 1998). The constructs were transformed into Arabidopsis Columbia wild-type plants via vacuum infiltration. At least 100 independent resistant lines were obtained for both the sense and antisense constructs in the T1 generation. The seed produced by gentamycin-resistant (100 mg/L gentamycin) T1 plants were harvested from individual plants. A total of 53 antisense and 45 sense lines were selected for gentamycin resistance on plates in the T2 generation and examined for the number of T-DNA insertion loci. To facilitate the selection, seed were germinated in darkness for 3 days on the selecting medium and then transferred to a white light growth chamber. The greening process of the sensitive plants was blocked completely, whereas the resistant plant still turned green. Among the tested lines, 19 antisense and 16 sense lines exhibited obvious seedling phenotypes and were transferred to soil for adult phenotype observation. Tissues from the plants with interesting phenotypes were collected, and proteins were extracted for protein blot analyses. To ensure that the phenotypes observed in the T2 generation were not caused by the side effects of antibiotics or other growth conditions, seed from those representative lines also were germinated on antibiotic-free medium and transferred to soil for phenotype observations. The genotypes of the T2 plants were confirmed by examining the resistance of the T3 seedlings to gentamycin. Selected T3 and T4 populations also were grown in soil for phenotype observation and protein gel blot analyses.
Yeast Two-Hybrid Assay
The full-length cDNA clone of CSN6A was cloned in frame into the pEG202 and pJG4-5 vectors (Gyuris et al., 1993; Ausubel et al., 1995). Other constructs used have been described (Serino et al., 1999). All of the LexA fusion constructs were transformed to yeast strain EGY48 (Ausubel et al., 1995). The activation domain fusion constructs were transformed in yeast strain L40 (Invitrogen, Carlsbad, CA). The transformants were selected and mated (Bendixen et al., 1994). The mated colonies were incubated with 2 mL of liquid medium lacking histidine and tryptophan and supplemented with 2% galactose and 1% raffinose. The β-galactosidase activity was assayed as described previously (Kwok et al., 1998).
Accession Numbers
The accession numbers for the two distinct Arabidopsis genes encoding the CSN6 subunit are AY048692 (CSN6A) and AF434762 (CSN6B).
Acknowledgments
We are grateful to Arthur Galston, Magnus Holm, Ning Wei, and David Liptak for commenting on the manuscript. This research was supported by National Science Foundation Grant No. MCD-0077217 and Binational Agricultural Research and Development Fund Grant No. IS-3123-99 to X.-W.D. X.-W.D. is a National Science Foundation Presidential Faculty Fellow. Z.P. was the recipient of a National Institutes of Health postdoctoral fellowship.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010248.
References
- Asano, K., Vornlocher, H.P., Richter-Cook, N.J., Merrick, W.C., Hinnebusch, A.G., and Hershey, W.B. (1997). Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. J. Biol. Chem. 272 27042–27052. [DOI] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds (1995). Short Protocols in Molecular Biology. (New York: John Wiley).
- Bech-Otschir, D., Kraft, R., Huang, X., Henklein, P., Kapelari, B., Pollmann, C., and Dubiel, W. (2001). COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 20 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendixen, C., Gangloff, S., and Rothstein, R. (1994). A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 22 1778–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, S.R.M., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8 505–520. [Google Scholar]
- Castle, L., and Meinke, D. (1994). A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.-W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121. [DOI] [PubMed] [Google Scholar]
- Chory, J. (1997). Light modulation of vegetative development. Plant Cell 9 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret, F.-X., Hibi, M., Dhu, S., Toda, T., and Karin, M. (1996). A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 383 453–457. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., et al. (2000). Unified nomenclature for the COP9 signalosome: An essential regulator of development. Trends Genet. 16 289. [DOI] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C. (1987). Auxin-resistant mutants in Arabidopsis with an altered morphology. Mol. Gen. Genet. 206 200–206. [Google Scholar]
- Freilich, S., Oron, E., Kapp, Y., Nevo-Caspi, Y., Orgad, S., Segal, D., and Chamovitz, D.A. (1999). The COP9 signalosome is essential for development of Drosophila melanogaster. Curr. Biol. 9 1187–1190. [DOI] [PubMed] [Google Scholar]
- Goto, N., Starke, M., and Kranz, A.R. (1987). Effect of gibberellins on flower development of the pin-formed mutant of Arabidopsis thaliana. Arabidopsis Inf. Serv. 23 66–71. [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25 133–138. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E.A., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 7 791–803. [DOI] [PubMed] [Google Scholar]
- Ingram, G.C., Goodrich, J., Wilkinson, M.D., Simon, R., Haughn, G.W., and Coen, E.S. (1995). Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish, V.F., and Sussex, I.M. (1990). Function of the apetala1 gene during Arabidopsis floral development. Plant Cell 2 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki, T., Deng, T., Hibi, M., and Karin, M. (1996). c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87 929–939. [DOI] [PubMed] [Google Scholar]
- Kapelari, B., Bech-Otschir, D., Hegerl, R., Schade, R., Dumdey, R., and Dubiel, W. (2000). Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J. Mol. Biol. 300 1169–1178. [DOI] [PubMed] [Google Scholar]
- Karniol, B., Malec, P., and Chamovitz, D.A. (1999). Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of COP9 complex. Plant Cell 11 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Misera, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/ FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Matsui, M., Ecker, J.R., and Deng, X.-W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Wolfe, D.S., Nilsson, O., and Weigel, D. (1997). A LEAFY co-regulator encoded by unusual floral organs. Curr. Biol. 7 95–104. [DOI] [PubMed] [Google Scholar]
- Levin, J.Z., and Meyerowitz, E.M. (1995). UFO: An Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7 529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, J.Z., Fletcher, J.C., Chen, X., and Meyerowitz, E.M. (1998). A genetic screen for modifiers of UFO meristem activity identifies three novel FUSED ORGAN genes required for early flower development in Arabidopsis. Genetics 149 579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Liu, X., and Ascoli, M. (2000). p38JAB1 binds to the intracellular precursor of the lutropin/choriogonadotropin receptor and promotes its degradation. J. Biol. Chem. 275 13386–13393. [DOI] [PubMed] [Google Scholar]
- Lolle, S.L., Hsu, W., and Pruitt, R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshiaes, R.J. (2001). Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292 1382–1385. [DOI] [PubMed] [Google Scholar]
- Mahalingam, S.A.V., Patel, M., Kieber-Emmons, T., Koo, G.D., Muschel, R.J., and Weiner, D.B. (1998). HIV-1 Vpr interacts with a human 34-kDa Mov34 homologue, a cellular factor linked to the G2/M phase transition of the mammalian cell cycle. Proc. Natl. Acad. Sci. USA 95 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- Miséra, S., Müller, A.J., Weiland-Heidecker, U., and Jürgens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol Gen. Genet. 244 242–252. [DOI] [PubMed] [Google Scholar]
- Mundt, K.E., Porte, J., Murray, J.H., Brikos, C., Christensen, P.U., Caspari, T., Haagan, I.M., Millar, J.B.A., Simanis, V., Hofmann, K., and Carr, A.M. (1999). The COP9 signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 9 1427–1430. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.H., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated Arabidopsis development. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Staub, J.M., Serino, G., Kwok, S.F., Kurepa, J., Bruce, B.D., Vierstra, R.D., Wei, N., and Deng, X.-W. (2001. a). The cellular level of PR500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol. Biol. Cell. 12 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.-W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development, in press. [DOI] [PubMed]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The Tir1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1P. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, G.W.H., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20 433–445. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X-W. (2001). Interaction of the COP9 signalosome with the E3 ubiquitin ligase SCFTir1 in mediating auxin-response. Science 292 1379–1382. [DOI] [PubMed] [Google Scholar]
- Seeger, M., Kraft, R., Ferrell, K., Bech-Otschir, D., Dumdey, R., and Schade, R. (1998). A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 12 469–478. [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S.F., Matsui, M., Wei, W., and Deng, X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same locus that encodes subunit 4 of the COP9 signalosome. Plant Cell 11 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain, B.H., Bowdish, K.S., Pacal, A.R., Staub, S.F., Koo, D., Chang, C.Y., Xie, W., and Colicelli, J. (1996). Two human cDNAs, including a homolog of Arabidopsis FUS6(COP11), suppress G-protein- and mitogen-activated protein kinase-mediated signal transduction in yeast and mammals. Mol. Cell. Biol. 16 6698–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., Wei, N., and Deng, X.-W. (1996). Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., and Poethig, R.S. (1994). Leaf development in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 379–401.
- Tomoda, K., Kuboto, Y., and Kato, J.-Y. (1999). Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398 160–165. [DOI] [PubMed] [Google Scholar]
- Tsuge, T., Matsui, M., and Wei, N. (2001). The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol. 305 1–9. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1992). COP9: A new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1996). The role of pleiotropic COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1999). Making sense of the COP9 signalosome, a conserved regulatory protein complex from Arabidopsis to human. Trends Genet. 15 98–103. [DOI] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.-W. (1994. a). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, N., Kwok, S.F., von Arnim, A.G., and Deng, X.-W. (1994. b). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Tomohiko, T., Serino, G., Dohmae, N., Takio, K., Matsui, M., and Deng, X.-W. (1998). The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8 919–922. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.D., and Haughn, G.W. (1995). UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell 7 1485–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J. 21 553–562. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., Matsui, M., Ang, L.H., and Deng, X.W. (1998). Role of COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101 25–33. [DOI] [PubMed] [Google Scholar]