Abstract
Interactions between TALE (three–amino acid loop extension) homeodomain proteins play important roles in the development of both fungi and animals. Although in plants, two different subclasses of TALE proteins include important developmental regulators, the existence of interactions between plant TALE proteins has remained unexplored. We have used the yeast two-hybrid system to demonstrate that the Arabidopsis BELL1 (BEL1) homeodomain protein can selectively heterodimerize with specific KNAT homeodomain proteins. Interaction is mediated by BEL1 sequences N terminal to the homeodomain and KNAT sequences including the MEINOX domain. These findings validate the hypothesis that the MEINOX domain has been conserved between plants and animals as an interaction domain for developmental regulators. In yeast, BEL1 and KNAT proteins can activate transcription only as a heterodimeric complex, suggesting a role for such complexes in planta. Finally, overlapping patterns of BEL1 and SHOOT MERISTEMLESS (STM) expression within the inflorescence meristem suggest a role for the BEL1–STM complex in maintaining the indeterminacy of the inflorescence meristem.
INTRODUCTION
The shoot of a plant arises through the reiterative formation of lateral primordia from the main axis of growth. These primordia, in turn, may develop as leaves, axillary shoots, or floral organs (Lyndon, 1998). Subtle variations in primordium initiation can produce remarkable differences in shoot architecture. Consequently, the control of lateral primordium formation and identity remains a focal point for studies of plant development.
The BELL1 (BEL1) gene of Arabidopsis is crucial to the production of lateral primordia within developing ovules, the reproductive structures that contain the female gametophyte and develop into seed after fertilization. Recessive mutations in BEL1 result in the inability to properly initiate the formation of the lateral integuments that would normally develop into the seed coat (Robinson-Beers et al., 1992; Modrusan et al., 1994). BEL1 encodes a TALE (three–amino acid loop extension) homeodomain transcription factor, an atypical superclass of homeodomain protein that is conserved among eukaryotes and characterized by a three–amino acid extension to the loop connecting the first and second helices of the homeodomain (Reiser et al., 1995; Bürglin, 1997). Consistent with its function in ovule development, the pattern of BEL1 transcription predicts the initiation of integument formation in the ovule primordium (Reiser et al., 1995). BEL1 appears to belong to a small subclass of plant TALE homeodomain proteins that includes the light-responsive Arabidopsis thaliana homeodomain 1 (ATH1; Quaedvlieg et al., 1995) and Malus domestica homeodomain 1 (MDH1; Dong et al., 2000).
Beyond the inference that it regulates the transcription of other genes, nothing is known of the molecular mechanisms by which BEL1 controls integument development. Computer modeling, however, suggests that a sequence of amino acids N terminal to the BEL1 homeodomain can form an amphipathic α-helix that might be involved in protein–protein interactions (Reiser et al., 1995). Heterodimerization between transcription factors is common and likely represents a way to optimize the potential of an individual transcription factor to differentially regulate multiple genes. Indeed, interactions between TALE homeodomain proteins have been well documented in fungi and animals and are known to regulate developmental processes, including mating type determination, embryogenesis, and limb development (Knoepfler et al., 1997; Rieckhof et al., 1997; Berthelsen et al., 1998b). Therefore, it is possible that BEL1 homodimerizes or heterodimerizes with other TALE homeodomain proteins of Arabidopsis to regulate the transcription of genes important to developmental processes.
In animals, the PBC subclass of TALE homeodomain proteins includes the vertebrate pre–B cell leukemia homeodomain proteins (PBX1, PBX2, and PBX3) and fruit fly EXTRADENTICLE (EXD). PBC proteins are named for two conserved N-terminal domains that are collectively referred to as the PBC domain (Bürglin and Ruvkun, 1992). Interestingly, an N-terminal portion of the PBC domain is essential for heterodimerization with a second subclass of TALE homeodomain proteins, the MEINOX proteins (Bürglin, 1997; Knoepfler et al., 1997; Berthelsen et al., 1998b). MEINOX proteins, including the vertebrate MEIS1 and PREP1 and fruit fly HOMOTHORAX (HTH), also are named for two conserved N-terminal domains that have been collectively referred to as the MEINOX domain (Bürglin, 1997). Experiments have demonstrated that the MEINOX domain is required for interaction with PBC proteins (Knoepfler et al., 1997; Berthelsen et al., 1998b). Although the interaction between PBC and MEINOX proteins results in cooperative binding to DNA (Knoepfler et al., 1997; Berthelsen et al., 1998b), heterodimerization between PBC and MEINOX proteins does not require their homeodomains or the presence of a DNA target (Berthelsen et al., 1998a). Apparently, interaction between the N-terminal domains of PBC and MEINOX proteins is required to mask a nuclear export sequence in the PBC protein and thereby facilitate the nuclear localization of the heterodimer (Rieckhof et al., 1997; Abu-Shaar et al., 1999; Berthelsen et al., 1999). PBC proteins and PBC–MEINOX heterodimers also can bind cooperatively to DNA target sequences with vertebrate HOX homeodomain proteins (ENGRAILED homeodomain protein in fruit fly) and thereby modify the target specificity, binding affinity, and regulatory activity of these typical homeodomain proteins (reviewed by Chariot et al., 1999). These interactions are mediated by C-terminal sequences of both PBC and HOX proteins and require DNA binding.
In plants, MEINOX proteins are represented by the KNOTTED1-like homeobox (KNOX) protein family (MEINOX being derived from MEIS and KNOX; Bürglin, 1997) and are easily distinguished from other TALE proteins, including BEL1, by their conserved homeodomains, MEINOX domains, and ELK domains (Kerstetter et al., 1994). KNOX proteins can be further classified into two subgroups on the basis of sequence similarity (Kerstetter et al., 1994; Bürglin, 1997). Several molecular and genetic analyses associate class 1 KNOX proteins with important roles in meristem formation and maintenance (for a comprehensive review, see Reiser et al., 2000). This is best exemplified by recessive mutations in the SHOOT MERISTEMLESS (STM) gene, which result in the inability to form or sustain the Arabidopsis shoot apical meristem (SAM; Barton and Poethig, 1993; Clark et al., 1996; Endrizzi et al., 1996). Consistent with its function in meristem activity, STM is expressed throughout the shoot meristems of Arabidopsis (Long et al., 1996; Long and Barton, 1998). Ectopic expression of class 1 KNOX proteins can cause ectopic meristem development (Sinha et al., 1993; Chuck et al., 1996). Class 1 KNOX proteins also may be important in lateral organ formation because loss-of-function mutations in KNOTTED1 (KN1) result in abnormal leaf and flower development in maize (Kerstetter et al., 1997), whereas gain-of-function mutations lead to knotted or lobed leaves in maize, Arabidopsis, tobacco, and tomato (reviewed by Reiser et al., 2000).
Because they contain the MEINOX domain, which is clearly required for interactions between animal TALE proteins, Bürglin (1998) has predicted that KNOX proteins dimerize with other plant TALE proteins to control transcription. This hypothesis implies homodimerization or heterodimerization between KNOX proteins and/or heterodimerization between KNOX and BEL1-like proteins. To test these possibilities, we used BEL1 as bait in a yeast two-hybrid screen to identify Arabidopsis BEL1-interacting proteins. In this article, we report the ability of BEL1 to interact with KNOX proteins from A. thaliana (KNATs). The data indicate that only specific combinations of BEL1 and KNAT proteins can heterodimerize. Although an N-terminal domain of each protein mediates the general interactions, the C-terminal region of BEL1 appears to be particularly important in discriminating between potential KNAT partners. The transcription patterns of BEL1 and STM overlap in the SAM and floral meristem, which suggests the potential for their protein products to heterodimerize. The identification of BEL1 and KNAT proteins as interacting partners has important implications not only for the study of their respective functions in plants but also for the study of PBC–MEINOX complexes in animals.
RESULTS
Identification of BEL1-Interacting Proteins
Reiser et al. (1995) identified an amino acid sequence N terminal to the BEL1 homeodomain that had the potential to form an amphipathic α-helix, a motif known to mediate protein–protein interactions through so-called coiled coil structures. This suggested that BEL1 may function within a complex of transcription factors. Interestingly, sequence comparisons suggest that this putative amphipathic α-helix is actually part of a larger domain, stretching from P262 to E350 of BEL1, that is highly conserved among 14 BEL1-LIKE HOMEODOMAIN (BLH) proteins identified in Arabidopsis, apple, and rice (Quaedvlieg et al., 1995; Reiser et al., 1995; The Arabidopsis Genome Initiative, 2000; Dong et al., 2000; A. Samach, M.S. Pidkowich, Z. Modrusan, and G.W. Haughn, unpublished results). We refer to this sequence as the BELL domain (Figure 1A). In addition, a short sequence N terminal to the BELL domain, from I193 to G213, also is conserved among at least 13 BLH proteins and also may form an amphipathic α-helix. We refer to this region as the SKY domain for the nearly invariable sequence of serine, lysine, and tyrosine residues found within this sequence (Figure 1A). Thus, BEL1 contains two sequences that possibly mediate protein–protein interactions.
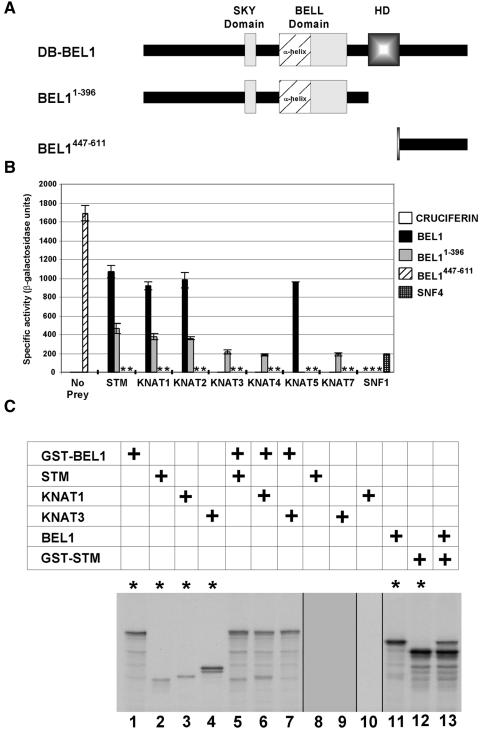
Figure 1.
Interactions between BEL1 and KNAT Proteins via the Yeast Two-Hybrid System.
(A) Scheme of intact and truncated BEL1 sequences used as bait in two-hybrid screens and quantitative assays. The patterned box (HD) represents the homeodomain, whereas the other boxes represent the SKY and BELL domains. The putative amphipathic α-helix, represented by the cross-hatched box, comprises the N-terminal portion of the BELL domain. Numbers in superscript indicate the BEL1 amino acids encoded by each clone. Intact BEL1 was used for two-hybrid screen A, whereas BEL1-396 was used for two-hybrid screen B.
(B) β-Galactosidase activity resulting from the combinations of BEL1 bait constructs and pTA-KNAT prey clones. pTA-based “prey” clones are indicated at the bottom of the histogram, and pDB-based “bait” clones are indicated to the right of the histogram. Enzyme activity resulting from the combination of the yeast pDB-SFN4 and pTA-SFN1 clones is included as a positive control for interacting proteins, combinations with pDB-CRUCIFERIN are included as a negative control, and pDB-BEL1447-611 is included as a clone that induces β-galactosidase activity autonomously. Asterisks denote tests that were not performed. For each prey category, the bait combinations are presented from left to right as follows: pDB-CRUCIFERIN, pDB-BEL1, pDB-BEL11-396, pDB-BEL1447-611, and pDB-SNF4.
(C) GST pulldown experiments demonstrating specific BEL1/KNAT interactions in vitro. The table indicates which combinations of translation products were used in each lane. The bottom panel shows an SDS-PAGE gel of the various translation products before or after incubation with glutathione–agarose beads as indicated (asterisks denote lanes in which translation products were not incubated with beads). In vitro translated GST-BEL1 displays a major, presumably full-length product and several minor truncated products (lane 1). STM and KNAT1 display single products (lanes 2 and 3, respectively), whereas KNAT3 displays two major products (lane 4). No STM, KNAT3, or KNAT1 products are retained by the beads alone (lanes 8 to 10, respectively). GST-BEL1, but not KNAT3, is retained by the beads when these translation products are incubated together (lane 7). STM and KNAT1, however, are retained by the beads in combination with GST-BEL1 (lanes 5 and 6, respectively), indicating specific interactions between BEL1 and STM or KNAT1. To confirm that BEL1 interacts with STM, the experiment was conducted in the reverse orientation. In vitro translated BEL1 (lane 11) and GST-STM (lane 12) display major, presumably full-length products and several minor truncated products. BEL1 was not retained by the glutathione–agarose beads alone (data not shown, but comparable to the results shown for STM, KNAT1, and KNAT3), but it was retained in combination with GST-STM (lane 13).
Because the identity of protein partners could reveal further clues regarding the role of BEL1 in development, we used a GAL4-based yeast two-hybrid screen to discover proteins that are capable of interacting with BEL1 (Kohalmi et al., 1997). The complete BEL1 coding sequence was cloned into the vector pBI-770 (Kohalmi et al., 1997) to give pDB-BEL1 (Figure 1A), by which BEL1 could be expressed as a C-terminal fusion to the GAL4 DNA binding domain (DB). This bait construct was introduced into the yeast two-hybrid host strain YPB2 (Bartel et al., 1993) and then used to screen a two-hybrid cDNA expression library encoding Arabidopsis proteins as C-terminal fusions to the GAL4 transcription activation domain (TA; Samach et al., 1999). Colonies expressing putative BEL1-interacting proteins were selected on the basis of histidine prototrophy and further screened on the basis of 3-amino-1′,2′,4′-triazol (3-AT) resistance and lacZ expression. Fourteen colonies were recovered in this manner (Table 1). To ensure that the histidine prototrophy depended on the combined presence of the pDB-BEL1 bait plasmid and a prey plasmid, TA-containing plasmids (pTA; see Methods) were recovered and reintroduced into YPB2 containing pDB-BEL1 or pDB-CRUCIFERIN (negative control). Of the original 14 prey clones, only 11 continued to activate both YPB2 marker genes in specific combination with pDB-BEL1. Remarkably, sequence analysis revealed that these 11 cDNA clones represented just two genes, each of which encodes a member of the KNAT family of homeodomain proteins; 8 clones represented KNAT1 (Lincoln et al., 1994), and 3 clones represented KNAT5 (Serikawa et al., 1996).
Table 1.
Results of Two-Hybrid System Screens
| Bait | Transformants Screened | His+ Colonies | LacZ+ | Specific Interactions | KNAT Clones |
|---|---|---|---|---|---|
| pDB-BEL1 | 1.8 × 107 | 14 | 14 | 11 | 11 |
| pDB-bel11-396 | 1.8 × 107 | 122 | 118 | 80 | 63 |
BEL1 Interacts with a Specific Subset of KNAT Proteins
At least eight individual KNAT genes exist in the Arabidopsis genome (Lincoln et al., 1994; Dockx et al., 1995; Long et al., 1996; Serikawa et al., 1996; Semiarti et al., 2001; this article). Interestingly, only two different KNAT cDNA clones were recovered in the two-hybrid screen using BEL1 as bait. This may indicate that BEL1 interacts with a limited set of KNAT protein partners. Alternately, this result may reflect only the relative abundance of KNAT cDNA clones within the cDNA library. To distinguish between these two possibilities, we used the two-hybrid system to test directly the ability of each remaining known KNAT protein to interact with BEL1 (Figure 1B). The coding sequences of STM, KNAT2, KNAT3, KNAT4, and KNAT7 (see Methods) were cloned into pTA to express the corresponding proteins as C-terminal fusion proteins with the GAL4 TA. Each plasmid was then used to transform YPB2 in combination with pDB-BEL1 or pDB-CRUCIFERIN (negative control). Strains were then assayed for β-galactosidase activity as a measure of their ability to activate the lacZ marker gene. As a positive control for interaction, we included a known pair of yeast proteins, SNF1 and SNF4, that have been demonstrated previously to interact via the two-hybrid system (Fields and Song, 1989).
pDB-CRUCIFERIN did not affect the activation of lacZ in combination with any pTA-KNAT bait constructs, indicating that the KNAT proteins neither interact nonspecifically with either the GAL4 DB or CRUCIFERIN nor activate transcription autonomously. In addition to combinations with pTA-KNAT1 and pTA-KNAT5, pDB-BEL1 also affected lacZ activation in combination with pTA-STM and pTA-KNAT2. However, combinations between pDB-BEL1 and pTA-KNAT3, pTA-KNAT4, or pTA-KNAT7 did not result in lacZ activation. Therefore, these results suggest that the BEL1 protein can discriminate between KNAT family members and interact with only a specific subset of KNAT proteins, namely STM, KNAT1, KNAT2, and KNAT5.
Interestingly, the two-hybrid results imply that BEL1 can interact equally well with KNAT1, KNAT2, KNAT5, and STM, yet no KNAT2 or STM clones were isolated in the two-hybrid screen using BEL1 as bait. This anomaly most likely reflects the relative representation of each gene in the cDNA library, with KNAT1 and KNAT5 clones being more abundant than KNAT2 or STM.
To demonstrate that the two-hybrid system results were not dependent on additional yeast-specific factors, we also tested the ability of BEL1 to interact with specific KNAT proteins in vitro (Figure 1C). We in vitro translated STM, KNAT1, KNAT3, and glutathione S-transferase–tagged BEL1 (GST-BEL1), alone and in combination using 35S-methionine, and then incubated the extracts with glutathione–agarose beads. Upon washing the beads, STM, KNAT1, and KNAT3 alone were not retained (Figure 1C, lanes 8 to 10). As expected, STM and KNAT1, but not KNAT3, were retained by the beads in combination with GST-BEL1 (Figure 1C, lanes 5 to 7). The retention of STM and KNAT1 is not likely to result from nonspecific interaction with the GST tag because STM, KNAT1, and KNAT3 were not retained in combination with GST alone (data not shown). The retention of STM in combination with GST-BEL1 was slightly ambiguous because one truncated GST-BEL1 translation product had approximately the same mobility as STM. Therefore, we repeated this experiment in the opposite orientation using GST-STM and BEL1 (Figure 1C, lanes 11 and 12). As expected, BEL1 alone was not retained by the beads (data not shown). BEL was retained only in combination with GST-STM (Figure 1C, lane 13). In summary, our results indicate specific interactions of BEL1 with STM and KNAT1.
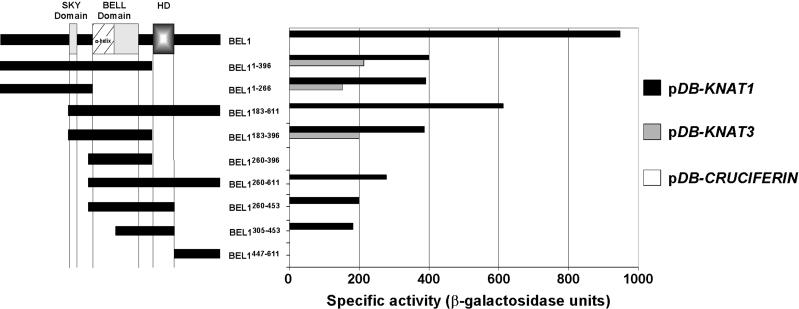
Identification of BEL1 Domains Mediating Interactions with KNAT Proteins
The presence of a putative amphipathic α-helix in BEL1 provided impetus for the two-hybrid system screen reported here. Therefore, we wished to determine if this domain, or other sequences within the BELL or SKY domains, was responsible for mediating the interactions between BEL1 and KNAT proteins. To this end, we tested the ability of a series of BEL1 deletions to interact with KNAT1 via the two-hybrid system (Figure 2). Briefly, sequences encoding the various BEL1 deletions were cloned into pTA and introduced into YPB2 strains containing pDB-KNAT1. Transformants were then assayed for β-galactosidase activity as a measure of lacZ activation. As a negative control, we tested these deletions against pDB-CRUCIFERIN.
Figure 2.
Deletion Analysis to Identify Protein–Protein Interaction Domains of BEL1.
Schemes of the BEL1 sequences expressed as C-terminal fusions to the GAL4 TA are depicted as black boxes at left. The patterned box (HD) represents the homeodomain, whereas the other boxes represent the SKY and BELL domains. Numbers in superscript indicate the BEL1 amino acids encoded by the deletion construct. β-Galactosidase activities resulting from the combinations of individual BEL1 deletion constructs with pDB-CRUCIFERIN, pDB-KNAT1, and pDB-KNAT3 are presented in the histogram at right.
No pTA-based BEL1 deletion constructs stimulated β-galactosidase activity in combination with pDB-CRUCIFERIN (Figure 2). As expected, pTA-BEL1 stimulated β-galactosidase activity in combination with pDB-KNAT1. Deletion of sequences from the C terminus of BEL1 diminished, but did not eliminate, the ability to stimulate β-galactosidase activity in combination with pDB-KNAT1. Remarkably, a clone expressing only the N-terminal 266 amino acids of BEL1 (pTA-BEL11-266) was sufficient to promote β-galactosidase activity in combination with pDB-KNAT1, whereas a clone expressing a 137–amino acid region spanning the putative amphipathic α-helix (pTA-BEL1260-396) was not. These results indicate that sequences N terminal to the putative amphipathic α-helix are sufficient for interaction with KNAT1 and that the amphipathic α-helix is neither sufficient nor necessary for interaction with KNAT1. Because pTA-BEL11-266 promoted the same level of β-galactosidase activity as pTA-BEL1183-396, it is tempting to speculate that the SKY domain mediates interactions with KNAT proteins. The SKY domain was not essential for interaction, however, because a C-terminal region of BEL1 from amino acids 260 to 611 (pDB-BEL1260-611) also could stimulate β-galactosidase activity in combination with pDB-KNAT1. All pTA-BEL1–based clones that retain the homeodomain still stimulated β-galactosidase activity in combination with pDB-KNAT1, indicating that a C-terminal interaction domain lies in the region of the homeodomain. Neither the N-terminal nor the C-terminal interaction domain alone affected β-galactosidase activity to the same extent as did intact BEL1, suggesting that these two domains function cooperatively to affect an interaction with KNAT1.
As a negative control, we initially used pDB-KNAT3 as a representative noninteracting KNAT protein. Surprisingly, deletion of the C-terminal portion of BEL1 including the homeodomain (pTA-BEL11-396) allowed for β-galactosidase activity in combination with pDB-KNAT3 (Figure 2). These data suggest that sequences in this C-terminal region, possibly the same sequences that are capable of interacting with KNAT1, are necessary to mediate the specificity of BEL1 for potential KNAT partners. The fact that pTA-BEL1260-611 was incapable of promoting β-galactosidase activity with pDB-KNAT3 is consistent with this hypothesis.
Together, these results identify at least two domains of BEL1 that are important for protein–protein interactions. One is located in the N-terminal 266 amino acids of BEL1 and is possibly capable of mediating general interactions between BEL1 and all KNAT proteins. A second region including the homeodomain is both necessary and sufficient to confer the specificity of BEL1 for interactions with KNAT1 relative to KNAT3. We predict that this region also is essential for specific interactions between BEL1 and KNAT2, STM, and KNAT5. Our results also are consistent with a hypothesis that both domains work cooperatively to allow the strongest and most selective protein–protein interactions.
Two-Hybrid System Screen Using the N-Terminal Region of BEL1
The previous results suggested that a C-terminal domain of BEL1 may confer specificity to the BEL1–KNAT interactions. Conceivably, the N-terminal region of BEL1 is capable of interacting with all KNAT proteins. Furthermore, strong interactions between the complete BEL1 protein and certain KNAT proteins may have precluded the identification of weaker, yet relevant, interactions with additional proteins in the yeast two-hybrid screen. To test this possibility, we constructed pDB-based plasmids to express the N-terminal 396 amino acids and the C-terminal 164 amino acids of BEL1 (Figure 1A) and used these as bait constructs in two-hybrid screens.
YPB2 colonies harboring only pDB-BEL1447-611 were His+ with high levels of β-galactosidase activity (Figure 1B). This finding suggests that the C-terminal region of BEL1 can function as a TA in yeast and precludes its use in a two-hybrid screen. This result, however, is consistent with the hypothesis that acidic residues C terminal to the BEL1 homeodomain may function as an activation domain (Reiser et al., 1995). In contrast, colonies harboring only pDB-BEL11-396 were His− with no detectable β-galactosidase activity and therefore suitable for use in a two-hybrid system screen.
We isolated 80 pTA-based clones that stimulated histidine prototrophy and β-galactosidase activity only in combination with pDB-BEL11-396 (Table 1). A combination of DNA sequencing and restriction digest analyses revealed that 63 of these clones encoded KNAT proteins, including STM (2 clones), KNAT1 (20 clones), KNAT3 (38 clones), KNAT4 (1 clone), and KNAT7 (2 clones). In general, direct tests indicated that each combination of pDB-BEL11-396 with a pTA-KNAT promoted comparable levels of β-galactosidase activity (Figure 1B), which suggests that the N-terminal interaction domain of BEL1 is competent to bind to each KNAT protein with similar affinity. In this respect, it is again interesting that no KNAT2 clones were isolated in this screen, and again we attribute this to the relative abundance of functional KNAT2 clones in the library. β-Galactosidase activity was notably greater in combinations of pTA-STM, pTA-KNAT1, and pTA-KNAT2 with pDB-BEL1 relative to DB-BEL11-396, a result that is consistent with N-terminal and C-terminal domains functioning cooperatively to affect an interaction.
KNAT5 proved to be an unexpected, yet interesting, exception. We were surprised not to isolate a single KNAT5 clone in the screen with pDB-BEL11-396, which suggests that KNAT5 is not capable of interacting with the N-terminal domain of BEL11-396. Consistent with the two-hybrid screen results, pTA-KNAT5 did not promote β-galactosidase activity in combination with pDB-BEL11-396 (Figure 1B). Therefore, we infer that KNAT5 interacts specifically with the C-terminal domain of BEL1.
The remaining 17 clones corresponded to five different Arabidopsis cDNAs that encode proteins with no homology with any protein of known function in the databases and no identifiable motifs. Furthermore, crude filter assays indicated that the β-galactosidase activities affected by combinations of these clones with pDB-BEL11-396 were very low compared with pDB-BEL11-396/pTA-KNAT combinations (blue precipitate detected after several hours compared with after 20 min for the combination of pDB-BEL11-396/pTA-KNAT1; data not shown). Therefore, the interactions of BEL1 with these proteins were not examined further in this study.
Conserved MEINOX Domain Mediates Interactions between BEL1 and KNAT Proteins
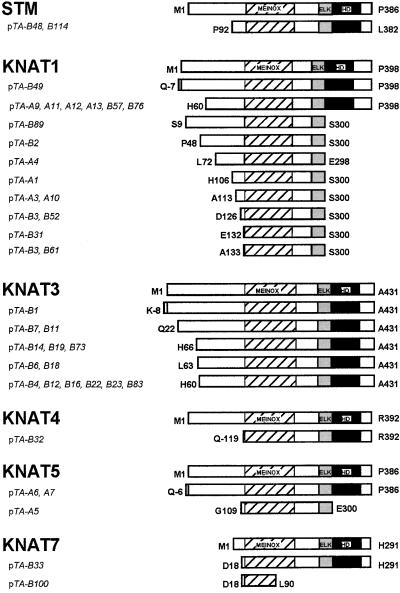
The KNAT cDNA clones isolated in both two-hybrid screens vary considerably in length (Figure 3). Consequently, their sequences prove informative with regard to the domain through which KNAT proteins interact with BEL1.
Figure 3.
Sequence Analyses of KNAT cDNA Clones Isolated in Two-Hybrid Screens Using BEL1 as Bait.
Prey plasmids were isolated with either pDB-BEL1 (pTA-A clones) or pDB-BEL1-396 (pTA-B clones) as bait. Predicted polypeptides are depicted schematically with N-terminal and C-terminal amino acids indicated at left and right of the schemes, respectively. Boxes with diagonal hatches represent the MEINOX domain, gray boxes represent the ELK domain, and black boxes represent the homeodomain (HD). Only a subset of the isolated KNAT1 and KNAT3 clones are depicted because several were identified by restriction digest analysis.
The pTA-KNAT1 and pTA-KNAT5 clones isolated using pDB-BEL1 as bait fell into four and two classes, respectively (Figure 3, A group). For both KNAT1 and KNAT5, the shortest cDNA clones encoded polypeptides just spanning the MEINOX and ELK domains. More specifically, the KNAT1 clones pTA-A3 and pTA-A10 encoded a 188–amino acid polypeptide from A113 to S300, whereas the KNAT5 clone pTA-A5 encoded a 192–amino acid polypeptide from G109 to E300. The structure of these KNAT clones implicates the MEINOX domain in interactions with BEL1. Consistent with this possibility, recent studies have demonstrated that the MEINOX domain mediates interactions between two subclasses of TALE homeodomain proteins in fruit flies and humans (Chang et al., 1997; Knoepfler et al., 1997; Berthelsen et al., 1998a; Abu-Shaar et al., 1999).
A subset of the KNAT clones isolated using pDB-BEL11-396 as bait also fell into multiple classes (Figure 3, B group). With one exception, every clone encoded a polypeptide containing both the MEINOX and ELK domains. The exception (pTA-B100) encoded a 71–amino acid N-terminal section of the KNAT7 MEINOX domain, which confirms that the KNAT MEINOX domain is sufficient for interaction with BEL1.
It is important to note that the KNAT1 and KNAT5 clones that terminated before the homeobox were polyadenylated. These data suggest that KNAT1 and KNAT5 genes may give rise to two different transcripts: one transcript that encodes a full-length KNAT1(5) protein, and a second truncated transcript that does not encode a homeodomain. It is unclear to what extent these transcripts accumulate or whether they correspond to the shorter class of KNAT1 transcripts reported by Lincoln et al. (1994). In any case, it is interesting to speculate that this truncated class of cDNA reflects a role for KNAT proteins independent of homeodomain-mediated DNA binding.
BEL1–KNAT Interactions Activate Transcription in Yeast
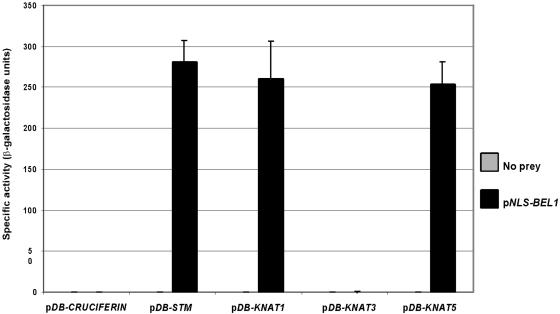
Our two-hybrid assay results suggest that the C-terminal 164 amino acids of BEL1 include a TA that functions efficiently in yeast when fused to the GAL4 DB. The intact BEL1, however, did not activate the transcription of reporter genes when fused to the GAL4 DB. The simplest explanation for this discrepancy is that the BEL1 protein assumes a tertiary structure that masks the TA activity of its C terminus. Subsequent interaction with another protein, such as a KNAT, could modify the BEL1 conformation to unmask the C-terminal TA.
To evaluate the merit of this hypothesis, we tested the ability of KNAT1 and BEL1 to activate lacZ without the aid of the GAL4 TA. We constructed a plasmid (pNLS-BEL1) to express the intact BEL1 as a fusion protein with a yeast nuclear localization signal (NLS). This construct was introduced into yeast strains harboring pDB-CRUCIFERIN, pDB-KNAT1, pDB-STM, pDB-KNAT3, and pDB-KNAT5. As anticipated, the combinations of pNLS-BEL1 with pDB-CRUCIFERIN or pDB-KNAT3 did not affect β-galactosidase activity (Figure 4). However, combinations of pNLS-BEL1 with pDB-KNAT1, pDB-STM, or pDB-KNAT5 were able to affect β-galactosidase activity in the absence of a GAL4 TA. These data strongly suggest that complexes between BEL1 and certain KNAT proteins are able to activate the transcription of target genes and that the interactions are necessary for activation domain function. Our data indicate that the C terminus of BEL1 most likely provides this activation domain. At this time, however, we cannot reject the possibility that the activation domain is provided, at least in part, by KNAT proteins or a yeast protein that interacts with the BEL1–KNAT complex. For example, pDB-KNAT2 alone was capable of activating lacZ in YPB2 (data not shown), which suggests that KNAT2 also contains an effective TA.
Figure 4.
Activation of Transcription by BEL1–KNAT Heterodimers.
β-Galactosidase activity resulting from the combinations of STM, KNAT1, KNAT3, and KNAT5 bait constructs with NLS-BEL1 are shown.
Bars indicate ±sd.
BEL1 and STM Transcript Patterns Overlap in the Inflorescence Apex
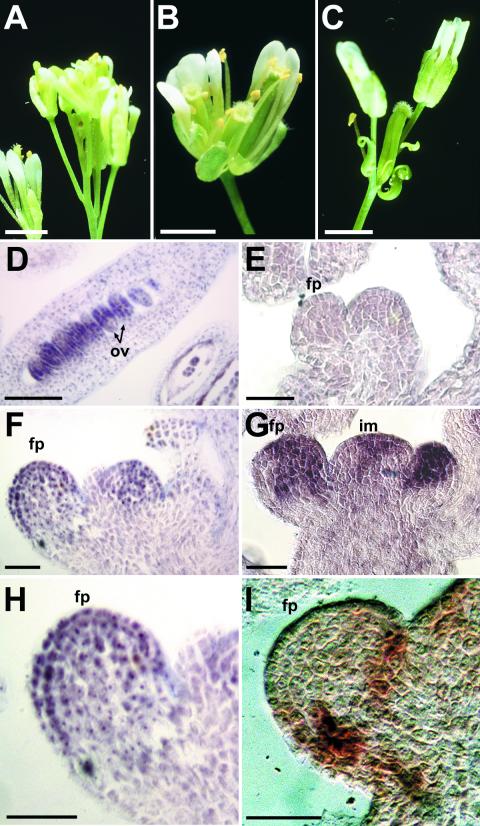
Consistent with the ovule defects of BEL1 mutants, BEL1 is transcribed in developing ovules (Reiser et al., 1995). Interestingly, BEL1 mutants also display an inflorescence phenotype resembling Terminal Flower 1 (Tfl1; Shannon and Meeks- Wagner, 1991) in which the inflorescence meristem terminates in a floral structure (compare Figures 5B and 5C with Figure 5A). This phenotype suggests that BEL1 is expressed and functions in the inflorescence SAM to maintain indeterminate growth. Therefore, we used in situ RNA hybridization to closely examine the pattern of BEL1 transcription in the Arabidopsis inflorescence. In agreement with Reiser et al. (1995), we detected BEL1 transcripts in the developing ovules in a manner that predicts the emergence of the integuments (Figure 5D). Consistent with a function in the inflorescence meristem, significant levels of BEL1 transcript also were detectable in the inflorescence apex and developing flowers (Figure 5F). This is unlikely to result from nonspecific hybridization because no such signal is detected in the inflorescence of bel1-3 homozygotes (Figure 5E), which are devoid of BEL1 transcripts (Reiser et al., 1995). BEL1 message also could be observed in the floral apex immediately before sepal formation (Figure 5H). A short time later, signal was detected in the sepal primordia (data not shown). Subsequently, transcripts were not detected until the emergence of ovule primordia. Notably, BEL1 expression was never observed in the developing pedicel (floral stem).
Figure 5.
BEL1 Function in Situ Hybridization Analyses of the Expression Patterns of BEL1, STM, and KNAT1 in the Inflorescence Apex of Arabidopsis.
(A) Wild-type Arabidopsis inflorescence apex from which lateral flowers are produced indeterminately.
(B) Bel1 mutant apex that has terminated in a flower.
(C) Bel1 terminal flower at a slightly later stage than in (B).
(D) BEL1 transcript is detected in developing ovules in a pattern that predicts the position of integument formation.
(E) No BEL1 transcript is detected in the inflorescence of a plant homozygous for the bel1-3 allele.
(F) Low levels of BEL1 mRNA can be detected in the inflorescence apex and developing floral primordia.
(G) STM transcript is detected across the apical meristem and in the apex of the developing floral primordium.
(H) Close-up of floral primordium shown at left in (F).
(I) KNAT1 is transcribed in the early floral primordium in what will become the pedicel. As the pedicel develops, this expression persists in the cortex next to the vascular tissue.
ov, ovule; fp, floral primordium; im, inflorescence meristem. Bars in (A) to (C) = 1 mm; bars in (D) to (I) = 30 μm.
Like BEL1, STM transcripts also were detected throughout the inflorescence SAM, in agreement with Long and Barton (1998) (Figure 5G). Also like BEL1, STM expression was observed in the early floral primordium but persists in the floral meristem until carpel development. Clearly, the transcript patterns of BEL1 and STM demonstrate the potential for interactions between their protein products in at least two stages of development. As in previous studies (Long and Barton, 1998), however, STM was not detected in ovules, which suggests that BEL1–STM interactions are not involved in ovule development.
Unlike BEL1 and STM, KNAT1 expression was not detected in the inflorescence apical meristem. In the inflorescence stem, message was observed in the cortex next to the vasculature (data not shown), as reported previously by Lincoln et al. (1994). In the flower, KNAT1 transcript was initially limited to what developed as the pedicel (Figure 5I). Later, KNAT1 expression was observed in the stigma. Thus, it is not obvious from the KNAT1 expression pattern that BEL1 has the potential to interact with KNAT1 protein in the inflorescence. It is worth noting, however, that the BEL1 transcript pattern juxtaposed that of KNAT1 in the inflorescence apex and in what becomes the flower receptacle (the junction of the pedicel and the remaining parts of the flower). Consequently, it is possible that KNAT1 and BEL1 proteins interact at these borders. As with STM, KNAT1 transcript was not detected in the developing ovule, which indicates that BEL1–KNAT1 interactions also are not involved in ovule development.
DISCUSSION
Previous mutant analyses have identified both BEL1 and KNOX genes as important regulators of morphogenesis in Arabidopsis (Barton and Poethig, 1993; Modrusan et al., 1994). The BEL1 and KNOX proteins belong to the TALE superfamily of homeodomain transcription factors. Until now, little has been determined about the molecular mechanisms by which they regulate development. In this study, we have used the yeast two-hybrid system to demonstrate that BEL1 can interact with Arabidopsis KNOX proteins (KNATs) to form heterodimeric complexes. Moreover, our results suggest that BEL1 can dimerize only with a specific subset of KNAT proteins and that a C-terminal domain of BEL1 including the homeodomain mediates this specificity. These findings are significant because the properties of these heterodimers may regulate the function of BEL1 and KNOX proteins in terms of target selectivity, affinity, and activation.
Specificity of Interactions between BEL1 and KNAT Proteins
Our yeast two-hybrid system experiments reveal that BEL1 can interact with Arabidopsis KNOX homeodomain proteins, and we believe that BEL1–KNAT interactions are both specific and meaningful for multiple reasons. First, the two-hybrid screen using a complete BEL1 protein as bait exclusively identified members of the KNOX family as BEL1-interacting proteins, and subsequent experiments confirmed that only a subset of KNAT proteins are capable of interacting with BEL1. Second, in vitro pulldown experiments provide physical evidence that BEL1 can interact with KNAT proteins with the same specificity demonstrated by two-hybrid system experiments. Third, the interactions between BEL1 and KNAT proteins are comparable to the interaction observed between a well-documented pair of interacting proteins, as inferred from their ability to activate a marker gene in the context of the two-hybrid system. Fourth, interactions between TALE homeodomain proteins are well documented in fungi and animals (Knoepfler et al., 1997; Berthelsen et al., 1998a). Fifth, the MEINOX domain mediates interactions of KNAT proteins with BEL1, as it does between TALE homeodomain proteins in animals (Knoepfler et al., 1997; Berthelsen et al., 1999). Finally, the transcription patterns of BEL1 and STM overlap, providing the potential for their protein products to interact in planta.
BEL1–KNAT Heterodimer Formation
Several studies have demonstrated that the MEINOX domain is essential for interactions between TALE proteins in animals (Knoepfler et al., 1997; Berthelsen et al., 1998a). In our two-hybrid screen, we identified a truncated KNAT7 cDNA clone that encoded only an N-terminal portion of the MEINOX domain yet retained the ability to interact with BEL1. These data indicate that the MEINOX domain is sufficient to mediate interactions with other TALE homeodomain proteins and confirm that this is a distinct interaction domain conserved between plants and animals.
The truncation analysis presented here indicates that BEL1 interacts with KNAT partners through an N-terminal domain. The SKY domain, a stretch of amino acids from I193 to G213 that is highly conserved among at least 12 other BLH proteins, represents the prime candidate for an N-terminal domain of BEL1 that is capable of interacting with most KNAT proteins. C-terminal sequences of BEL1 including the homeodomain also are sufficient for inter-action with certain KNAT partners. We suggest that this C-terminal interaction allows BEL1 to discriminate between potential KNAT partners, although it is possible that distinct domains impart C-terminal interaction and KNAT specificity.
We are intrigued that the putative amphipathic α-helix is neither sufficient nor necessary for interaction with KNAT proteins. The function of this sequence, and the rest of the BELL domain, remains unknown, but it is undoubtedly important because these sequences are highly conserved between BEL1 and BLH proteins. It is still possible that this domain contributes to the interactions with KNAT proteins. Alternately, this region may mediate interactions with additional factors not identified in the two-hybrid screens.
Parallels between TALE Homeodomain Protein Properties in Plants and Animals
Striking parallels exist between BEL1–KNOX associations and PBC–MEIS associations. Both involve interactions between members of two distinct subclasses of TALE homeodomain protein, and both involve important regulators of morphogenesis. These similarities not only support the validity of our conclusions that BEL1 and KNAT proteins act as transcription factor partners in plants but also could provide further insight into how these proteins function.
As in BEL1–KNAT interactions, MEIS and PBC proteins interact through the N-terminal domains of each protein. Similar to our observations with KNAT7, multiple studies have demonstrated that the N-terminal portion of the MEINOX domain is essential for the interactions of MEIS proteins with PBC proteins (Knoepfler et al., 1997; Berthelsen et al., 1998a). In addition to establishing a transcription factor complex, the interactions between these N-terminal domains regulate the subcellular localization, and thereby the function, of PBC and MEIS proteins. In animal cells, PBX1 (EXD) localizes to the nucleus only upon interaction with PREP1 (HTH) in the cytoplasm, presumably because PREP1 (HTH) masks a nuclear export sequence present in PBX1 (EXD; Abu-Shaar et al., 1999; Berthelsen et al., 1999). Analogous to PBX1 and EXD, BEL1 contains a sequence resembling a leucine-rich nuclear export sequence (Fischer et al., 1995; Wen et al., 1995) within the N-terminal domain that is sufficient for interaction with KNAT partners. Therefore, we predict that interactions between BEL1 and KNAT proteins also regulate their subcellular localization, and thereby their function, in a manner similar to PBX1–PREP1 (EXD–HTH) interactions.
In multiple cases, the sequences immediately C terminal to the homeodomain of a TALE protein contribute to interactions with a second homeodomain protein partner. In yeast, the C-terminal tail of α2 contacts the homeodomain of α1 (Mak and Johnson, 1993). In human cells, deletion of the region C terminal to the homeodomain reduces the ability of PBX1 to interact with MEIS1 (Knoepfler et al., 1997). Similarly, deletion of the C-terminal sequences of BEL1 including the homeodomain reduces its ability to interact with KNAT1. More importantly, this truncation eliminates the ability of BEL1 to discriminate between KNAT partners. These results suggest that the C-terminal regions of PBX1 and BEL1 make important contacts with their respective MEINOX partners, and because this contact is important to mediate partner specificity in BEL1–KNAT interactions, it will be interesting to learn if this region functions similarly in PBC–MEIS interactions.
The downstream target genes of BEL1 and KNOX proteins remain to be determined, and the identification of target sequences for BEL1–KNOX complexes could conceivably facilitate this process. DNA target sequences of PBC–MEIS heterodimers have been reported, and these elements also are found in the promoter region of key genes that regulate development in Arabidopsis. We are currently testing the ability of BEL1–KNAT heterodimers to recognize these sequences.
In animals, PBC and MEIS proteins function in ternary complexes with the typical HOX homeodomain proteins. Consequently, Bürglin (1998) predicted that KNOX proteins, and hence possibly BEL1-like proteins, heterodimerize with typical homeodomain proteins such as those belonging to the HD-zip family. Although no proteins other than KNATs were identified as BEL1-interacting proteins, it is possible that additional factors interact specifically with a BEL1–KNAT heterodimer, but such ternary complexes would not be identified in a yeast two-hybrid screen.
Role of Interactions between BEL1 and KNAT Proteins
Consistent with its function in ovule development, the BEL1 expression pattern predicts the position of integument initiation (Reiser et al., 1995; this article). That BEL1 also regulates processes distinct from integument formation is strongly implied by its expression in other tissues (Reiser et al., 1995), including the inflorescence SAM and emerging floral apices. Interestingly, several important regulators of ovule morphogenesis also are known to function in other plant tissues (Gasser et al., 1998).
Our results indicate that the BEL1 and STM expression patterns overlap within the inflorescence and floral apices. Interestingly, a subtle defect in Bel1 inflorescence SAM development has received little attention (Modrusan et al., 1994). Unlike wild-type plants, in which the primary inflorescence grows indeterminately up until senescence, Bel1 shoot apices terminate in a flower. This phenotype and the expression of BEL1 in the shoot apex suggest that BEL1 has a direct role in maintaining the indeterminate growth of the inflorescence meristem, possibly by inhibiting the floral development program. One attractive hypothesis would have BEL1 interacting with STM to maintain indeterminate growth programs in the inflorescence SAM.
Our expression analysis also indicates the potential for BEL1 to interact with KNAT1 in the young floral primordium. The juxtaposition of the BEL1 and KNAT expression domains makes it tempting to speculate that BEL1–KNAT1 heterodimers establish a boundary between the apical and basal portions of the primordium. Development of the young floral primordium, however, is not obviously affected in Bel1 mutants, which suggests that BEL1–KNAT1 interactions are not specifically required in early flower development. Nevertheless, this apparent lack of BEL1 function could be explained by redundancy conferred by additional BLH genes whose expression patterns overlap with BEL1 (A. Samach, M.S. Pidkowich, K. Kushalappa, and G.W. Haughn, unpublished results).
Consistent with previous findings (Lincoln et al., 1994; Long and Barton, 1998), we did not detect STM or KNAT1 expression in developing ovules, and preliminary experiments indicate that KNAT5 is not expressed in ovules either. It is worth noting, however, that STM and KNAT2 are transcribed in placental tissues that are immediately adjacent to the domain of BEL1 expression in young ovule primordia (Long and Barton, 1998; Bowman et al., 1999). It is possible that BEL1 interacts with KNAT proteins to establish the boundary between placenta and developing ovule and/or that additional KNAT proteins with expression patterns in the ovule remain to be discovered. Alternately, BEL1 may function independently of KNAT proteins within the ovule. Finally, we expect that more complete expression analyses of BEL1, KNAT1, KNAT2, STM, and KNAT5 will identify additional tissues in which BEL1–KNAT interactions play a role. We have begun to use reporter genes under the control of BEL1 and KNAT promoters to assist us in such identification.
BEL1 and KNAT Proteins Form Complexes That Can Activate Transcription
Our results indicate that although neither BEL1 nor various KNAT proteins can do so alone, BEL1–KNAT complexes are capable of activating transcription in yeast. Because the C-terminal 164 amino acids of BEL1 can activate transcription in isolation from the rest of the protein, we suggest that BEL1 possesses a latent activation domain that can be triggered through interaction with certain KNAT proteins.
The ability of a transcription factor to activate or repress transcription depending on the identity of its interacting partner is a common theme in developmental biology. Hence, the interactions of BEL1 with multiple KNAT partners, some of which contain activation domains themselves, may reflect a dynamic capacity of BEL1 to activate or repress, depending on the specific KNAT partner.
Implications for the Study of KNOX Protein Function
Studies in various systems have demonstrated that ectopic expression of some, but not all, KNOX proteins results in abnormal leaf, stem, and flower morphology (reviewed by Reiser et al., 2000). In Arabidopsis, for example, KNAT1 but not KNAT3 is capable of inducing defects in leaf and flower development (Lincoln et al., 1994; Serikawa and Zambryski, 1997).
Two related pieces of evidence hint that interactions with a BEL1-like protein may be important for the generation of this phenotype. First, domain exchange experiments between class 1 and class 2 KNOX proteins suggest that the major determinant of the abnormal phenotype rests within an N-terminal region containing the BEL1-interacting MEINOX domain (Serikawa and Zambryski, 1997; Sakamoto et al., 1999). Second, it is a striking correlation that BEL1 can interact with KNAT1, the ectopic expression of which can affect the abnormal phenotype, but cannot interact with KNAT3, which is incapable of affecting an abnormal phenotype. Because multiple BLH genes are expressed simultaneously in leaves, stems, and flowers, we hypothesized that loss-of-function mutations in BEL1 and/or additional BLH proteins would attenuate the KNAT1 ectopic expression phenotype. Removal of BEL1 function alone, however, does not achieve this effect (A. Samach, K. Kushalappa, and G.W. Haughn, unpublished results).
Coevolution of TALE Homeodomain Proteins
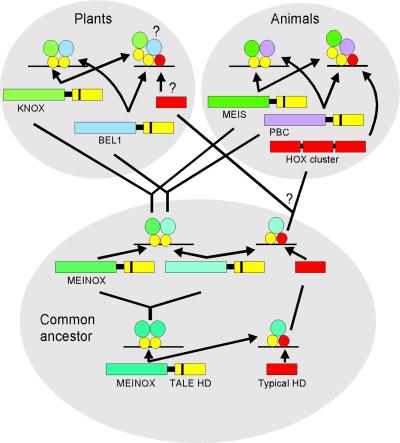
A model has been proposed previously for the evolution of TALE homeodomain proteins in which an ancestral MEINOX–TALE protein was able to form homodimers (Bürglin, 1998). In animals, this MEINOX gene duplicated and diverged to give rise to the MEIS- and PBC-class proteins, which retained the ability to dimerize with each other. Because no obvious PBC-like proteins exist in plants, Bürglin (1998) predicted that KNOX proteins homodimerize or heterodimerize among each other. The results presented here strongly suggest that KNOX proteins interact with BEL1. In light of these findings, we suggest a modification of this model such that the ancestral MEINOX–TALE gene duplicated and diverged before the emergence of plants and animals (Figure 6). One of the MEINOX ancestors gave rise to the KNOX and MEIS families in plants and animals, respectively, whereas the second ancestral gene gave rise to the BEL1-like and PBC genes. Although the N-terminal, MEINOX-interacting sequences of BEL1 and PBC proteins do not share sequence similarity, it is possible that they have diverged beyond recognition. The merit of this hypothesis will be easier to evaluate once the three-dimensional structures of these complexes are determined.
Figure 6.
Model Depicting the Evolution of Interactions between TALE Homeodomain Proteins (after Bürglin, 1998).
It has been hypothesized that an ancestral MEINOX–TALE homeodomain (HD) protein was capable of forming homodimers and interacting with another, typical homeodomain protein. Before the emergence of plants and animals, this ancestral MEINOX gene duplicated, and the diverging proteins retained the ability to interact with each other. In animals, MEINOX proteins (represented by the MEIS class) retained the ability to interact with TALE homeodomain proteins that evolved into the PBC family and typical homeodomain proteins that evolved into the HOX family. In plants, the MEINOX proteins have been conserved as KNOX proteins that have retained the ability to interact with the BEL1 homeodomain protein. The existence of BEL1–KNAT interactions with typical homeodomain proteins is hypothetical.
METHODS
Plasmid Construction
For the purposes of this article, pBI-770– and pBI-771–based plasmids (Kohalmi et al., 1997) are designated pDB-X and pTA-X, respectively, where X represents a coding sequence of interest.
For many of the constructs used, coding sequences were amplified from the appropriate cDNA source by polymerase chain reaction (PCR) using the Expand High-Fidelity PCR System (Hoffmann-La Roche) and oligonucleotides that introduced a SalI site at the 5′ end and a NotI site at the 3′ end of the open reading frame. This fragment was digested subsequently with SalI and NotI and inserted into pDB and/or pTA. The following plasmids were constructed in this way: pDB/TA-BEL1 (BF1, 5′-ACGCGTCGACACATGGCAAGAGATCAG-3′; BR1, 5′-ATAAGAATGCGGCCGCGTTACATAACCGAATCG-3′), pTA-BEL11-396 (BF1; BR2, 5′-ATAAGAATGCGGCCGCGTTGTGGA-CGCCCAAGG-3′), pTA-BEL11-266 (BF1; BR3, 5′-ATAAGAATGCGG-CCGCTTGAGAGTGAAGTGGTGG-3′), pTA-BEL1183-611 (BF2, 5′-ACG-CGTCGACATCAGCATCATAATCATC-3′; BR1), pTA-BEL1183-396 (BF2; BR2), pTA-BEL1260-396 (BF3, 5′-ACGCGTCGACAATCTGCGACT-ACTTCTTC-3′; BR2), pTA-BEL1260-611 (BF3; BR1), pTA-BEL1260-453 (BF3; BR4, 5′-ATAAGAATGCGGCCGCTCAAATCATTGGTTTCCATA-G-3′), pTA-BEL1305-453 (BF4, 5′-ACGCGTCGACACATGGAAGCGGCGGTTGGACT-3′; BR4), pDB/TA-BEL1447-611 (BF5, 5′-GTCGACTAGGCTATGGAAACC-3′; BR1), pDB/TA-STM (5′-ACGCGTCGA-CGTATGGAGAGTGGTTCCAAC-3′; 5′-ATAAGAATGCGGCCGCCC-AAGTATACCGAGAACC-3′), pDB/TA-KNAT2 (5′-ACGCGTCGACTA-ATGGATAGAATGTGTG-3′; 5′-ATAAGAATGCGGCCGCCTCTT-TTCATTACTCGG-3′), and pDB/TA-KNAT4 (5′-ACGCGTCGACCG-ATGGCGTTTCATAAC-3′; 5′-ATAAGAATGCGGCCGCGGATCTCT-ACATGCAAGC-3′). pLR113 (Reiser et al., 1995) was used as a template in BEL1-related PCRs, an ATK1 cDNA (Dockx et al., 1995) was used as a template in KNAT2-related PCRs, and an Arabidopsis thaliana cDNA library (see below, Rapid Amplification of cDNA Ends) was used as a template in STM- and KNAT4-related PCRs.
pTA-KNAT1 corresponds to pTA-B49 (Figure 3), which contains the entire KNAT1 coding sequence and includes the first glutamine upstream of the predicted methionine start codon. pTA-KNAT3 corresponds to pTA-B3, which contains the entire KNAT3 coding sequence. pTA-KNAT5 corresponds to pTA-A6. That this truly represents the complete KNAT5 coding region is unclear because GenBank accession number D43962 reports the “complete” coding sequence, but no stop codons are observed upstream of the most 5′ ATG. Therefore, the coding may actually be longer. pTA-KNAT7 corresponds to pTA-B33, which contains a partial KNAT7 cDNA. The cDNA inserts within pTA-KNAT1, pTA-KNAT3, pTA-KNAT5, and pTA-KNAT7 were liberated from pBI-771 as SalI–NotI fragments and cloned into pBI-770 to produce pDB-KNAT1, pDB-KNAT3, pDB-KNAT5, and pDB-KNAT7, respectively.
To construct pNLS-BEL1, pTA-BEL1 was first digested with SalI and XhoI. This produced three fragments: a 383-bp fragment including the transcription activation domain (TA) coding sequence, a 1.4-kb fragment including the yeast nuclear export sequence coding sequences, and the 7.2-kb fragment including the remaining plasmid and BEL1 sequences. The 1.4- and 7.2-kb fragments were then ligated to produce an in-frame fusion between the yeast nuclear localization signal (NLS) sequences and the BEL1 cDNA.
For in vitro glutathione S-transferase (GST) pulldown experiments, BEL1, STM, KNAT1, and KNAT3 fragments were excised from pTA-BEL1, pTA-STM, pTA-KNAT1, and pTA-KNAT3, respectively, with SalI and NotI and then inserted into pET28A (Novagen, Madison, WI) to yield pHIS-BEL1, pHIS-STM, pHIS-KNAT1, and pHIS-KNAT3. To create a plasmid for GST fusion proteins, a GST fragment was amplified from pGEX-4T (Amersham Pharmacia Biotech) using primers 5′-GCCTCTAGATCACACAGGAAACAG-3′ and 5′-TCCTGGGGATCC-ACGCGG-3′ and then inserted into pET28A to produce pET28A-GST. BEL1 and STM fragments were then excised from pTA-BEL1 and pTA-STM and cloned into pET28A-GST as SalI–NotI fragments to produce pGST-BEL1 and pGST-STM, respectively, which encoded BEL1 and STM as C-terminal fusions to GST.
For in situ hybridization probe synthesis, 5′ STM and KNAT1 fragments that did not include the homeobox were amplified from pTA-STM and pTA-KNAT1 and cloned into pCR2.1 (Invitrogen, Carlsbad, CA) to produce pSTMT7 and pK1T7, such that antisense transcripts could be generated using T7 RNA polymerase. The oligonucleotides used for STM amplification were 5′-GACGACGACAAGGGTCG-3′ and 5′-CTAATACGACTCACTATAGGGTTTGGAGAGCTCTTGC-3′, whereas those used for KNAT1 amplification were 5′-GACGACGACAAG-GGTCG-3′ and 5′-CTAATACGACTCACTATAGGGTCACAGTATGCTTCC-3′.
Yeast Two-Hybrid Library Screening
Arabidopsis cDNA clones encoding BEL1-interacting proteins were identified using a modified GAL4-based yeast two-hybrid system and library that have been described previously (Fields and Song, 1989; Kohalmi et al., 1997; Samach et al., 1999). Briefly, the appropriate bait construct was introduced into the yeast two-hybrid host strain YPB2 (Bartel et al., 1993). If the resulting colonies were histidine auxotrophs, sensitive to 3-amino-1′,2′,4′-triazol (3-AT), and lacked lacZ expression, this strain was used to screen the Arabidopsis two-hybrid cDNA expression library. Colonies expressing putative BEL1-interacting proteins were selected on the basis of the activation of two marker genes, HIS3, which conferred both histidine prototrophy and resistance to 3-AT, and lacZ. pTA-based plasmids were isolated from yeast, transformed into Escherichia coli, and purified again. Upon retransformation of these plasmids into YPB2, we sequenced plasmids that were found to activate a lacZ only in specific combination with the original bait plasmid.
β-Galactosidase Assays
β-Galactosidase assays were performed as described by Miller (1972), except that the yeast cells were permeabilized using glass beads, and soluble protein was determined using the Bradford (1976) method. Cultures were grown in an appropriate liquid medium to OD600 = 1 before being sedimented, resuspended in breaking buffer, and frozen at −80°C. The sample then was vortexed at 4°C for at least 5 min. The lysate was centrifuged for 15 min, and the supernatant was collected and used for the β-galactosidase assay. Specific activity was calculated using the formula β-galactosidase units = 1000(OD420/OD600)·min−1·mL−1. Values represent the averages of three separate assays for each treatment.
Rapid Amplification of cDNA Ends
5′ sequences of the KNAT7 cDNA were amplified using the Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA). Briefly, a cDNA library was constructed according to the manufacturer's instructions with Arabidopsis mRNA isolated from floral bud tissue with RNeasy Plant and Oligotex mRNA kits (Qiagen, Valencia, CA). This library provided the template for PCRs with the kit-provided adaptor-specific primer and the KNAT7-specific primer 5′-ATCTGACGA-GAAATCCATCGGAA-3′. Candidate PCR products were then gel purified and subcloned using the TA-Cloning System (Invitrogen). Finally, the inserts were sequenced to determine the 5′ sequence of the cDNA of interest. A minimum of two independently amplified PCR products was sequenced to confirm the sequence of the KNAT7 cDNA.
In Vitro GST Pulldown Experiments
The pet28a-BEL1, -STM, -KNAT1, -KNAT3, -GST-BEL1, and -GST-STM plasmids were transcribed and translated in vitro, alone or in combination, using the TNT Coupled Wheat Germ Extract System (Promega) according to the manufacturer's instructions using T7 RNA polymerase and 35S-methionine (Amersham Pharmacia Biotech). Twenty-five microliters of the translation products were mixed with 20 μL of glutathione–agarose beads (Sigma) in binding buffer (PBS containing 0.1% BSA, 0.1% naphthylphthalamic acid, 10 mM phenylmethylsulfonyl fluoride, and 1 mM DTT) and incubated at 4°C for 2 hr with shaking. Beads were then washed four times with binding buffer. Transcription/translation extracts or the glutathione–agarose beads were run on a 12% SDS-PAGE gel to size fractionate the retained translation products. The gel was fixed and treated with Amplify (Amersham Pharmacia Biotech) to enhance the signal. Translation products were then visualized by autoradiography.
Plant Material and Growth Conditions
Arabidopsis wild type, ecotypes Columbia-2 (Col-2) and Wassilewskija (Ws), and Bel1 mutant (Ws ecotype) plants were grown at 20°C under continuous light (90 to 120 μE·m−2·sec−1 PAR) on prepared soil mix (Terra-Lite Redi Earth; W.R. Grace and Company of Canada Ltd., Ajax, Ontario, Canada).
In Situ Analysis
Tissue fixation, sectioning, hybridization, and detection were conducted as described previously by Samach et al. (1997). Sections were photographed under differential interference contrast optics. The resulting images were digitized and manipulated using Photoshop 5 (Adobe Systems, San Jose, CA).
To generate digoxigenin-labeled antisense transcripts, pSTMT7 and pK1T7 were linearized by digestion with BamHI and HindIII, respectively, and then transcribed as described previously (Wilkinson and Haughn, 1995).
Accession Numbers
The GenBank accession numbers for the sequences described in this article are AF306661 (KNAT5) and AF308451 (KNAT7).
Acknowledgments
We thank Jacek Nowak (National Research Council of Canada–Plant Biotechnology Institute) for his expertise in yeast two-hybrid system screening, Véronique Pautot (Institut National de la Recherche Agronomique) for her gift of the ATK1 template used for KNAT2-related PCR, Ravi S. Kumar for help with the figures, and two anonymous reviewers for their constructive criticism of the manuscript. This work was funded in part through National Science and Engineering Research Council of Canada (NSERC) postgraduate scholarships and a University of British Columbia University Graduate Fellowship to M.S.P., an NSERC research grant to G.W.H. and W.L.C., and the National Research Council of Canada.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010161.
References
- Abu-Shaar, M., Ryoo, D.H., and Mann, R.S. (1999). Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bartel, P., Chien, C.T., Sternglanz, R., and Fields, S. (1993). Elimination of false positives that arise in using the 2-hybrid system. Biotechniques 14 920–924. [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of the development in the wild-type and shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Berthelsen, J., Zappavigna, V., Mavilio, F., and Blasi, F. (1998. a). PREP1, a novel functional partner of PBX proteins. EMBO J. 17 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen, J., Zappavigna, V., Ferretti, E., Mavilio, F., and Blasi, F. (1998. b). The novel homeoprotein PREP1 modulates PBX-Hox protein cooperativity. EMBO J. 17 1434–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen, J., Kilstrup-Nielsen, C., Blasi, F., Mavilio, F., and Zappavigna, V. (1999). The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45 155–205. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Bürglin, T.R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin, T.R. (1998). The PBC domain contains a MEINOX domain: Coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 208 113–116. [DOI] [PubMed] [Google Scholar]
- Bürglin, T.R., and Ruvkun, G. (1992). New motif in PBX genes. Nat. Genet. 1 319–320. [DOI] [PubMed] [Google Scholar]
- Chang, C.-P., Jacobs, Y., Nakamura, T., Jenkins, N.A., Copeland, N.G., and Cleary, N.L. (1997). MEIS proteins are major in vivo DNA binding partners for wild-type but not chimeric PBX proteins. Mol. Cell. Biol. 17 5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chariot, A., Gielen, J., Merville, M.-P., and Bours, V. (1999). The homeodomain-containing proteins: An update on their interacting partners. Biochem. Pharmacol. 58 1851–1857. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122 1567–1575. [DOI] [PubMed] [Google Scholar]
- Dockx, J., Quaedvlieg, N., Keultjes, G., Kock, P., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28 723–737. [DOI] [PubMed] [Google Scholar]
- Dong, Y.H., Yao, J.L., Atkinson, R.G., Putterill, J.J., Morris, B.A., and Gardner, R.C. (2000). MDH1, an apple homeobox gene belonging to the BEL1 family. Plant Mol. Biol. 42 623–633. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in the Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O.K. (1989). A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Fischer, U.L., Huber, J., Boelens, W.C., Mataj, I.W., and Luhrmann, R. (1995). The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82 443–449. [DOI] [PubMed] [Google Scholar]
- Gasser, C.S., Broadhvest, J., and Hauser, B.A. (1998). Genetic analysis of ovule development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 1–24. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R., Vollbrecht, E., Lowe, B., Veit, B., Yamaguchi, J., and Hake, S. (1994). Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter, R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124 3045–3054. [DOI] [PubMed] [Google Scholar]
- Knoepfler, P.S., Calvo, K.R., Chen, H., Antonarakis, S.E., and Kamps, M.P. (1997). Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. USA 94 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohalmi, S.E., Nowak, J., and Crosby, W.L. (1997). A practical guide to using the yeast 2-hybrid system. In Differentially Expressed Genes in Plants: A Bench Manual, E. Hansen and G. Harper, eds (London: Taylor and Francis), pp. 63–82.
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Lyndon, R.F. (1998). The Shoot Apical Meristem: Its Growth and Development. (New York: Cambridge University Press).
- Mak, A., and Johnson, A.D. (1993). The carboxy-terminal tail of the homeo domain protein α2 is required for function with a second homeo domain protein. Genes Dev. 7 1862–1870. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg, N., Dockx, J., Rook, F., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det. Plant Cell 7 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L.M., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule. Cell 83 735–742. [DOI] [PubMed] [Google Scholar]
- Reiser, L., Sanchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42 151–166. [PubMed] [Google Scholar]
- Rieckhof, G.E., Casares, F., Ryoo, H.D., Abu-Shaar, M., and Mann, R.S. (1997). Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91 171–183. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruit, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Nishimura, A., Tamaoki, M., Kuba, M., Tanaka, H., Iwahori, S., and Matsuoka, M. (1999). The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell 11 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Kohalmi, S.E., Motte, P., Datla, R., and Haughn, G.W. (1997). Divergence of function and regulation of class B floral organ identity genes. Plant Cell 9 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20 433–445. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 10 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serikawa, K.A., and Zambryski, P.C. (1997). Domain exchanges between KNAT3 and KNAT1 suggest specificity of the kn1-like homeodomains requires sequences outside of the third helix and the N-terminal arm of the homeodomain. Plant J. 11 863–869. [DOI] [PubMed] [Google Scholar]
- Serikawa, K.A., Martinez-Laborda, A., and Zambryski, P. (1996). Three knotted1-like homeobox genes in Arabidopsis. Plant Mol. Biol. 32 673–683. [DOI] [PubMed] [Google Scholar]
- Shannon, S., and Meeks-Wagner, D.R. (1991). A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7 787–795. [DOI] [PubMed] [Google Scholar]
- Wen, W., Meinkoth, J.L., Tsien, R.Y., and Taylor, S.S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82 463–473. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.D., and Haughn, G.W. (1995). UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell 7 1485–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]