Abstract
In higher plants, developmental phase changes are regulated by a complex gene network. Loss-of-function mutations in the EMBRYONIC FLOWER genes (EMF1 and EMF2) cause Arabidopsis to flower directly, bypassing vegetative shoot growth. This phenotype suggests that the EMF genes play a major role in repression of the reproductive program. Positional cloning of EMF2 revealed that it encodes a zinc finger protein similar to FERTILIZATION-INDEPENDENT SEED2 and VERNALIZATION2 of Arabidopsis. These genes are characterized as structural homologs of Suppressor of zeste 12 [Su(z)12], a novel Polycomb group gene currently identified in Drosophila. In situ hybridization studies have demonstrated that EMF2 RNA is found in developing embryos, in both the vegetative and the reproductive shoot meristems, and in lateral organ primordia. Transgenic suppression of EMF2 produced a spectrum of early-flowering phenotypes, including emf2 mutant–like phenotype. This result confirms the role of EMF2 in phase transitions by repressing reproductive development.
INTRODUCTION
Postembryonic plant development undergoes phase changes (Poethig, 1990). Each phase is characterized by lateral organs with distinct morphological features that are produced by the shoot apical meristem (SAM). The major phase change involves the transition from vegetative to reproductive development (floral transition), which results in flowering. In Arabidopsis, the floral transition occurs when the vegetative SAM converts to the inflorescence SAM. During reproductive development, the main inflorescence shoot produces lateral buds that develop into additional inflorescences or flowers.
The floral transition is regulated by endogenous factors such as plant hormones and environmental factors such as photoperiod and temperature (Martínez-Zapater et al., 1994; Koornneef et al., 1998). These developmental regulations are mediated by a complex network of flowering time genes (Levy and Dean, 1998; Simpson et al., 1999; Blázquez, 2000). APETALA1 (AP1) and LEAFY (LFY), which encode transcription factors, are necessary for flower initiation (Mandel et al., 1992; Weigel et al., 1992). They accelerate the floral transition when ectopically expressed under the constitutive 35S promoter of Cauliflower mosaic virus (Mandel and Yanofsky, 1995; Weigel and Nilsson, 1995). Gibberellin and long days promote flowering in part through the upregulation of LFY (Simon et al., 1996; Blázquez et al., 1998), suggesting that LFY integrates environmental and endogenous signals. Other flowering time genes, such as TERMINAL FLOWER1 (TFL1) and FLOWERING LOCUS T (FT), which encode similar phosphatidylethanolamine binding protein homologs, act antagonistically in flowering time regulation. FT promotes and TFL1 delays flowering (Ratcliffe et al., 1998; Kardailsky et al., 1999; Kobayashi et al., 1999). Plants that overexpress both FT and LFY flower immediately after germination, indicating that these two genes are sufficient to bypass vegetative shoot development (Kardailsky et al., 1999; Kobayashi et al., 1999). In Arabidopsis, long-day photoperiods promote flowering. CONSTANS (CO) is a major mediator of the photoperiod-dependent regulation pathway that encodes the mammalian GATA factor–like zinc finger protein (Putterill et al., 1995). Evidence indicates that CO promotes flowering through the upregulation of LFY, FT (Simon et al., 1996; Kobayashi et al., 1999), and another flowering time gene, AGAMOUS-LIKE 20 (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). However, winter annual ecotypes of Arabidopsis are vernalization sensitive with respect to flowering. In these ecotypes, active alleles of FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) repress flowering through the negative regulation of FT and other flowering time genes. Vernalization promotes flowering by suppressing FLC activity in these ecotypes (Sheldon et al., 2000). Molecular cloning of FRI and FLC revealed that they encode a novel protein with a coiled-coil domain and a MADS box protein, respectively (Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). Several other genes, including VERNALIZATION1 (VRN1), VRN2, and VRN3, were identified as vernalization signal mediators from their mutant phenotypes (Chandler et al., 1996).
The EMBRYONIC FLOWER (EMF) genes also are involved in shoot development and phase transitions in Arabidopsis. Loss-of-function emf mutants (emf1 and emf2) skip rosette growth, producing small inflorescences whose lateral buds produce only flowers and no additional inflorescences (Sung et al., 1992). Genetic analysis using double mutations demonstrated that the EMF genes are epistatic to flowering time genes such as CO, AP1, FT, and FWA with respect to the rosette-to-inflorescence transition (Chen et al., 1997; Haung and Yang, 1998). These phenotypes suggest that the EMF genes play major roles in the repression of reproductive development. Moreover, the emf mutants express AP1 precociously and AGAMOUS ectopically in young seedlings, suggesting negative regulation of these genes by EMF genes (Chen et al., 1997).
Here, we report the molecular cloning of EMF2 and its functional analysis. EMF2 encodes a novel zinc finger protein that is related to regulatory proteins in Arabidopsis and animals. EMF2 transcripts were detected throughout the life cycle of Arabidopsis, especially in actively proliferating tissues. Altering EMF2 expression by transgenic approaches caused phenotypic changes in flowering time and shoot morphogenesis, supporting the idea that EMF2 activity regulates phase transitions and shoot development. The possible molecular function of EMF2 in Arabidopsis development also is discussed based on functional similarity to EMF2-related genes.
RESULTS
Map-Based Cloning of EMF2
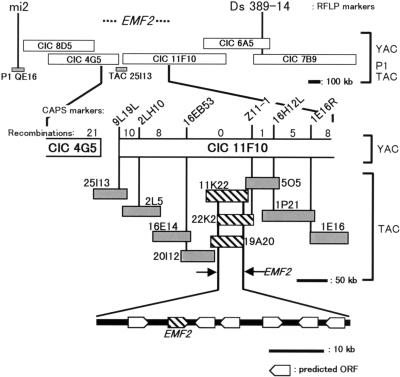
We isolated a new emf2 mutant, emf2-9. Like other emf2 mutants (Yang et al., 1995), emf2-9 displays the characteristic features of the weak emf1 mutant (Sung et al., 1992), namely, petioleless cotyledons and leaves, abbreviated inflorescence bearing terminal flowers, and impairment of petal development. Using emf2-9, we cloned EMF2 using a map-based strategy (Figure 1). The EMF2 locus was mapped between the restriction fragment length polymorphism markers mi2 (Lister and Dean, 1995) and Ds 389-14 (Smith et al., 1996) on chromosome 5. The DNA contig covering this 5.7-centimorgan region was established with DNA clones from the CEPH, INRA, and CNRS (CIC)-yeast artificial chromosome library (Creusot et al., 1995), the P1 library (Liu et al., 1995), and the transformation-competent artificial chromosome (TAC) library (Liu et al., 1999). Fine mapping with cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993; Glazebrook et al., 1998) narrowed the locus to ∼80 kb. The TAC clones, which cover the 80-kb region, were tested to rescue the emf2 mutant phenotype by Agrobacterium-mediated root transformation (Akama et al., 1992). As a result, three complementary clones, 11K22, 22K2, and 19A20, were identified. Using a 50-kb fragment common to these three clones as a probe, we isolated six independent cDNA clones from a library that was made from various tissues and developmental stages (Newman et al., 1994). In addition, one open reading frame (ORF) was predicted from the genomic sequence. To identify the gene complementing the emf2 mutation, a series of deletion subclones covering the 50-kb region was constructed and introduced into the mutants. Consequently, the region corresponding to the cDNA, named 58-1, was identified as responsible for the complementation. A nearly full- length clone of the cDNA was obtained through rescreening of the library. Comparison of genomic (GenBank accession number AB053262) and cDNA sequences revealed that the ORF consists of 21 exons spanning 5813 bp. Molecular lesions of four emf2 alleles were mapped in the ORF (Figure 2A). Thus, we concluded that the ORF is EMF2.
Figure 1.
Map-Based Cloning of the EMF2 Locus.
Scheme of the physical map of the EMF2 locus. CEPH, INRA, and CNRS (CIC)-yeast artificial chromosome (YAC) clones are shown as open bars, and TAC and P1 clones are shown as gray or striped bars. The three striped TAC clones can complement the emf mutation. The positions of restriction fragment length polymorphism (RFLP) and CAPS markers are represented by vertical lines. Recombination numbers in the 2000 tested chromosomes are indicated between each pair of CAPS markers. Predicted ORFs are mapped in the region common to the three complementary TAC clones.
Figure 2.
Structure of the EMF2 Gene.
(A) Scheme of the EMF2 gene. Solid boxes and lines represent exons and introns, respectively. Mutations of four emf2 alleles are indicated at the corresponding sites.
(B) Deduced amino acid sequence of the EMF2 protein. A C2H2-type zinc finger motif is boxed. Two putative nuclear localization signals and an acidic cluster are underlined with single solid lines and a double solid line, respectively. The sequence marked with a dotted line is rich in tryptophan and methionine.
EMF2 Encodes a Novel Polycomb Group Protein Homolog
The predicted EMF2 ORF encodes a novel protein with 631 amino acid residues whose molecular mass is estimated to be 71.7 kD (Figure 2B). The EMF2 protein has a single C2H2 zinc finger motif as well as putative nuclear localization signals. The C-terminal region of the protein is characterized by an acidic cluster and a tryptophan/methionine-rich sequence. We named this region the acidic-W/M domain. Molecular lesions in the two emf2 alleles (emf2-3 and emf2-9) were mapped to this domain (Figure 2A); thus, the domain is crucial for EMF2 function.
Through a survey of Arabidopsis genome databases, at least three EMF2-related genes were detected: FIS2 (GenBank accession number AF096096), VRN2 (GenBank accession number AX03288), and one hypothetical ORF. VRN2, which is involved in the vernalization-dependent promotion of flowering, was isolated and characterized recently (A.R. Gendall, personal communication). The VRN2 gene encodes a protein containing a C2H2 zinc finger and an acidic-W/M domain, showing 67 and 76% identity with corresponding domains of EMF2, respectively, although it lacks the sequence corresponding to six exons in the N-terminal half of EMF2. The second gene, FIS2, was characterized originally by its loss-of-function mutation phenotype of partial seed development in the absence of fertilization (Luo et al., 1999). FIS2 also is similar to EMF2 in the zinc finger and the acidic-W/M domain, showing 55 and 49% identity, respectively, but it differs from EMF2 in the unique repetitive sequences that constitute a putative protein–protein interaction domain (Figure 3) (Luo et al., 1999). The third gene, a hypothetical ORF, was tentatively termed EMF2-LIKE 1 (EML1; GenBank accession number AB053265). It seems to be a pseudogene because the transcripts were not detected by reverse transcriptase–mediated polymerase chain reaction (RT-PCR) using the total RNA from seedlings as templates (data not shown). This gene can potentially encode an acidic-W/M domain, showing 54% identity with EMF2, but it lacks zinc fingers. The exon-intron organization is conserved among these EMF2-related genes, suggesting that they have evolved from the same ancestral sequence (data not shown).
Figure 3.
Structural Similarity in the EMF2/Su(z)12 Protein Family.
(A) Schematic comparison of EMF2, VRN2, FIS2, Su(z)12, and KIAA1610 protein structures. Conserved N-terminal basic domains, C2H2-type zinc finger domains, and C-terminal acidic-W/M domains are colored blue, green, and pink, respectively.
(B) to (D) Sequence alignment of the proteins. Identical amino acids and similar amino acids are boxed in red and yellow, respectively. con, consensus sequence.
(B) Sequence alignment in N-terminal basic domains. Basic amino acid clusters are indicated by a blue bar.
(C) Sequence alignment in zinc finger regions. Conserved cysteine and histidine residues involved in zinc finger formation are indicated with green circles.
(D) Sequence alignment in acidic-W/M domains. Acidic amino acid clusters are indicated by a pink bar.
Interestingly, these EMF2-related proteins show homology with a newly identified Polycomb group (PcG) protein, Su(z)12, of Drosophila (Figure 3) (Birve et al., 2001). Mutations of the Su(z)12 gene cause typical phenotypes of PcG mutants, such as homeotic transformations and misexpression of homeobox genes, in the developmental process of Drosophila. Until now, three classes of PcG gene homologs have been reported in Arabidopsis. Those are CURLY LEAF (CLF) (Goodrich et al., 1997), FIS1/MEDEA (MEA) (Grossniklaus et al., 1998; Luo et al., 1999), and FIS3/FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Ohad et al., 1999). Thus, EMF2, VRN2, and FIS2 constitute the fourth class of PcG gene homologs in Arabidopsis. A cDNA clone, KIAA1060 (Nagase et al., 1995), should be the human homolog of Su(z)12, because these proteins are conserved over the entire sequence. The Arabidopsis and animal genes share three conserved domains, the N-terminal basic domains, zinc finger domains, and acidic-W/M domains, except that FIS2 lacks the N-terminal basic domain (Figure 3).
Expression of EMF2 in Wild-Type Arabidopsis
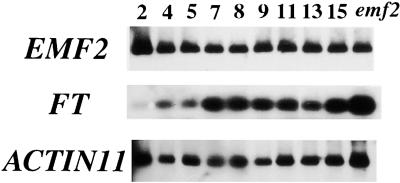
It has been hypothesized that the EMF genes play a major role in repressing floral transition and that a decrease in EMF activity leads to flowering. To determine whether the transcriptional regulation of EMF2 is involved in floral transition, semiquantitative RT-PCR analysis was performed at different stages of growth. The time course of RNA accumulation in seedlings showed that the EMF2 transcript level did not change significantly during vegetative and reproductive development, whereas the FT transcript level was upregulated during flower initiation (Figure 4). Thus, transcriptional regulation of EMF2 appeared not to be involved in the floral transition.
Figure 4.
Expression Time-Course of EMF2 during Arabidopsis Growth.
Wild-type Columbia plants were cultivated in vitro under LD conditions (see Methods), and seedlings were harvested at 2, 4, 5, 7, 8, 9, 11, 13, and 15 days after sowing. Semiquantitative RT-PCR was performed using total RNA as a template for EMF2, FT, and ACTIN11 RNA amplifications. PCR products were detected by DNA gel blot analysis. Numbers indicate days of cultivation before harvesting. Floral buds are visible after 10 to 14 days of cultivation in this condition. emf2, emf2-3 plants harvested after 14 days of cultivation.
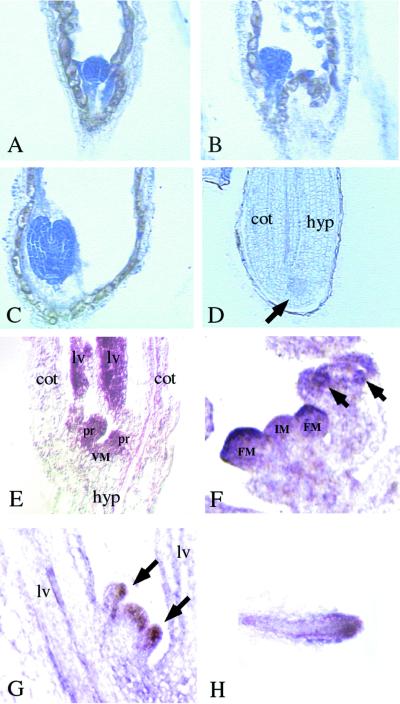
We further studied the spatial expression pattern of EMF2 by in situ RNA hybridization. The results demonstrated that EMF2 is expressed throughout the life cycle of Arabidopsis with preferential localization in actively proliferating tissues. In early seed development, EMF2 mRNA was observed in the developing embryos and endosperm. EMF2 mRNA was detected in the entire embryos as early as the globular stage. When embryos were about to mature, EMF2 mRNA decreased to an undetectable level, tending to remain for a longer period in the embryonic SAMs (Figures 5A to 5D). In endosperm, EMF2 mRNA was observed in cytoplasm around the free nuclei in syncytium and in early cellularized endosperm (Figure 5B), but it was reduced to an undetectable level during later endosperm development. After germination, EMF2 mRNA was again detected in the SAMs, leaf primordia, and young leaves (Figure 5E). In the reproductive shoots, transcripts were detected in both the inflorescence and the floral meristems (Figure 5F). At later stages of flower development, EMF2 mRNA appeared to accumulate in floral organ primordia (Figure 5F). In coflorescences, the transcripts were detected in SAMs and lateral organs, as seen in the main shoots (Figure 5G). In roots, higher levels of the transcripts were detected in root tips (Figure 5H).
Figure 5.
In Situ Localization of EMF2 mRNA in Wild-Type Arabidopsis.
(A) to (D) EMF2 mRNA signals are detected as dark blue areas with bright-field optics. Longitudinal sections of developing embryos are shown.
(A) Globular stage.
(B) Early heart stage.
(C) Early torpedo stage.
(D) Maturing stage. Arrow indicates the embryonic shoot apex, which showed slightly higher signals than other parts of the embryo. cot, cotyledon; hyp, hypocotyl.
(E) to (H) EMF2 mRNA signals are detected as purple areas with differential interference contrast optics. Longitudinal sections of shoot and root apical meristems are shown.
(E) Shoot apex of a 6-day-old seedling. Stronger signals are detected at the meristem (VM), leaf primordia (pr), and young leaves (lv).
(F) Inflorescence apex. Stronger signals are detected on the inflorescence meristem (IM), floral meristem (FM), and floral organ primordia (arrows).
(G) Coflorescence shoot emerging from the axil of a rosette leaf. Stronger signals are detected on the meristem and leaf primordia. Arrows indicate two leaf primordia emerging from the meristem.
(H) Longitudinal section of a root tip. Stronger signals are detected at the meristem and the provascular tissue.
Suppression of EMF2 in Transgenic Plants
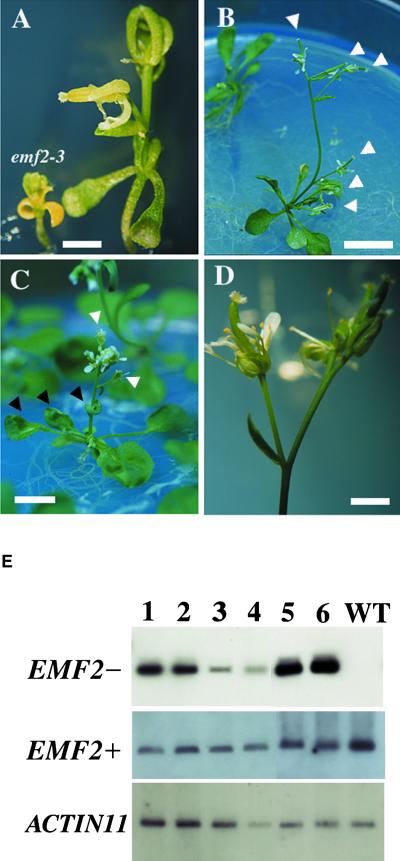
To confirm that EMF2 activity regulates flowering, we generated transgenic Arabidopsis plants expressing antisense or sense EMF2 under the control of the 35S promoter of Cauliflower mosaic virus. For the antisense experiment, the construct containing 2280 bp of the entire cDNA in the antisense orientation was introduced into wild-type plants. Among 69 T1 transformants, 46 plants (67%) exhibited early flowering phenotypes with formation of terminal flowers, whereas 18 plants (26%) showed the wild-type–like phenotype under long-day (LD) conditions. Five plants (7%) had other morphological abnormalities. A gradient of the early- flowering phenotype was observed, which can be grouped into two categories: emf2–like (Figure 6A) and terminal flower1 (tfl1)–like (Figures 6B to 6D). In emf2-like plants, the main and lateral shoots are terminated by infertile flowers. They have petiolated cotyledons but formed two to six small sessile leaves instead of rosette leaves. On the other hand, tfl1-like plants had petiolated rosette leaves and cotyledons and retained some fertility. Under LD conditions, these plants produced six to seven rosette leaves before flowering, the same as wild-type plants, but they are distinguished by terminal flowers and curled leaves (Figure 6C), like clf mutants (Goodrich et al., 1997). emf2-like plants accumulated higher levels of the antisense EMF2 RNA compared with those of the tfl1-like plants (Figure 6E, EMF2−), although the two categories were not significantly different with respect to accumulation levels of the endogenous EMF2 (Figure 6E, EMF2+). This finding suggests that the phenotypic difference may result from varying the amounts of the antisense RNA, which directs the suppression of endogenous EMF2 activity. These results indicate that the suppression of EMF2 affects flowering time by converting inflorescence to floral meristems prematurely, as in 35S::LFY plants (Weigel and Nilsson, 1995) or tfl1 mutants (Shannon and Meeks-Wagner, 1991), and that further suppression causes the plants to bypass the rosette phase, as in emf2 mutants.
Figure 6.
Phenotypes and RNA Levels of Antisense EMF2 Transgenic Plants.
(A) An emf2-like plant. Both the main and lateral shoots develop into terminal flowers. An emf2-3 plant is shown for comparison of size. Bar = 2 mm.
(B) A typical tfl1-like plant. Both the main and all lateral shoots develop into terminal flowers (white arrowheads). Bar = 1 cm.
(C) The clf-like phenotype of a tfl1-like plant. Rosette and cauline leaves are curled (black arrowheads). Both the main and all lateral shoots develop into terminal flowers (white arrowheads). Bar = 5 mm.
(D) Close-up view of terminal flowers of a tfl1-like plant. There is no visible structural abnormality. Bar = 2 mm.
(E) Antisense (EMF2−) and endogenous (EMF2+) RNA levels in transgenic plants. Semiquantitative RT-PCR analysis was performed using total RNA as a template. ACTIN11 RNA was amplified as a control. Four independent tfl1-like plants (lanes 1 to 4) and two emf2-like plants (lanes 5 and 6) were tested. WT, wild type, ecotype Columbia.
In the case of sense transgenic plants, 138 plants among 292 T1 transformants (47%) showed the early-flowering/terminal flower phenotype like the antisense plants (data not shown), whereas 148 (51%) were wild type–like and six (2%) were other mutants. Unexpectedly, no late-flowering phenotype was observed in the sense transformants. Enhancement of only EMF2 expression might be insufficient to repress the reproductive program in Arabidopsis development. Alternatively, ectopic expression of EMF2 might cause suppression of the endogenous gene function in a cosuppression mechanism.
The T2 generation of the antisense transformants was cultivated under LD or short-day (SD) conditions (Table 1). The tfl1-like phenotype was not affected by daylength (Table 1). Some wild-type–like lines segregated the early-flowering phenotype only under SD conditions (Table 1). They bolted after developing 10 to 12 rosette leaves, in contrast to 26.7 leaves for wild-type plants. They produced no terminal flowers and were completely fertile. This early-flowering phenotype under SD conditions appears to be mediated by moderate suppression of EMF2, suggesting that the LD signal might promote flowering partly by suppressing the endogenous EMF2 in wild-type Arabidopsis.
Table 1.
Flowering Times of the Antisense EMF2 Transgenic Plants
| Line, Photoperioda | Ratio of Hygromycin-Resistant Plants | Number of Rosette Leavesb | n |
|---|---|---|---|
| WTL-1, LD | 0.75 | 6.6 ± 0.6 | 21 |
| WTL-1, SD | 0.75 | 11.6 ± 1.6 | 38 |
| WTL-2, LD | 1.00 | 6.4 ± 1.0 | 25 |
| WTL-2, SD | 1.00 | 11.5 ± 1.3 | 23 |
| TFL-1, LD | N. d.c | 6.8 ± 1.2 | 14 |
| TFL-1, SD | N. d.c | 6.0 ± 0.8 | 42 |
| Co-0, LD | 0 | 6.5 ± 1.0 | 23 |
| Co-0, SD | 0 | 26.7 ± 3.5 | 7 |
a WTL and TFL represent wild-type–like and terminal flower1–like transgenic lines, respectively. Co-0 represents wild-type ecotype Columbia plants.
b Average leaf numbers ±sd at the time of bolting. Leaf numbers of hygromycin-resistant plants and tfl1-like plants were counted for the WTL and TFL lines, respectively.
c N. d., not determined.
DISCUSSION
Structural and Functional Homology between EMF2 and PcG Genes
EMF2, together with FIS2 and VRN2, shows significant homology with Su(z)12, a newly identified class of PcG gene from Drosophila (A.R. Gendall, personal communication; Birve et al., 2001). In Drosophila, PcG proteins repress certain sets of target genes and maintain the repression state through cell division (Elgin, 1996; Pirrotta, 1997). The loss-of-function mutations of the EMF genes skip a large part of vegetative growth and flower precociously. This phenotype suggests that the EMF genes could repress the reproductive program, which is likely to be a default program of Arabidopsis development. Similarly, the FIS genes (FIS1/MEA, FIS2, and FIS3/FIE) are thought to repress seed development until fertilization occurs (Grossniklaus et al., 1998; Luo et al., 1999; Ohad et al., 1999). As a result of the discovery of Su(z)12, all of the FIS genes can be characterized as PcG gene homologs. EMF2 and the FIS genes appear to share structural and functional homology, although the EMF1 protein has no similarity to previously characterized proteins (Aubert et al., 2001).
CLF, another PcG gene homolog in Arabidopsis, functions as a repressor of the floral homeotic genes AGAMOUS (AG) and PISTILLATA (PI) in vegetative tissue (Goodrich et al., 1997). clf mutants display early-flowering and curled leaf phenotypes, which also are observed in antisense EMF2 transgenic plants (Figure 6C). Moreover, a previous study showed precocious expression of AP1 and ectopic expression of AG in emf2 mutants (Chen et al., 1997). From these facts, it can be hypothesized that EMF2 and CLF act in the same pathway of phase transitions by regulating proper expression of the floral homeotic genes.
In animals, PcG proteins form large protein complexes (Tie et al., 1998, 2001; Shao et al., 1999) and act to remodel chromatin structures, altering the accessibility of DNA to factors required for transcription. In plants, protein–protein interactions were examined among the FIS genes, and only the MEA–FIE interaction was detected by the yeast two-hybrid system (Luo et al., 2000; Yadegari et al., 2000). Protein interaction between CLF and EMF2 is intriguing. The role of the Arabidopsis PcG homologs in chromatin remodeling remains to be investigated, which should lead to further understanding of the molecular mechanisms of plant development and the evolutionary correlation of developmental regulation between the animal and plant kingdoms.
Involvement of EMF2 in Shoot Development and Flowering
It has been hypothesized that the EMF genes repress reproductive development by delaying the vegetative-to-inflorescence (V/IF) and inflorescence-to-flower (IF/F) transitions. The early-flowering/terminal flower phenotypes of the transgenic plants harboring the antisense EMF2 support this hypothesis. emf2-like and tfl1-like phenotypes demonstrate the role of EMF2 in the repression of the V/IF and IF/F transitions, whereas early flowering under SD conditions suggests EMF2-mediated, photoperiod-dependent regulation of the V/IF transition. However, our findings indicate that transcriptional regulation of endogenous EMF2 is not likely to direct the phase transitions. The possibility of other regulatory mechanisms, such as modification of the protein, remains to be investigated. The expression pattern of EMF1, another EMF gene, is similar to that of EMF2 (Aubert et al., 2001). Like the 35S::sense EMF2 transgenic plants, ectopic 35S::sense EMF1 transgenic plants do not exhibit late-flowering phenotypes (Aubert et al., 2001). These findings indicate that each EMF gene is strictly required but not sufficient for repression of the transitions. The two genes should act in the same pathway, but their molecular interaction remains to be determined. A previous study suggested that the EMF genes interact with LFY and AP1 genes in a negative, reciprocal manner (Chen et al., 1997). Thus, modification of EMF activity by LFY and AP1 may contribute to the phase transitions. EMF2 shows structural similarity to PcG genes, which regulate the expression of homeotic genes that interact reciprocally with Trithorax group (TrxG) genes. An important question is whether a regulatory system similar to the PcG/TrxG system in animals exists in plants.
In inflorescence development, the TFL1 gene interacts with AP1/LFY genes in a reciprocal, negative manner to specify meristem identity (Liljegren et al., 1999). Arabidopsis shoot development is thought to be a highly integrated process that is controlled by a common mechanism, named the controller of phase switch (COPS) (Schultz and Haughn, 1993). In tfl1 mutants, all phases are shortened, whereas ectopic expression of TFL1 causes the extension of all phases (Ratcliffe et al., 1998). These observations support the COPS hypothesis and suggest that TFL1 affects COPS activity. The phenotype of emf mutants also can be interpreted by compressing all of the developmental phases, and the EMF2 transcripts accumulate in all phases that we analyzed. Thus, it can be speculated that EMF2 may be involved in COPS by acting in a manner similar to TFL1. Otherwise, EMF2 activity, possibly along with EMF1 activity, may be required for TFL1 activity. The phenotypic similarity between tfl1 and tfl1-like transgenic plants supports this speculation. In the reproductive phase, EMF2 mRNA was detected throughout the inflorescence and floral meristems (Figure 5G), whereas the TFL1 mRNA accumulated specifically in a group of cells just below the apical dome of inflorescence meristems (Bradley et al., 1997). Further investigations of gene interaction between EMF1/EMF2 and TFL1 will help explain this diverse expression pattern. In addition to these investigations, discovery of the genes regulated directly by EMF2 should provide further insights into the gene network that controls Arabidopsis shoot development.
METHODS
Plant Material and Mutant Screening
All Arabidopsis thaliana plants were grown under white light at 24°C. The photoperiod was 16 hr of light and 8 hr of dark for long-day (LD) conditions and 8 hr of light and 16 hr of dark for short-day (SD) conditions. Plants were planted on rock fiber blocks and supplied Hyponex (1:1000 dilution; Hyponex, Inc., Tokyo, Japan) as liquid nutrient. For in vitro culture, 0.8% agar plates containing half-strength B5 salts (Sigma), 1% sugar, and Hyponex diluted 1:2000 (pH adjusted to 5.8) were used. M2 seed of ethyl methanesulfonate–mutagenized Landsberg ecotype were purchased from Lehle Seed (Round Rock, TX). Approximately 5000 M2 seed from each of 10 parental groups, totaling 50,000 seed, were sowed on the agar medium and screened for emf2-like seedlings. Five thousand M3 lines were established from the parental group in which the emf2-like mutant was obtained. Consequently, the line segregating the mutant was isolated. We confirmed the allelism between this line and emf2-3 by reciprocal crossing. The line was identified as the ninth allele of emf2 (emf2-9).
Map-Based Cloning
Genetic and physical mapping of emf2-9 was conducted as described by Schmidt and Dean (1992). All genetic complementation analysis was performed using the root transformation method (Akama et al., 1992). Agrobacterium tumefaciens strain MP90 was transformed with transformation-competent artificial chromosome (TAC) plasmids (Liu et al., 1999), and then root sections from emf2-9 plants were infected with the transformed bacteria. Complementation was judged from regeneration of the normal inflorescence stalks from transformed calli.
In Situ RNA Hybridization
Wild-type Columbia plants were grown under LD conditions. Shoots, flowers, roots, and siliques at different developmental stages were harvested and fixed as described (Chen et al., 1997). Methods for digoxigenin labeling of RNA probes, slide preparation, and in situ hybridization were as described on the World Wide Web (http://www.arabidopsis.org/cshl-course/5-in_situ.html; http://www.wisc.edu/genetics/CATG/barton/protocols.html). For synthesis of the EMF2-specific probe, the 441-bp PstI fragment of EMF2 cDNA, the region deleted in VRN2, was subcloned into pBluescript KS+ and transcribed in vitro with digoxigenin-UTP using T3 and T7 RNA polymerase for the antisense and sense probes, respectively.
Semiquantitative Reverse Transcriptase–Mediated Polymerase Chain Reaction
Total RNA was extracted from seedlings of wild-type, mutant, or transgenic plants using RNeasy (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Reverse transcription–mediated polymerase chain reaction (RT-PCR) was performed using the mRNA Selective PCR kit (TaKaRa; Kyoto, Japan) according to the manufacturer's instructions using 0.1 μg of total RNA as starting material. PCR was stopped in the exponential phase of amplification, and the products were detected by DNA gel blot analysis. Primers used to amplify the endogenous EMF2 mRNA were 5′-GGCAAGACTCGTTTCTCCTAAGC-3′ (EMF-F) and 5′-GCAACAAGGAAGAGG-AAGGATGT-3′ (EMF-R), designed for amplification of the EMF2-specific region deleted in VRN2. For amplification of FT mRNA, previously described primers (Kobayashi et al., 1999) were used. As a control reaction, the Arabidopsis ACTIN11 gene was amplified using primers 5′-ATGGCAGATGGTGAAGACATTCAG-3′ and 5′-GAAGCA-CTTCCTGTGGACTATTGATG-3′. For amplification of the antisense EMF2 RNA, 5′-GTTTGAACGATCGGGGAAATTC-3′, the sequence upstream of the polyadenylation signals in the nopaline synthase terminator, was used in combination with EMF-R or other EMF2-specific primers to check for reproducibility. For amplification of endogenous EMF2 mRNA from the antisense transformants, DNase I–treated RNA samples were reverse transcribed by ReverTraAce (Toyobo, Osaka, Japan) using the EMF-R primer. PCR was performed successively using KOD Dash polymerase (Toyobo) with the following program: 94°C for 3 min followed by 18 cycles of 94°C for 30 sec, 60°C for 10 sec, and 74°C for 1 min.
Generation of Transgenic Arabidopsis
The antisense EMF2 construct was generated as follows. The cDNA clone was cut with SalI and NotI and blunted with the Klenow fragment. The cDNA region was separated by agarose gel electrophoresis and ligated with XbaI-digested and blunted KH2 vector, the pBI121-based binary plasmid with a hygromycin selection marker, courtesy of Dr. Kenzo Nakamura (Nagoya University, Nagoya, Japan). The clones containing the antisense-oriented EMF2 coding sequence were selected by enzymatic digestion of their plasmids. The antisense constructs were introduced into Agrobacterium strain MP90 by electroporation. Wild-type Columbia plants were infected with the bacteria harboring the constructs by means of the vacuum infiltration method (Bechtold and Pelletier, 1998). T1 transformants were selected on the agar medium containing 30 mg/L hygromycin B.
Accession Number
The GenBank accession number for the EMF2 cDNA clone is AB053171.
Acknowledgments
We particularly thank Jürg Müller (Max Plank Institute, Tubingen, Germany) and Tony Gendall (John Innes Centre, Norwich, UK) for sharing unpublished results. We thank the former researchers of the Mitsui Plant Biotechnology Research Institute, especially Daisuke Shibata, for supplying mi markers, the P1 library, and the TAC library, and Robert Wittier, Yao-Gang Liu, and Norihiro Mitsukawa for useful advice and suggestions on the cloning of EMF2. We acknowledge the Arabidopsis Biological Resource Center (Columbus, OH) for providing the cDNA library, the yeast artificial chromosome library, and restriction fragment length polymorphism markers. We thank Takashi Araki (Kyoto University) for reviewing the manuscript. We especially thank Takayuki Nakano, the former head of the bioscience laboratory at Mitsui Petrochemical Industries, Ltd., for encouraging this study. This work was supported by United States Department of Agriculture Grant No. 99-35301-7984 to Z.R.S.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010227.
References
- Akama, K., Shiraishi, H., Ohta, S., Nakamura, K., Okada, K., and Shimura, Y. (1992). Efficient transformation of Arabidopsis thaliana: Comparison of efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep. 12 7–11. [DOI] [PubMed] [Google Scholar]
- Aubert, D., Chen, L., Moon, Y.H., Martin, D., Castle, L.A., Yang, C.H., and Sung, Z.R. (2001). EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Birve, A., Sengupta, A., Beuchle, D., Larsson, J., Kennison, J.A., Rasmuson-Lestander, Å., and Müller, J. (2001). Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128 3371–3379. [DOI] [PubMed] [Google Scholar]
- Blázquez, M. (2000). Flower development pathways. J. Cell Sci. 113 3547–3548. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Green, R., Nilsson, O., Sussman, M.R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, R., Kampmann, G., Chandler, J., Gleissner, R., Wisman, E., Apel, K., and Melzer, S. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24 591–599. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275 80–83. [DOI] [PubMed] [Google Scholar]
- Chandler, J., Wilson, A., and Dean, C. (1996). Arabidopsis mutants showing an altered response to vernalization. Plant J. 10 637–644. [DOI] [PubMed] [Google Scholar]
- Chen, L., Cheng, J.C., Castle, L., and Sung, Z.R. (1997). EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot, F., et al. (1995). The CIC library: A large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 8 763–770. [DOI] [PubMed] [Google Scholar]
- Elgin, S.C. (1996). Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 6 193–202. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Drenkard, E., Preuss, D., and Ausubel, F.M. (1998). Use of cleaved amplified polymorphic sequences (CAPS) as genetic markers in Arabidopsis thaliana. Methods Mol. Biol. 82 173–182. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44–51. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Haung, M.D., and Yang, C.H. (1998). EMF genes interact with late-flowering genes to regulate Arabidopsis shoot development. Plant Cell Physiol. 39 382–393. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 345–370. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10 1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., and Yanofsky, M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, C., and Dean, C. (1995). Lister and Dean RI map. Weeds World 2 11–18. [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Vazquez-Tello, A., and Whittier, R.F. (1995). Generation of a high-quality P1 library of Arabidopsis suitable for chromosome walking. Plant J. 7 351–358. [Google Scholar]
- Liu, Y.G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., and Shibata, D. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377 522–524. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 377 273–277. [DOI] [PubMed] [Google Scholar]
- Martínez-Zapater, J.M., Coupland, G., Dean, C., and Koornneef, M. (1994). The transition to flowering in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 403–433.
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase, T., Seki, N., Tanaka, A., Ishikawa, K., and Nomura, N. (1995). Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121–KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 2 167–174. [DOI] [PubMed] [Google Scholar]
- Newman, T., de Bruijn, F.J., Green, P., Keegstra, K., Kende, H., Mclntosh, L., Ohlrogge, J., Raikhel, N., Somerville, S., Thomashow, M., Rentzel, E., and Somerville, C. (1994). Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta, V. (1997). Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 13 314–318. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250 923–930. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Amaya, I., Vincent, C.A., Rothstein, S., Carpenter, R., Coen, E.S., and Bradley, D.J. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125 1609–1615. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., and Dean, C. (1992). Physical mapping of the Arabidopsis thaliana genome. In Strategies for Physical Mapping, K.E. Davis and S.M. Tilghman, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 71–98.
- Schultz, E.A., and Haughn, G.W. (1993). Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119 745–765. [Google Scholar]
- Shannon, S., and Meeks-Wagner, D.R. (1991). A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Z., Raible, F., Mollaaghababa, R., Guyon, J.R., Wu, C.T., Bender, W., and Kingston, R.E. (1999). Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98 37–46. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Rouse, D.T., Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R., Igeno, M.I., and Coupland, G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384 59–62. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., Gendall, A.R., and Dean, C. (1999). When to switch to flowering. Annu. Rev. Cell Dev. Biol. 15 519–550. [DOI] [PubMed] [Google Scholar]
- Smith, D., Yanai, Y., Liu, Y.G., Ishiguro, S., Okada, K., Shibata, D., Whittier, R.F., and Fedroff, N.V. (1996). Characterization and mapping of Ds-GUS-T-DNA lines for targeted insertional mutagenesis. Plant J. 10 721–732. [DOI] [PubMed] [Google Scholar]
- Sung, Z.R., Belachew, L., Shunong, B., and Bertrand-Garcia, R. (1992). EMF, an Arabidopsis gene required for vegetative shoot development. Science 258 1645–1647. [DOI] [PubMed] [Google Scholar]
- Tie, F., Furuyama, T., and Harte, P.J. (1998). The Drosophila Polycomb group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 125 3483–3496. [DOI] [PubMed] [Google Scholar]
- Tie, F., Furuyama, T., Prasad-Sinha, J., Jane, E., and Harte, P.J. (2001). The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128 275–286. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377 495–500. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Katz, A., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.H., Chen, L.J., and Sung, Z.R. (1995). Genetic regulation of shoot development in Arabidopsis: Role of the EMF genes. Dev. Biol. 169 421–435. [DOI] [PubMed] [Google Scholar]