Abstract
The potato pathogenesis-related gene PR-10a is transcriptionally activated in response to pathogen infection or elicitor treatment. Characterization of the cis-acting elements of the PR-10a promoter revealed the presence of a silencing element between residues −52 and −27 that contributes to transcriptional regulation. In this study, we have isolated a silencing element binding factor (SEBF) from potato tuber nuclei that binds to the coding strand of the silencing element in a sequence-specific manner. The consensus binding site of SEBF, PyTGTCNC, is present in a number of PR genes and shows striking similarity to the auxin response element. Mutational analysis of the PR-10a promoter revealed an inverse correlation between the in vitro binding of SEBF and the expression of PR-10a. SEBF was purified to homogeneity from potato tubers, and sequencing of the N terminus of the protein led to the isolation of a cDNA clone. Sequence analysis revealed that SEBF is homologous with chloroplast RNA binding proteins that possess consensus sequence–type RNA binding domains characteristic of heterogenous nuclear ribonucleoproteins (hnRNPs). Overexpression of SEBF in protoplasts repressed the activity of a PR-10a reporter construct in a silencing element–dependent manner, confirming the role of SEBF as a transcriptional repressor.
INTRODUCTION
A variety of defense-specific events are induced in plants in response to pathogen infection, including the production of reactive oxygen species, activation of G-proteins, reinforcement of the cell wall, and induction of signal transduction cascades leading to the transcriptional activation of defense genes (Dixon et al., 1994; Blumwald et al., 1998). Although key components of the signaling cascades are being discovered, few transcription factors have been identified that integrate these signals at the transcriptional level.
The well-characterized PR genes induced by pathogen invasion provide excellent models to study the transcriptional regulation of defense genes. PR genes are subdivided into 11 classes based on sequence homology (Van Loon et al., 1994). The PR-10 gene family is subdivided into two groups and encodes small, acidic intracellular proteins of 15 to 18 kD (Osmark et al., 1998). Although RNase activity has been demonstrated for PR-10 proteins, they do not possess any sequence similarity to classic RNases (Moiseyev et al., 1994; Swoboda et al., 1996). In potato, three members of this family have been identified, of which PR-10a is the best characterized.
Expression studies have identified cis-acting elements involved in PR-10a gene regulation. An elicitor response element (ERE) located between nucleotides −135 and −105 is essential and sufficient for elicitor-induced gene expression (Matton et al., 1993; Després et al., 1995; Desveaux et al., 2000). PBF-2, a single-stranded DNA binding factor, appears to play a role in the activation of PR-10a from the ERE (Després et al., 1995; Desveaux et al., 2000). Although the presence of the ERE is sufficient for PR-10a activation, removal of the silencing element (SE), which is located between −52 and −27, leads to further activation, suggesting that this element participates with the ERE in the regulation of PR-10a (Matton et al., 1993; Després et al., 1995). Therefore, a thorough understanding of PR-10a regulation requires that we understand the contribution of the SE and its trans-acting factors.
In this article, we demonstrate, using transient expression studies, that only nine nucleotides containing the sequence GACTGTCAC are required for full SE activity. We have identified a nuclear factor (silencing element binding factor [SEBF]) that shows sequence-specific binding to the sequence PyTGTCNC. Database searches revealed the presence of this sequence in the promoter of numerous PR genes. SEBF was purified to homogeneity from potato tubers, and sequencing of the N terminus of the protein led to the isolation of a cDNA encoding SEBF. The deduced amino acid sequence shows a high degree of sequence similarity to chloroplast RNA binding proteins. Subcellular partitioning of leaf cells demonstrated that SEBF is located in both chloroplasts and nuclei, suggesting functions in both cellular compartments. Finally, overexpression of recombinant SEBF in a transient expression system resulted in SE-dependent transcriptional repression of a PR-10a–uidA reporter fusion, confirming the role of SEBF as a transcriptional repressor.
RESULTS
Identification of SEBF
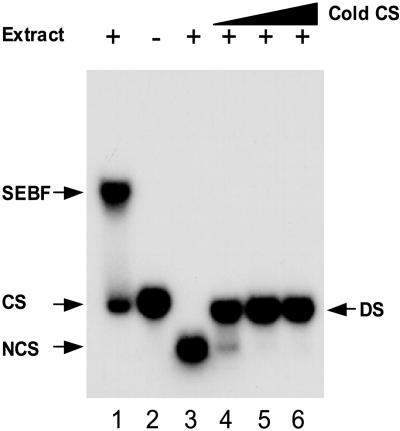
Previous studies identified a negative regulatory sequence (SE) in the promoter of the potato PR-10a gene (Matton et al., 1993; Després et al., 1995). Therefore, we sought to identify a nuclear protein able to regulate PR-10a expression through binding to this sequence. Because PR-10a appears to be regulated positively at the ERE by a single- stranded DNA (ssDNA) binding protein (Desveaux et al., 2000), we searched for proteins that could bind either single- or double-stranded forms of the SE. A crude nuclear extract of potato tubers was incubated with single- or double-stranded radiolabeled SE and analyzed by electrophoretic mobility shift assay (EMSA). Figure 1 demonstrates that a nuclear factor (SEBF) bound the coding strand of the SE (lane 1). SEBF was not able to bind the noncoding strand (lane 3) or double-stranded DNA (lanes 4 to 6). These results show that SEBF is a ssDNA binding protein that recognizes only one strand of the SE.
Figure 1.
SEBF Binds Single-Stranded SE.
EMSA was performed with 10 μg of crude nuclear preparation and 20,000 cpm of 32P-labeled SE coding strand (CS; lane 1) or noncoding strand (NCS; lane 3). The double-stranded probe (DS) was made by annealing radiolabeled noncoding and nonlabeled (cold) coding strands. The ratio of CS to NCS is 0.75:1, 1.5:1, and 3:1 for lanes 4, 5, and 6, respectively. No extract was added in lane 2. Arrows indicate the positions of the CS, NCS, and DS probes and of the SEBF shift in the gel.
Binding of SEBF to the SE Correlates with the Transcriptional Repression of PR-10a
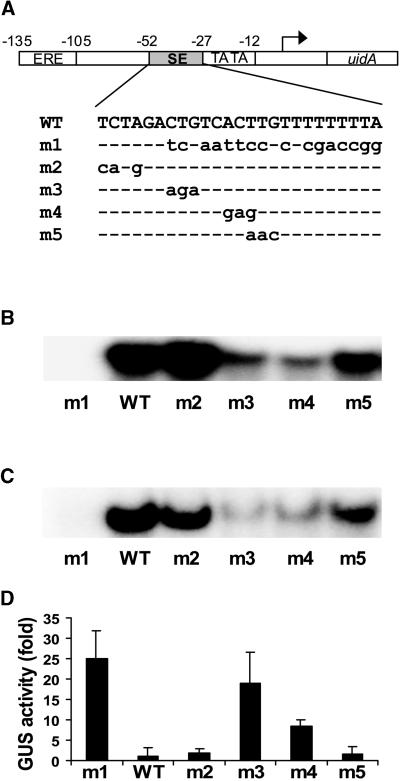
Mutational analysis of the SE was performed to determine which nucleotides constitute the SEBF binding site and whether these nucleotides are important in vivo for the regulation of PR-10a expression. Mutated forms of the SE coding strand were synthesized (Figure 2A) and used as probes in EMSA studies (Figure 2B). The same mutations also were introduced into the PR-10a promoter fused to the uidA reporter gene encoding β-glucuronidase, and the resulting transcriptional activity was measured in a transient expression system (Figure 2D). As shown in Figure 2B, binding of SEBF to mutants m3 and m4 was reduced significantly compared with that of the wild type. In contrast, introduction of these mutations in the promoter of PR-10a led to an increased activity of the reporter gene in transient studies (Figure 2D, m3 and m4). Mutations that showed either a slight increase (Figure 2B, m2) or decrease (Figure 2B, m5) in binding did not show altered transcriptional activities (Figure 2D, m2 and m5). Binding of SEBF was abolished completely using mutant m1, in which 80% of the sequence has been changed (Figure 2B, m1). This absence of binding correlated with the highest level of transcriptional activity observed in transient studies (Figure 2D, m1). Therefore, the transcriptional activity of the PR-10a promoter is inversely correlated with the in vitro binding of SEBF at the SE. This finding suggests that SEBF is the trans-acting factor responsible for SE-mediated repression of PR-10a.
Figure 2.
Mutational Analysis of the SE.
(A) Scheme of the reporter constructs and mutant oligonucleotides used in this study. Reporter constructs contain the PR-10a promoter region from −135 to +136 fused to the bacterial gene uidA encoding β-glucuronidase. The positions of the ERE, the SE, and the TATA box are shown. The transcriptional start site is indicated with an arrow. The common sequence between the wild-type (WT) and mutant oligonucleotides is indicated by dashed lines, and mutated nucleotides are represented by lowercase letters.
(B) EMSA studies using 10 μg of crude potato tuber nuclear preparation and 20,000 cpm of the 32P-labeled single-stranded CS oligonucleotides presented in (A).
(C) EMSA studies using 10 ng of purified recombinant mature SEBF and 20,000 cpm of the 32P-labeled single-stranded CS oligonucleotides presented in (A).
(D) The sequence from −52 to −27 of the PR-10a promoter fused to the uidA gene was replaced with the sequences presented in (A). The resulting plasmids were electroporated in potato leaf protoplasts, and the β-glucuronidase (GUS) activity was measured. The histogram represents fold activity to wild type (WT = 1). Transfection efficiencies were corrected by coelectroporating a luciferase reporter gene. Results represent the mean from a minimum of six individual electroporations. Error bars indicate ±sem.
Sequence PyTGTCNC Defines the DNA Binding Site of SEBF
The results presented in Figure 2 indicate that the SEBF binding site is located within the sequence GACTGTCAC. To further delineate this element, additional mutations were introduced into this region, and their effect on SEBF binding was monitored by EMSA. As indicated in Figure 3, mutations affecting the sequence CTGTCAC reduced the binding of SEBF dramatically (mutants m8, m9, m11, m13, m14, m15, m16, m17, and m19), whereas mutations outside of this region did not reduce binding (mutants m6, m7, m12, and m20). Mutations of the only A in this sequence did not affect binding (mutants m10 and m18). In fact, any nucleotide could replace the A without affecting SEBF binding (data not shown). Substitution of the first C by an A (m13) or a G in the sequence CTGTCAC led to a strong reduction in SEBF binding, whereas substitution by T led to a small increase in binding (data not shown). Therefore, the consensus SEBF binding site is PyTGTCNC.
Figure 3.
Determination of the Consensus SEBF Binding Site.
(A) Two-by-two mutational analysis of the SEBF binding site.
(B) Single-nucleotide analysis of the SEBF binding site.
Oligonucleotides used for fine mapping of the SEBF binding site are listed in each panel. The common sequence between wild-type (WT) and mutant oligonucleotides is indicated by dashed lines, and mutated nucleotides are represented by lowercase letters. EMSA results show the binding of SEBF to oligonucleotides containing the mutated nucleotides. Studies were performed using 10 μg of crude nuclear preparation and 20,000 cpm of the 32P-labeled single- stranded oligonucleotides listed in each panel.
Increased binding of SEBF was observed with mutants m6 and m10 (Figure 3). Transcription factors are known to make secondary hydrogen bonds with either the phosphate backbone or nucleotides outside of their binding sites, and changes of nucleotides involved in these secondary contacts often modify binding affinity (for review, see Pabo and Sauer, 1992). Therefore, mutants m6 and m10 may favor secondary contacts, leading to an increase in SEBF binding.
Purification of SEBF
Further characterization of SEBF required the cloning of its gene. Toward this goal, SEBF was purified from tuber nuclei using a combination of anion exchange chromatography and affinity purification. Table 1 shows that a 20,700-fold purification of SEBF was achieved. Analysis of the chromatographic fractions by SDS-PAGE and Coomassie blue staining revealed two distinct bands of 29 and 28 kD in the most purified fraction (Figure 4, lane 4). To determine which of these two bands possessed DNA binding activity, proteins from the second affinity purification were transferred to nitrocellulose and probed with the wild-type SE oligonucleotide. The results indicate that the two purified proteins can interact independently with the SE (Figure 4, lane 5). Both of these proteins were subjected to N-terminal sequence analysis.
Table 1.
Purification of SEBF from Potato Tubers
| Fraction | Total Protein (mg) | Total Activitya (pg DNA) | Specific Activity (pg DNA/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude nuclear | 112 | 3.3 × 106 | 2.9 × 104 | 1 | 100 |
| Q-Sephb | 1.15 | 3.0 × 106 | 2.6 × 106 | 90 | 91 |
| Aff.1c | 4.0 × 10−3 d | 2.3 × 106 | 5.8 × 108 | 20,000 | 70 |
| Aff.2e | 3.5 × 10−3 d | 2.1 × 106 | 6.0 × 108 | 20,700 | 64 |
a Total activity was determined by measuring labeled probe bound by SEBF in EMSA and calculating total picograms of DNA bound in each fraction.
b Q-Seph, Q-Sepharose Fast-Flow anion exchange chromatography.
c Aff.1, first round of DNA affinity chromatography.
d Estimated from stained gels.
Aff.2, second round of DNA affinity chromatography.
Figure 4.
Purification of SEBF.
Coomassie blue staining of proteins from each step of the purification of SEBF (lanes 1 to 4). The crude nuclear extract (Crude; lane 1) was loaded onto a Q-Sepharose column. SEBF was eluted at 400 mM NaCl (Q-Seph; lane 2) before two rounds of affinity purification (Aff.1 and Aff.2; lanes 3 and 4). Lane 5 shows the results of a DNA probed protein gel blot (Dp) experiment performed with affinity 2–purified SEBF and the wild-type coding strand as a radiolabeled probe. Arrows indicate the two purified bands. Molecular mass markers are indicated at left.
Cloning of SEBF
A 50–amino acid sequence was obtained from N-terminal sequencing of the 29-kD protein. N-terminal sequencing of the 28-kD protein showed more than one amino acid at each cycle. However, a clear sequence similarity with that of the 29-kD protein could be observed, suggesting that the 28-kD band contained degraded forms of the 29-kD protein. Using the amino acid sequence obtained from the 29-kD protein, a polymerase chain reaction (PCR) strategy was elaborated to clone a cDNA (see Methods). The amino acid sequence derived from the cDNA clone is presented in Figure 5. The 50 residues determined by N-terminal sequencing (amino acids 60 to 109, indicated by stars) are preceded by a 59–amino acid sequence, suggesting that SEBF is synthesized as a precursor (pre-SEBF). The TargetP program, which predicts the cellular location of a protein (Emanuelson et al., 2000), predicted that SEBF would be located at the chloroplast with reliability class 1 (Table 2). Ninety-nine percent of the proteins having reliability class 1 were predicted correctly when using TargetP on a nonredundant subset of plant proteins (Emanuelson et al., 2000). For comparison, the predicted locations of the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit and the chlorophyll a/b binding protein are given. TargetP also predicted that the putative transit sequence in SEBF would be cleaved after residue 58, suggesting that the 59 residues encoded in the cDNA but absent from the purified protein could act as a chloroplastic transit peptide. Similar results were obtained using the Predotar (http://www.inra.fr/Internet/Produits/Predotar/) and iPSORT (http://www.hypothesiscreator.net/iPSORT/) subcellular location prediction programs (data not shown).
Figure 5.
Sequence of SEBF and Alignment with a Family of Nucleus-Encoded Chloroplast RNA Binding Proteins.
The amino acid sequence derived from the SEBF cDNA is presented. A member of each group of nucleus-encoded chloroplast RNA binding proteins was chosen for sequence comparison (cp29A from group I, GenBank accession number Q08935; cp31 from group II, GenBank accession number P19683; cp33 from group III, GenBank accession number P19684). Amino acids obtained through N-terminal sequencing of the purified protein are indicated by stars. The beginning of the mature form of SEBF is indicated by the arrowhead above the sequence. The putative transit peptide (T), the acidic region (A), and the cs-RBDs (RI and RII) are underlined. The conserved amino acids that define the cs-RBD are indicated by triangles below the sequence. Black boxes indicate sequence identity, and gray shading indicates conservative substitutions.
Table 2.
TargetP Predictions of the Subcellular Localization of SEBF
| TargetP Scores
|
||||||
|---|---|---|---|---|---|---|
| Precursor (Accession Number) | cTPa | mTPb | SPc | Other | TargetP Predicted Location | RCd |
| SEBF (AF38941) | 0.987 | 0.015 | 0.010 | 0.010 | Chloroplast | 1 |
| Rubisco small subunit (X69759) | 0.874 | 0.044 | 0.096 | 0.099 | Chloroplast | 2 |
| Chlorophyll a/b binding protein (CAA78379) | 0.846 | 0.031 | 0.177 | 0.154 | Chloroplast | 2 |
a Score for chloroplastic transit peptide.
b Score for mitochondrial transit peptide.
Score for signal peptide.
d Reliability class (RC) is the difference (diff.) between the highest score and the second highest score. The classes are defined as follows: RC1, diff. > 0.8; RC2, 0.8 > diff. > 0.6; RC3, 0.6 > diff. > 0.4; RC4, 0.4 > diff. > 0.2; RC5, 0.2 > diff.
An acidic region present in the N-terminal region of the mature protein precedes two consensus sequence–type RNA binding domains (cs-RBDs; identified as RI and RII in Figure 5). These domains, also known as RNA recognition motifs, ribonucleoprotein consensus sequences (RNP-cs), and RNP motifs (Burd and Dreyfuss, 1994), are separated by a glycine-rich region. Database searches indicated a high degree of sequence similarity to a family of nucleus-encoded chloroplast RNA binding proteins (Figure 5). The greatest similarity is to tobacco cp29A (86% sequence identity), a member of the group I chloroplast RNA binding proteins (Ohta et al., 1995).
To confirm that the cDNA-encoded protein corresponds functionally to SEBF, the mature protein was expressed as a glutathione S-transferase fusion protein in Escherichia coli, purified, and tested for binding to the SE. Figure 2C shows that the recombinant mature SEBF binds to SE oligonucleotides with binding specificity similar to native SEBF (Figure 2B).
Cellular Distribution of SEBF
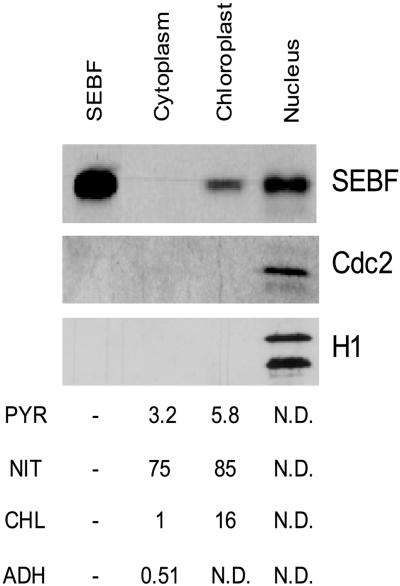
The purification of SEBF from isolated nuclei (Table 1) indicates that SEBF is a nuclear protein. However, the presence of a putative transit sequence in recombinant pre-SEBF suggests that the protein could be present in plastids as well. This led us to investigate the subcellular localization of SEBF. Antibodies were raised against recombinant mature SEBF, and subcellular fractions were obtained from potato leaves. The integrity of each fraction was verified immunologically and by enzymatic assays. Figure 6 shows that as expected, alcohol dehydrogenase activity was restricted to the cytosolic fraction. The nuclei were free of the chloroplastic marker chlorophyll, and little contamination was observed in the cytosolic fraction. No detectable alkaline pyrophosphatase or nitrite reductase activities, which are used as plastid markers (MacDonald and ap Rees, 1983; Smith et al., 1993), were found in the nuclear fraction. The chloroplastic fraction was clear of nuclear contamination, as shown by protein gel blot analysis using antibodies against the nuclear proteins histone H1 and Cdc2 (Figure 6). Protein gel blot analysis performed on these fractions using an anti-SEBF antibody revealed that a protein immunologically related to SEBF was present in chloroplasts and nuclei (Figure 6). A single 29-kD band is present in both cellular compartments and comigrates with the mature form of recombinant SEBF (in which the 59–amino acid putative transit peptide has been removed) (Figure 6, SEBF). No proteins of higher molecular mass were detected in any fraction, suggesting that if SEBF is synthesized as a precursor, it is processed efficiently by peptidases before nuclear import.
Figure 6.
Cellular Localization of SEBF.
Potato leaves were fractionated into cytoplasm, nuclei, and chloroplasts. Ten micrograms of each fraction were separated by 12% SDS-PAGE. Proteins were transferred to nitrocellulose, and the presence of SEBF and the nuclear proteins histone H1 and Cdc2 was revealed with specific antibodies. The first lane (10 ng of recombinant mature SEBF) shows the protein that served to immunize the rabbits. Chlorophyll (CHL), nitrite reductase (NIT), and alkaline pyrophosphatase (PYR) were used as chloroplastic markers (data are presented as μg chlorophyll·mg−1 protein, nmol nitrite·mg−1 protein·min−1, and μmol PO4·mg−1 protein·min−1, respectively), and alcohol dehydrogenase (ADH) was used as a cytoplasmic marker (data are presented as increased OD·mg−1 protein·min−1). N.D., not detectable; −, not determined.
SEBF Is a Single-Copy Gene
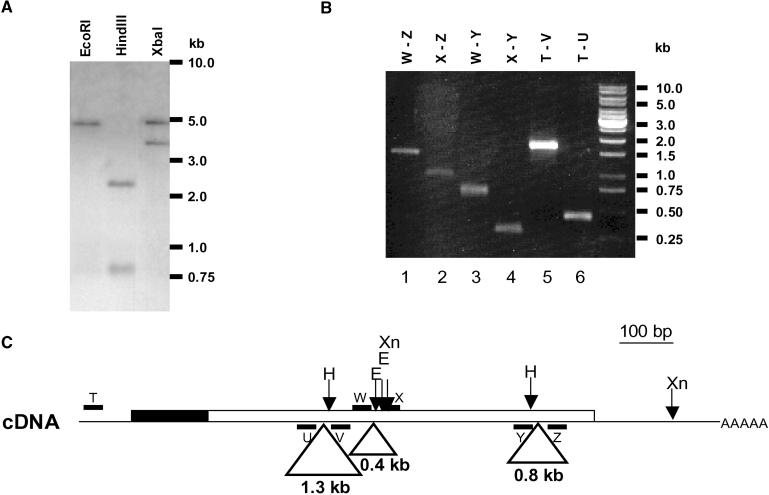
To exclude the possibility that nuclear and chloroplastic SEBF could be the products of different genes, genomic DNA was digested with restriction enzymes and used for DNA gel blot analysis. As shown in Figure 7A, probing the genomic DNA with an SEBF cDNA fragment resulted in a single EcoRI band and two HindIII bands. These were the expected results for a single-copy gene because no EcoRI sites and a single HindIII site are found within the probe. The presence of two XbaI fragments is attributable to the presence of an XbaI site in the third intron of the gene (data not shown). Analysis of potato genomic DNA by PCR led to the identification of three introns in the gene coding for SEBF (Figure 7B). These three introns are present at the same position in homologs of SEBF in Arabidopsis and tobacco (Ye et al., 1991; Ohta et al., 1995). These results clearly indicate that there is only one copy of the SEBF gene in the potato genome. Because no introns are present at the 5′ end of the gene (Figure 7C), the absence of a transit peptide on the purified protein is not caused by alternative splicing. This was confirmed by sequencing 5′ rapid amplification of cDNA ends products, all of which showed sequences identical to that of SEBF (data not shown). These results are supported by the conservation of the genomic organization of this gene in tobacco and Arabidopsis. Together, these results indicate that the nucleus-localized SEBF is encoded by this gene.
Figure 7.
Genomic Organization of SEBF.
(A) DNA gel blot analysis of SEBF. Five micrograms of digested genomic DNA was loaded per lane and probed with a randomly labeled XmnI fragment from the SEBF cDNA shown in (C). Molecular mass markers are indicated at right.
(B) PCR analysis of the genomic DNA. The positions of the oligonucleotides (T to Z; lanes 1 to 6) on the cDNA are presented in (C). The differences in size between the amplification products of lanes 5 and 6, lanes 3 and 4, and lanes 2 and 4 define the sizes of introns 1, 2, and 3, respectively. Molecular mass markers are indicated at right.
(C) Deduced genomic organization of SEBF. The cDNA is represented as a line, and the coding region is represented as a box. The N-terminal transit sequence is presented in black. Introns are indicated as triangles emerging from the cDNA. The oligonucleotides used in the PCR reactions are designated by letters (T, U, V, W, X, Y, Z). The positions and sizes of the introns are deduced from the PCR analysis (B). Restriction sites are indicated by arrows: H, HindIII; E, EcoRI; Xn, XmnI.
SEBF Can Repress Transcription
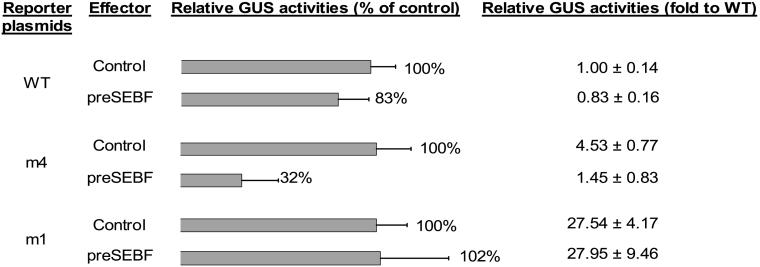
The correlation between the binding of SEBF to mutated forms of the SE and their activity as negative regulatory elements in protoplasts suggests that SEBF is the factor mediating SE-dependent repression. This was confirmed by coexpressing pre-SEBF and various reporter constructs in potato protoplasts. Although repression was observed using the wild-type promoter construct, reporter activity was too low and the results were not statistically significant (Figure 8, WT). This result was expected, because promoter activity with the wild-type SE is already fully repressed. To circumvent this problem, we overexpressed pre-SEBF in conjunction with a construction carrying the partially derepressed mutant m4 (Figure 2D), which is bound with less affinity than is wild type by recombinant and purified SEBF (Figures 2B and 2C). We expected that overexpression of SEBF would compensate for the lower binding affinity to this mutant. Figure 8 demonstrates that in fact, a 68% decrease of promoter activity was attained when pre-SEBF was overexpressed, compared with the activity observed with the overexpression of FK506 binding protein, a control protein (Figure 8, m4). Repression was not seen when pre-SEBF was overexpressed with mutant m1 (Figure 8, m1), to which it does not bind, confirming that the repression observed requires a cis element bound by SEBF and is not the result of tethering of an activator or translational blocking. These results demonstrate that SEBF participates in SE-mediated PR-10a repression and that expression of the pre-SEBF results in nucleus-localized activity.
Figure 8.
Overexpression of SEBF Represses PR-10a Expression.
The coding sequence of pre-SEBF and the coding sequence of a control protein were each inserted into plasmid pBI223D (effector plasmids). These plasmids were coelectroporated in potato leaf protoplasts with the reporter plasmids described in Figure 2D (reporter plasmids). The histogram represents the effect of pre-SEBF overexpression on reporter activity compared with the overexpression of the control protein (control = 100). Fold activity to wild-type SE (WT = 1) is presented for easier reference to Figure 2D. Transfection efficiencies were corrected by coelectroporating a luciferase reporter gene. Results represent means from a minimum of three individual electroporations. GUS, β-glucuronidase. Error bars indicate ±sem.
DISCUSSION
In this study, we identified a ssDNA binding factor that binds with sequence specificity to the coding strand of the SE present in the promoter of the potato PR-10a gene. A strong inverse correlation was shown between the binding of SEBF in vitro and the transcriptional activity of the PR-10a promoter. N-terminal sequencing of the purified factor allowed the cloning of a cDNA that encodes a protein with RNA binding domains similar to those found in mammalian hnRNPs. Recombinant and purified SEBF displayed similar binding specificity toward the SE. Overexpression of the recombinant protein in protoplasts confirmed its implication in the transcriptional repression of the PR-10a gene. Subcellular partitioning followed by immunoblotting with anti-SEBF antibodies revealed the presence of a 29-kD protein in chloroplasts and nuclei, suggesting that SEBF is present in both cellular compartments.
ssDNA Binding Proteins and Regulation of Expression by hnRNP
We demonstrated previously that another cis element of the PR-10a promoter also interacts with a ssDNA binding protein. This element, located between −135 and −105, interacts with the ssDNA binding factor PBF-2 (Després et al., 1995; Desveaux et al., 2000), which has been shown to activate the transcription of a PR-10a–uidA reporter gene construct in protoplasts (D. Desveaux and N. Brisson, unpublished data). Therefore, our data suggest that a large region of the PR-10a promoter is contacted by ssDNA binding proteins. A similar situation has been reported for several genes in animals (reviewed by Rothman-Denes et al., 1998) and is illustrated by the promoter of the c-myc proto-oncogene, in which several classes of single-stranded cis elements occur in vivo and are bound by various specific ssDNA binding factors (Michelotti et al., 1996). The formation of ssDNA regions is known to occur as a consequence of transcription (Giaever and Wang, 1988), which generates torsional stress on the DNA, resulting in an upstream negative supercoiling that forces strands to separate. This also can affect neighboring genes, because the effects of negative supercoiling can be perceived as far as 4 kb from a transcribed gene (Wang and Dröge, 1997). Localized negative supercoiling also has been shown to be produced by the binding of the non-sequence-specific HMG proteins, which generate a bend in the DNA (reviewed by Grasser, 1998). DNA supercoiling then may lead to the formation or stabilization of noncanonical DNA structures that possess ssDNA regions, providing a way for ssDNA binding proteins to gain access to their target sequence (reviewed by Rothman-Denes et al., 1998). On the other hand, the transcriptional repressor YB-1, a cold shock domain protein, has intrinsic strand-melting activity that allows it to bind to its high-affinity ssDNA site (MacDonald et al., 1995). Both phenomena are mechanisms that could generate ssDNA and allow SEBF binding to the SE.
hnRNPs are among the best-characterized ssDNA binding proteins that play a role in the regulation of gene expression in eukaryotes. For example, hnRNP D and its homologs have been shown to activate (Tay et al., 1992; Lau et al., 2000; Tolnay et al., 2000) or repress (Kamada and Miwa, 1992; Smidt et al., 1995; Chen et al., 1998) transcription from a variety of promoters. Another cs-RBD–containing protein, human hnRNP A1, has been shown to repress transcription of the thymidine kinase gene (Lau et al., 2000). Therefore, it is not surprising that SEBF, a plant protein containing cs-RBDs, can act as a transcriptional repressor. Other plant proteins containing two cs-RBDs, such as Arabidopsis FMV-3b, carnation CEBP-1, and tobacco ACBF, also were able to bind specific regulatory cis elements (Didier and Klee, 1992; Maxson and Woodson, 1996; Séguin et al., 1997) and therefore represent potential transcriptional regulators.
The presence of RNA binding domains in SEBF, and the multiple functions associated with this domain, suggests that SEBF also could play a role in RNA processing. Tobacco chloroplast homologs of SEBF have been shown to bind mRNA and intron-containing pre-tRNAs (Nakamura et al., 1999). They also have been shown to confer stability and ribonuclease protection to mRNA in the chloroplasts (Nakamura et al., 2001). In addition, RNA editing in chloroplasts, like mammalian nuclear editing, also involves the presence of hnRNPs (Lau et al., 1997; Hirose and Sugiura, 2001).
Dual Localization of SEBF
The presence of a protein of similar size and immunologically related to SEBF in chloroplasts suggests that this protein is targeted efficiently to this cellular compartment. This is supported by the presence of a putative transit peptide encoded in the cDNA of SEBF and by studies in other species that have shown that SEBF homologs are present in chloroplasts (Ye et al., 1991; Mieszczak et al., 1992; Ohta et al., 1995). Our results, however, indicate that SEBF is encoded by a single-copy gene and that the transit peptide is not eliminated by differential splicing. This suggests that both the nuclear SEBF and the chloroplast protein are derived from the same precursor. Because only the mature form of SEBF is detected in the nucleus, this also suggests that the precursor can be processed outside the chloroplast. Similar results were found in potato for the enzyme starch phosphorylase, which is synthesized as a precursor and targeted to the amyloplast in young tubers but accumulates in a processed form in the cytoplasm of older tubers (Brisson et al., 1989). In agreement with these data, Dahlin and Cline (1991) reported that the import of proteins into plastids is regulated developmentally, with the plastids losing their ability to import precursors in older tissues. This differential import of proteins could be controlled by the phosphorylation of transit peptides, which has been shown to reduce chloroplast import rates (Waegemann and Soll, 1996). It will be interesting in the future to determine whether the subcellular localization of SEBF changes during development. Nonetheless, the dual localization of SEBF in chloroplasts and the nucleus makes this protein an ideal candidate for regulating metabolism in both subcellular compartments during the defense response. It is well established that in addition to the activation of nuclear defense genes, infection leads to major changes in the primary metabolism of the plant, including reductions in the rate of photosynthesis and synthesis of Rubisco (Kombrink and Hahlbrock, 1990; Somssich and Hahlbrock, 1998). SEBF, therefore, could play a role in coordinating defense-induced changes in both compartments.
SEBF Binding Site Is Present in Other Defense Genes and Is Similar to the Auxin Response Element (AuxRE)
DNA database searches revealed that the SEBF binding site is present in the promoter of a number of genes known to be induced by pathogens (Table 3). Many more sites will be added to this list when an exhaustive study of SEBF binding to naturally occurring sites found in the promoter of PR genes is performed. The SEBF binding site also was found in numerous wound- or stress-inducible genes. This suggests an important and general role for SEBF in plant defense responses. It will be interesting to determine whether the SEBF binding site actually contributes to the regulation of these genes and if SEBF homologs in other species are involved in this regulation.
Table 3.
Occurrence of the SEBF Binding Site in the Promoter of PR Genes
| Gene | Organism | Accession Number |
Sequence |
|---|---|---|---|
| PR-10a | Potato | M29041 | CTGTCAC |
| PR-10b | Potato | M29042 | CTGTCAC |
| PR-10 (ypr10b) | Birch | AJ289770 | CTGTCTC |
| PR-10 (ypr10a) | Birch | AJ289771 | CTGTCAC |
| PR-10 | Apple | AF020542 | CTGTCAC |
| PR-10 | Parsley | U48862 | TTGTCTC |
| PR-10 (MLP)a | Opium poppy | L06467 | CTGTCAC |
| PR-10 (MLP)a | Peach | AF239177 | CTGTCAC |
| Chitinase | Potato | AF153195 | TTGTCAC |
| Chitinase | Bermuda grass | AF105426 | TTGTCTC |
| Chitinase | Arabidopsis | Y14590 | TTGTCTC |
| Glucanase | Rice | X58877 | CTGTCAC |
| Glucanase | Tomato | AF077340 | CTGTCAC |
| PR-1 | Tobacco | X66942 | CTGTCAC |
| PR-1 | Barley | Z48728 | TTGTCAC |
| Osmotin (PR-5) | Tobacco | S68111 | TTGTCAC |
| Peroxidase | Wheat | X85230 | CTGTCAC |
| Alternative oxidase | Voodoo lily | Z15117 | CTGTCAC |
| Antifungal protein | Gastrodia elata | AF334813 | CTGTCTC |
Major latex protein.
Comparison of the SEBF binding site (PyTGTCNC) with other regulatory elements revealed a strong similarity to the auxin response element (AuxRE; TGTCTC) present in composite AuxRE. In the absence of auxin, this element represses the action of a constitutive enhancer element (Ulmasov et al., 1995) and appears to be occupied, regardless of auxin status, by a yet unidentified factor that can interact with auxin response factors (Ulmasov et al., 1999). Auxin is known to negatively regulate several defense genes (Grosset et al., 1990; Jouanneau et al., 1991). Recently, an Arabidopsis pleiotropic mutant was isolated that shows both increased susceptibility to pathogens and auxin insensitivity, demonstrating complex interactions between the pathways leading to these phenotypes (Mayda et al., 2000). These results, and the strong similarity between the SEBF binding site and the AuxRE, raise the possibility that SEBF also could be involved in the hormonal control of gene expression.
Conclusion
It is difficult at this stage to formulate a general model explaining the role of SEBF in the regulation of PR-10a expression. Promoter fusion studies conducted in leaf protoplasts indicate that deletion of the SE is required for optimal ERE-mediated expression (Matton et al., 1993; this study). This demonstrates the importance of repression for the proper regulation of PR-10a expression in this system in which SEBF could play a major role. Although SE-mediated repression also is observed in transgenic tubers, an enhancer element positioned between −155 and −135 suppresses the action of the silencer after elicitor treatment (Després et al., 1995). This could be the result of a specific interplay between tuber-specific enhancer binding factors and SEBF, thus allowing maximal gene expression through the action of the ERE. In this context, post-translational modifications of SEBF could be required for specific protein–protein interactions to occur. Yeast two-hybrid studies are being conducted to identify proteins that interact with SEBF. In addition, our future experiments will aim at determining whether SEBF activity and localization are regulated by phosphorylation or other post-translational modifications and whether this factor controls other genes involved in defense responses.
METHODS
All standard molecular biology procedures were performed according to Sambrook et al. (1989).
Plant Material
Potato tubers (Solanum tuberosum cv Kennebec) were obtained from the Quebec Ministry of Agriculture Les Buissons Research Station (Pointe-aux-Outardes, Canada). Tubers were stored in the dark at 4°C and brought to room temperature 24 hr before use. Leaf mesophyll protoplasts were isolated from 4- to 5-week-old potato plants grown in growth chambers as described by Magnien et al. (1980). Potato leaves were isolated from plants grown in an environmental growth chamber (Conviron, Winnipeg, Canada) under long-day photoperiod conditions.
β-Glucuronidase Reporter Constructs and Analysis
The wild-type construct was created by polymerase chain reaction (PCR) amplification of the PR-10a promoter fragment of plasmid p-135 (Matton et al., 1993) using PCR oligonucleotide 1 (5′-GCC-AAGCTTTAGATAAAATGACACAAATGTCAAAAATGG-3′) and oligonucleotide 2 (5′-CCACCCGGGGATCCAGCTTTGAAC-3′) to replace the XbaI site at position −135 with a HindIII site. After digestion with HindIII and BamHI, the fragment was inserted into pBI201. Mutant m1 was described previously (pLP9; Desveaux et al., 2000). Mutants m3, m4, and m5 were created by PCR using mutated oligonucleotides (Figure 2A) and PCR oligonucleotide 2. The PCR fragments were cleaved with XbaI and BamHI and inserted into the wild-type plasmid described above. For mutant m2, a HindIII-NcoI PCR fragment was synthesized using oligonucleotide 1 and oligonucleotide 3 (5′-CAGTCCATGGTTAAATCAAC-3′). Then, a NcoI-BamHI PCR fragment was synthesized using oligonucleotide 2 and oligonucleotide 4 (5′-TAACCATGGACTGTCACTTG-3′). Ligation of these fragments into pBI201 cleaved with HindIII and BamHI created mutant m2. Amplifications were performed in a Whatman/Biometra TGradient thermal cycler. All constructs were sequenced to ensure that no mutations were inserted by PCR. The transient expression system was described previously (Matton et al., 1993). All values were corrected for the efficiency of electroporation using luciferase activity resulting from the coelectroporation of 1 μg of plasmid pWB216 (Barnes, 1990).
The coding sequence for the precursor silencing element binding factor (pre-SEBF) was inserted into pBI223D, which contains a double 35S enhancer of Cauliflower mosaic virus. For transrepression studies, 5 μg of expression vector was added to the assays. The control vector expressed a human fusion protein containing FK506 binding protein (Pelletier et al., 1998) under the same promoter.
Preparation of Extracts
Crude nuclear extracts were prepared as follows: 200 g of potato tubers or 75 g of leaves was homogenized in a blender at maximum speed for 1 min in 250 mL of cold NEBH (10 mM Pipes-KOH, pH 6.0, 1 M 2-methyl-2,4-pentanediol, 0.15 mM spermine, 0.5 mM spermidine, 10 mM MgCl2, 14.3 mM β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride). After decantation at 4°C for 5 min, the homogenate was filtered through five layers of Miracloth under vacuum. The filtrate was centrifuged at 4000g for 5 min in a Sorvall (Newtown, CT) GSA rotor to pellet the nuclei. The supernatant served as the cytosolic fraction. Nuclei were washed three times in Nonidet P-40 buffer (10 mM Mes-NaOH, pH 6.0, 260 mM sucrose, 10 mM NaCl, 1 mM EDTA, 0.15 mM spermine, 0.5 mM spermidine, 14.3 mM β-mercaptoethanol, 0.1% BSA, and 1% Nonidet P-40) to remove organelle and cytoskeleton contamination. The final pellet was resuspended in 5 mL of 200 mM NaCl SEBF binding buffer (SBB; 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 14.3 mM β-mercaptoethanol, and variable NaCl concentrations). Nuclei were ruptured by sonication, and the lysate was centrifuged at 19,000g for 45 min. The supernatant was either used immediately for SEBF purification or stored at −70°C in 10% glycerol. In such conditions, SEBF binding activity was stable for months. Chloroplasts were prepared by the method of Gegenheimer (1990).
Determination of Enzymatic Activities in Extracts
Chlorophyll content was measured according to Arnon (1949). Alkaline pyrophosphatase, nitrite reductase, and alcohol dehydrogenase were measured according to Gross and ap Rees (1986), Hucklesby et al. (1972), and Smith and ap Rees (1979), respectively.
Electrophoretic Mobility Shift Assays
Protein extracts (in 200 mM NaCl SBB) were mixed with 20,000 cpm of 32P end-labeled probe. The binding reaction (40 μL) contained 100 ng of poly(dI-dC) to eliminate nonspecific binding. The reaction was left on ice for 15 min. Electrophoretic separation of the complexes was achieved by loading the mixture on a 5.7% polyacrylamide gel prepared in Tris-glycine buffer (25 mM Tris and 195 mM glycine) and applying 200 V for 2.5 hr at 4°C. The gel was exposed overnight to Kodak X-Omat AR film at −70°C.
Purification of SEBF from Nuclear Extracts
Crude nuclear extracts were loaded onto a 3 mL Q-Sepharose fast protein liquid chromatography column (Amersham Pharmacia Biotech) equilibrated in 200 mM NaCl SBB. The column was washed with 10 volumes of 200 mM NaCl SBB followed by 15 volumes of 300 mM NaCl SBB. SEBF was eluted in 4 mL of 400 mM NaCl SBB. The eluate then was submitted to two rounds of affinity purification. The affinity beads were prepared by coupling a biotinylated wild-type SE oligonucleotide to streptavidin-coated paramagnetic beads (Sigma) according to Desveaux et al. (2000). The beads were washed twice with 400 mM NaCl SBB before use. To 1 mL of eluate was added 20 μL of 500 mM EDTA, pH 8.0, 10 μg of poly(dI-dC), and 5 μL of Nonidet P-40. The mixture was added to the affinity beads and left on ice for 15 min with sporadic mixing. After separation of the beads from the solution using a magnet, the beads were washed in 1 mL of 200 mM NaCl SBB followed by a 1-mL wash with 1 M NaCl SBB. SEBF was eluted twice from the beads with 0.1 mL of 2 M NaCl SBB and incubated at 37°C for 5 min. The eluate containing SEBF was desalted using Ultrafree 4 centrifugal filter 10 K BioMax (Millipore, Bedford, MA). Proteins from the equivalent of 1 kg of tubers were pooled and acetone precipitated. The pellet was resuspended in Laemmli solubilizing buffer (Laemmli, 1970) and loaded onto a 12% SDS-polyacrylamide gel. The proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad) and stained with Coomassie Brilliant Blue R 250. Bands were cut and sent for N-terminal analysis at the Eastern Quebec Proteomics Core Facility (Ste-Foy, Canada). A 50–amino acid sequence was obtained from N-terminal analysis of the 29-kD band (see Figure 5). The last 10 amino acids of this sequence (residues 41 to 50) eluted with minor contaminating amino acids. DNA-probed protein gel blot analysis was performed according to Vinson et al. (1988).
Cloning of SEBF
Degenerate oligonucleotides (5′-GTKACWYTITCIGATTTYGAYCA-3′ and 5′-ARRTTWCCIACRAARATYTT-3′) deduced from residues 1 to 8 and 41 to 46, respectively, of the N-terminal sequence of the purified protein were used to generate a 141-bp genomic PCR fragment of SEBF. A specific primer (5′-GTTTGAGTGATGAAGGTGC-3′) was derived from this fragment and used to probe a potato cDNA bank (Matton and Brisson, 1989) using the method of Israel (1993). Twenty-three pools of 18,000 phage were used in PCR reactions using the gene-specific primer and the M13 universal primer. Two pools gave a PCR fragment of ∼1000 bp. These pools were further diluted, and the procedure was repeated until it was possible to isolate a single plaque. A positive clone was excised using the ExAssist system (Stratagene) according to the manufacturer's instructions. The clone was sequenced on both strands with Thermosequenase (Amersham Pharmacia Biotech) in a cycle sequencing reaction that was then processed on a Li-Cor automated sequencer.
DNA Gel Blot Analysis and Genomic Analysis
Procedures for the extraction of genomic DNA and DNA gel blot analysis were as described by Ausubel et al. (2001). The following primers were designed according to the predicted positions of the introns in the tobacco and Arabidopsis SEBF homologs (Ye et al., 1991; Ohta et al., 1995) and used for PCR analysis of the genomic DNA: T, 5′-GCCGTTCTGTCTTCACAATTCTTTTGCTTC-3′; U, 5′-CAACATCTCCAGCACGCTCAAAAAGCTCAG-3′; V, 5′-CCTCTGCTT-CTTCCTGTAAGCTTGTCATAG-3′; W, 5′-CAGTTGAAGCCGCCTGTC-AACAATTTAATG-3′; X, 5′-TTGACGGGAGGGCACTGAGGGTGAATT-CTG-3′; Y, 5′-GCTTTCAATTGCATCGTTGACCTCCTTAG-3′; and Z, 5′-GCTTCAGCAGGACTTACACGGATGGCCCTG-3′.
Antibody Production and Protein Gel Blot Analysis
The cDNA encoding SEBF was fused to the glutathione S-transferase (GST) gene in plasmid pGEX-4T1 (Amersham Pharmacia Biotech). The fusion protein (GST-SEBF) was expressed in BL21 cells. After lysis of the cells, the fusion protein was purified on a glutathione column (Smith and Johnson, 1988). The GST tag was cleaved with thrombin (Amersham Pharmacia Biotech) and removed from the extract by a second pass on the glutathione column. Rabbit immunization was performed according to Harlow and Lane (1988). Briefly, 50 μg of recombinant SEBF was used to immunize rabbits, followed by a second injection of 150 μg 30 days later to boost the immunological reaction. The rabbits were killed 14 days after the second injection. For protein gel blot studies, the SEBF antiserum was used at a dilution 1:5000, and the antibody–antigen interaction was revealed with the enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Antibodies to histone H1 (sc-8030) and Cdc2 (sc-53) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used at a dilution of 1:1000.
Accession Number
The GenBank accession number for SEBF is AF389431.
Acknowledgments
We thank Rajagopal Subramanian for helpful discussions and the members of the laboratory, particularly Darrell Desveaux, for reviewing the manuscript. We also thank Louise Cournoyer for preparing in vitro grown plants and Barbara Otrysko for the gift of certified potato tubers. We also are grateful to Franz Lang, Gertraud Burger, and their group for sequencing SEBF. This research was supported by research grants from the Natural Sciences and Engineering Research Council of Canada and from the Fonds pour la Formation de Chercheurs et l'Aide à la Recherche, Québec.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010231.
References
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts: Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2001). Current Protocols in Molecular Biology. (New York: Wiley).
- Barnes, W.M. (1990). Variable patterns of expression of luciferase in transgenic tobacco leaves. Proc. Natl. Acad. Sci. USA 87 9183–9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald, E., Gilad, S.A., and Lam, B.C.H. (1998). Early signal transduction pathways in plant–pathogen interactions. Trends Plant Sci. 3 342–346. [Google Scholar]
- Brisson, N., Giroux, H., Zollinger, M., Camirand, A., and Simard, C. (1989). Maturation and subcellular compartmentation of potato starch phosphorylase. Plant Cell 1 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G., and Dreyfuss, G. (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265 615–621. [DOI] [PubMed] [Google Scholar]
- Chen, H., Hu, B., Gacad, M.A., and Adams, J.S. (1998). Cloning and expression of a novel dominant-negative-acting estrogen response element-binding protein in the heterogeneous nuclear ribonucleoprotein family. J. Biol. Chem. 273 31352–31357. [DOI] [PubMed] [Google Scholar]
- Dahlin, C., and Cline, K. (1991). Developmental regulation of the plastid protein import apparatus. Plant Cell 3 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., Subramaniam, R., Matton, D.P., and Brisson, N. (1995). The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux, D., Després, C., Joyeux, A., Subramaniam, R., and Brisson, N. (2000). PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12 1477–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier, D.K., and Klee, H.J. (1992). Identification of an Arabidopsis DNA-binding protein with homology to nucleolin. Plant Mol. Biol. 18 977–979. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., Harisson, M.J., and Lamb, C.J. (1994). Early events in the activation of plant defense responses. Annu. Rev. Phytopathol. 32 479–501. [Google Scholar]
- Emanuelson, O., Nielsen, H., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequences. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Gegenheimer, P. (1990). Preparation of extracts from plants. Methods Enzymol. 182 174–193. [DOI] [PubMed] [Google Scholar]
- Giaever, G.N., and Wang, J.C. (1988). Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell 55 849–856. [DOI] [PubMed] [Google Scholar]
- Grasser, K.D. (1998). HMG1 and HU proteins: Architectural elements in plant chromatin. Trends Plant Sci. 3 260–265. [Google Scholar]
- Gross, P., and ap Rees, T. (1986). Alkaline inorganic pyrophosphatase and starch synthesis in amyloplasts. Planta 167 140–145. [DOI] [PubMed] [Google Scholar]
- Grosset, J., Meyer, Y., Chartier, Y., Kauffmann, S., Legrand, M., and Fritig, B. (1990). Tobacco mesophyll protoplasts synthesize 1,3-β-glucanase, chitinases, and “osmotins” during in vitro culture. Plant Physiol. 92 520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hirose, T., and Sugiura, M. (2001). Involvement of a site specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: Development of a chloroplast in vitro RNA editing system. EMBO J. 20 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucklesby, D.P., Dalling, M.J., and Hageman, R.H. (1972). Some properties of two forms of nitrite reductase from corn (Zea mays L.) scutellum. Planta 104 220–233. [DOI] [PubMed] [Google Scholar]
- Israel, D.I. (1993). A PCR-based method for high stringency screening of DNA libraries. Nucleic Acids Res. 21 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau, J.-P., Lapous, D., and Guern, J. (1991). In plant protoplasts, the spontaneous expression of defense reactions and the responsiveness to exogenous elicitors are under auxin control. Plant Physiol. 96 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, S., and Miwa, T. (1992). A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene 119 229–236. [DOI] [PubMed] [Google Scholar]
- Kombrink, E., and Hahlbrock, K. (1990). Rapid, systemic repression of the synthesis of ribulose 1,5-biphosphate carboxylase small-subunit mRNA in fungus-infected or elicitor-treated potato leaves. Planta 181 216–219. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lau, J.S., Baumeister, P., Kim, E., Roy, B., Hsieh, T.-Y., Lai, M., and Lee, A.S. (2000). Heterogenous nuclear ribonucleoproteins as regulators of gene expression through interactions with the human thymidine kinase promoter. J. Cell. Biochem. 79 395–406. [DOI] [PubMed] [Google Scholar]
- Lau, P.P., Zhu, H.-J., Nakamuta, M., and Chan, L. (1997). Cloning of an apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J. Biol. Chem. 272 1452–1455. [DOI] [PubMed] [Google Scholar]
- MacDonald, F.D., and ap Rees, T. (1983). Enzymatic properties of amyloplasts from suspension cultures of soybean. Biochim. Biophys. Acta 755 81–89. [Google Scholar]
- MacDonald, G.H., Itoh-Lindstrom, Y., and Ting, J.P.-Y. (1995). The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J. Biol. Chem. 270 3527–3533. [DOI] [PubMed] [Google Scholar]
- Magnien, E., Dalschaert, X., and Devreux, M. (1980). Different radiosensitivities of Nicotiana plumbaginifolia leaves and regenerating protoplasts. Plant Sci. Lett. 19 231–241. [Google Scholar]
- Matton, D.P., and Brisson, N. (1989). Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. Plant-Microbe Interact. 2 325–331. [DOI] [PubMed] [Google Scholar]
- Matton, D.P., Prescott, G., Bertrand, C., Camirand, A., and Brisson, N. (1993). Identification of cis-acting elements involved in the regulation of the pathogenesis-related gene STH-2 in potato. Plant Mol. Biol. 22 279–291. [DOI] [PubMed] [Google Scholar]
- Maxson, J.M., and Woodson, W.R. (1996). Cloning of a DNA-binding protein that interacts with the ethylene-responsive enhancer element of the carnation GST1 gene. Plant Mol. Biol. 31 751–759. [DOI] [PubMed] [Google Scholar]
- Mayda, E., Mauch-Mani, B., and Vera, P. (2000). Arabidopsis dth9 mutation identifies a gene involved in regulating disease susceptibility without affecting salicylic acid-dependent responses. Plant Cell 12 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti, G.A., Michelotti, E.F., Pullner, A., Duncan, R.C., Eick, D., and Levens, D. (1996). Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16 2656–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieszczak, M., Klahre, U., Levy, J.H., Goodall, G.J., and Filipowicz, W. (1992). Multiple plant RNA binding proteins identified by PCR: Expression of cDNAs encoding RNA binding proteins targeted to chloroplasts in Nicotiana plumbaginifolia. Mol. Gen. Genet. 234 390–400. [DOI] [PubMed] [Google Scholar]
- Moiseyev, G.P., Beintema, J.J., Federoyeva, L.I., and Yakovlev, G.I. (1994). High sequence similarity between a ribonuclease from ginseng calluses and fungus-elicited proteins from parsley indicates that intracellular pathogenesis-related proteins are ribonucleases. Planta 193 470–472. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Ohta, M., Sugiura, M., and Sugita, M. (1999). Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett. 460 437–441. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Ohta, M., Sugiura, M., and Sugita, M. (2001). Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276 147–152. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Sugita, M., and Sugiura, M. (1995). Three types of nuclear encoding chloroplast RNA-binding proteins (cp29, cp31, cp33) are present in Arabidopsis thaliana: Presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant. Mol. Biol. 27 529–539. [DOI] [PubMed] [Google Scholar]
- Osmark, P., Boyle, B., and Brisson, N. (1998). Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol. Biol. 38 1243–1246. [DOI] [PubMed] [Google Scholar]
- Pabo, C.O., and Sauer, R.T. (1992). Transcription factors: Structural families and principles of DNA recognition. Annu. Rev. Biochem. 61 1053–1095. [DOI] [PubMed] [Google Scholar]
- Pelletier, J.N., Campbell-Valois, F.-X., and Michnick, S.W. (1998). Oligomerization domain–directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. USA 95 12141–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman-Denes, L.B., Dai, X., Davydova, E., Carter, R., and Kazmierczak, K. (1998). Transcriptional regulation by DNA structural transitions and single-stranded DNA-binding proteins. Cold Spring Harbor Symp. Quant. Biol. 63, 63–73. [DOI] [PubMed]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Séguin, A., Laible, G., Leyva, A., Dixon, R.A., and Lamb, C.J. (1997). Characterization of a gene encoding a DNA-binding protein that interacts in vitro with vascular specific cis elements of the phenylalanine ammonia-lyase promoter. Plant Mol. Biol. 35 281–291. [DOI] [PubMed] [Google Scholar]
- Smidt, M.P., Russchen, B., Snippe, L., Wijnholds, J., and Ab, G. (1995). Cloning and characterisation of a nuclear, site specific ssDNA binding protein. Nucleic Acids Res. 23 2389–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.M., and ap Rees, T. (1979). Pathways of carbohydrate fermentation in the roots of marsh plants. Planta 146 327–334. [DOI] [PubMed] [Google Scholar]
- Smith, A.G., Marsh, O., and Elder, G.H. (1993). Investigation of the subcellular location of the tetrapyrrole-biosynthesis enzyme coproporphyrinogen oxidase in higher plants. Biochem. J. 292 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.B., and Johnson, K.S. (1988). Single-step purification of polypeptides expressed in Escherichia coli as fusions with the glutathione S-transferase. Gene 67 31–40. [DOI] [PubMed] [Google Scholar]
- Somssich, I.E., and Hahlbrock, K. (1998). Pathogen defence in plants: A paradigm of biological complexity. Trends Plant Sci. 3 86–90. [Google Scholar]
- Swoboda, I., Hoffmannsommergruber, K., Oriordain, G., Schneiner, O., Heberlebors, E., and Vincente, O. (1996). Bet v1 proteins, the major birch pollen allergens and members of a family of conserved pathogenesis-related proteins, show ribonuclease activity in vitro. Physiol. Plant. 96 433–438. [Google Scholar]
- Tay, N., Chan, S.-H., and Ren, E.-C. (1992). Identification of a novel heterogeneous nuclear ribonucleoprotein C-like protein that functions as transcriptional activator of the hepatitis B virus enhancer II. J. Virol. 66 6841–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolnay, M., Barany, L., and Tsokos, G.C. (2000). Heterogeneous nuclear ribonucleoprotein D0 contains transactivator and DNA-binding domains. Biochem. J. 348 151–158. [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z.-B., Hagen, G., and Guilfoyle, T.J. (1995). Composite structure of auxin response elements. Plant Cell 7 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C., Pierpoint, W.S., Boller, T., and Conejero, V. (1994). Recommendations for naming plant pathogenesis-related proteins. Plant Mol. Biol. Rep. 12 245–264. [Google Scholar]
- Vinson, C.R., LaMarco, K.L., Johnson, P.F., Landschulz, W.H., and McKnight, S.L. (1988). In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 2 801–806. [DOI] [PubMed] [Google Scholar]
- Waegemann, K., and Soll, J. (1996). Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem. 271 6545–6554. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and Dröge, P. (1997). Long-range effects in a supercoiled DNA domain generated by transcription in vitro. J. Mol. Biol. 271 499–510. [DOI] [PubMed] [Google Scholar]
- Ye, L., Li, Y., Fukami-Kobayashi, K., Go, M., Konishi, T., Watanabe, A., and Sugiura, M. (1991). Diversity of a ribonucleoprotein family in tobacco chloroplasts: Two new chloroplast ribonucleoproteins and a phylogenetic tree of ten chloroplast RNA-binding domains. Nucleic Acids Res. 19 6485–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]