Abstract
A 128-bp insertion into the maize waxy-B2 allele led to the discovery of Tourist, a family of miniature inverted repeat transposable elements (MITEs). As a special category of nonautonomous elements, MITEs are distinguished by their high copy number, small size, and close association with plant genes. In maize, some Tourist elements (named Tourist-Zm) are present as adjacent or nested insertions. To determine whether the formation of multimers is a common feature of MITEs, we performed a more thorough survey, including an estimation of the proportion of multimers, with 30.2 Mb of publicly available rice genome sequence. Among the 6600 MITEs identified, >10% were present as multimers. The proportion of multimers differs for different MITE families. For some MITE families, a high frequency of self-insertions was found. The fact that all 340 multimers are unique indicates that the multimers are not capable of further amplification.
INTRODUCTION
Transposable elements usually are divided into two classes. Class 1, the retroelements, including the long terminal repeat (LTR) retrotransposons, makes up the largest fraction of most plant genomes (reviewed by Kumar and Bennetzen, 1999). Retroelements are capable of attaining very high copy numbers in a relatively short period because the element-encoded mRNA, and not the element itself, forms the transposition intermediate. Class 2, the DNA elements, is characterized by short terminal inverted repeats (TIRs) and transposition via a DNA intermediate (reviewed by Kunze et al., 1997). Plant DNA elements (such as Ac/Ds, Spm/dSpm, and Mutator) generally excise from one site and reinsert elsewhere in the genome. Class 2 elements can be further divided into two groups. Autonomous elements, such as Ac and Spm from maize, encode the products (transposase) necessary for their transposition (Baker et al., 1986; Yoder, 1990; Kunze et al., 1997). Nonautonomous elements, such as Ds and dSpm, usually are internally deleted versions of autonomous elements. As a result, they require the presence of the autonomous elements Ac and Spm, respectively, for their transposition.
As a result of their conservative mechanism of transposition, the copy number of class 2 element families is usually <100 per haploid genome. One exception to this generalization is miniature inverted repeat transposable elements (MITEs), a special category of nonautonomous elements that display very high copy number (in the thousands) and are uniformly short (usually <500 bp). In addition, most MITEs in plants have TIRs and insert into the TA dinucleotide or into a 3-bp trinucleotide (Bureau and Wessler, 1992, 1994a, 1994b; Mao et al., 2000; Zhang et al., 2000). Although first identified in several plant species, including maize (Bureau and Wessler, 1992, 1994a, 1994b), rice (Bureau and Wessler, 1994a, 1994b; Bureau et al., 1996), green pepper (Pozueta-Romero et al., 1996), and Arabidopsis (Casacuberta et al., 1998; Le et al., 2000), MITEs also are abundant in several animal genomes, including Caenorhabditis elegans (Oosumi et al., 1995a, 1995b; Surzycki and Belknap, 2000), mosquito (Tu, 1997, 2001; Feschotte and Mouchès, 2000), fish (Izsvák et al., 1999), and human (Morgan, 1995; Smit and Riggs, 1996).
Another important feature of MITEs is their preference for insertion into low copy number sequences or genic regions (Tikhonov et al., 1999; Mao et al., 2000; Zhang et al., 2000). In addition, MITEs also were found, in several cases, to insert into each other. For instance, the first Stowaway element was found as an insertion in a sorghum Tourist element (Bureau and Wessler, 1994b), whereas in another case a Tourist dimer was found in the same organism (Tikhonov et al., 1999). Such MITE multimers also were reported in other organisms, including rice (Tarchini et al., 2000) and mosquito (Tu, 1997; Feschotte and Mouchès, 2000). Therefore, it was proposed that MITEs could be preferential targets for other MITEs (Feschotte and Mouchès, 2000).
Given the previously identified target site preference of MITEs and the frequent detection of MITE multimers, we wondered about the propensity of MITE insertion into other MITEs. Such a determination is possible only with a systematic comparison between the insertion frequency of MITEs into MITEs and of MITEs into other sequences. In this article, we report a detailed characterization of maize Tourist multimers and a comprehensive analysis of MITE multimers in rice, a species known to be particularly rich in MITEs (Bureau et al., 1996; Mao et al., 2000; Tarchini et al., 2000). The availability of 30.2 Mb of rice genomic sequence has enabled us to address questions about MITE multimers that could not be answered with the available maize sequence. The analysis of rice genomic sequence not only allows us to evaluate the prevalence of MITE multimers but also may provide new insight into the temporal order of amplification of different transposable elements in the rice genome.
RESULTS
Tourist Multimers in Maize
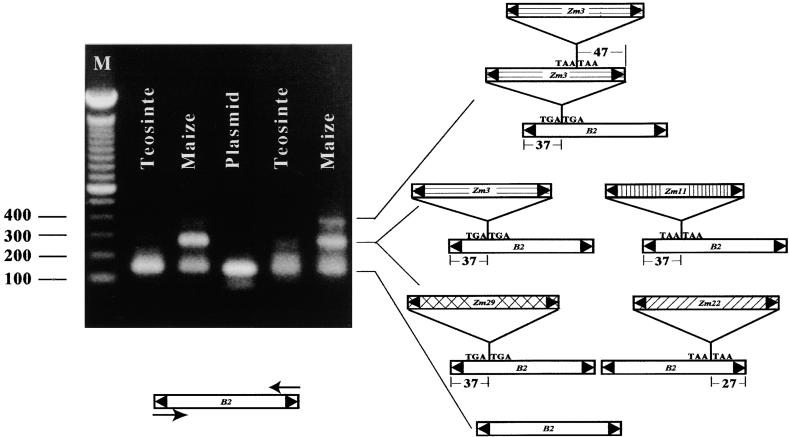
The first reported MITE was the B2 element, found as a 128-bp insertion into the maize waxy (wx) gene in the mutant wxB2 allele (Wessler and Varagona, 1985; Bureau and Wessler, 1992). Subsequent database searches revealed that this element belongs to a large family of related elements, called Tourist, whose members are associated with the noncoding regions of genes from maize, sorghum, barley, and rice (Bureau and Wessler, 1992, 1994a). Tourist multimers were discovered initially in a polymerase chain reaction (PCR) assay that was intended to identify additional B2-like (Tourist) elements in maize. Genomic DNA from maize inbred B79 was amplified with primers derived from the B2 TIR (14 bp) and 11 bp of internal element sequence. In addition to a product corresponding to the size of the B2 element (128 bp), we observed larger fragments that varied in size depending on the annealing temperature (Figure 1). PCR products from all size classes were cloned, and several were sequenced, revealing monomers, dimers, and a trimer (Figure 1). Four different multimers were found among the six dimer-sized clones that were sequenced. In contrast, the largest PCR fragment corresponded to a single trimer. All multimers contained a variety of elements that, like B2, are members of Tourist subfamily A (Bureau and Wessler, 1994a).
Figure 1.
The Identity of Multimers Resulting from PCR Amplification of B2-Tourist Elements in Maize Line B79.
PCR products resolved by agarose gel electrophoresis and visualized by ethidium bromide staining were purified, cloned, and sequenced (see Methods). The annealing temperature for PCR was 60°C for the two samples at left and 55°C for the three samples at right. The identities of PCR bands are diagrammed at right. The positions of the insertion sites of the various Tourist subfamily members (Zm3, Zm11, Zm22, and Zm29) (Bureau and Wessler, 1992, 1994a; N. Jiang and S. Wessler, unpublished data) into B2 elements are shown along with the TSD. M, DNA molecular weight standard (bp).
To exclude the possibility that the Tourist multimers were artifacts of PCR amplification, we used dimer and trimer products to probe a small insert library derived from B79 genomic DNA. Three of 11 sequenced clones contained Tourist multimers, thus confirming the presence of Tourist multimers in the genome.
Insertion into Preexisting MITEs
Among maize inbred lines, the insertion sites of MITEs frequently were polymorphic with respect to the presence or absence of an element at a particular locus (Casa et al., 2000; Zhang et al., 2000). Polymorphism of this type usually is associated with the recent spread of transposon families through the genome. In light of these findings, we designed a PCR assay to detect insertion site polymorphism within Tourist multimers. In this way, evidence might be obtained for the sequential insertion of one element into another.
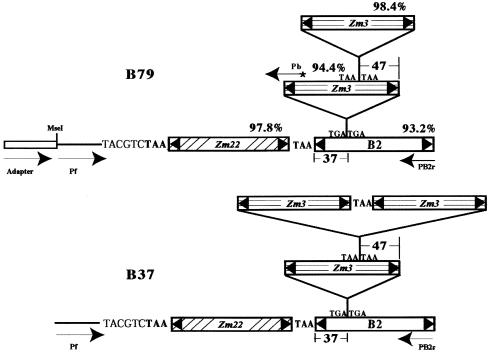
The locus harboring the Tourist trimer (Figure 1) was investigated for possible insertion polymorphism among different maize lines. Following the methodology described in Methods, we obtained B79 genomic sequence adjacent to one end of the trimer, revealing that another Tourist element (Tourist-Zm22) had inserted adjacent to the trimer with only an intervening target site duplication (TSD) (Figure 2). A locus-specific primer was designed from the sequence flanking Tourist-Zm22 and used to amplify B37 genomic DNA together with a B2 terminal primer (PB2r). The resulting PCR product, which harbored an additional Tourist element (Tourist-Zm3) (Figure 2), provided evidence for the progressive formation of multimers (tetramers from trimers).
Figure 2.
Diagram of a MITE Trimer and Tetramer Found at the Same Locus in Different Maize Lines.
As in Figure 1, the positions of the insertion sites of the various Tourist subfamily members into Tourist-B2 and other Tourist-Zm elements are shown along with the TSD. The sequence flanking the trimer in B79 was amplified with primer Pb and an adapter primer (see Methods for details). For B37, the tetramer was amplified using a locus-specific primer, Pf, and the B2 primer PB2r. The number above each element in B79 indicates the similarity between the element in B79 and its counterpart in B37. The asterisk indicates that the primer was labeled with 33P.
Nonrandom Insertion Sites
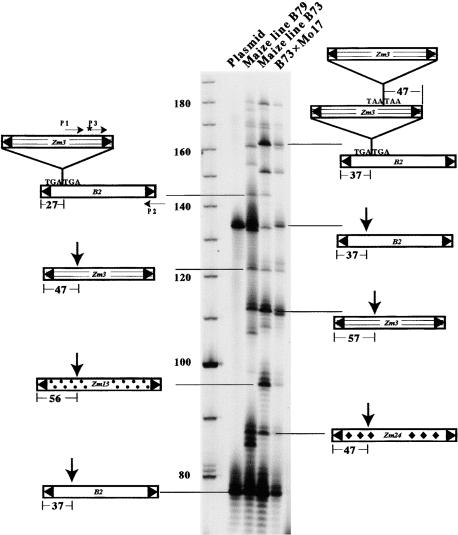
Insertion sites within the sequenced multimers clearly were nonrandom. For ease of comparison, insertion sites have been calculated as the number of base pairs from the closest end of the target element to the first nucleotide of the TIR of the insertion element. For all insertions examined, this value corresponded to 27, 37, or 47 bp (Figure 1). To determine whether this periodicity was representative of the multimers in the maize genome, a two-step PCR assay was used to isolate additional multimers. In this assay (see Methods), the length of the PCR products reflects the position of the insertion sites within the multimers. That is, if the insertion sites are 10 bp apart, the PCR products will appear, more or less, as a 10-bp “ladder” on the gel. Such a ladder was observed (Figure 3). Furthermore, sequencing of selected PCR products revealed that all contained a Tourist-Zm3 element inserted into another Tourist element at ∼10-bp intervals. The composition of some of the multimers is diagrammed in Figure 3. In addition, these data and the data from all previous multimer sequences are summarized in Table 1.
Figure 3.
Autoradiograph of PCR Products Resolved on an Acrylamide Gel Showing an ∼10-bp Ladder.
The positions of primers used to obtain the PCR products from maize lines or plasmid DNA are indicated by horizontal arrows; the composition of multimers represented by the indicated PCR products also is diagrammed. Vertical arrows over some multimers represent Zm3 insertions. Lane 1, Zm3-B2 dimer containing plasmid; lane 2, B79 genomic DNA; lane 3, B73 genomic DNA; lane 4, recombinant inbred DNA from a B73 × Mo17 mapping population. The asterisk indicates that the primer was labeled with 33P.
Table 1.
Insertion Sites within Tourist Dimers and Trimers
| Maize Line | Preexisting Element | Insertion Element | Insertion Sitea (bp) | TSD | Source |
|---|---|---|---|---|---|
| B79 | B2 | Zm3 | 37 | TGA | B2 PCR |
| B79 | B2 | Zm11 | 37 | TAA | B2 PCR |
| B79 | B2 | Zm29 | 37 | TGA | B2 PCR |
| B79 | B2 | Zm22 | 27 | TTA | B2 PCR |
| B79 | B2b (1) | Zm3b (2) | 37 | TGA | B2 PCR |
| B79 | Zmb (2) | Zm3b (3) | 47 | TTA | B2 PCR |
| B73 | B2 | Zm3 | 27 | TTA | Two-step PCR |
| RIL22c | Zm13 | Zm3 | 46 | TAA | Two-step PCR |
| B73 | B2 | Zm3 | 37 | TTA | Two-step PCR |
| B73 | Zm3 | Zm3 | 47 | TAA | Two-step PCR |
| B73 | Zm3 | Zm3 | 57 | TTA | Two-step PCR |
| B73 | Zm13 | Zm3 | 67 | TTA | Two-step PCR |
| B73 | Zm24 | Zm3 | 47 | TCA | Two-step PCR |
| B79 | B2 | Zm3 | 37 | TGA | Two-step PCR |
| RIL7 | Zm24 | Zm3 | 37 | TGA | Two-step PCR |
| Spanco | Zm13 | Zm22 | 67 | TTA | Two-step PCR |
| Spanco | Zm24 | Zm22 | 59 | TAA | Two-step PCR |
| B79 | B2 | Zm28 | 58 | TAA | Genomic library |
| B79 | Zm13 | Zm25 | 46 | TGA | Genomic library |
| B79 | Zm27b (1) | Zm3b (2) | 47 | TGA | Genomic library |
| B79 | Zm3b (2) | Unknown elementb (3) | 47 | TAA | Genomic library |
Insertion site is defined as the number of base pairs from the closest end of the target element to the first nucleotide of the TIR of the insertion element.
Elements that form trimers. The numbers in parentheses indicate the order of insertion. Element 1 is the first preexisting element; element 2 is the element that inserted into element 1; element 3 is the element that inserted into element 2.
RIL represents recombinant inbred lines from the B73 × Mo17 cross.
MITE Multimers in Rice
In the absence of a significant amount of maize genomic sequence, analysis of maize multimers is restricted to a description of the phenomenon and the characterization of a small fraction of the existing elements. A more thorough survey, including an estimate of the proportion of the multimers present, is possible for rice because a large amount of rice genomic sequence is available publicly (Yuan et al., 2001) and the rice genome contains thousands of MITEs (Mao et al., 2000; Tarchini et al., 2000).
Prevalence of Multimers
Computer searches were restricted to 30.2 Mb of complete bacterial artificial chromosome and P1-derived artificial chromosomes (PAC) sequences, of which 6.6 Mb was derived from pericentromeric regions (based on the most recent data on the location of rice centromeres; Harushima et al., 1998; Cheng et al., 2001). No significant differences were observed in the insertion patterns of MITEs between the sequences from chromosomal arms and those from pericentromeric regions (data not shown). Of the rice sequences queried, 6641 MITEs were detected (Table 2) with the RepeatMasker program (see Methods for details). This corresponds to 0.22 MITEs per kb of genomic DNA or 1 MITE per 4.5 kb. MITEs account for 1.54 Mb of DNA or 5.1% of the genomic sequence analyzed. These values are very close to those found in a previous study of MITEs from a 350-kb contig (Tarchini et al., 2000). MITEs grouped into 41 different families, of which 26 were reported previously and 15 were identified in this study (Bureau and Wessler, 1994a, 1994b; Bureau et al., 1996; Song et al., 1998; Zhang and Kochert, 1998; Tarchini et al., 2000; Turcotte et al., 2001) (see supplemental data for sequences of the new identified MITE families).
Table 2.
Multimers Containing Rice MITEs
| Insertion of MITEs into
|
Insertion of Other Elements into
|
|||||
|---|---|---|---|---|---|---|
| MITEs | LTR Elements |
Other DNA Elements |
All Genomic Sequences Analyzed |
MITEs | All Genomic Sequences Analyzed |
|
| Number of elements |
6,641 | 1,171 | 646 | —c | 6,641 | —c |
| (kb) DNA | 1,540 | 4,650 | 1,780 | 30,183 | 1,540 | 30,183 |
| Insertion events |
387 | 14 | 14 | 6,641 | 93 | 2,185 |
| Insertion (kb) | 0.251a (1/4.0 kb) | 0.003b (1/330 kb) | 0.008b (1/127 kb) | 0.220 (1/4.5 kb) | 0.060 (1/17 kb) | 0.072 (1/14 kb) |
P < 0.05 compared with that for all genomic sequences by χ2 test.
P < 0.01 compared with that for all genomic sequences by χ2 test.
Indicates all sequences contain both elements and non-element sequences.
Of the 6641 MITEs, 732 (or ∼11%) are part of 340 multimers. These include 293 dimers, 35 trimers, nine tetramers, and three pentamers (the trimers and tetramers also contain non-MITE elements). These 387 MITEs inserted into other MITEs correspond to 387 MITEs per 1540 kb of MITEs, or an insertion frequency of MITEs into MITEs of 0.25 per kb or one MITE per 4 kb (Table 2). In contrast, there are very few insertions of MITEs into class 1 elements or into other class 2 elements, despite the fact that these elements constitute a much larger fraction of the genome. Although there is one MITE inserted per 4 kb of MITE DNA, there is only one MITE inserted per 330 kb of LTR retrotransposons and per 127 kb of other class 2 elements. These data indicate either a target site preference of MITEs for other MITEs or that MITE amplification preceded the amplification of the other elements in the genome. In the latter situation, it is envisioned that the bulk of the class 1 and non-MITE class 2 elements were not in the genome when most of the MITE families were undergoing amplification. In contrast, non-MITE elements show no discrimination for insertion into MITEs (Table 2); while the frequency of insertion into MITEs is one per 17 kb of MITEs, the insertion frequency into all genomic DNA is slightly higher at one per 14 kb.
Self-Insertions
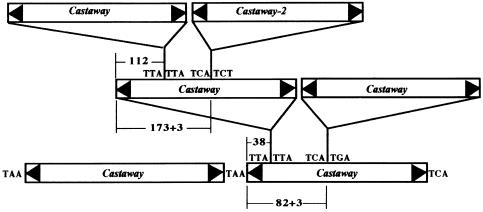
The data presented in Table 2 reveal a slight preference for insertion of MITEs into MITEs. However, analysis of these data for individual MITE families indicates that this preference is not displayed by all families and is attributable largely to self-insertions. Four MITE families were analyzed in detail (Table 3). These families were chosen because they are abundant and they represent different groups of MITEs. Among the four families analyzed, Castaway, Gaijin, and Ditto are related to Tourist elements in maize, whereas Stowaway elements belong to another superfamily (see Discussion). As shown in Table 3, all of the Tourist-related elements have sustained more insertions per kb of DNA than has the genome as a whole (insertion frequencies of 0.38, 0.32, and 0.60, respectively, versus 0.22 for all DNA; Tables 2 and 3). In contrast, Stowaway has sustained insertions at approximately the same frequency as the rest of the genome. Although the increased insertions into Castaway and Gaijin elements can be accounted for completely by self-insertions, Ditto elements appear to attract a variety of MITEs (see Discussion). The cluster of six elements from chromosome 1 (Figure 4) illustrates the propensity for self-insertion among Castaway family members.
Table 3.
Self-Insertion Preference of Some MITEs in Rice
| Elements | Castaway | Gaijin | Ditto | Stowaway |
|---|---|---|---|---|
| Copies of the element in 30.2 Mb of genomic sequence | 225 | 486 | 345 | 2690 |
| Total size (kb) | 76 | 71 | 89 | 561 |
| Insertions by MITEs | 29 | 23 | 53 | 104 |
| Self-insertions | 20 (69%a) | 16 (70%a) | 13 (25%a) | 77 (71%a) |
| Insertion frequency by all MITEsb | 0.38c | 0.32 | 0.60c | 0.19 |
| Self-insertion frequencyb | 0.26 | 0.23 | 0.15 | 0.14 |
Percentage of self-insertions.
Insertion frequency equals insertions into the element divided by the total size (kb) of the element.
P < 0.01 compared with the average insertion frequency (0.223) in the genome by χ2 test.
Figure 4.
A Cluster of Six Castaway Elements on a Rice P1-derived Artificial Chromosomes (PACs) Clone (GenBank Accession Number ap002844) from Chromosome 1.
The distance from the end of the element to the insertion site is shown. Castaway-2 is a member of a subfamily of Castaway. The +3 indicates the inclusion of the 3-bp TSD that was generated upon insertion.
One could argue that the observed higher self-insertion frequency of MITEs reflects a preference of MITEs for particular regions of the genome rather than a preference for other members of the same family. If this is the case, for a certain family of MITEs there would be a comparable number of insertions into sequences flanking MITEs as there are into MITEs. Fortunately, the availability of 30.2 Mb of rice contigs permits an analysis of the insertions into MITEs and their flanking sequences. On the basis of the data presented in Figure 5, it is evident that for all families examined, except Stowaway, the self-insertion frequencies of MITEs are significantly higher than the insertion frequencies into their flanking sequences (P < 0.01 by χ2 test).
Figure 5.
The Frequencies of Self-Insertions and Insertions into Flanking Sequences for Four MITE Families in Rice.
Frequencies were calculated in the same way as in Table 2 (see Methods for details).
MITE Multimers Cannot Transpose
A MITE multimer can arise in at least two ways. The first is by the insertion of a MITE into another MITE, and the second is by amplification of a multimer. If a multimer is capable of transposition, several copies of the same multimer should be detected and the multimers should evolve similarly as single elements. Furthermore, these copies should be composed of the same elements in the same relative orientation and with the same insertion site and TSD. Among the 340 MITE multimers identified in this study, only three pairs of dimers share these structural features. However, the sequence similarity between the members of each dimer pair ranges from 65 to 72%, whereas at least one of the insertion elements in each dimer pair has homologs with >90% similarity in the same database. This striking discrepancy suggests that these dimer pairs resulted from independent insertions instead of amplification of dimers.
There was one exception involving a dimer composed of a MITE and a DNA element. This element (called Midway), initially found as an 850-bp insertion in a Stowaway-Os1 element, has 11-bp TIRs and an 8-bp TSD. A closer examination indicates that Midway harbors another Stowaway element (Stowaway-Os25). That there are three Midway/Stowaway composite elements in the database sharing 93 to 96% overall DNA sequence identity suggests that Midway can still transpose despite (or because of) the Stowaway-Os25 insertion.
DISCUSSION
Here, we report the characterization and quantification of MITE multimers in maize and rice. Although MITE multimers were first discovered in maize, limited genomic sequence precluded further analysis of these multimers. However, the high density of MITEs in the rice genome (Bureau et al., 1996; Mao et al., 2000; Tarchini et al., 2000) coupled with the availability of large amounts of genomic sequence facilitated a more comprehensive analysis of multimers in rice and has led to the following conclusions: (1) MITEs are numerically the most abundant transposable elements in the rice genome (one MITE per 4.5 kb); (2) >10% of rice MITEs are part of multimers, thus suggesting a preference for MITE insertion into MITEs; (3) an insertion preference is displayed by some, but not all, MITE families; (4) for the Castaway and Gaijin families, this preference is caused by a high frequency of self-insertions; in contrast, Ditto elements are targeted by many element families; (5) the frequency of MITE insertions into class 1 or other class 2 elements is surprisingly low; and (6) on the basis of our analysis of 30.2 Mb of rice sequence, nested MITE multimers arise from independent insertion events.
Self-Insertion Preference for Some MITE Families
As calculated in Table 2, the insertion frequency of all MITEs into other MITEs is slightly higher than the average value into the whole genome. However, there is a threefold variation in the frequency of MITE insertions into MITEs when individual families are examined (Table 3). More significantly, self-insertions constituted a major part of the multimers for several families. For Castaway, Gaijin, and Stowaway, self-insertions account for two-thirds of all insertions. These data indicate that the preferential insertion of MITEs into MITEs that is displayed by some families can be attributed, to a great extent, to self-insertions. One exception is the Ditto element. Among the rice MITEs, Ditto elements are targeted frequently by various types of elements, including other Ditto elements. In addition to being targeted 53 times by 12 families of MITEs, we detected five cases of insertions by four different LTR retrotransposons and 22 examples of MITEs inserted in adjacent (with an intervening TSD) sequences.
Composite elements, arising from self-insertion, have been reported previously in maize, in which double Ds and Ac elements were shown to be responsible for chromosome breakage and more complex rearrangements (McClintock, 1949; Courage-Tebbe et al., 1983; Döring and Starlinger, 1984; Weck et al., 1984; Döring et al., 1989; Michel et al., 1994). It was later hypothesized that chromosome breakage resulted from aberrant transposition of composite or adjacent Ds elements (English et al., 1993; Weil and Wessler, 1993). In contrast to the composite Ds elements that are still capable of transposition, the uniqueness of each MITE multimer suggests that self-insertion creates an inactive composite element. Inactivating self-insertions of the Tp1 element of Physarum polycephalum have been observed previously (Rothnie et al., 1991). It has been proposed that a preference for inactivating self-insertions minimizes deleterious effects on the host by providing a safe haven for insertion while simultaneously limiting the overall transposition frequency (Rothnie et al., 1991).
Regional versus Self-Insertion Preference
Previous studies indicate that some MITE families insert preferentially into genic regions (Mao et al., 2000; Zhang et al., 2000). A preference for genic regions also has been observed for the maize class 2 families Ac/Ds and Mutator (Chen et al., 1992; Cresse et al., 1995). Regional preferences have been demonstrated for many elements in a wide variety of species. For example, yeast Ty5 elements integrate preferentially into regions of silent chromatin at the telomeres and the mating loci (Zou et al., 1996), and for P elements, euchromatic sites, especially 5′ regions of genes, are targeted more often than heterochromatin (Berg and Spradling, 1991; Liao et al., 2000).
Regardless of the mechanism responsible, an element with a regional preference is more likely to have a higher frequency of self-insertion than an element with no such preference. If the regional preference is the major factor leading to a high self-insertion frequency, comparable insertion frequencies are expected into elements and into their flanking genomic sequences. The availability of 30.2 Mb of rice sequence allowed us to test this assumption (Figure 5). For Castaway, Gaijin, and Ditto, the self-insertion preference is more likely to be caused by the targeting of preexisting elements than by a regional preference. In contrast, Stowaway elements show no significant difference between insertion into preexisting elements and insertion into flanking DNA, thus suggesting that the high ratio of self-insertions results from a regional preference. Alternatively, the presence of one Stowaway element may alter the flanking DNA in some manner, thereby creating a better target for future insertions. A similar effect was observed for the in vitro transposition of the C. elegans Tc1 element (Ketting et al., 1997). Interestingly, Stowaway elements, like Tc1, use TA dinucleotide targets.
The difference between Stowaway and the three other MITE families may indicate distinct integration mechanisms for different MITE families. Like the Tourist elements in maize, Castaway, Ditto, and Gaijin all create a 3-bp TSD upon insertion (Bureau et al., 1996). More importantly, the TIRs of Castaway, Ditto, and Gaijin are related to the TIR of Tourist elements in maize, suggesting that they may belong to the same superfamily. In contrast, Stowaway elements appear to belong to another superfamily based on their TIR and TSD (Bureau and Wessler, 1994b). Therefore, it is likely that these two superfamilies rely on distinct sources of transposases.
Target Site Preference in Maize Multimers
The potential to form secondary structures has been noted for several MITE families since the discovery of the Tourist family in maize (Bureau and Wessler, 1992; Izsvák et al., 1999). Given the occurrence of multimers among maize Tourist-B2 elements, we hypothesized that secondary structures might play a role in targeting. Consistent with this notion is the inability to detect MITE multimers involving two other maize MITE families (Hbr and mPIF) lacking significant secondary structures (N. Jiang, Q. Zhang, X. Zhang, and S.R. Wessler, unpublished data). However, in the rice genome, multimer formation does not correlate with the potential to form significant secondary structures. In rice, the MITEs that sustained the most insertions, Castaway and Ditto, are those without significant secondary structures (Bureau et al., 1996). In contrast, Stowaway elements usually have significant secondary structures but do not show a targeting bias. However, these data cannot rule out the possibility that small, local stem loops, such as the 14-bp palindrome targeted by P elements (Liao et al., 2000), might influence targeting of MITEs.
The analysis of MITE multimers in rice also was prompted by the discovery of nonrandom insertion sites among Tourist multimers in maize (Figures 1 and 2, Table 1). The 10-bp periodicity observed for Tourist multimers is reminiscent of the integration of human immunodeficiency virus. Integration of human immunodeficiency virus in vitro occurs preferentially into bent DNA in which the major groove is on the exposed face of the nucleosome (Pryciak and Varmus, 1992; Pruss et al., 1994). The 10-bp periodicity for Tourist multimers could be produced in a similar pattern (i.e., the transposition machinery attacks only major or minor grooves of the DNA double helix).
In rice, some “hot” spots for insertion were observed inside the sequence of some MITEs, and some of the insertion sites are ∼10 bp apart. However, insertions that are not 10 bp apart also were observed. Because of the fact that the rice MITEs that sustained most insertions are much larger than maize Tourist elements (maize Tourist, 130 bp; Ditto, 244 bp; Castaway, 364 bp), the distribution of insertion sites appears to be sporadic within rice MITEs. Thus, more rice multimers need to be examined to determine whether or not the 10-bp pattern is statistically significant. Alternatively, this feature may belong only to Tourist elements in maize.
To date, no autonomous element responsible for the transposition of MITEs has been available. The isolation of such elements and their associated protein(s) will ultimately facilitate the biochemical analysis of the various levels of targeting exhibited by MITE families.
Deficiency of MITE Insertions into Non-MITE Elements: Targeting Preference or Temporal Differences in Amplification?
A surprising and dramatic conclusion of the data presented in Table 2 is that MITEs have inserted into MITEs 80 times more often than they have inserted into LTR retrotransposons and 32 times more often than they have inserted into other DNA elements (one MITE insertion versus 4, 330, and 127 kb, respectively). In contrast, the frequency of insertion of LTR retrotransposons and other DNA elements into MITEs is only slightly lower than the overall frequency of insertion of these elements into rice genomic DNA (one insertion per 17 kb of MITEs versus one insertion per 14 kb of genomic DNA).
Previous studies have noted a genic preference for maize class 2 elements, including members of the Ac/Ds and Mutator families (Chen et al., 1992; Cresse et al., 1995). Differences in chromatin density and/or the extent of DNA methylation between gene-rich and other regions of the genome have been proposed as possible target recognition mechanisms (Chen et al., 1992). A similar preference for genic regions has been demonstrated for members of the MITE families Hbr and mPIF (Casa et al., 2000; Zhang et al., 2000; X. Zhang, N. Jiang, and S.R. Wessler, unpublished data). In contrast, MITEs appear to be underrepresented in regions of the maize and barley genomes containing nested or clustered LTR retrotransposons (Tikhonov et al., 1999; Dubcovsky et al., 2001). Although MITEs may target gene-rich regions by the same or similar mechanisms as other class 2 elements, the analysis of MITE multimers in rice provides at least two alternate explanations for the observed (skewed) distribution. Enrichment for MITEs in genic regions and their apparent absence from retrotransposon clusters or domains could reflect a self-insertion preference coupled with avoidance of retrotransposon targets. Alternatively, a dearth of MITE insertions into non-MITE transposons also would result if the bulk of MITE amplification occurred before the amplification of LTR retrotransposons and other class 2 elements. To unambiguously distinguish between these seemingly mutually exclusive hypotheses, it will be necessary to identify an active MITE system that can be exploited to experimentally determine MITE target preference(s). In the mean time, we must rely on the comparative analysis of related genomes to provide clues to the mechanisms underlying the observed distributions of transposable elements.
METHODS
Plant Material, DNA Extraction, and Library Construction
Maize (Zea mays) lines B79 and B37 were obtained from the U.S. Department of Agriculture, Agricultural Research Service Plant Introduction Station at Ames, Iowa. Maize line B73 and recombinant inbred lines from a cross between B73 and Mo17 were provided by Michael Lee (Iowa State University, Ames). Maize line Spanco was provided by Andy Tull (University of Georgia, Athens). Plant DNA was extracted as described (McCouch et al., 1988). The small insert genomic library from B79 genomic DNA was constructed as described (Zhang et al., 2000).
Polymerase Chain Reaction and Gel Electrophoresis
Polymerase chain reaction (PCR) was performed as described (Bureau and Wessler, 1992) with annealing temperature ranging from 55 to 60°C, depending on the primers. Sequences of primers are available on request.
To clone the flanking sequence of the Tourist trimer in Figure 1, B79 genomic DNA was digested with MseI and ligated with adapters. The DNA then was amplified with a primer complementary to the adapter and primer Pb, which contains the sequence at the junction of (Tourist) Zm3 and the B2-like element (Figure 2). To separate PCR products that resulted only from adapters and PCR products from the two primers, primer Pb was labeled with 33P, and the PCR products were loaded on 6% denaturing acrylamide-bisacrylamide gels and electrophoresed as described previously (Casa et al., 2000).
The two-step PCR assay described in Figure 3 involved amplification of genomic DNA with primers P1 and P2, followed by amplification of the PCR products with primers P2 and P3 (P3 was labeled with 33P). PCR products were resolved by PAGE, as described above.
Recovery of Gel Bands
DNA fragments were excised from radioactive gels by scratching the dried gel with yellow tips (Stumm et al., 1997; Elsevier Trends Journals Technical Tips online, http://tto.biomednet.com/cgi-bin/tto/pr), placing the tip in 20 μL of PCR reaction mix with relevant primers for 1 min before discarding, and reamplifying with the same cycling parameters as that of the original reaction. PCR products were resolved on 0.8% agarose gels, and fragments were excised, purified (QIAquick; Qiagen, Chatsworth, CA), and cloned (TA cloning kit; Invitrogen, Carlsbad, CA). DNA templates were sequenced at the Molecular Genetics Instrumentation Facility (University of Georgia).
DNA Sequence Analysis
DNA sequence analysis (pairwise comparisons, multiple sequence alignments, and sequence assembling and formatting) was performed with programs in the University of Wisconsin Genetics Computer Group program suite (GCG, version 10.1) accessed through Research Computing Resources (University of Georgia).
Retrieval of Sequences
Completely sequenced rice (Oryza sativa) bacterial artificial chromosomes and P1-derived artificial chromosomes (PACs) were retrieved from the World Wide Web sites of different rice genomic projects, including groups in the United States (http://www.usricegenome.org/), Japan (http://rgp.dna.affrc.go.jp/), Korea (http://bioserve.myongji.ac.kr/ricemac.html), the People's Republic of China (http://www.ncgr.ac.cn/Ls/index.html), and Taiwan (http://genome.sinica.edu.tw/).
Screening for Transposable Elements
Transposable elements in rice sequences were searched with RepeatMasker (http://ftp.genome.washington.edu/RM/webrepeatmaskerhelp.html). The grass repeats database in RepeatMasker was modified by adding sequences of other previously characterized transposable elements in maize and rice (references not listed in Results: Hirochika et al., 1992, 1996; SanMiguel et al., 1996; Kumekawa et al., 1999; Ohtsubo et al., 1999) and new transposable elements identified in this study. New elements were found either by their similarity to known elements or by insertion into known elements. The rice genome sequences described above were used as query sequences in analysis with RepeatMasker using the modified grass repeats database at default settings. In the output of RepeatMasker, the annotation files display all of the matches and the positions of matches between the query sequences and any of the sequences in the repeats database.
Identification of Multimers
Potential multimers were first selected from the query sequences on the basis of the distance between two elements in the annotation files. For example, if one element is flanked by another element on both sides, the two elements probably form a dimer. The sequences of potential multimers were further analyzed manually with programs in GCG. If the ends of one element were located inside the sequence of another element, the elements were deduced to constitute a multimer; otherwise, elements were deduced to be monomeric.
Calculations
The insertion frequency was calculated by dividing the number of insertions by the size (in kilobases) of available sequences. For individual MITE families, the amount of DNA equals the size of the consensus element multiplied by the number of elements. If the length of the match was less than half of the consensus element, it was considered as half an element in calculating the amount of DNA. If a match was <30 bp, it was eliminated from consideration. The total amount of long terminal repeat (LTR) retrotransposons was approximated by multiplying the number of elements and solo-LTRs by their average lengths, which are 6.9 and 1.8 kb, respectively. The total amount of DNA representing DNA elements was estimated similarly, with an average size of 1.9 kb. The average size of LTR elements and other DNA elements was obtained by sampling an 880-kb region in chromosome 1 (71.8 to 73.5 cm).
In Figure 5, the length of flanking sequences was estimated by the number of elements multiplied by 2 and then by the range of flanking sequences, where 2 represents the fact that for each element there are flanking sequences on both sides. For example, 2690 Stowaway elements were detected in the 30.2-Mb rice genomic sequence, and 359 Stowaway insertions were observed in the range of 1.0 to 2.0 kb from another Stowaway element. In this case, the total length of available sequences = 2690 × 2 × (2.0 − 1.0) = 5380 kb, and the insertion frequency in this range of flanking sequences = 359 ÷ 5380 = 0.067 insertion per kb. Because the purpose of the analysis is to determine whether the high self-insertion frequency for some MITE families is caused by the targeting of preexisting elements or by a regional preference, adjacent insertions (only one target site duplication [TSD] between two elements) were not included. This type of insertion was not considered because it is not clear whether it is caused by the targeting for preexisting elements or for flanking sequences.
Supplementary Material
Acknowledgments
We thank Arian Smit (Institute for Systematic Biology, Seattle, WA) and Phil Green (Washington University, St. Louis, MO) for providing the RepeatMasker and cross_match programs, Zhirong Bao (Washington University) for valuable suggestions and discussions, Cedric Feschotte and Xiaoyu Zhang for critical reading of the manuscript, Alexander Nagel for communicating unpublished data, and Qiang Zhang and Liangjiang Wang for technical assistance. This study was supported by grants from the National Institutes of Health, the U.S. Department of Energy, and the National Science Foundation to S.R.W.
Article, publication date, and citation information can be found at www.aspb.org/cgi/doi/10.1105/tpc.010235.
Footnotes
Online version contains Web-only data.
References
- Baker, B., Schell, J., Lorz, H., and Fedoroff, N. (1986). Transposition of the maize controlling element “Activator” in tobacco. Proc. Natl. Acad. Sci. USA 83 4844–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, C.A., and Spradling, A.C. (1991). Studies on the rate and site-specificity of P-element transposition. Genetics 127 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1992). Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1994. a). Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1994. b). Stowaway: A new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., Ronald, P.C., and Wessler, S.R. (1996). A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc. Natl. Acad. Sci. USA 93 8524–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa, A.M., Brouwer, C., Nagel, A., Wang, L., Zhang, Q., Kresovich, S., and Wessler, S.R. (2000). The MITE family Heartbreaker (Hbr): Molecular markers in maize. Proc. Natl. Acad. Sci. USA 97 10083–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta, E., Casacuberta, J.M., Puigdomenech, P., and Monfort, A. (1998). Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: Characterization of the Emigrant family of elements. Plant J. 16 79–85. [DOI] [PubMed] [Google Scholar]
- Chen, J., Greenblatt, I.M., and Dellaporta, S.L. (1992). Molecular analysis of Ac transposition and DNA replication. Genetics 130 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Presting, G.G., Buell, C.R., Wing, R.A., and Jiang, J. (2001). High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courage-Tebbe, U., Döring, H.P., Fedoroff, N.V., and Starlinger, P. (1983). The controlling element Ds at the Shrunken locus in Zea mays: Structure of the unstable sh-m5933 allele and several revertants. Cell 34 383–393. [DOI] [PubMed] [Google Scholar]
- Cresse, A.D., Hulbert, S.H., Brown, W.E., Lucas, J.R., and Bennetzen, J.L. (1995). Mu1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics 140 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring, H.P., and Starlinger, P. (1984). Barbara McClintock's controlling elements: Now at the DNA level. Cell 39 253–260. [DOI] [PubMed] [Google Scholar]
- Döring, H.P., Nelsensalz, B., Garber, R., and Tillmann, E. (1989). Double Ds elements are involved in specific chromosome breakage. Mol. Gen. Genet. 219 299–305. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J., Ramakrishna, W., SanMiguel, P.J., Busso, C.S., Yan, L., Shiloff, B.A., and Bennetzen, J.L. (2001). Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol. 125 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J., Harrison, K., and Jones, J. (1993). A genetic analysis of DNA sequence requirements for Dissociation state I activity in tobacco cells. Plant Cell 5 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., and Mouchès, C. (2000). Recent amplification of miniature inverted-repeat transposable elements in the vector mosquito Culex pipiens: Characterization of the Mimo family. Gene 250 109–116. [DOI] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Fukuchi, A., and Kikuchi, F. (1992). Retrotransposon families in rice. Mol. Gen. Genet. 233 209–216. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimito, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposon of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák, Z., Ivics, Z., Shimoda, N., Mohn, D., Okamoto, H., and Hackett, P.B. (1999). Short inverted-repeat transposable elements in teleost fish and implications for a mechanism of their amplification. J. Mol. Evol. 48 13–21. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E.J., and Plasterk, R.A. (1997). Target choice determinants of the Tc1 transposon of Caenorhabditis elegans. Nucleic Acids Res. 25 4041–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., and Bennetzen, J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33 479–532. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Ohtsubo, H., Horiuchi, T., and Ohtsubo, E. (1999). Identification and characterization of novel retrotransposons of the gypsy type in rice. Mol. Gen. Genet. 260 593–602. [DOI] [PubMed] [Google Scholar]
- Kunze, R., Saedler, H., and Lönnig, W.E. (1997). Plant transposable elements. Adv. Bot. Res. 27 331–470. [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, G.C., Rehm, E.J., and Rubin, G.M. (2000). Insertion site preference of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97 3347–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., Wood, T.C., Yeisoo, Y., Budiman, M.A., Tomkins, J., Woo, S., Sasinowski, M., Presting, G., Frisch, D., Goff, S., Dean, R.A., and Wing, R.A. (2000). Rice transposable elements: A survey of 73,000 sequence-tagged-connectors. Genome Res. 10 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1949). Mutable loci in maize. Carnegie Inst. Washington Year Book 48 142–154. [PubMed] [Google Scholar]
- McCouch, S.R., Kochert, G., Yu, Z.H., Khush, G.S., Coffman, W.R., and Tanksley, S.D. (1988). Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76 815–829. [DOI] [PubMed] [Google Scholar]
- Michel, D., Salamini, F., Motto, M., and Döring, H.P. (1994). An unstable allele at the maize opaque2 locus is caused by the insertion of a double Ac element. Mol. Gen. Genet. 243 334–342. [DOI] [PubMed] [Google Scholar]
- Morgan G.T. (1995). Identification in the human genome of mobile elements spread by DNA-mediated transposition. J. Mol. Biol. 254 1–5. [DOI] [PubMed] [Google Scholar]
- Ohtsubo, H., Kumekawa, N., and Ohtsubo, E. (1999). RIRE2, a novel gypsy-type retrotransposon from rice. Genes Genet. Syst. 74 83–91. [DOI] [PubMed] [Google Scholar]
- Oosumi, T., Belknap, W.R., and Garlick, B. (1995. a). Mariner transposons in humans. Nature 378 672. [DOI] [PubMed] [Google Scholar]
- Oosumi, T., Garlick, B., and Belknap, W.R. (1995. b). Identification and characterization of putative transposable DNA elements in solanaceous plants and Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 92 8886–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozueta-Romero, J., Houlne, G., and Schantz, R. (1996). Non-autonomous inverted repeat Alien transposable elements are associated with genes of both monocotyledonous and dicotyledonous plants. Gene 171 147–153. [DOI] [PubMed] [Google Scholar]
- Pruss, D., Reeves, R., Bushman, F.D., and Wolfe, A.P. (1994). The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biochem. 269 25031–25041. [PubMed] [Google Scholar]
- Pryciak, P.M., and Varmus, H.E. (1992). Nucleosomes, DNA-binding proteins and DNA sequence modulate retroviral integration target site selection. Cell 69 769–780. [DOI] [PubMed] [Google Scholar]
- Rothnie, H.M., McCurrach, K.J., Glover, L.A., and Hardman, N. (1991). Retrotransposon-like nature of Tp1 elements: Implications for the organisation of highly repetitive, hypermethylated DNA in the genome of Physarum polycephalum. Nucleic Acids Res. 19 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.-K., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic region of the maize genome. Science 274 765–768. [DOI] [PubMed] [Google Scholar]
- Smit, A.F., and Riggs, A.D. (1996). Tiggers and other transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA 93 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Pi, L.Y., Bureau, T.E., and Ronald, P.C. (1998). Identification and characterization of 14 transposon-like elements in the non-coding regions of the members of the Xa21 family of disease resistance genes in rice. Mol. Gen. Genet. 258 449–456. [DOI] [PubMed] [Google Scholar]
- Stumm, G.B., Vedder, H., and Schlegel, J. (1997). A simple method for isolation of PCR fragments from silver-stained polyacrylamide gels by scratching with a fine needle. Elsevier trends journals technique tips online http://tto.biomednet.com/cgi-bin/tto/pr.
- Surzycki, S.A., and Belknap, W.R. (2000). Repetitive-DNA elements are similarly distributed on Caenorhabditis elegans autosomes. Proc. Natl. Acad. Sci. USA 97 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z. (1997). Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 94 7475–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z. (2001). Eight novel families of miniature inverted-repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. USA 98 1699–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte, K.T., Srinivasan, S., and Bureau, T. (2001). Survey of transposable elements from rice genomic sequences. Plant J. 25 169–179. [DOI] [PubMed] [Google Scholar]
- Weck, E., Courage, U., Doring, H.-P., Fedoroff, N., and Starlinger, P. (1984). Analysis of sh-m6233, a mutation induced by the transposable element Ds in the sucrose synthase gene of Zea mays. EMBO J. 3 1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil, C.F., and Wessler, S.R. (1993). Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell 5 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S.R., and Varagona, M. (1985). Molecular basis of mutations at the waxy locus of maize: Correlation with the fine structure genetic map. Proc. Natl. Acad. Sci. USA 82 4177–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, J.I. (1990). Rapid proliferation of the maize transposable element Activator in transgenic tomato. Plant Cell 2 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q., Quackenbush, J., Sultana, R., Pertea, M., Salzberg, S.L., and Buell, C.R.. (2001). Rice bioinformatics: Analysis of rice sequences data and leveraging the data to other plant species. Plant Physiol. 125 1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., and Kochert, G. (1998). Independent amplification of two classes of Tourists in some Oryza species. Genetica 101 145–152. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Arbuckle, J., and Wessler, S.R. (2000). Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker (Hbr) into genic regions of maize. Proc. Natl. Acad. Sci. USA 97 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, S., Ke, N., Kim, J.M., and Voytas, D.F. (1996). The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 10 634–645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.