Abstract
Functional screening of an Arabidopsis cDNA library enabled the identification of a novel cDNA, ESR1 (for Enhancer of Shoot Regeneration), that can confer cytokinin-independent shoot formation when overexpressed in Arabidopsis root explants. Neither callus induction nor root formation was affected by ESR1 overexpression. ESR1 encodes a putative transcription factor with an AP2/EREBP domain. Surprisingly, ESR1 overexpression also greatly increased the efficiency of shoot regeneration from root explants in the presence of cytokinin, with a shift in the optimal cytokinin concentration required for this process. The effects of ESR1 overexpression on shoot regeneration are synergistic with those of cytokinin. Overexpression of ESR1 cannot induce callus formation or root formation, suggesting that its effects are specific to shoot formation. In wild-type Arabidopsis plants, ESR1 expression was induced by cytokinin. ESR1 transcript levels also increased transiently during shoot regeneration from root explants, most probably in response to cytokinin in the shoot-inducing medium. This transient increase occurred after the acquisition of competence for regeneration and before shoot formation, which is consistent with the physiological effects of ESR1 overexpression. Our results suggest that ESR1 may regulate the induction of shoot regeneration after the acquisition of competence for organogenesis.
INTRODUCTION
Despite the considerable potential of de novo organogenesis as a model experimental system to elucidate the intricacies of plant cell differentiation, at present there is little insight regarding the primary determinants of the capacity of undifferentiated callus cells to regenerate shoot buds. The requirement for a defined auxin/cytokinin ratio in the regeneration medium has long been appreciated (Reinert and Bajaj, 1977). High auxin/cytokinin ratios usually induce root formation, whereas lower auxin/cytokinin ratios promote shoot formation. Media with intermediate auxin/cytokinin ratios promote disorganized cellular proliferation as calli. However, little is understood concerning the mechanisms by which the auxin/cytokinin balance exerts these effects. The wide variation between plant species and explant types in terms of the ease with which they regenerate shoots in vitro remains to be explained. In particular, the duration of the regeneration response and the inability to predict which cells within a multicellular explant may undergo organogenesis have retarded progress in understanding the physiological basis of shoot organogenesis.
In general, it is thought that organogenesis is composed of three sequential phases (Hicks, 1994; Sugiyama, 1999). In the first phase, cells acquire developmental competence, which is the ability to recognize hormonal or other signals that commit them to a particular developmental program. Subsequently, the signals induce a process that causes competent cells to alter their developmental fate. The last phase is the differentiation to form determined organs, during which the inductive signals are no longer required. Although experimental procedures for in vitro organogenesis vary among species, in Arabidopsis tissue culture the first phase is initiated by culturing on a callus-inducing medium (CIM) containing 2,4-D (Koncz et al., 1989). Then explants are cultured on a shoot-inducing medium (SIM) or root-inducing medium (RIM) that contains a specific ratio of auxin to cytokinin.
Using an Arabidopsis tissue culture system, three temperature-sensitive mutants, srd1, srd2, and srd3, with defects in shoot regeneration (Yasutani et al., 1994; Ozawa et al., 1998) were isolated and characterized. Given that cytokinins act both synergistically with auxin to promote cell division and antagonistically to auxin to promote shoot and root initiation from callus cultures (Skoog and Miller, 1957), it seems reasonable to anticipate that genes involved in cytokinin signaling might regulate in vitro shoot bud formation. Recently, a cytokinin receptor gene, CRE1/AHK4, which encodes a histidine kinase, was identified (Inoue et al., 2001; Ueguchi et al., 2001). The roles that other genes potentially involved in cytokinin signaling (Kakimoto, 1998; D'Agostino and Kieber, 1999) may play in regulating organogenesis remain obscure. This most likely arises, at least in part, from the participation of cytokinins in aspects of plant growth and development (e.g., senescence and photomorphogenesis) that may not overlap directly with their role in regulating cell division and shoot initiation and growth.
Here, we describe the cloning of a novel cDNA, Arabidopsis ESR1 (for Enhancer of Shoot Regeneration), whose overexpression confers cytokinin-independent shoot generation from Arabidopsis root cultures. ESR1 overexpression also greatly increased shoot regeneration efficiency in the presence of cytokinin, although neither callus induction nor root regeneration was affected. Expression of ESR1 in wild-type Arabidopsis is induced by cytokinins. We also investigated the timing of ESR1 expression during the shoot regeneration process. A transient increase in ESR1 expression during shoot regeneration is consistent with its participation in this developmental transition.
RESULTS
Screening of cDNAs Whose Overexpression Confers Cytokinin-Independent Shoot Formation
To identify Arabidopsis genes involved in shoot regeneration, we screened for cDNAs that can confer cytokinin-independent shoot formation from root cultures when overexpressed. For this purpose, a cDNA library was constructed by cloning cDNAs downstream of a 35S promoter of Cauliflower mosaic virus in the binary vector pSK34. Arabidopsis root segments were transformed with the cDNA library using Agrobacterium, and the root cultures were incubated on media lacking cytokinin. On the basis of the efficiency of shoot regeneration from a portion of the total transformed roots incubated on SIM supplemented with kanamycin and carbenicillin, we estimated that ∼100,000 independent transformation events were interrogated for their ability to confer cytokinin-independent shoot formation. After 4 weeks, nine shoots and one dark green callus were recovered from the plates. Genomic DNA was prepared from the transformed explants, and the cDNA transgenes that were recovered by polymerase chain reaction (PCR)–mediated amplification were reinserted into the binary vector pSK34 for retransformation. Among the recovered cDNAs, constitutive expression of cDNA 9 (recovered from a dark green callus) clearly conferred cytokinin autonomy. The transformed calli were darker green and unable to produce any shoots. cDNA 9 was cloned into a 17β-estradiol–inducible expression vector, pER10 (Zuo et al., 2000). The construct pER10-ESR1 was introduced into Arabidopsis roots by Agrobacterium-mediated transformation, and the roots were incubated on plates with or without cytokinin and in the presence or absence of the inducer.
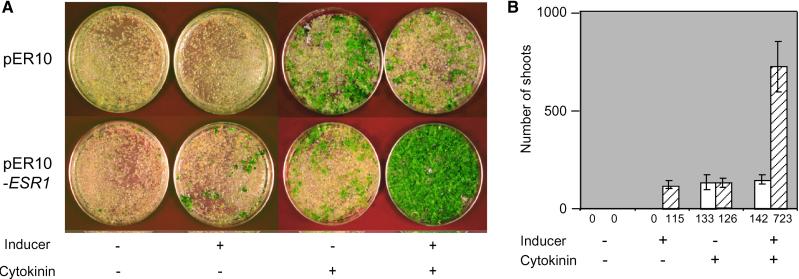
Figure 1 shows that transformants carrying the empty vector pER10 generated shoots only in the presence of cytokinin. On the other hand, in the absence of cytokinin, transformants that overexpressed cDNA 9 displayed a shoot regeneration response comparable to that observed in root cultures transformed with the empty vector in the presence of cytokinin. Root cultures transformed with pER10-ESR1 occasionally generated green calli in the absence of cytokinin and the 17β-estradiol inducer, but shoot formation was not observed (Figure 1A). Surprisingly, in the presence of exogenous cytokinin, overexpression of cDNA 9 also significantly increased the number of regenerated shoots (on average 5.7-fold) compared with explants transformed with the vector alone (Figure 1B). Because of its ability to promote shoot regeneration when overexpressed, this cDNA was named ESR1.
Figure 1.
Effects of ESR1 Overexpression on Shoot Regeneration from Root Explants.
(A) Representative plates 4 weeks after transformation. Arabidopsis root cultures transformed with the vector alone (pER10) or pER10-ESR1 were cultured on SIM with or without cytokinin (25 μM 2-ip) or an inducer of transcription (5 μM 17β-estradiol).
(B) Numbers of transformed shoots from 0.1 g fresh weight of root culture. Data represent the average of four independent experiments, and error bars indicate standard deviations. Open columns show transformants with the vector alone (pER10), and cross-hatched columns show transformants with pER10-ESR1.
Sequence Analysis of ESR1
Sequence analysis predicted that ESR1 encodes a protein of 328 amino acids with a molecular mass of 36.27 kD (Figure 2A). The ESR1 protein contains a domain with sequence homology with the AP2/EREBP domain found in a group of transcriptional factors in higher plants (Okamuro et al., 1997; Riechmann and Meyerowitz, 1998; Chang and Shockey, 1999) (Figure 2B). Whereas the AP2/EREBP domain of ESR1 is most similar to that of tobacco EREBP3, ESR1 did not display sequence homology with any known protein outside of the AP2 domain. Therefore, ESR1 does not appear to be an Arabidopsis ortholog of EREBP3, although the DNA sequences of their target binding sites may be similar.
Figure 2.
Deduced Amino Acid Sequence of ESR1 and Comparison of the AP2/EREBP Domains of ESR1 with Those of Other AP2/EREBP Proteins.
(A) Predicted amino acid sequence encoded by ESR1 cDNA. Amino acid numbers are indicated at right, and the AP2/EREBP domain is underlined.
(B) Alignment of the AP2/EREBP domains of ESR1 and other representative AP2/EREBP proteins by the multiple alignment function of Lasergene (DNASTAR, Madison, WI). GenBank accession numbers are T02433 (EREBP3), BAA32421(AtERF4), Y09942 (AtEBP), AAD25937 (ABI4), CAB80440 (Aintegumenta), AAC13770 (APETALA2), and BAB11119 (Leafy petiole).
Sensitivity of ESR1-Overexpressing Cells to Cytokinin
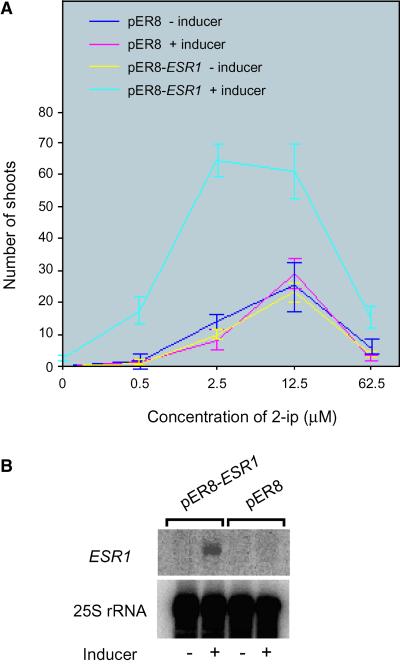
We examined the effects of ESR1 overexpression on sensitivity to cytokinin using root cultures derived from a transgenic plant line carrying a 17β-estradiol–inducible ESR1 cDNA (pER8-ESR1) (Figure 3A). Transgenic root cultures induced to overexpress ESR1 were hypersensitive to cytokinin compared with those under noninducing conditions or with control root cultures carrying the empty vector under inducing or noninducing conditions. The number of regenerated shoots from ESR1-overexpressing root cultures was much greater than that of root cultures without induced ESR1 expression or that of root cultures carrying the empty vector alone under inducing or noninducing conditions. In addition, the optimum concentration of N6-Δ2-isopentenyladenine (2-ip) was shifted to a lower value. Figure 3B shows the ESR1 transcript levels in transgenic lines under inducing and noninducing conditions. ESR1 transcripts were induced specifically with 17β-estradiol in the transgenic line carrying a pER8-ESR1 construct. Three other lines with high ESR1 inducibility also showed the same effects (data not shown).
Figure 3.
Sensitivity of ESR1-Overexpressing Cells to Cytokinin.
(A) Numbers of regenerated shoots in different concentrations of cytokinin. Ten root segments from transgenic seedlings carrying either pER8 or pER8-ESR1 were cultured on plates with MS medium supplemented with the indicated concentrations of 2-ip with or without an inducer (10 μM 17β-estradiol). Each value represents the average of three independent experiments, and bars indicate standard deviations.
(B) RNA gel blot analysis of the ESR1 transcript. Four micrograms of total RNA was loaded on each lane and hybridized with an ESR1 cDNA probe or a 25S rRNA probe.
Effects of ESR1 Overexpression on Callus Development, Root Regeneration, and Shoot Regeneration
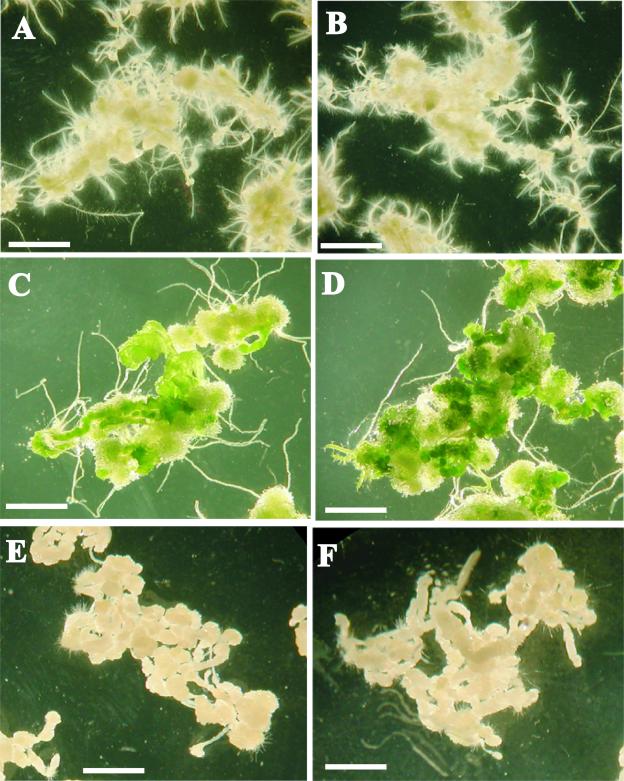
We also examined whether ESR1 overexpression affected callus development or root regeneration (Figure 4). Root segments from the transgenic line used in the experiment described above were precultured on CIM for 4 days before being transferred onto SIM, RIM, or CIM in the presence or absence of the inducer for 4 weeks. Whereas shoot formation was enhanced by ESR1 overexpression, neither callus formation nor root formation was affected. The effects of ESR1 overexpression were specific to shoot regeneration. ESR1-induced explants not only generated more shoots on SIM but also produced darker green calli than did explants incubated without the inducer (Figures 4C and 4D). Extended incubation on RIM occasionally resulted in shoot formation, although the number was much lower than that on SIM with the inducer (data not shown). Without the inducer, explants did not generate any shoots on RIM even after an 8-week incubation. These results indicate that ESR1 overexpression specifically enhances shoot regeneration and greening rather than callus formation or root formation.
Figure 4.
Effects of ESR1 Overexpression on the Formation of Calli, Shoots, and Roots.
(A) and (B) Root segments on RIM with (B) or without (A) inducer.
(C) and (D) Root segments on SIM with (D) or without (C) inducer.
(E) and (F) Root segments on CIM with (F) or without (E) inducer.
17β-Estradiol (10 μM) was used as an inducer in (B), (D), and (F). Root segments from a transgenic Arabidopsis line carrying a pER8-ESR1 construct were preincubated on CIM for 3 days and then transferred onto the media described above. Photographs were taken 4 weeks after transfer onto individual media. Bars = 0.5 cm.
Effects of the Constitutive Expression of ESR1 on Plants
Root explants were transformed with a 35S:ESR1 construct, and 100 independent transformed calli were recovered. These transformed calli were cultured on Murashige and Skoog (MS) medium or SIM, but none developed normal leaves. In contrast, ∼80% of calli carrying the empty vector developed into transgenic plants with normal leaves and flowers. Figure 5A shows representative independent calli carrying the 35S:ESR1 construct. Overexpression of ESR1 strongly inhibited normal leaf formation and resulted in dark green calli. We also attempted to transform Arabidopsis with the 35S:ESR1 construct via vacuum transformation, but only one transgenic plant was obtained (Clough and Bent, 1998). This plant did not develop normally; instead, it produced dark green calli similar to calli carrying the 35S:ESR1 construct obtained by root transformation (Figure 5B). These results suggest that ESR1 overexpression induces the initiation of shoot regeneration but interferes with the subsequent differentiation of plant cells.
Figure 5.
Inhibition of Shoot Regeneration Caused by Constitutive ESR1 Expression.
(A) Phenotype of transformants obtained by root transformation. Root segments were transformed with pSK-ESR1, a 35S:ESR1 construct, after incubation on CIM for 3 days and then transferred onto SIM containing kanamycin (50 mg/L) and carbenicillin (300 mg/L) for 4 weeks. Six representative explants are shown.
(B) Phenotype of seedling obtained by in planta transformation. Seedlings harboring a 35S:ESR1 construct were germinated on MS medium containing kanamycin and carbenicillin. The photograph was taken 6 weeks after germination.
(C) Magnified root part of (B).
(D) Magnified aerial part of (B).
Bars = 1 cm.
ESR1 Expression during Shoot Regeneration
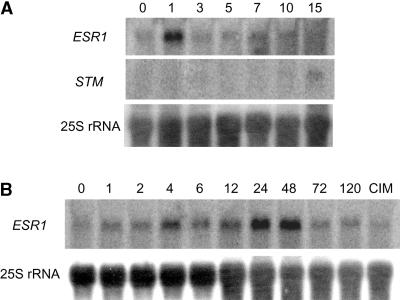
We used RNA gel blot hybridization to investigate the time course of ESR1 expression in wild-type root cultures during shoot regeneration. After preincubation on CIM, root segments were transferred onto SIM and total RNAs were prepared. Proliferating cells formed small clusters inside root cortex when the root cultures were transferred onto SIM. Subsequently, the clusters formed green calli 1 week after the transfer to SIM, and shoot structures were observed 1 week later. A 5′ nucleotide sequence encoding the N-terminal region located before the ESR1 AP2 domain was used as a specific RNA probe to prevent cross-hybridization with other AP2/EREBP transcripts. We found that ESR1 transcript levels increased 1 hr after transfer onto SIM, reached a maximum after 24 hr, and then remained at this level until ∼48 hr before they declined (Figure 6B). The induction was not caused by the stress associated with the transfer of the roots or by supplying unconditioned medium, because incubation on fresh CIM did not affect ESR1 transcript levels (Figure 6B). Whereas expression of shoot meristemless (STM), which is required for the formation of shoot apical meristem (SAM) (Endrizzi et al., 1996; Long et al., 1996), was detected 15 days after transfer onto SIM, ESR1 transcripts accumulated much earlier than STM transcripts (Figure 6A). These results suggest that the expression of ESR1 occurs at early stages during the shoot regeneration process.
Figure 6.
Time Course of ESR1 Expression during Shoot Regeneration.
RNA gel blot analysis of the ESR1 transcript. Root segments were preincubated on CIM for 4 days and then transferred onto SIM. Total RNAs were prepared from root explants at the times indicated, and 10 μg of total RNA was loaded. RNA gel blots were hybridized to different cDNA probes. Numbers above the lanes indicate days (A) or hours (B) after transfer onto SIM. In (B), in the first lane from the right (CIM), after pretreatment on CIM, root segments were incubated on fresh CIM for 24 hr.
Induction of ESR1 Transcript Accumulation by Cytokinin
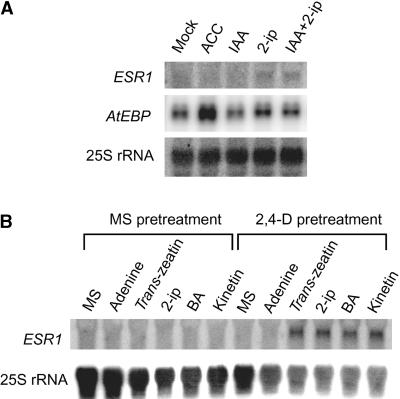
We investigated ESR1 transcript levels in wild-type plants after treatment with various phytohormones (Figure 7A). Whereas the transcript levels of AtEBP (Buttner and Singh, 1997), a member of the AP2/EREBP gene family (Figure 7A), were induced by 1-aminocyclopropane-1-carboxylic acid treatment, those of ESR1 were increased specifically after incubation on medium containing 2-ip. To verify the induction of ESR1 expression by cytokinin, various cytokinins (5 μM) were tested using root cultures because of their accessibility to phytohormone (Figure 7B). Adenine was used as a control because most cytokinins are adenine derivatives. Roots from liquid cultures of Arabidopsis plants were cut into ∼1-cm segments and transferred onto MS medium with or without 2,4-D, because the CIM used in the pretreatment for shoot regeneration contained 2,4-D and the pretreatment was essential for the highest shoot regeneration efficiency. We found that pretreatment on MS medium containing only 2,4-D could replace CIM for efficient shoot regeneration (unpublished data). Accordingly, roots were incubated on MS medium containing 2,4-D for 3 days and then transferred onto MS media containing various cytokinins. ESR1 transcript levels were induced by all cytokinin treatments, but only after preincubation with 2,4-D. The induction was not caused by 2,4-D removal, because transfer to MS medium without 2,4-D did not induce ESR1 expression. These results demonstrate that ESR1 expression is induced by cytokinins and that the induction requires a 2,4-D preincubation, suggesting that ESR1 expression can be switched on after the acquisition of competence for shoot differentiation.
Figure 7.
Induction of ESR1 Expression by Cytokinin.
(A) RNA gel blot analysis in seedlings. Two-week-old Arabidopsis plants grown on MS medium were transferred onto MS medium containing various phytohormones. After 24 hr, total RNAs were prepared and analyzed on a gel. Ten micrograms of total RNA was loaded and hybridized to different cDNA probes. Mock, fresh MS medium without any phytohormones; ACC, 1-aminocyclopropane-1-carboxylic acid (50 μM); IAA, indole-3-acetic acid (2.5 μM). 2-ip was used at a concentration of 5 μM.
(B) Expression of ESR1 in response to cytokinin with or without 2,4-D pretreatment in root explants. Root segments were pretreated on MS medium with or without 2,4-D (2 μM) for 4 days (indicated as MS pretreatment or 2,4-D pretreatment, respectively) and then transferred onto MS medium containing the compound(s) indicated. After 12 hr of incubation, total RNAs were prepared. Ten micrograms of total RNA was loaded on each lane and hybridized to different cDNA probes. Concentrations were as follows: adenine, 50 μM; trans-zeatin, 5 μM; 2-ip, 5 μM; 6-benzyladenine (BA), 5 μM; kinetin, 5 μM.
DISCUSSION
Functions of ESR1 in Cytokinin Signaling
We have isolated and characterized a novel cDNA whose overexpression conferred cytokinin-independent shoot regeneration on Arabidopsis root explants. Because our screening was based on the ability of overexpressed cDNAs to substitute for cytokinin required for shoot regeneration, the cDNAs obtained may encode factors involved in cytokinin signaling. However, overexpression of ESR1 under the control of the estradiol-inducible XVE system (Zuo et al., 2000) also increased the efficiency of shoot regeneration in the presence of cytokinin (Figure 1). Experiments with a cytokinin concentration series showed that ESR1 overexpression increased the sensitivity not only for cytokinin but also for the maximum number of regenerated shoots (Figure 3); this finding suggests that ESR1 and cytokinin act synergistically. Shoot formation by ESR1 overexpression in the absence of cytokinin may be attributable to an increased sensitivity to a low level of endogenous cytokinin that by itself is not sufficient to induce shoot formation in wild-type root cultures. Considering its late induction by cytokinin, ESR1 is unlikely to be a primary response gene in cytokinin signaling, although it may be a downstream effector for cytokinin-induced shoot regeneration. ESR1 overexpression was not sufficient to induce the highest efficiency of shoot formation because the latter still required cytokinin (Figures 1 and 3), which may induce or activate other factors in addition to ESR1. ESR1 most likely functions in a branch of the cytokinin signaling pathway that directs shoot regeneration.
We have recovered a T-DNA–tagged mutant of Arabidopsis with insertion in the ESR1 coding sequence. This mutant is indistinguishable from the wild-type plants, suggesting the presence of related genes with redundant function (data not shown). Indeed, analysis of the Arabidopsis genome uncovered at least one gene with sequence homology with ESR1.
Biochemical Function of ESR1
ESR1 encodes a putative transcriptional factor belonging to the AP2/EREBP family. The AP2 domain of ESR1 is most similar in sequence to those found in EREBPs, AtEBP, or AtERFs (Figure 2B). These transcriptional factors are thought to be involved in ethylene signaling because they can bind to the GCC box found in ethylene-responsive elements, and with the exception of AtERF3, their expression is induced by ethylene (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997; Allen et al., 1998; Hao et al., 1998; Fujimoto et al., 2000). We consider it unlikely that ESR1 is an ortholog of EREBPs or can substitute for AtERFs or AtEBP because ESR1 expression is induced by cytokinin but not ethylene (Figure 7). The sequence homology of the ESR1 AP2 domain with those of EREBPs, AtEBP, or AtERFs suggests that ESR1 may bind to DNA sequences containing a GCC box; however, the binding sequence remains to be determined biochemically. If this is the case, how can these protein factors differentially regulate promoters with GCC boxes? It is known that ethylene delays shoot regeneration in mangosteen (Lakshmanan et al., 1997), and overex-pression of AtEBP strongly inhibited shoot regeneration (unpublished results). One simple interpretation is that ESR1 functions as a transcriptional repressor. However, this possibility is unlikely because overexpression of a fusion protein of ESR1 and VP16, a peptide that functions as a strong transactivator in plants (Aoyama and Chua, 1997; Zuo et al., 2000), gave the same effect as ESR1 alone on shoot regeneration (unpublished results). Another possibility is that a combination of an AP2/EREBP protein and another transcriptional factor recognizes an extended sequence containing a GCC box. In fact, AtEBP was identified as a protein interacting with another transcriptional factor, OBP3 (Buttner and Singh, 1997), and some AP2/EREBP proteins have two DNA binding domains (Klucher et al., 1996; Moose and Sisco, 1996; Kagaya et al., 1999). A single AP2/EREBP domain may be insufficient for selection of a specific binding sequence from plant genomic DNA.
Insights for ESR1 Functions in the Shoot Regeneration Process
In Arabidopsis tissue culture, CIM pretreatment is essential for efficient de novo organogenesis, in which it is thought that differentiated cells acquire competence for dedifferentiation and subsequent redifferentiation (Hicks, 1994; Sugiyama, 1999). After the acquisition of developmental competence, cells proceed to alter their developmental fate directed by signals from SIM or RIM. ESR1 transcripts accumulated rapidly after transfer onto SIM, and the levels reached a maximum 1 to 2 days after the transfer (Figure 6), but ESR1 expression in root explants required pretreatment on CIM (Figure 7) before transfer onto SIM. At the time when ESR1 expression was induced, shoot formation was not observed, although proliferating cells had formed small clusters. ESR1 appears to function sometime before SAM formation, because expression of STM, a marker gene for SAM formation (Endrizzi et al., 1996; Long et al., 1996), could be detected only 15 days after the transfer. These results imply that ESR1 transcripts accumulate after the acquisition of competence on CIM but before shoot formation. Although inducible expression of ESR1 under the control of the XVE system (Zuo et al., 2000) could increase shoot formation on SIM dramatically, constitutive overexpression of ESR1 strongly inhibited normal leaf development (Figure 5). We found that expression under the control of the XVE system declines within several days after induction (Zuo et al., 2000; unpublished results). Thus, transient ESR1 overexpression under the control of the XVE system appears to enhance shoot regeneration efficiency without inhibiting shoot formation after its initiation. This hypothesis is supported by the observation that ESR1 is expressed transiently soon after transfer onto SIM. ESR1 may regulate the initiation of shoot regeneration after the acquisition of competence for organogenesis.
Induction of ESR1 expression required a pretreatment on CIM (Figure 7B). Some factors required for ESR1 expression appear to be induced or activated by CIM pretreatment, during which cells acquire competence for shoot regeneration. We noted that overexpression of ESR1 itself was not sufficient for efficient shoot regeneration. The latter appears to require some other factors that are activated or induced by cytokinin during incubation on SIM. In 2-week-old plants, ESR1 was induced by cytokinin without pretreatment with 2,4-D (Figure 7A), although the induced transcript level was much lower than that obtained in root cultures after 2,4-D pretreatment (Figure 7B). Vegetative tissues appear to contain a substantial number of cells that are competent for ESR1 induction.
Our results support the hypothesis (Christianson and Warnick, 1983, 1984, 1985) that de novo organogenesis can be divided into three phases. Physiological studies with Convolvulus leaf explants led to the proposal that cells in leaf explants first acquire competence for induction and organogenesis (phase 1). The competent cells then are induced to undergo differentiation (phase 2) before subsequent morphological differentiation and growth (phase 3). Because ESR1 expression on cytokinin-containing SIM required previous incubation on CIM (a preorganogenesis treatment corresponding to phase 1), it is likely to act after the acquisition of competence for regeneration. The identification of proteins that interact with ESR1 and the genes under ESR1 regulation should enable us to determine the subsequent chain of events that culminates in shoot regeneration.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Wassilewskija was used for root cultures. Seed were sterilized and sown on Murashige and Skoog (MS)(1962) medium (MS salts [Sigma], Gamborg's B5 vitamins [Sigma], 1% sucrose, and 0.25% Phytagel [Sigma]) and grown for 7 days at 22°C under continuous light. Seedlings were transferred to B5 liquid medium (Gamborg's B5 salts [Sigma], Gamborg's B5 vitamins, and 2% glucose) and incubated for 15 days with shaking at 125 rpm. Aerial parts were removed from the hydroponic culture, and the harvested roots were cultured. Cultured roots were cut into 3- to 6-mm segments and used for transformation. Compositions of the callus-inducing medium (CIM), the shoot-inducing medium (SIM), and the root-inducing medium (RIM) were as follows (Yasutani et al., 1994): CIM: Gamborg's B5 salts, 2% glucose, Gamborg's B5 vitamins, 2 μM 2,4-D, 0.25 μM kinetin, and 0.25% Phytagar; SIM: MS salts, 1% sucrose, Gamborg's B5 vitamins, 0.8 μM indole-3-acetic acid, 12.5 μM N6-Δ2-isopentenyladenine (2-ip), and 0.25% Phytagel; RIM: Gamborg's B5 salts, 2% glucose, Gamborg's B5 vitamins, 2.5 μM indole-3-acetic acid, and 0.25% Phytagel. C medium is SIM without 2-ip but supplemented with 0.4 g/L carbenicillin and 50 mg/L kanamycin.
Construction of a cDNA Library
Poly(A) RNAs were prepared from equal amounts of total RNAs extracted from several different sources using Straight A's mRNA Isolation System (Novagen, Madison, WI). The sources used were 7-day-old seedlings, mature plants before flowering, root cultures incubated on CIM for 3 days, and root cultures incubated on CIM for 3 days followed by incubation on SIM for 1 day, 3 days, or 5 days. Double-stranded cDNAs were synthesized from 1 μg of the poly(A) RNAs using Anchor oligo(dT) primer (5′-AAGCAGTGGTAACAACGC-AGAGTGCGGCCGCTTTTTTTTTTTTTTTTA/G/C-3′), second strand primer (5′-AAGCAGTGGTAACAACGCAGAGTGGCGCGCCGGG-3′), SuperScript II RNase H reverse transcriptase (Gibco BRL), and the buffer supplied by Gibco BRL. cDNAs were amplified by five cycles of polymerase chain reaction (PCR) using AMP primer (5′-AAGCAGTGGTAACAACGCAGAGTG-3′) after RNase A treatment and subsequent gel filtration. The cDNAs were normalized by the self-subtraction method (Foote et al., 1994). After three cycles of subtraction, the cDNAs were amplified by eight cycles of PCR using the AMP primer and then digested with AscI and NotI. The digested cDNAs were cloned between a 35S promoter of Cauliflower mosaic virus and the poly(A) addition sequence of the nopaline synthetase gene in the plasmid pSK34 (a derivative of pSK1 [Kojima et al., 1999]). The plasmid cDNA library was amplified once in Escherichia coli DH10B (Gibco BRL) and then transformed into Agrobacterium tumefaciens EHA105.
Screening of Cytokinin-Independent Shoots
Root transformation was performed as described by Koncz et al. (1989). Root cultures transformed by the cDNA expression library were incubated on C medium at 22°C for 4 weeks under continuous light. cDNA inserts were amplified by PCR using the vector primers 5′-CGTTCCAACCACGTCTTCAAAGCAAGTGG-3′ and 5′-AACGAC-GGGGAAATTCGAGCTGCGG-3′ from shoots or a dark green callus produced on C medium that lacks cytokinin. Amplified fragments digested with AscI and NotI were cloned into pSK34 digested with AscI and NotI, resulting in pSK-ESR1. The plasmid construct was transformed into root cultures mediated by Agrobacterium. pER10-ESR1 was constructed by ligating the AscI-NotI fragment of the ESR1 cDNA to linearized pER10. pER10 is identical to pER8 (Zuo et al., 2000) except that the hygromycin resistance gene is replaced by a kanamycin resistance gene.
Cytokinin Sensitivity Assay
T2 plants transformed with pER8 or pER8-ESR1 were used. pER8-ESR1 was constructed by ligating the AscI-NotI fragment of the ESR1 cDNA to linearized pER8. Roots were cut into ∼1-cm segments. Ten root segments were first incubated on CIM and then transferred onto SIM containing various concentrations of 2-ip.
RNA Gel Blot Analysis
Total RNAs were prepared using RNeasy Plant Mini Kits (Qiagen, Valencia, CA). All procedures for electrophoresis and blotting were performed as described by Sambrook et al. (1989).
Accession Numbers
A gene corresponding to the ESR1 cDNA is found in the Arabidopsis genomic database and is assigned to chromosome 1 (GenBank accession number AC007357). The accession number for the ESR1 cDNA described in this article is AF353577.
Acknowledgments
We thank Dr. Peter Hare and Dr. Mathias Zeidler for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010234.
References
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Buttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., and Shockey, J.A. (1999). The ethylene-response pathway: Signal perception to gene regulation. Curr. Opin. Plant Biol. 2, 352–358. [DOI] [PubMed] [Google Scholar]
- Christianson, M.L., and Warnick, D.A. (1983). Competence and determination in the process of in vitro shoot organogenesis. Dev. Biol. 95, 288–293. [DOI] [PubMed] [Google Scholar]
- Christianson, M.L., and Warnick, D.A. (1984). Phenocritical times in the process of in vitro shoot organogenesis. Dev. Biol. 101, 382–390. [DOI] [PubMed] [Google Scholar]
- Christianson, M.L., and Warnick, D.A. (1985). Temporal requirement for phytohormone balance in the control of organogenesis in vitro. Dev. Biol. 112, 494–497. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I.B., and Kieber, J.J. (1999). Molecular mechanisms of cytokinin action. Curr. Opin. Plant Biol. 2, 359–364. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 967–979. [DOI] [PubMed] [Google Scholar]
- Foote, H.C.C., Brady, G., Thorlby, G.J., and Flanklin, C.H. (1994). Subtractive hybridization of different mRNA populations. In Plant Molecular Biology: A Laboratory Manual, M.S. Clark, ed (Berlin: Springer-Verlag), pp. 201–220.
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hicks, G.S. (1994). Shoot induction and organogenesis in vitro: A developmental perspective. In Vitro Cell Dev. Biol. 30, 10–15. [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto, T. (1998). Cytokinin signaling. Curr. Opin. Plant Biol. 1, 399–403. [DOI] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, S., Banno, H., Yoshioka, Y., Oka, A., Machida, C., and Machida, Y. (1999). A binary vector plasmid for gene expression in plant cells that is stably maintained in Agrobacterium cells. DNA Res. 6, 407–410. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Mayerhofer, R., Koncz-Kalman, Z., Korber, H., Redei, G.P., and Schell, J. (1989). High-frequency T-DNA-mediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 86, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan, P., Ng, S.K., Loh, C.S., and Gof, C.J. (1997). Auxin, cytokinin and ethylene differentially regulate specific developmental states associated with shoot bud morphogenesis in leaf tissues of mangosteen (Garcinia mangostana L.) cultured in vitro. Plant Cell Physiol. 38, 59–64. [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins en-coded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Moose, S.P., and Sisco, P.H. (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10, 3018–3027. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa, S., Yasutani, I., Fukuda, H., Komamine, A., and Sugiyama, M. (1998). Organogenic responses in tissue culture of srd mutants of Arabidopsis thaliana. Development 125, 135–142. [DOI] [PubMed] [Google Scholar]
- Reinert, J., and Bajaj, Y.P.S. (1977). Plant Cell, Tissue and Organ Culture. (New York: Springer-Verlag).
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Skoog, F., and Miller, C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11, 118–140. [PubMed] [Google Scholar]
- Sugiyama, M. (1999). Organogenesis in vitro. Curr. Opin. Plant Biol. 2, 61–64. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Sato, S., Kato, T., and Tabata, S. (2001). The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42, 751–755. [DOI] [PubMed] [Google Scholar]
- Yasutani, I., Ozawa, S., Nishida, T., Sugiyama, M., and Komamine, A. (1994). Isolation of temperature-sensitive mutants of Arabidopsis thaliana that are defective in the redifferentiation of shoots. Plant Physiol. 105, 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). An estrogen receptor–based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273. [DOI] [PubMed] [Google Scholar]