Abstract
We cloned a cDNA encoding an Arabidopsis Ran binding protein, AtRanBP1c, and generated transgenic Arabidopsis expressing the antisense strand of the AtRanBP1c gene to understand the in vivo functions of the Ran/RanBP signal pathway. The transgenic plants showed enhanced primary root growth but suppressed growth of lateral roots. Auxin significantly increased lateral root initiation and inhibited primary root growth in the transformants at 10 pM, several orders of magnitude lower than required to induce these responses in wild-type roots. This induction was followed by a blockage of mitosis in both newly emerged lateral roots and in the primary root, ultimately resulting in the selective death of cells in the tips of both lateral and primary roots. Given the established role of Ran binding proteins in the transport of proteins into the nucleus, these findings are consistent with a model in which AtRanBP1c plays a key role in the nuclear delivery of proteins that suppress auxin action and that regulate mitotic progress in root tips.
INTRODUCTION
Ran/TC4 is a nuclear small GTP binding protein whose GTP- or GDP-bound state is modulated by the action of interacting proteins such as RCC1 (a Ran nuclear guanine nucleotide exchange factor), Ran-GTPase–activating protein (RanGAP), Ran binding protein 1 (RanBP1; a RanGAP cofactor), and Mog1 (a guanine nucleotide release factor) (Klebe et al., 1993; Bischoff et al., 1994, 1995; Steggerda and Paschal, 2000). The modulation of Ran status in concert with the actions of other importin/karyopherin and exportin superfamily members in the nuclear pore complex regulates many cellular processes, including nuclear protein transport and cell cycle progression (Allen et al., 2000; Dasso, 2001). Ran hydrolyzes GTP to GDP with the aid of a cytosolic RanGAP or SUMO-1–conjugated RanGAP1 (Becker et al., 1995; Matunis et al., 1996; Saitoh et al., 1998) and RanBP1 (Bischoff et al., 1995). Replacement of GDP with GTP is achieved by the action of RCC1 (Bischoff and Ponstingl, 1991). Free Ran then is recharged by the excess amount of GTP in cells, where the concentration of GTP is 30-fold greater than that of GDP (Rush et al., 1996).
Modulation of the GTP/GDP-bound state of Ran by RanBPs is essential for nuclear protein transport in interphase cells. Ran-GDP is predominant on the cytoplasmic side of the nuclear pore complex, whereas Ran-GTP is most abundant on the nucleoplasmic side of the complex (Kahana and Cleveland, 1999). This unequal distribution of Ran-GTP/Ran-GDP is achieved mostly by the compartmentation of the Ran binding proteins that regulate the ratio of Ran-GTP to Ran-GDP. RanGAP and RanBP1 occur mainly on the cytoplasmic side and help maintain cytosolic Ran in a GDP-bound form, whereas RCC1, which is nuclear in localization, keeps nuclear Ran mainly in a GTP-bound form (Koepp and Silver, 1996). This asymmetric distribution of Ran-GTP and Ran-GDP is thought to be a major driving force giving directionality in nuclear transport.
In addition to their role in nuclear transport, Ran and Ran binding proteins also are involved in the regulation of nuclear assembly and cell cycle progression, including mitotic spindle formation in mitotic cells (Nakamura et al., 1998; Kahana and Cleveland, 1999; Heald and Weis, 2000). Some of the strongest evidence for this comes from studies using Xenopus egg extracts. Carazo-Salas et al. (1999) showed that chromatin-associated RCC1 in these extracts generates a high local concentration of Ran-GTP around the chromatin, which, they proposed, is needed for the local nucleation of microtubules and spindle formation. Consistent with this model, Ran-GTP added to Xenopus egg extracts promotes spindle assembly by organizing microtubules in the vicinity of chromosomes (Dasso, 2001). Added Ran-GTP also releases TPX2 protein or APA (aster-promoting activity) protein from importin α (Gruss et al., 2001) or NuMA microtubule-associated protein from importin β (Nachury et al., 2001; Wiese et al., 2001), and it is the release of these proteins from the importing cargos by Ran-GTP that then induces a spindle-like structure.
There is evidence that RanBP1 may be an especially important partner of Ran in mediating cell cycle progression. Although most genes encoding Ran-interacting partners are constitutively active, transcription of the mRNA for RanBP1 is repressed in nonproliferating cells, is activated at the G1/S transition in cycling cells, peaks during S-phase or metaphase of mitosis, and declines abruptly in late telophase (Battistoni et al., 1997; Guarguaglini et al., 2000). Furthermore, Kalab et al. (1999) and Guarguaglini et al. (2000) have shown that the overexpression or addition of exogenous RanBP1 yields severe defects in spindle formation, whereas depletion of the protein in mitotic cells induces mitotic delay and abnormal nuclear division.
Valuable insights regarding how RanBP1 could interfere with cell cycle progression have come from yeast studies. Depletion of a yeast RanBP1, Yrb1p, arrests cells in late anaphase/telophase. This phenotype is attributed to the suppression of two important proteolysis events: the anaphase-promoting complex (APC)-mediated proteolysis of the anaphase inhibitor Pds1p and the Skp1/Cullin/F-box (SCF) ubiquitin ligase–mediated proteolysis of cyclin-dependent kinase inhibitor Sic1p protein (Baumer et al., 2000). The yeast results underscore the role of Ran binding proteins in directing mitosis-controlling protease complexes to the nucleus.
In summary, increasing data suggest that the Ran-GDP/Ran-GTP switch and the generation of Ran-GTP around the mitotic apparatus play just as important a role in directing cell cycle progression through mitosis as they do in regulating nuclear transport in interphase cells. This advance in our understanding of the in vivo functions of Ran and Ran binding proteins has come primarily from studies in animals and yeast. In contrast to the rapid progress that has been made in clarifying the roles of Ran and its binding partners in these organisms, relatively little is known about them in plants.
Ach and Gruissem (1994) showed that the tomato Ran protein was functionally homologous with a yeast Ran-like protein in suppressing the effects of a mutation in a yeast homolog of RCC1 on mitosis. Xia et al. (1996) found a RanBP1 homolog in their expression screen for Arabidopsis genes that could alter cell cycle progression in yeast. Subsequently, Haizel et al. (1997) isolated genes encoding RanBPs (AtRanBP1a and AtRanBP1b) in Arabidopsis, characterized their binding specificity to Ran, and demonstrated coordinated expression of the genes with Arabidopsis Ran. Recently, Meier (2000) proposed that Ran signal transduction is linked to the control of nuclear assembly, postulating that the association of plant RanGAP with nuclear envelope vesicles was needed to ensure the completion of mitosis before the assembly of the nuclear envelope.
To gain a better understanding of the functions of Ran and Ran binding proteins in plants, we obtained initial results showing that at least five different Ran binding proteins are detectable in total pea plumule extracts, as judged by an overlay assay using labeled Ran-GTP as a probe (unpublished observations). We then cloned a cDNA encoding an Arabidopsis RanBP1 homolog, AtRanBP1c, using the yeast two-hybrid technique, and generated transgenic Arabidopsis expressing antisense transcripts of the gene. Here, we report that the suppression of AtRanBP1c expression in Arabidopsis results in a unique phenotype, which includes altered growth and development of primary roots and hypersensitivity of these roots to auxin. Our results lead us to hypothesize that the regulation of the Ran-GDP/Ran-GTP cycle by AtRanBP1c is critically involved in the control of proteins that regulate auxin sensitivity and cell cycle progression, especially the transition from metaphase to telophase.
RESULTS
An Arabidopsis Ran Binding Protein, AtRanBP1c, Interacts with a Pea Ran/TC4, PsRan1, in Cotransformed Yeast and Preferentially Binds to the GTP-Bound Conformation of PsRan1
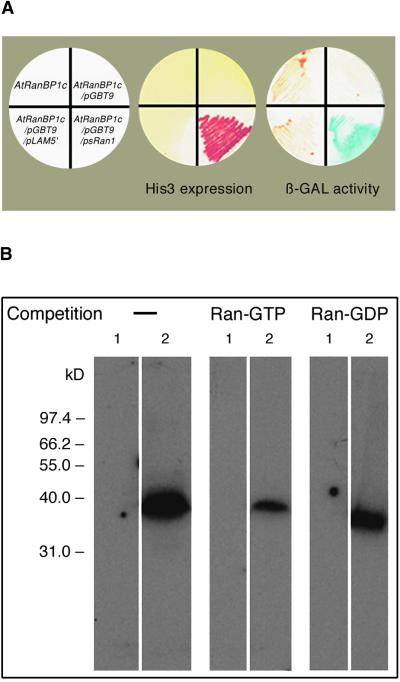
Thirty putative PsRan1 binding clones were isolated from 1.3 × 106 transformants by the entrapment technique, and one of them (named AtRanBP1c) was characterized further. Figure 1 shows that only the yeast cotransformed with AtRanBP1c fused to pACT, and PsRan1 fused to pGBT9 survived in minimal medium lacking histidine and showed β-galactosidase activity. AtRanBP1c did not activate the yeast reporters, either alone or in combination with a bait plasmid that is not directed to PsRan1. This result implies that AtRanBP1c interacted specifically with PsRan1 in transformed yeast.
Figure 1.
AtRanBP1c Is an Arabidopsis Ran Binding Protein.
(A) In vivo test of the specific interaction between PsRan1 and AtRanBP1c in yeast. A cDNA clone encoding the AtRanBP1c fusion to the GAL4 activation domain vector pACT was isolated from a cDNA expression library and was used to transform the reporter strain Saccharomyces cerevisiae Y190 either alone or along with psRan1 or PLAM5′ (a bait unrelated to Ran used as a negative control) fused to the GAL4 DNA binding domain vector pGBT9 or to the DNA binding domain vector without an insert. Cotransformed yeast then were streaked on amino acid dropout medium (SD-Trp-Leu-His) containing 50 mM 3-aminotriazole to test the expression of the His3 reporter and on complete medium to test the expression of the LacZ reporter gene by a β-galactosidase (β-GAL) assay.
(B) Binding of AtRanBP1c to PsRan1 and binding preference for Ran-GTP. Total bacterial extracts from the cells containing the expression construct pET15-b (lane 1) and pET15-b/AtRanBP1c (lane 2) were fractionated on 12% SDS-PAGE, transferred to a nitrocellulose membrane, renatured, and overlaid with α-32P-GTP–PsRan1 in the absence or presence of competition by an excess amount of cold Ran-GTP or Ran-GDP.
The derived protein of the cDNA for AtRanBP1c is a 219–amino acid polypeptide with a calculated molecular weight of 24,741 (Figure 2), whereas bacterially expressed protein runs as 33 kD on SDS-PAGE (data not shown). The protein has a conserved Ran binding domain (RanBD) in the middle of the peptide that is known to serve as a Ran binding site for the gene product of the human AB1 clone (Beddow et al., 1995). Computer analysis performed with the PSORT program also revealed a putative RNA binding motif inside RanBD. AtRanBP1c is a very acidic protein (pI = 4.52), with glutamate accounting for 22% of the total amino acids, much of it concentrated in the C terminus of the sequence. The protein is essentially identical in its amino acid sequence to the Arabidopsis Ran binding protein homolog, which has been classified as one of the cell cycle modulators in yeast (Xia et al., 1996), and it is 70% identical to the two other known plant RanBPs, AtRanBP1a and AtRanBP1b (Haizel et al., 1997), but it differs significantly from both of these in its C-terminal sequence (Figure 2).
Figure 2.
Amino Acid Sequence Alignment of Three Arabidopsis Ran BPs.
Identical residues among three RanBPs are highlighted and considered as consensus sequences. The putative Ran binding domain is marked with asterisks. The RNA binding motif stretches from amino acid 72 to 78 (GAGTVKL). Uninterrupted stretches of 10 or more identical amino acids among the proteins are underlined. Dots represent gaps in the sequence. Dashes in the consensus (Con.) rows indicate unconserved amino acids.
Using an overlay assay performed with labeled PsRan1, we confirmed the binding of PsRan1 to a 38-kD protein whose size corresponds to that of His-tagged AtRanBP1c (Figure 1B, left). The binding was inhibited greatly by competition with an excess amount of cold Ran-GTP (Figure 1B, middle) but not significantly by an excess amount of Ran-GDP (Figure 1B, right). This result implies that AtRanBP1c binds to the GTP-bound conformation of PsRan1 preferentially over that of the GDP-bound conformation.
Expression of Antisense AtRanBP1c in Transgenic Arabidopsis Enhances the Growth of Primary Roots but Suppresses Lateral Root Growth and Alters the Pattern of Root Hair Development
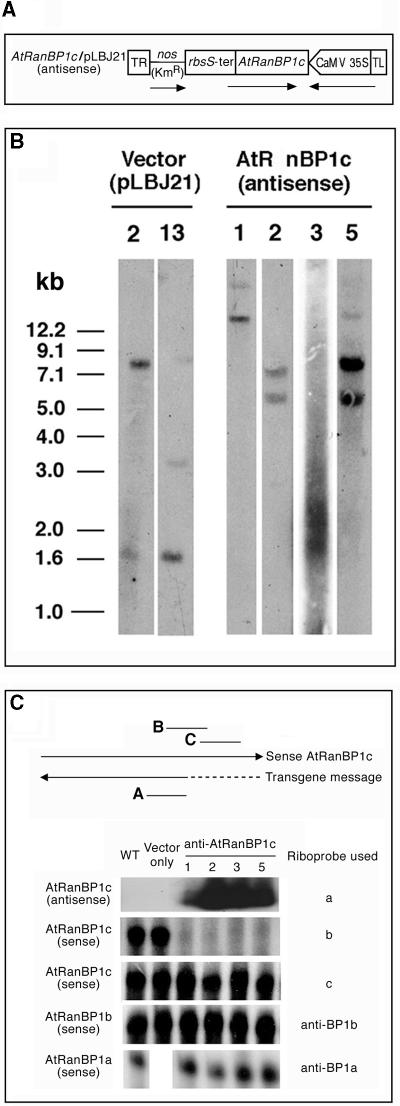
Independent transformation events of the antisense fusion construct into Arabidopsis were confirmed by demonstrating different restriction fragment patterns in DNA gel blots of transgenic lines (Figure 3B). Four independent transgenic lines transformed with antisense AtRanBP1c were produced, judging from both their ability to survive kanamycin selection (see Methods) and the fact that their DNA generated different restriction fragment patterns when the 35S promoter region of Cauliflower mosaic virus (CaMV) was used as a probe for DNA gel blot analysis. All four of the transgenic lines expressed antisense AtRanBP1c, as determined by RNA gel blot analysis using a strand-specific riboprobe (Figure 3C).
Figure 3.
Construction and Characterization of anti-AtRanBP1c Transgenic Plants.
(A) Construction of antisense AtRanBP1c in pLBJ21. Two-thirds of the AtRanBP1c gene was subcloned into pLBJ21 in an antisense direction to the orientation of a 35S promoter of CaMV. Arrows indicate the transcriptional direction of the AtRanBP1c gene. TR and TL, right and left border of the T-DNA; nos, nopaline synthase; KmR, kanamycin resistance gene.
(B) Identification of independent transgenic lines by DNA gel blot hybridization analysis. Ten micrograms of genomic DNA isolated from each transgenic line was digested with EcoRI and fractionated by agarose gel electrophoresis. After the DNA was transferred to a nylon membrane, the blot was hybridized with a 32P-dATP–labeled 35S promoter region of CaMV and washed as described in Methods. The numbers above the gels represent independent transgenic lines.
(C) Expression of known Arabidopsis AtRanBP1 genes in anti-AtRanBP1c transgenic plants. Total RNAs were isolated from wild-type (WT) or transgenic Arabidopsis plants, and the expression of the endogenous AtRanBP1a, AtRanBP1b, AtRanBP1c, or anti-AtRanBP1c transgene was examined by RPA or RNA gel blot analysis as described in Methods. Lines designated a, b, and c represent regions of the anti-AtRanBP1c (a) or AtRanBP1c gene (b and c) protected by riboprobes a, b, and c, respectively. Arrows indicate the transcriptional direction of the sense and antisense message for the AtRanBP1c gene. The dotted line represents the region of the AtRanBP1c cDNA that is missing in the antisense transgene.
We performed RNase protection assays (RPA) with total RNA obtained from different transgenic lines. Different riboprobes were used to detect all known Arabidopsis RanBP1 genes or to check the abundance of endogenous AtRanBP1c messages and the integrity of different regions of endogenous AtRanBP1c messages. Results (Figure 3C) showed that region B of endogenous AtRanBP1c message was not protected from RNase digestion, whereas the 3′ end of the messages (region C) was protected fully by riboprobe c (Figure 3C). This implies that the 3′ truncated cytoplasmic transgene actually forms an RNA duplex with the corresponding complementary endogene in the cell.
It does not appear that the formation of an RNA duplex induces transcriptional gene silencing because, as seen in RPA using riboprobe c, the transgenic plants still express AtRanBP1c messages in the seedlings of the T2 generation we used as a source of total RNA. The expression of other known AtRanBP1 genes, AtRanBP1b (Figure 3C) and AtRanBP1a (Figure 3C), was unaffected by the presence of the transgene. Thus, it is reasonable to conclude that the mutant phenotype is attributable mostly to post-transcriptional suppression of endogenous AtRanBP1c expression (Van Houdt et al., 2000).
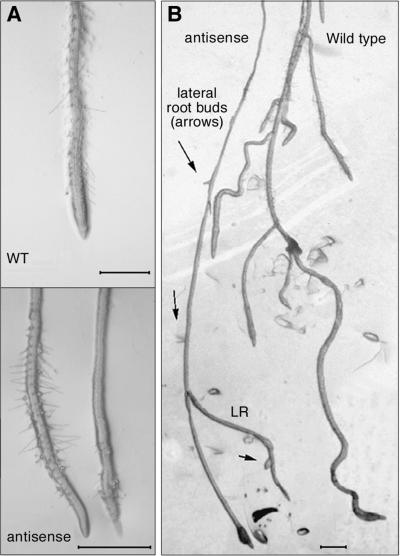
Three of the transgenic lines (anti-AtRanBP1c-1, -2, and -3) showed a long-rooted phenotype, ∼1.6 to four times longer than that of wild type, on a medium containing germination medium without sucrose (data not shown), and all of them showed suppressed lateral root growth and abnormal patterns of root hair development (Figure 4). An increased rate of growth of apical root cells was the main reason for the long-rooted phenotype of anti-AtRanBP1c-1 plants (Table 1). An initial test of whether the increased root length was caused primarily by increased cell number or by increased cell length was performed by analyzing longitudinal sections in one of the transgenic lines (anti-AtRanBP1c-1). This study revealed that mature epidermal, cortex, and endodermal root cells in the transformants were significantly longer than in wild-type plants but that they did not differ from the wild type in mean cell diameter (Table 1). Transverse sections of transgenic plant roots showed no difference in structure or arrangement compared with those of the wild-type plants. Lateral root initiation in transgenic plants was normal, but growth beyond the epidermis of the primary root was <2 mm in >90% of the lateral roots in these plants compared with being longer than 10 mm in >90% of the wild-type lateral roots. In anti-AtRanBP1c-1, only eight of 51 plants had any lateral roots at all, and of these, all had only one lateral root. This was in sharp contrast to wild-type plants, the vast majority of which had multiple lateral roots, averaging 8.5 per plant (Table 1).
Figure 4.
Microscopic Analysis of Root Phenotypes of Anti-AtRanBP1c Transgenic Plants.
(A) At top, root of wild-type (WT) Arabidopsis showing normal even distribution of root hairs throughout the region extending 20 to 500 μm behind the meristematic zone. At bottom, two representative roots from transgenic plants expressing anti-AtRanBP1c, each showing only patchy distribution of root hairs, with bare spots visible either near the distal end of the root hair zone (left root) or all along most of the root length shown (right root).
(B) Roots of wild-type (right) and anti-AtRanBP1c plants (left) showing the typical pattern of multiple lateral roots growing out from the primary root of the wild-type plant and only one lateral root (LR) growing out from the transformant, with additional lateral root buds forming in a normal pattern but arrested in their growth (arrows).
Bars = 500 μm.
Table 1.
Comparison of Transgenic and Wild-Type Roots
| Plant Line | Mean Root Length (mm) |
Percentage of Wild Type |
sd (mm) | n | ||||
|---|---|---|---|---|---|---|---|---|
| Transgenic | 9.0 | 164a | 6.4 | 473 | ||||
| Wild type | 5.5 | — | 1.8 | 54 | ||||
| Mean Cell Length (μm)b
|
Mean Cell Diameter (μm)b
|
|||||||
| Cell Type | Transgenic | Percentage of Wild Type |
sd | Wild Type | sd | Transgenic | Percentage of Wild Type | Wild Type |
| Epidermal | 157.0a | 173 | 24.0 | 91.0 | 21.0 | 14.8 | 103 | 14.0 |
| Cortex | 146.0a | 151 | 21.0 | 97.0 | 16.0 | 28.0 | 116 | 24.0 |
| Endodermal | 107.0a | 130 | 21.0 | 82.0 | 17.0 | 15.9 | 98 | 16.0 |
| Mean Lateral Rootsc
|
Mean Root Apical Growthc
|
Mean Lateral Roots Initiated/Plantd
|
||||||
| Plant Line | Percentage with Lateral Roots |
Lateral Roots/ Plant |
Rate (μm/day) | Percentage of Wild Type | +IAAe | −IAA | ||
| Wild type | 77 | 8.5 | 505 | 100 | N.D.f | 8.75 | ||
| Transgenic | 16a | 1.0a | 750a | 149a | 10.0 | 2.33a | ||
Significantly different from the wild type at P ≤ 0.05.
n, wild type = 8 plants; transgenic = 6 plants; ≥5 cells each plant.
n, wild type and transgenic ≥ 22; these lateral roots are all fully developed.
n, wild type = 8; transgenic = 15 (−IAA) or 4 (+IAA).
IAA concentration = 10−11 M.
N.D., not determined.
Regarding root hair development, in transgenic plants the circumferential arrangement of root hairs was the same as in wild-type plants (i.e., in columns separated by one or two non-root-hair–bearing epidermal cells). However, the longitudinal arrangement of root hairs in transgenic lines varied dramatically from that of wild-type plants in that it was very erratic and uneven (Figure 4). In >90% of the transgenic plants, in the region in which there was even spacing of root hairs along the entire longitudinal axis of wild-type plant roots, there were irregularly spaced patches up to 200 μm long in which there were no root hairs at all.
Antisense Transgenic Plants Show a Hypersensitive Response to Indoleacetic Acid for Induction of Lateral Root Initiation and for Growth Inhibition of Primary Roots
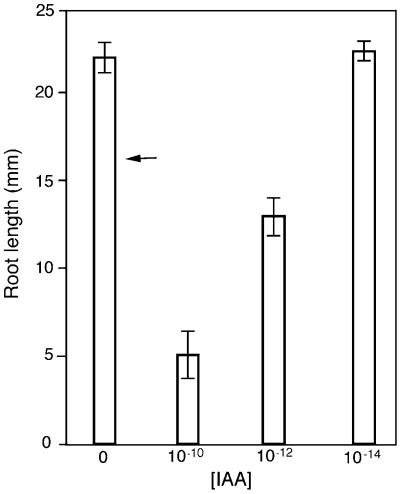
The addition of only 1 to 10 pM indoleacetic acid (IAA) to the medium stimulated lateral root formation in the transgenic plants (Table 1), and it severely inhibited primary root growth in these plants (Figure 5). A dose–response analysis of the inhibitory response showed that the length of primary roots in the transgenic plants was inhibited significantly at concentrations down to 10−12 M compared with that in plants grown on medium without IAA. The amount of growth inhibition by IAA in transgenic plants was proportional to the dose of IAA, with lower concentrations causing less inhibition (Figure 5). Wild-type plants were unaffected by concentrations of IAA at or below 10−10 M, and the presence or absence of 10−14 M IAA in the medium made no difference in the root length of transgenic plants.
Figure 5.
IAA Dose–Response for Primary Root Length in Arabidopsis Plants Transformed with Antisense Orientation of AtRanBP1c.
Data shown are means ±sd of primary roots (n ≥ 5). The arrow indicates the mean length of the primary root of wild-type plants (17.1 ± 2.7 mm). This length did not change significantly in wild-type plants treated with IAA concentrations ≤10−10 M. Mean root lengths of transgenic plants treated with 10−10 and 10−12 M IAA differed significantly from each other and from the lengths of plants not treated with IAA or treated with 10−14 IAA (P ≤ 0.05), but the lengths of roots not treated with IAA or treated with 10−14 M IAA did not differ significantly when significance was analyzed by Student's t test.
Subnanomolar Auxin Induces a Higher Mitotic Index and Mitotic Arrest at Metaphase or Anaphase in Transgenic Roots and Leads to the Death of Cells in the Meristematic Zone of the Roots
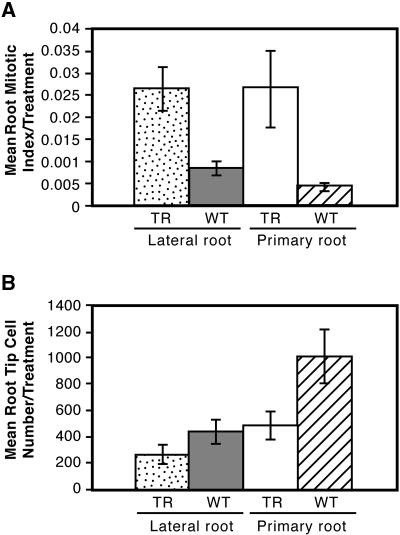
Even low concentrations of auxin (10−11 M) induced an increase in the mitotic index in both the lateral and primary roots of the transformants (Figure 6A). Most of the mitotic figures found in the transgenic lateral root tips were in metaphase (91%, with only 9% being in early anaphase and none in telophase), whereas those in wild-type lateral roots were distributed more widely among different phases (metaphase, 47%; anaphase, 13%; telophase, 40%). The mitotic figures in primary root tips of transgenic plants showed 77% in metaphase and 23% in early anaphase, whereas those in wild-type plants showed 46% in metaphase, 23% in anaphase, and 31% in telophase (data not shown). The arrest of mitosis at metaphase, which accounted for the increased mitotic index, was accompanied eventually by death of the cells in the meristematic zone of the root (Figures 7A and 7B). Cells basal to the meristematic zone generally were not affected by the IAA application because they were produced and differentiated before the onset of mitotic disruption (Figure 7A). Cells produced before the onset of mitotic disruption continued to expand normally in the absence of newly produced cells from the meristem. This led to a truncation of the root tip, with fully differentiated and expanded cells being directly adjacent to the meristem, as seen in Figures 7A and 7B, by the reduction in the number of cells in the meristematic zone (Figure 6B). Once cells in the meristematic zone died, root cap cells were not replaced, which ultimately led to the death of the entire root tip. In contrast, the growth and early development of those lateral roots that formed in the transgenic plants were normal in the absence of exogenous IAA (Figure 7C).
Figure 6.
Mitotic Index (A) and Cell Number (B) in Tips of Both Lateral and Primary Roots Treated with 10−11 M IAA.
In both (A) and (B), the differences between wild-type and anti-AtRanBP1c transgenic plants are statistically significant as judged by Student's t test (P ≤ 0.05). TR, anti-AtRanBP1c transgenic plants; WT, wild-type plants.
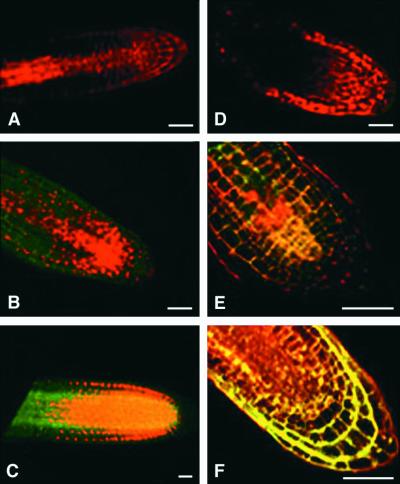
Figure 7.
Fluorescein Diacetate and Propidium Iodide Test of Cell Viability in Root Tips of Arabidopsis Plants Treated with Various Levels of IAA and Untreated Controls.
Plants were either antisense ([A] to [C]) or wild-type plants ([D] to [F]). Because fluorescein diacetate must be transported into the nucleus and processed before it fluoresces green and propidium iodide always fluoresces red, this combination is excellent for determining cell viability. A living cell will show a combination of the red and green (yellow nucleus), and a dead cell will show only the red from the propidium iodide.
(A) Primary root of a seedling germinated on germination medium plus sucrose (GM-S) plus IAA (10−11 M). The root grew only 1 mm before it stopped growing and died.
(B) Lateral root of a seedling germinated on GM-S and transferred at 10 days to a plate containing GM-S plus IAA (10−11 M). Roots were stained with fluorescein diacetate and propidium iodide 4 days later. A root initiated during IAA treatment died soon after emergence from the epidermis.
(C) Lateral root from a seedling treated as in (A) except that no IAA was in the medium. This same pattern of staining was seen in primary root tips from transgenic seedlings grown in the absence of IAA (data not shown).
(D) Lateral root of a seedling germinated on GM-S and transferred at 10 days to a plate containing GM-S plus IAA (10−5 M). Roots were stained with fluorescein diacetate and propidium iodide 4 days later. A lateral root initiated during treatment died soon after emergence from the epidermis.
(E) Lateral root of a seedling germinated on GM-S and transferred at 10 days to a plate containing GM-S + IAA (10−11 M). Roots were stained with fluorescein diacetate and propidium iodide 4 days later. A lateral root initiated during treatment remained alive after the treatment.
(F) Wild-type root tip from a seedling treated as in (C).
Bars = 0.2 mm.
Exogenous auxin also could induce mitotic arrest and the ultimate death of meristematic cells in wild-type plants as well, but only at much higher concentrations. When wild-type plants were treated with 10−5 M IAA, lateral roots were initiated but died immediately after emergence from the epidermis (Figures 7D to 7F), as seen in transgenic plants treated with 10−9 M IAA. Likewise, the inhibitory response of the roots of wild-type plants to 10−7 M IAA closely mimicked the response seen in the transgenic plants at 10−11 M, including having enhanced numbers of meristematic cells in the metaphase stage of mitosis (data not shown). Wild-type roots showed neither positive nor negative growth responses to IAA when the applied concentration decreased to 10−10 M IAA, whereas the transgenic plants failed to show a response only when the applied IAA concentration decreased to 10−14 M.
Lateral root initiation and early development in both transgenic and wild-type plants were qualitatively the same on medium with and without IAA at concentrations at or below 0.1 nM. However, in transgenic plants, unlike in wild-type plants, the growth of the lateral roots ceased once they extended >1 mm out from the epidermis.
DISCUSSION
Overexpression of tomato and tobacco Ran genes in Schizosaccharomyces pombe suppresses the temperature-sensitive RCC1 phenotypes that undergo mitotic chromosomal condensation and spindle formation without completion of the S-phase (Ach and Gruissem, 1994; Merkle et al., 1994). Overexpression of an Arabidopsis RanBP1 in yeast, which is virtually identical to AtRanBP1c, yields a phenotype similar to that of the yeast pim1 mutant, which is defective in its normal progression through the cell cycle (Xia et al., 1996). However, little information has been available regarding the in vivo functions of the RanBPs in plant growth and development, and we are unaware of any other analysis of the effect of a perturbation to the Ran system in plants. Our results show clear evidence that Ran/RanBP signal pathways play a critical role in normal root growth and development by affecting root sensitivity to auxin and regulating the mitosis-to-interphase transition process in mitotic root tips.
Celenza et al. (1995) described an alf3-1 mutant in Arabidopsis that was defective in its maturation of lateral roots and, in this respect, greatly resembled the transgenic plants expressing an antisense orientation of AtRanBP1c. However, the alf3-1 mutant, unlike the anti-AtRanBP1c plants, had arrested primary root growth in the absence of auxin, initiated more than twice the number of lateral root primordia per length of primary root as wild-type plants, and had apparently normal root hair distribution. Moreover, the lateral root growth defect in alf3-1, but not that in the antisense transformants, could be rescued by growing the mutants in IAA.
Roots of Anti-AtRanBP1c Plants Show Hypersensitive Responses to Auxin Treatment
The roots of transgenic plants are hypersensitive to IAA effects on growth inhibition, lateral root initiation, and cell cycle progression in the mitotic zones of primary and lateral roots. Regarding the growth inhibition response, exogenous IAA typically does not inhibit the growth of wild-type Arabidopsis roots until its concentration reaches 10−9 M (Knee and Hangarter, 1996). In contrast, this response is induced at 10−12 M IAA in transgenic plants. In the absence of added auxin, transgenic plants have longer cells in their primary roots than do wild-type plants, which is consistent with the interpretation that in the roots of transgenic plants, endogenous levels of IAA induce relatively more growth in them than the endogenous levels in wild-type roots induce in them.
In wild-type plants, the level of IAA needed to induce lateral root initiation is ≥10−7 M, significantly higher than that needed to inhibit root growth (Knee and Hangarter, 1996). Thus, wild-type plants grown in 10−7 M IAA initiate more lateral roots, but those roots are inhibited from further growth soon after they emerge through the epidermis, similar to the effects of 10−11 M IAA on the AtRanBP1c antisense plants described here.
The induction of lateral roots by IAA and the inhibition of root elongation by IAA use distinct signal transduction pathways (Celenza et al., 1995), although these pathways may share some steps in common. The fact that roots suppressed in their expression of AtRanBP1c are hypersensitive to the effects of IAA in both of these pathways could indicate that this protein plays a key role in one or more of the steps shared by both pathways. Alternatively, roots suppressed in AtRanBP1c production may accumulate higher levels of endogenous IAA and thus generically require less exogenous IAA to show any characteristic response.
Regarding auxin-induced effects on mitosis, in transgenic plants, IAA selectively blocked new cell production in root tip cells at greater than or equal to picomolar concentrations. This effect also was seen in wild-type plants treated with 10−5 M IAA (Figures 7D to 7F), but in wild-type plants, 10−11 M IAA had no effect. In both transgenic and wild-type plants, the inhibition of mitosis was accompanied by the loss of viability of cells in the mitotic zones of lateral and primary roots. Recent findings on the cellular function of RanBP1 suggest a hypothetical mechanism for how this protein could regulate mitotic cell cycle progression, and as discussed below, this model could help explain how the lack of AtRanBP1c renders cells more sensitive to auxin.
A Model Explaining How AtRanBP1c Is Involved in the Regulation of Mitotic Cell Cycle Progression and Auxin Hypersensitivity
Yeast deficient in Yrb1p, a homolog of RanBP1, are defective in both APC- and SCF-mediated proteolysis of important cell cycle regulatory proteins, in which APC promotes sister chromatid separation by targeting an anaphase inhibitor, Pds1p, for destruction (Baumer et al., 2000). One interpretation of the yeast results is that the Yrb1p1 mutant is defective in the transport of the APC complex to the nucleus.
Using the yeast results as a starting point for a model, one could predict that in transgenic Arabidopsis the suppression of AtRanBP1c would result in an inappropriate increase in the ratio of Ran-GTP to Ran-GDP in the cytoplasm. This increase would decrease the affinity of importin α and importin β for their cargo and decrease the rate of delivery of key proteins involved in the mitotic cell cycle to the nucleus. Specifically, because cell cycle–dependent proteolytic activity has been demonstrated in plants (Genschik et al., 1998; Inze et al., 1999), if the transgenic plants were defective in their rate of import of APC-like complexes (or other ubiquitin ligase–like complexes), this would lead to the failure of sister chromatid separation, freezing many cells at metaphase. Given the critical role of RanBP1 in the nuclear transport of TPX2 or APA protein needed to initiate spindle assembly in Xenopus egg extracts (Gruss et al., 2001), one also could predict that disrupting the rate of nuclear delivery of proteins required for spindle assembly in the transgenic plants would result in the failure of cells to complete the cell cycle.
Could this same model help explain the apparent hypersensitivity of roots to auxin? Increased levels of auxin could induce cell growth and the G2/M transition in roots by removing a suppressor protein that functions in the nucleus and must be delivered there by an importin-dependent mechanism. If this is the case, a decrease in the efficiency of this delivery system resulting from the lack of AtRanBP1c could mimic an increased auxin concentration in cells. This would make the cells appear to be hypersensitive to auxin. Examples of suppressors of auxin action that must be delivered to the nucleus to block the expression of auxin-induced genes are members of the IAA family of proteins, which have been implicated in both the suppression and induction of auxin responses (Ulmasov et al., 1999). Although in principal the hypersensitivity to exogenous auxin exhibited by transgenic plants could result from these plants having higher endogenous levels of auxin, the model would favor the alternative that the sensitivity is caused by reduced activity of auxin suppressors.
Much remains to be clarified regarding the biological functions of plant Ran and RanBPs and the way in which these proteins work together in plant growth and development. Detailed analyses of the effects of AtRanBP1c suppression on the nuclear abundance of auxin-induced transcription factors and plant mitotic regulators may reveal additional roles for Ran and RanBPs in plant growth and development and may clarify the molecular steps in which AtRanBP1c is involved.
METHODS
Two-Hybrid Screening
All procedures used for the two-hybrid screening were performed according to Clontech's Matchmaker manual (Palo Alto, CA). For generation of a bait construct for screening, a cDNA of PsRan1 containing the full coding region of Ran was amplified by polymerase chain reaction and subcloned into pGBT9 (a GAL4 DNA binding domain vector). For screening, the yeast (Saccharomyces cerevisiae Y190) containing the pGBT9/PsRan1 hybrid (bait) was cotransformed with Arabidopsis cDNAs fused to pACT (a GAL4 DNA activation domain vector). The cDNAs were provided as an expression library by the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The transformants then were grown for 1 week on selective medium (synthetic medium lacking His, Trp, and Leu) containing 50 mM 3-aminotriazole to identify putative positive clones. The positive colonies then were streaked on the same selective medium to confirm the expression of His3, and on a complete medium to test the expression of LacZ using a β-galactosidase assay. From these positive colonies, the plasmids containing cDNAs for PsRan1 binding proteins as fusions with the pACT vector were rescued into Escherichia coli HB101 for further characterization and sequencing.
PsRan1 Overlay Assay
The overlay assays for PsRan1 binding were performed as described by Lounsbury et al. (1994) with some modifications. Noninduced total bacterial extracts were fractionated by 12% SDS-PAGE and transferred electrophoretically to a nitrocellulose membrane. The blot was soaked in renaturation buffer (20 mM 3-[N-morpholino]-propanesulfonic acid, pH 7.1, 100 mM sodium acetate, 5 mM magnesium acetate, 0.25% Tween 20, 0.5% BSA, and 5 mM DTT) for 24 hr at 4°C and then overlaid with α-32P-GTP–PsRan1 in Ran binding buffer (20 mM 3-[N-morpholino]-propanesulfonic acid, pH 7.1, 100 mM potassium acetate, 5 mM magnesium acetate, 0.1% Tween 20, 0.5% BSA, 5 mM DTT, and 100 μM GTP) in the presence or absence of a 12-fold excess amount of unlabeled PsRan1-GTP or PsRan1-GDP protein. After washing the membrane with the binding buffer, the blot was exposed to a phosphorimage screen and shown using a phosphorimager. The α-32P-GTP–PsRan1 protein used in the overlay was generated by charging 1 μg of PsRan1 protein with 30 μCi of α-32P-GTP (3000 Ci/mmol; DuPont) for 20 min at room temperature in the presence of GTP binding buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.2 mg/mL BSA, 2.5 mM EDTA, 1 mM DTT, and 10 μM GTP. The binding was stabilized by adding MgCl2 to a final concentration of 10 mM at the end of the reaction.
Construction of an Antisense AtRanBP1c in pLBJ21 and Transformation into Agrobacterium tumefaciens
The binary vector pLBJ21 used as the transformation vehicle is a derivative plasmid of pKYLX71 (Schardl et al., 1987; Lloyd et al., 1992) with a unique EcoRI site engineered in place of the HindIII site of the multicloning region of the expression cassette. For the construction of a fusion vector between antisense AtRanBP1c and pLBJ21, a cDNA encoding two-thirds of AtRanBP1c was cloned as a SacI-XbaI fragment into the corresponding sites of pLBJ21, resulting in the fusion construct anti-AtRanBP1c/pLBJ21 with the 35S promoter of Cauliflower mosaic virus directing expression of the antisense orientation of the cDNA. The fusion plasmid was transformed into A. tumefaciens GV3101 harboring pMP90 plasmid by electroporation (Koncz and Schell, 1986).
Transformation into Arabidopsis
The pLBJ21-fused antisense AtRanBP1c was introduced into root explants of Arabidopsis using the Agrobacterium-mediated transformation method as described by Valvekens et al. (1988). T1 seed from primary transformants were planted on germination medium (GM) (Valvekens et al., 1988) containing 50 μg/L kanamycin to select for transgenic progeny. Transgenic T2 seed from individual T1 plants were plated on the same medium to identify homozygous T2 seed. T2 homozygous seed were used to generate all of the transgenic plants used for the described experiments.
Genomic DNA Gel Blot Analysis
Genomic DNA isolated from 3-week-old green transgenic plants was digested with EcoRI, separated electrophoretically on a 0.8% agarose gel, and blotted onto a zeta-probe membrane (Bio-Rad). The membrane was prehybridized in 0.25 M Na2HPO4, pH 7.2, and 7% SDS at 65°C for 30 min and hybridized in the same solution containing α-32P-dATP–labeled 35S promoter region of Cauliflower mosaic virus (XbaI-HindIII–digested fragment of pBI221; Clontech) for 20 hr at 65°C. The membrane was washed with 20 mM Na2HPO4, pH 7.2, and 5% SDS and finally with the same buffer containing 1% SDS for 1 hr in each solution at 65°C.
Total RNA Isolation and RNA Gel Blot Analysis
Total RNA (10 μg), isolated from 3-week-old green transgenic plants using Trizol reagent (Gibco BRL), was size fractionated on a 1.2% agarose gel containing 6% formaldehyde and transferred to a Hybond N+ membrane (Amersham) in 20 × SSPE (1 × SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4). The membrane then was prehybridized in a solution containing 5% SSPE, 50% formamide, 2 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.5% SDS, and 250 μg/mL denatured salmon sperm DNA for 12 hr at 56°C. Hybridization was performed for 20 hr at 56°C to detect antisense RNA of AtRanBP1c in the same solution supplemented with sense riboprobe. The probe was generated from the region encoding RanBD of AtRanBP1c using the MAXIscript T3/T7 in vitro transcription kit (Ambion, Austin, TX) as indicated by the manufacturer's protocol. The blot was washed with 1 × SSPE and 0.5% SDS at room temperature, 0.1% SSPE and 0.1% SDS at room temperature, and finally 0.1 × SSPE and 0.1% SDS at 60°C. Autoradiography was performed for 18 hr at −80°C using two intensifying screens.
Total RNA Isolation and Gene Expression Analysis by RNase Protection Assays
Total RNA was isolated from 3-week-old green transgenic plants using Trizol reagent (Gibco BRL) and was used for RNase protection assay (RPA) experiments. For the RPA experiments, each DNA fragment was amplified by polymerase chain reaction from AtRanBP1a, AtRanBP1b, or AtRanBP1c cDNA using the following primer sets: anti-BP1a, 5′-TGG-TTATCTGAAAGAGTACTTTGG-3′ and 5′-TTTTTGTCAAACAATTGG-CAATTACC-3′; anti-BP1b, 5′-TTAAATCCATCGAAGGAAGACTTGG-3′ and 5′-AAAGAGAACACGCGAATCATCCATG-3′; probe a, 5′-GAC-GGCGAGTTGAAAGATGAGCTTT-3′ and 5′-TTTCTCCTCACTTATATTTTCCTCA-3′; probe b, 5′-GCTGGTTTAATAGAGAATCTTTCGG-3′ and 5′-CCAAATTGACACAGTAAAGAGACGAC-3′; and probe c, 5′-AGGAAGCAGAAGAGAAAGAGCCTGC-3′ and 5′-CCAAATTGACACAGTAAAGAGACGAC-3′. The DNA fragment was ligated with T7 adaptor using the Lig'nScribe RNA polymerase promoter addition kit (Ambion), and actual DNA template for in vitro transcription was prepared according to the manufacturer's manual. Riboprobe was transcribed from the template using the MAXIscript T3/T7 in vitro transcription kit (Ambion) and size fractionated on a 5% acrylamide gel to determine the longest riboprobe. The RNase protections were performed using the RPA III kit (Ambion) according to the manufacturer's directions with 10 μg of total RNA and 105 cpm of labeled probe. In brief, sample RNA and labeled probe were mixed and precipitated to dissolve in the hybridization buffer. The RNA and labeled probe in the buffer were denatured and incubated overnight at 42°C. The RNA mixture then was digested with a 1:100 dilution of RNase stock solution for 30 min at 37°C. The protected RNA fragment was separated on a 5% denaturing polyacrylamide gel and detected by autoradiography using two intensifying screens for 5 to 6 hr at −70°C. Riboprobe anti-BP1a was used for the detection of AtRanBP1a sense messages, and anti-BP1b was used for the detection of AtRanBP1b. Riboprobe a was used for the detection of the antisense transgene of AtRanBP1c, and riboprobe b was used for the detection of the region on sense AtRanBP1c at the junction between the sequence duplexed by the transgene and the 3′ end that is missing in the transgene. Riboprobe c was used for the detection of the 3′ end of the AtRanBP1c sense message.
Analyses of Anatomy, Growth, and Auxin Effects
Plants used for anatomical analysis were fixed in a modified Navishins solution as described by Mauseth et al. (1984). After fixation, plant tissue was dehydrated through a standard ethanol series. Ethanol was exchanged with xylenes before embedding in paraffin. Tissue was microtomed at 7 μm, stained, and examined using standard bright-field microscopy techniques.
Statistics were computed using SAS software, release 6.12 (SAS Institute, Cary, NC). Measurements were made using an ocular micrometer calibrated to a ×6.3 objective.
Mean apical growth rate was calculated as the mean per plant of transgenic or wild-type plants. Measurements of each root were taken at 24-hr intervals using a Nikon (Tokyo, Japan) dissecting microscope at ×40 and a ruler.
Plates used for indoleacetic acid (IAA) dose–response experiments were identical to those used for all other experiments except for the addition of IAA to the medium (germination medium without sucrose [GM-S]). Seed were surface sterilized in 30% (v/v) commercial bleach solution for 15 min, rinsed in sterile water, and air dried. Seed were then placed on plates and vernalized for 72 hr to synchronize germination. After vernalization, all plates were placed in the same growth chamber and allowed to grow for 10 days to determine the effect of various concentrations of auxin on root length and lateral root production (modified from Knee and Hangarter, 1996).
For mitotic index studies, transgenic and wild-type plants were grown on GM-S plates as described previously. Ten days after germination, all plants were transferred to new plates that were identical to the germination plates except for the addition of 10−11 M IAA. After 10 days on plates containing IAA, all lateral and primary root tips were removed, washed in double-distilled water, and fixed using a 3:1 mixture of 95% ethanol and acetic acid. After fixation, samples were stored in 70% ethanol at 4°C; then 10 each of wild-type and transgenic primary and lateral root tips were selected randomly for examination by fluorescence microscopy. The tips were hydrolyzed in 1 N HCl for 90 sec, rinsed with double-distilled water, immersed in a solution of 5 μg/mL 4′,6-diamidino-2-phenylindole in PBS buffer for 30 min, placed on slides, covered with a cover slip, and squashed using a pencil eraser. The edges of the cover slips were sealed with clear, fast-drying nail polish. All slides were stored in the dark at 4°C until viewing. Slides were viewed using an Olympus (Tokyo, Japan) BH2 epifluorescence microscope with a ×100 Dplan Apo UV oil immersion objective and a standard UV filter cube. For each root tip, all mitotic figures and total numbers of cells in the meristematic zone were counted. Mitotic indices were calculated using the following formula: number of cells in mitosis/total number of cells.
For confocal laser microscopy studies, wild-type and transgenic plants were germinated and grown on GM-S for 10 days before being transferred to plates that were identical except for the addition of 10−10 M IAA. Five days after the initiation of lateral roots, all roots were removed, washed in double-distilled water, and double stained with fluorescein diacetate and propidium iodide (for details, see Celenza et al. [1995]). After staining and mounting, all root tips were viewed using a Leica (Wetzlar, Germany) TCS 4D confocal laser microscope. Each root tip was viewed using wavelengths specific for each stain, producing two images that were merged together using Adobe Photoshop 5.0 (Mountain View, CA).
Accession Numbers
The accession numbers for the genes described in this article are X97377 (AtRanBP1a), X97378 (AtRanBP1b), and U62742 (AtRanBP1c).
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the Arabidopsis cDNA expression library and Roger Hangarter and Mark Estelle for critical comments. This work was supported by grants from the National Science Foundation (IBN-9603884 and IBN-0080363) and the National Aeronautics and Space Administration (NAG5-3887 and NAG2-1347) to S.J.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010214.
References
- Ach, R.A., and Gruissem, W. (1994). A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell-cycle mutant. Proc. Natl. Acad. Sci. USA 91, 5863–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, T.D., Cronshaw, J.M., Bagley, S., Kiseleva, E., and Goldberg, M.W. (2000). The nuclear pore complex: Mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113, 1651–1659. [DOI] [PubMed] [Google Scholar]
- Battistoni, A., Guarguaglini, G., Degrassi, F., Pittoggi, C., Palena, A., Matteo, G.D., Pisano, C., Cundari, E., and Lavia, P.C. (1997). Deregulated expression of the RanBP1 gene alters cell cycle progression in murine fibroblasts. J. Cell Sci. 110, 2345–2357. [DOI] [PubMed] [Google Scholar]
- Baumer, M., Kunzler, M., Steigemann, P., Braus, G.H., and Irniger, S. (2000). Yeast Ran-binding protein Yrb1p is required for efficient proteolysis of cell cycle regulatory proteins Pds1p and Sic1p. J. Biol. Chem. 275, 38929–38937. [DOI] [PubMed] [Google Scholar]
- Becker, J., Melchior, F., Gerke, V., Bischoff, F.R., Ponstingl, H., and Wittinghofer, A. (1995). RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J. Biol. Chem. 270, 11860–11865. [DOI] [PubMed] [Google Scholar]
- Beddow, A.L., Richards, S.A., Orem, N.R., and Macara, I.G. (1995). The Ran/TC4 GTPase-binding domain: Identification by expression cloning and characterization of a conserved sequence motif. Proc. Natl. Acad. Sci. USA 92, 3328–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., and Ponstingl, H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., Klebe, C., Kretschmer, J., Wittinghofer, A., and Ponstingl, H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91, 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., Krebber, H., Smirnova, E., Dong, W., and Ponstingl, H. (1995). Co-activation of Ran-GTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 14, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Guarguaglini, G., Gruss, O.J., Segref, A., Karsenti, E., and Mattaj, I.W. (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181. [DOI] [PubMed] [Google Scholar]
- Celenza, J., Grisafi, P., and Fink, G. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9, 2131–2142. [DOI] [PubMed] [Google Scholar]
- Dasso, M. (2001). Running on Ran: Nuclear transport and the mitotic spindle. Cell 104, 321–324. [DOI] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y. Derevier, A., and Fleck, J. (1998). Cell cycle–dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteosome inhibitor MG132. Plant Cell 10, 2063–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, O.J., Carazo-Salas, R.E., Schatz, C.A., Guarguaglini, G., Kast, J., Wilm, M., Bot, N.L., Vernos, I., Karsenti, E., and Mattaj, I.W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin β on TPX2 activity. Cell 104, 83–93. [DOI] [PubMed] [Google Scholar]
- Guarguaglini, G., Renzi, L., Dottavio, F., DiFiore, B., Casenghi, M., Cundari, E., and Lavia, P. (2000). Regulated Ran-binding protein 1 activity is required for organization and function of the mitotic spindle in mammalian cells in vivo. Cell Growth Differ. 11, 455–465. [PubMed] [Google Scholar]
- Haizel, T., Merkle, T., Pay, A., Fejes, E., and Nagy, F. (1997). Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 11, 93–103. [DOI] [PubMed] [Google Scholar]
- Heald, R., and Weis, K. (2000). Spindles get the Ran around. Trends Cell Biol. 10, 1–4. [DOI] [PubMed] [Google Scholar]
- Inze, D., Gutierrez, C., and Chua, N.-H. (1999). Trends in plant cell cycle research. Plant Cell 11, 991–994. [PMC free article] [PubMed] [Google Scholar]
- Kahana, J.A., and Cleveland, D.W. (1999). Beyond nuclear transport: Ran-GTP as a determinant of spindle assembly. J. Cell Biol. 146, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P., Pu, R.T., and Dasso, M. (1999). The Ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9, 481–484. [DOI] [PubMed] [Google Scholar]
- Klebe, C., Nishimoto, T., and Wittinghofer, F. (1993). Functional expression in Escherichia coli of the mitotic regulator proteins p24ran and p45rcc1 and fluorescence measurements of their interaction. Biochemistry 32, 11923–11928. [DOI] [PubMed] [Google Scholar]
- Knee, E.M., and Hangarter, R.P. (1996). Differential IAA dose response of the axr1 and axr2 mutants of Arabidopsis. Physiol. Plant. 98, 320–324. [Google Scholar]
- Koepp, D.M., and Silver, P.A. (1996). A GTPase controlling nuclear trafficking: Running the right way or walking randomly? Cell 87, 1–4. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vectors. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Lounsbury, K.M., Beddow, A.L., and Macara, I.G. (1994). A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J. Biol. Chem. 269, 11285–11290. [PubMed] [Google Scholar]
- Matunis, M.J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauseth, J.D., Montenegro, G., and Walckowiak, A.M. (1984). Studies of the holoparasite Tristerix aphyllus (Loranthaceas) infecting Trichocereus chilensis (Cactaceae). Can. J. Bot. 62, 847–857. [Google Scholar]
- Meier, I. (2000). A novel link between Ran signal transduction and nuclear envelope proteins in plant. Plant Physiol. 124, 1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle, T., Haizel, T., Matsumoto, T., Harter, K., Dallmann, G., and Nagy, F. (1994). Phenotype of the fission yeast cell cycle regulating mutant pim1-46 is suppressed by a tobacco cDNA encoding a small, Ran-like GTP-binding protein. Plant J. 6, 555–565. [DOI] [PubMed] [Google Scholar]
- Nachury, M.V., Maresca, T.J., Salmon, W.C., Waterman-Storer, C.M., Heald, R., and Weis, K. (2001). Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., Masuda, H., Horii, J., Kuma, K.I., Yokoyama, N., Ohba, T., Nishitani, H., Miyata, T., Tanaka, M., and Nishimoto, T. (1998). When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to γ-tubulin. J. Cell Biol. 143, 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush, M.G., Drivas, G., and D'Eustachio, P. (1996). The small nuclear GTPase Ran: How much does it run? BioEssays 18, 103–112. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., Sparrow, D.B., Shiomi, T., Pu, R.T., Nishimoto, T., Mohun, T.J., and Dasso, M. (1998). Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8, 121–124. [DOI] [PubMed] [Google Scholar]
- Schardl, C.L., Byrd, A.D., Benzion, G., Altschuler, M.A., Hildebrand, D.F., and Hunt, A.G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61, 1–11. [DOI] [PubMed] [Google Scholar]
- Steggerda, S.M., and Paschal, B.M. (2000). The mammalian Mog1 protein is a guanine nucleotide release factor for Ran. J. Biol. Chem. 275, 23175–23180. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Montagu, M.V., and Lijsebettens, M.V. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt, H., Van Montagu, M., and Depicker, A. (2000). Both sense and antisense RNAs are targets for the sense transgene-induced posttranscriptional silencing mechanism. Mol. Gen. Genet. 263, 995–1002. [DOI] [PubMed] [Google Scholar]
- Wiese, C., Wilde, A., Moore, M.S., Adam, S.A., Merdes, A., and Zheng, Y. (2001). Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science 291, 653–656. [DOI] [PubMed] [Google Scholar]
- Xia, G., Ramachandran, S., Hong, Y., Chan, Y.-S., Simanis, V., and Chua, N.-H. (1996). Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 10, 761–769. [DOI] [PubMed] [Google Scholar]