Abstract
A third member of the ZTL gene family was identified in the Arabidopsis genome and was named LKP2 (for LOV kelch protein2). A cDNA was isolated corresponding to this gene, and plants overexpressing LKP2 were generated. The overexpression of LKP2 resulted in arrhythmic phenotypes for a number of circadian clock outputs in both constant light and constant darkness, long hypocotyls under multiple fluences of both red and blue light, and a loss of photoperiodic control of flowering time. The LKP2 mRNA is not regulated by the circadian clock and was detected in all tissues examined. These results suggest that LKP2 functions either within or very close to the circadian oscillator in Arabidopsis. A model is presented for its mode of action.
INTRODUCTION
Diverse organisms possess a circadian system that allows them to keep track of the time of day and to coordinate specific physiological events with environmental day/night cycles. In conceptual terms, an organism's circadian system consists of three broad domains: a central oscillator, input pathways used to entrain the oscillator, and output pathways used to convey temporal information from the clock to physiological events controlled by the clock. Although the boundaries between these broad domains have become less clear recently as the molecular components of the system have been identified, these fundamental concepts still apply and provide a useful framework for understanding circadian systems in a wide range of organisms.
At the core of the circadian system is the central oscillator, which has been the focus of intense research for many years. Studies of the central oscillator from several different organisms indicate that it consists of a transcriptional feedback loop in which constitutive positively acting factors promote the expression of autoregulatory negatively acting factors (Dunlap, 1999). This autoregulatory feedback loop oscillates with an ∼24-hr periodicity, and peripheral factors that can affect the pace of the oscillator are important for maintaining the cycle near 24 hr. A circadian oscillator that approximates the day-night cycles experienced by the organism has been shown to confer a competitive advantage to the organism (Ouyang et al., 1998). A major element of the circadian system is the input pathways that entrain the oscillator so that it maintains a stable phase relationship with the environment. Recently, the characterization of factors that are regulated by the clock (i.e., outputs) and feedback on the input pathways have made the distinction between input and central oscillator difficult to determine (Kume et al., 1999; Merrow et al., 1999; Covington et al., 2001).
Even though pioneering work on plants led to the discovery of circadian rhythms more than 250 years ago, identification of the molecular components of the oscillator in plants has lagged behind that in other organisms and has begun to emerge only recently. Characterization of a short-period Arabidopsis mutant resulted in the identification of TOC1, a clock-regulated gene containing response regulator and Constans-like domains, indicating a role in transcription (Strayer et al., 2000). Physiological analysis of the toc1 mutant strongly suggests that it plays a central role in the oscillator (Somers et al., 1998b), but further molecular characterization is needed. Two clock-regulated transcription factors containing single myb domains were identified (CCA1 and LHY), and their overexpression resulted in arrhythmia for a number of circadian outputs (Schaffer et al., 1998; Wang and Tobin, 1998). These two myb factors currently are prime candidates for transcription factors that function within the feedback loop. The nuclear-localized GI protein is involved in the photoperiodic control of flowering, and although it clearly can affect clock function, phenotypic analysis of gi mutants indicates that it most likely functions in the light input pathway to the clock (Fowler et al., 1999; Park et al., 1999; Huq et al., 2000). The elf3 mutant was isolated as an early flowering mutant exhibiting a conditional arrhythmic phenotype in continuous light (Zagotta et al., 1992; Hicks et al., 1996), and physiological analysis suggests a function for the ELF3 protein in the regulation of light input to the clock (McWatters et al., 2000; Covington et al., 2001). Clearly, each of these proteins functions in the circadian system of Arabidopsis, although further characterization is needed to determine their exact roles.
Two other genes, ZTL and FKF1, were described recently that appear to be involved in the photoperiodic control of flowering and, in the case of ZTL, the circadian clock. The ztl mutants were isolated in a genetic screen for mutations affecting period length in Arabidopsis (Millar et al., 1995). These mutants exhibited a late flowering phenotype in long days and short hypocotyls in red light (Somers et al., 2000). It also was shown that fluence rate strongly affected the period length phenotype in ztl mutants, an observation consistent with ZTL functioning on the light input pathway to the clock. A related gene (FKF1) was characterized at the same time based on its late flowering phenotype (Nelson et al., 2000). The fkf1 mutant was isolated as a large deletion on the bottom of chromosome 1 that was shown to cause both late flowering in long days and short hypocotyls in red and blue light. Sequence analysis of ZTL and FKF1 identified a unique combination of motifs, including a PAS domain at their N terminus followed by an F-box and six repeats at their C terminus, that form a kelch domain. This unique gene structure and the phenotypes associated with these mutants are consistent with them playing a role in the circadian system in Arabidopsis.
A gene containing a PAS domain was identified on top of chromosome 2 in the Arabidopsis genome, and further analysis of its sequence indicated that it also possesses an F-box and a kelch domain, very similar to ZTL and FKF1, making it the third member of the ZTL gene family. The gene was named LKP2 (for LOV kelch protein2). A cDNA was isolated corresponding to this gene, and plants overexpressing LKP2 were generated (LKP2-OX). The overexpression of LKP2 was shown to cause arrhythmia in multiple outputs under both constant light and constant darkness, an increase in hypocotyl length, and a delay in flowering under long days. The expression of endogenous LKP2 transcripts does not appear to be regulated by the circadian clock, and its mRNA was detected in a wide range of tissues. These results, particularly arrhythmia in both the light and dark, place LKP2 very close to the circadian oscillator. A model is presented for the role of the ZTL gene family in the Arabidopsis circadian system.
RESULTS
A third member of the ZTL gene family in Arabidopsis was identified using BLAST searches of the sequence databases and was named LKP2. Members of this gene family contain a unique combination of protein motifs. The N-terminal half of these proteins contains a PAS domain (described previously as the LOV domain in Arabidopsis) followed by an F-box motif, whereas the C terminus consists of six repeats forming a kelch domain. The PAS domain of LKP2 shares 77 and 67% identity with ZTL and FKF1, respectively; the F-box regions share 66 and 63% identity, and the kelch domains share 80 and 62% identity.
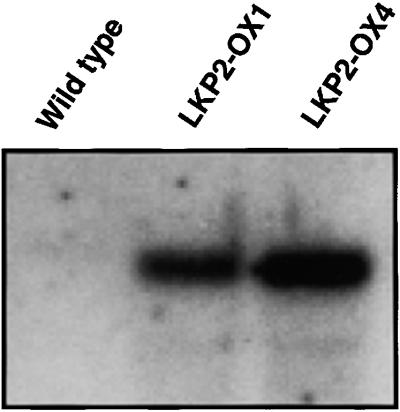
An Arabidopsis cDNA library was screened, and a cDNA containing the full-length LKP2 coding sequence was isolated. This cDNA was placed under the control of the 35S promoter, and transgenic plants overexpressing LKP2 were generated. RNA gel blot analysis of RNA isolated from T2 seedlings of multiple transgenic lines showed that LKP2 transcript levels were increased five- to 10-fold relative to wild-type plants, depending on the transgenic line (Figure 1). These LKP2-OX lines were analyzed for circadian clock, hypocotyl length, and flowering time defects, phenotypes shown previously to be associated with ztl and fkf1 mutants (Nelson et al., 2000; Somers et al., 2000). Line LKP2-OX4 was used for the results presented below.
Figure 1.
LKP2 Transcripts Are Overexpressed in LKP2-OX Lines.
RNA gel blot (10 μg of total RNA) showing LKP2 mRNA levels in wild-type control seedlings (ecotype Columbia) and two different LKP2-overexpressing lines (LKP2-OX1 and LKP2-OX4). Total RNA samples were prepared from 4-week-old plants grown in soil under 16-hr-light/8-hr-dark cycles.
LKP2-OX Seedlings Are Arrhythmic under Continuous Light
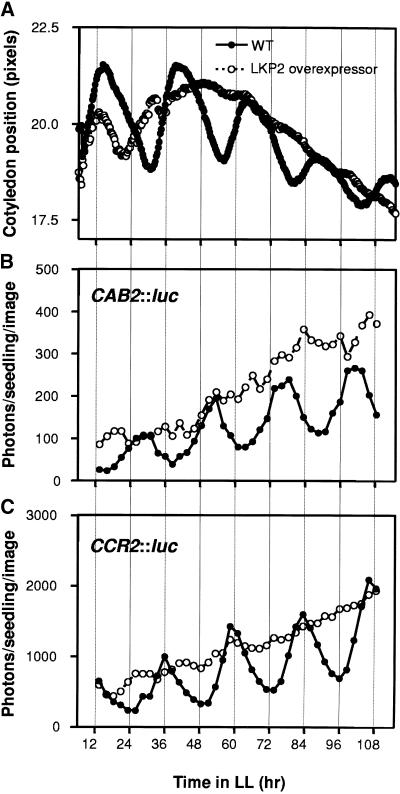
Circadian rhythms in cotyledon movement, CAB2::luc expression, and CCR2::luc expression were assayed in continuous white light (LL) for the LKP2-OX seedlings. Figure 2A shows circadian rhythms in cotyledon position in both wild-type and LKP2-OX plants. Wild-type seedlings exhibited a robust rhythm of cotyledon movement during the first 3 to 4 days under constant conditions, and these rhythms persisted for another 3 to 5 days, although a decrease of the rhythm was observed typically (Figure 2A). The LKP2-OX plants went through ∼1.5 cycles after release into LL but then rapidly became arrhythmic for the rest of the time course (Figure 2A). The first cycle observed in LKP2-OX seedlings, after release into LL, most likely was the result of a residual rhythm persisting from the light/dark (LD) conditions used to entrain the seedlings. The overexpression of LKP2 clearly has a strong effect on the circadian rhythm of cotyledon movements.
Figure 2.
Overexpression of LKP2 Results in Arrhythmicity of Multiple Circadian Outputs.
(A) Seedlings were germinated for 5 to 7 days under LD (12 hr/12 hr) cycles and then transferred to LL, and cotyledon position was monitored for 4 to 5 days. Cotyledon position along the vertical axis was plotted against time. Each trace represents the average of at least 50 seedlings from three independent experiments. WT, wild type.
(B) Bioluminescence was monitored from F1 seedlings carrying both the LKP2-OX and CAB2::luc constructs for 4 to 5 days under LL. Seedlings were germinated and entrained for 6 days in LD (12 hr/12 hr) and then transferred to LL (30 μmol·m−2·sec−1), and bioluminescence was monitored every 2.5 hr for 4 to 5 days. Traces represent averages from 16 seedlings from two independent experiments.
(C) Same as (B) except CCR2::luc was used as the reporter construct.
Circadian rhythms of both CAB2::luc and CCR2::luc also were assayed in LKP2-OX seedlings under LL. T2 plants from LKP2-OX lines were crossed to both CAB2::luc and CCR2::luc reporter lines, and progeny containing both the reporter gene and the overexpression cassette were assayed for rhythmic luciferase activity. In wild-type seedlings, both CAB2 and CCR2 expression patterns exhibited robust rhythmicity under LL, with CAB2 levels peaking ∼4 hr after subjective dawn and CCR2 levels peaking at approximately subjective dusk (Figures 2B and 2C). In LKP2-OX lines, both reporters were arrhythmic throughout the time course (Figures 2B and 2C). In contrast to the first 24 hr of cotyledon movement (Figure 2A), there was no residual effect of the conditions used to entrain the seedlings for both CAB2 and CCR2 expression (Figures 2B and 2C).
Rhythmic Gene Expression Patterns Occur under LD Cycles
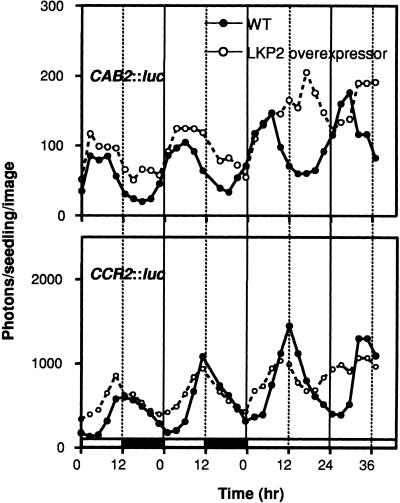
The remnants of a driven rhythm of cotyledon movement (first 24 hr in Figure 2A) indicated that the LKP2-OX plants are able to exhibit rhythmic behavior under LD (i.e., driven) conditions. CAB2::luc and CCR2::luc expression patterns in the LKP2-OX lines were assayed under driven conditions (LD cycles) to determine if these plants can be entrained by LD cycles or if they are completely arrhythmic. The expression of both reporters was clearly rhythmic under LD conditions, although subtle changes in waveform were observed. For CCR2::luc, luciferase activity began to accumulate in the LKP2-OX lines ∼4 hr earlier than in the wild type, but they declined at a similar rate (Figure 3). The peak of CCR2 expression after release into LL was not observed consistently (Figure 3). The expression of CAB2, on the other hand, appeared to accumulate late; these plants most likely were responding to the dark-to-light transition (i.e., lights on), which has been shown previously to drive CAB2 expression (Anderson et al., 1997). These results show that although LKP2-OX plants are arrhythmic under constant conditions, they are able to respond to LD conditions, exhibiting a diurnal mode of regulation for both CAB2 and CCR2.
Figure 3.
LKP2-OX Plants Are Rhythmic under Driven Conditions.
Seedlings containing both the LKP2 overexpression construct and either the CAB2::luc or the CCR2::luc reporter were germinated for 7 days under LD (12 hr/12 hr) conditions and then transferred to LL for 2 days. Bioluminescence was monitored beginning on day 5 and continued through day 9. Open bars along the x axis indicate light, and solid bars indicate dark. Traces represent averages of 10 seedlings. WT, wild type.
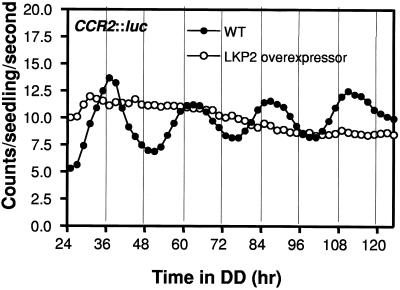
LKP2-OX Seedlings Are Arrhythmic under Continuous Darkness
The CCR2::luc reporter, unlike CAB2::luc, exhibits robust cycling in continuous darkness (DD) and therefore serves as a useful marker for circadian function in the dark (Strayer et al., 2000). LKP2-OX seedlings carrying the CCR2::luc reporter were assayed for rhythmic luciferase activity in DD. Figure 4 shows that CCR2::luc expression was completely arrhythmic under DD, in contrast to wild-type plants, which exhibited robust rhythmic expression of CCR2::luc. These results show that circadian rhythms of three distinct outputs (CAB2::luc, CCR2::luc, and cotyledon movement) were affected under various conditions by LKP2 overexpression, indicating that LKP2 acts within or very near the circadian clock in Arabidopsis.
Figure 4.
LKP2-OX Seedlings Are Arrhythmic in DD.
Seedlings containing both the CCR2::luc reporter and the LKP2 overexpression construct were germinated and entrained for 5 days and then transferred to DD for 5 days while bioluminescence was monitored. Traces represent averages from at least 16 seedlings from two independent experiments. WT, wild type.
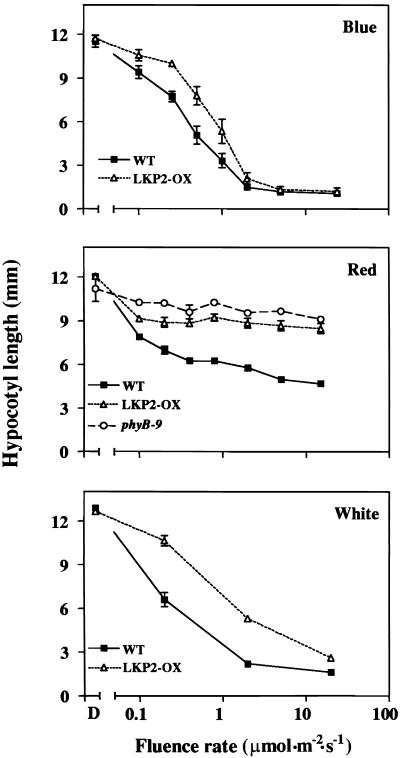
LKP2-OX Seedlings Exhibit Long Hypocotyls under both Red and Blue Light
Loss-of-function fkf1 mutants exhibited short hypocotyls under both red and blue light, whereas ztl mutants exhibited short hypocotyls specifically under red light conditions but not in blue light (Nelson et al., 2000; Somers et al., 2000). Hypocotyl lengths for LKP2-OX seedlings were determined under various fluences of red, blue, and white light (Figure 5). At all fluence rates tested, LKP2-OX seedlings had long hypocotyls in red light, a phenotype similar to loss-of-function phyB mutants (Figure 5). Under low to moderate fluence rates of blue light, LKP2-OX seedlings also exhibited long hypocotyls, but at higher fluences they appeared similar to wild-type controls. White light conditions were similar to blue light conditions, with long hypocotyls being observed at low to moderate fluences. Overexpression of LKP2 presumably results in a gain-of-function situation that causes hypocotyl lengthening. This result is consistent with the loss-of-function mutation in fkf1 and the point mutations in ztl, which caused a shortening of hypocotyl lengths.
Figure 5.
LKP2-OX Plants Exhibit Long Hypocotyl Phenotypes.
Wild-type (WT) and LKP2-OX seed (phyB-9 [null] seed were included in the red light experiment) were stratified in the dark, given a 2-hr white light pulse, transferred back to dark for 24 hr, and then transferred to the specific light conditions indicated. Hypocotyl lengths were recorded after 3 days, and each data point is the mean ±se of 20 hypocotyls from three independent experiments (n = 60).
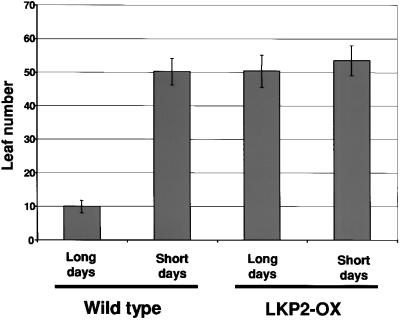
LKP2-OX Plants Flower Late under Long Days
Arabidopsis is a facultative long-day plant that flowers more rapidly under long day conditions than it does under short days. The fkf1 mutant was identified by virtue of its late flowering phenotype (Nelson et al., 2000), and the ztl mutants also exhibit late flowering (Somers et al., 2000). Leaf number at the time of bolting was determined for LKP2-OX plants grown under both long and short days (Figure 6). Flowering in LKP2-OX plants was delayed significantly under long days compared with flowering in wild-type plants, but it occurred at the same time as in wild-type plants under short days. Like both fkf1 and ztl mutants, LKP2-OX plants appear to have lost photoperiodic control of flowering time.
Figure 6.
LKP2-OX Plants Flower Late under Long Days.
Wild-type and LKP2-OX plants were grown under long days (16 hr of light/8 hr of dark; ∼60 μmol·m−2·sec−1) or short days (8 hr of light/16 hr of dark; ∼120 μmol·m−2·sec−1), and total leaf number was determined at the time of bolting (n = 5 to 17). Error bars represent standard deviations.
LKP2 Transcripts Are Expressed in a Variety of Tissues and Are Not Regulated by the Circadian Clock
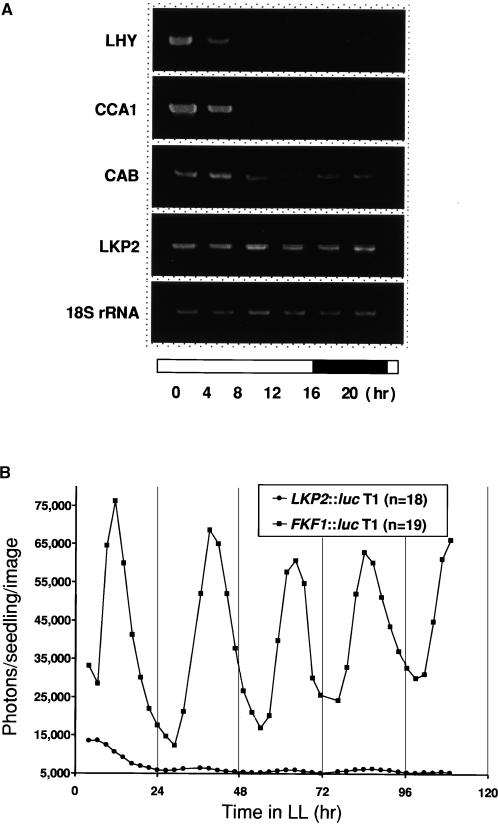
In contrast to ZTL mRNA levels, which do not appear to cycle, the transcripts for FKF1 cycle robustly, with peak levels occurring at approximately 8 hr after dawn. Therefore, it was of interest to determine whether LKP2 transcripts are regulated by the circadian clock. Because the LKP2 transcripts are of low abundance and difficult to detect by RNA gel blot analysis (Figure 1), reverse transcriptase–mediated polymerase chain reaction (RT-PCR) was used to detect the expression of LKP2 in total RNA samples isolated from plants at various times during an LD cycle. Transcripts corresponding to LHY, CCA1, and CAB have been shown to cycle and were used as cycling controls for this experiment (Figure 7A). The LKP2 transcript levels did not appear to change during the course of one LD cycle. To further support this result, bioluminescence was monitored from transgenic seedlings carrying both LKP2 and FKF1 promoters fused to the luciferase reporter gene (Figure 7B). The FKF1 promoter exhibited robust cycling, as was observed for steady state transcript levels, whereas the LKP2 promoter was expressed at a much lower level. The slight rhythmicity observed in the LKP2::luc seedlings had an amplitude of <1.2 and is most likely insignificant. Together, these results indicate that LKP2 is expressed at a low level and is not regulated by the circadian clock in Arabidopsis, similar to ZTL expression.
Figure 7.
LKP2 Transcripts Are Not Expressed Rhythmically under Both LD Cycles and LL.
(A) Gene-specific primers were designed and used in RT-PCR on total RNA isolated at the times indicated during one light (open bar)/dark (solid bar) cycle. PCR products were electrophoresed on agarose gels and visualized by ethidium bromide staining.
(B) Bioluminescence was monitored from transgenic T1 seedlings carrying LKP2::luc and FKF1::luc constructs as indicated. Seedlings were germinated and entrained for 6 days under LD (12 hr/12 hr) and then released into LL. Traces represent averages from multiple seedlings (n = 18 to 19).
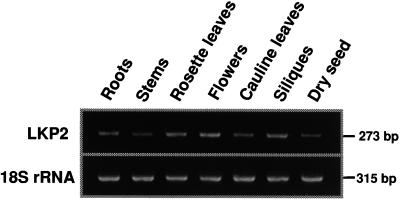
RT-PCR also was performed on total RNA isolated from a number of tissues to determine the tissue-specific expression of LKP2. Figure 8 shows that endogenous LKP2 transcripts were detected in all tissues assayed, including roots, stems, cauline and rosette leaves, flowers, siliques, and dry seed. This expression profile is very similar to those of ZTL and FKF1, with the exception that ZTL transcripts were not detected in dry seed and FKF1 transcripts were not detected in stems and seed (Kiyosue and Wada, 2000; Nelson et al., 2000).
Figure 8.
LKP2 Is Expressed in a Wide Variety of Tissues.
Gene-specific primers were designed and RT-PCR was used to detect LKP2 transcripts in total RNA isolated from a variety of tissues, as indicated.
Overexpression of LKP2 Results in the Arrhythmic Expression of Clock-Associated Factors
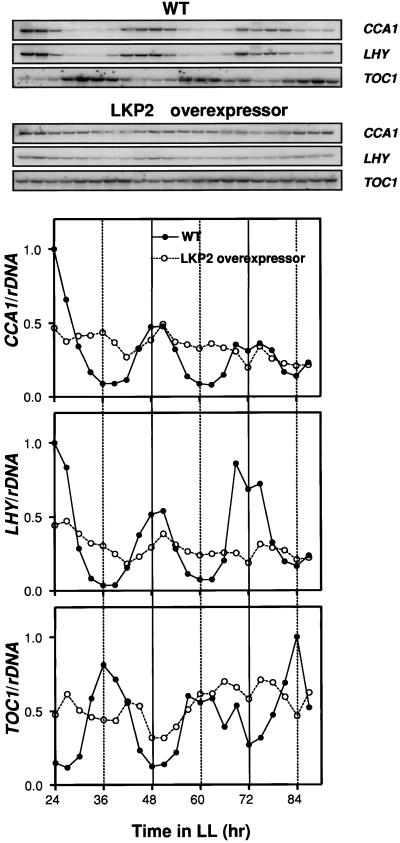
Two related MYB-like transcription factors (CCA1 and LHY) have been ascribed clock functions based on the arrhythmic phenotypes resulting from their overexpression. TOC1 is a putative clock component isolated from a genetic screen for clock mutants. One possible explanation for the arrhythmic phenotypes observed in LKP2-OX seedlings is that these transcription factors become overexpressed as a result of the degradation of a transcriptional repressor or activator. This hypothesis was tested by determining the expression levels of CCA1, LHY, and TOC1 during a 3-day RNA time course in continuous light (Figure 9). As reported previously, the RNA levels for each gene are circadian clock-regulated in wild-type plants, with peak expression occurring at subjective dawn for CCA1/LHY and at subjective dusk for TOC1. In LKP2-OX plants, the expression of all three genes was clearly altered, exhibiting arrhythmic expression patterns at intermediate levels compared with peak and trough levels for wild-type plants. These results do not support the hypothesis that LKP2 targets a direct repressor or activator for CCA1, LHY, or TOC1.
Figure 9.
Arrhythmic Expression of CCA1, LHY, and TOC1 Transcript Levels in LKP2-OX Plants.
RNA gel blot analysis was performed on total RNA isolated from wild-type (WT) and LKP2-OX plants during 3 days in LL. Seed were germinated under LD (12 hr/12 hr) for 14 days and then transferred to LL at time 0. Seedlings were harvested at the times indicated, and total RNA was extracted and probed for CCA1, LHY (5 μg/lane), and TOC1 (10 μg/lane) transcript levels.
DISCUSSION
ZTL Gene Family Consists of Three Members
LKP2 belongs to a small gene family in Arabidopsis consisting of three members, ZTL (for ZEITLUPE), FKF1, and LKP2. The ZTL gene was identified from a screen for mutants affecting period length in Arabidopsis (Millar et al., 1995), and ZTL is located on the bottom arm of chromosome 5 (P1 clone MSF19). Another group working independently characterized a deletion mutant causing late flowering, and this work resulted in the identification of FKF1, the second member of this gene family (Nelson et al., 2000), which is located on the bottom arm of chromosome 1 (bacterial artificial chromosome clone T23K23). Before publication of the ZTL sequence, Nelson et al. (2000) identified an FKF1-like sequence through homology searches and deposited it into GenBank as FKF1-like protein2 (FKL2). Kiyosue and Wada (2000) and Jarillo et al. (2001) also identified ZTL using homology searches of the databases, and each assigned it a different name (LKP1 and ADO1, respectively). FKL2, LKP1, and ADO1 all correspond to ZTL located on the P1 clone MSF19 from the bottom of chromosome 5. The focus of this article is on the third member of this gene family, which is called LKP2 and is located on the upper arm of chromosome 2 (bacterial artificial chromosome clone F19F24).
Overexpression of LKP2 Results in Arrhythmic Phenotypes for Multiple Circadian Outputs
Seedlings overexpressing LKP2 were constructed and assayed for three circadian phenotypes. LKP2-OX seedlings were arrhythmic for CAB2 and CCR2 expression and for cotyledon movement in LL. Although CAB2 and CCR2 expression became rapidly arrhythmic in LL, rhythms in cotyledon movement persisted for one to two cycles after release into free-run conditions (LL). The one to two cycles observed for cotyledon movement after release into free-run conditions most likely are attributable to remnants of a driven cycle (i.e., the LD conditions used to entrain the seedlings) and indicated that although the LKP2-OX plants are arrhythmic under free-run conditions, they may be able to respond to the light-to-dark transitions under LD. Luciferase activity was monitored for both CAB2::luc and CCR2::luc seedlings during two LD cycles followed by release into constant light, and the LKP2-OX seedlings exhibited a clear diurnal mode of regulation under driven conditions, indicating that they are able to respond to light.
Because CAB2 expression requires light, the utility of the CAB2::luc reporter for assaying circadian clock function has been limited to LL. The development of the CCR2::luc reporter line has provided a very powerful tool in Arabidopsis for assaying clock function under free-run conditions in the dark (Strayer et al., 2000). Overexpression of LKP2 results in the arrhythmic expression of CCR2::luc in DD (Figure 4). This is an important observation because it indicates that the circadian phenotype of LKP2-OX plants is not light dependent, which according to classic criteria places LKP2 either in or very near the clock. This conclusion is in contrast to the results obtained from the fluence rate response curves for the ztl mutant (Somers et al., 2000), which indicated that ZTL functions in the light input pathway to the clock. Because CAB2::luc was used for the fluence rate response curves for ztl, DD period length was not included; it will be interesting to determine the DD period length phenotype for CCR2::luc in the ztl mutant background. If the period length of the ztl mutant remains long in DD, then its role in the circadian system is not limited to light input and our model must include clock function as well. The arrhythmic phenotype of LKP2-OX plants in DD suggests that members of this gene family occupy a position very close to the central oscillator.
LKP2-OX Plants Exhibit Long Hypocotyls and Late Flowering Phenotypes
Two phenotypes commonly associated with the circadian clock in Arabidopsis are changes in hypocotyl length and alterations in flowering time. CCA1 and LHY are two MYB-like transcription factors closely associated with the circadian clock in Arabidopsis that when overexpressed cause arrhythmia as well as long hypocotyls and delayed flowering (Schaffer et al., 1998; Wang and Tobin, 1998). ELF3 and GI are two other proteins known to be associated with the clock in Arabidopsis and to have effects on both hypocotyl length and flowering time (Zagotta et al., 1996; Huq et al., 2000). Therefore, it is not surprising that members of the ZTL gene family show similar phenotypes when they are either mutated or expressed ectopically. The LKP2-OX seedlings are presumably gain-of-function, and this results in long hypocotyls in both red and blue light (Figure 5). This observation is consistent with the short hypocotyl phenotypes in both red and blue light exhibited by the deletion of FKF1 (Nelson et al., 2000). It is interesting that ztl mutants, which exhibit a semidominant phenotype resulting from point mutations in a conserved amino acid within the kelch domain, showed short hypocotyls in red light but not blue light (Somers et al., 2000). It has been shown that Arabidopsis exhibits a circadian rhythm in hypocotyl elongation (Dowson-Day and Millar, 1999), and this observation could explain the connection between hypocotyl length and circadian phenotypes.
Arabidopsis is a facultative long-day plant, meaning that it flowers more quickly under long days as opposed to short days, and the ZTL gene family clearly plays a role in the photoperiodic control of flowering time. fkf1 resulted from a deletion and was identified based on its late flowering phenotype when grown under long days; the ztl mutant has a similar late flowering phenotype (Nelson et al., 2000; Somers et al., 2000). These phenotypes are very similar to the gain-of-function phenotype exhibited by LKP2-OX plants, which also flower late (Figure 6). Each of these lines appears to be aphotoperiodic, a phenomenon that is known to be controlled by the circadian clock in Arabidopsis. How both loss-of-function and gain-of-function alleles of different members of the ZTL gene family cause late flowering is not clear, although a similar situation exists for PhyB, in which both overexpressors and null plants exhibit early flowering phenotypes (Bagnall et al., 1995). Further analysis of the effect ZTL family members have on flowering time will determine the nature of the interaction between the circadian clock, hypocotyl length, and flowering time.
ZTL Gene Family Mode of Action
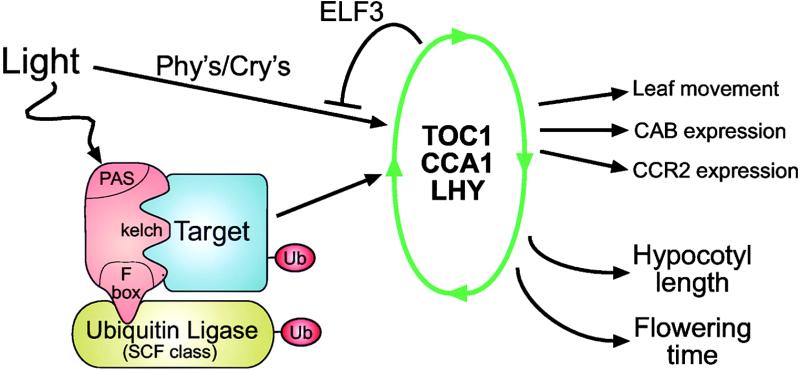
Members of the ZTL gene family possess a unique combination of protein motifs—a PAS domain, an F-box region, and kelch repeats—and this combination strongly suggests a model for their mode of action (Figure 10). The F-box motif in the ZTL protein family indicates a role in protein turnover. The stability of clock components is predicted to have a strong effect on the pace of the clock, so it is not surprising that a family of F-box proteins was found that can affect clock function. The function of the F-box motif is to interact with SKP proteins, which are a component of the SCF (for SKP, Cullin/Cdc53, F-box) class of ubiquitin ligases. This interaction then targets other proteins for ubiquitination and subsequent degradation. Components of the SCF class of ubiquitin ligases have been described in Arabidopsis, with the genome containing a staggering number (>500) of F-box proteins, >20 SKP-like proteins, and six Cullin-like proteins (http://smart.embl-heidelberg.de/; Schultz et al., 2000). Beyond sequence analysis of the Arabidopsis genome, the SCF class of ubiquitin ligases has been implicated functionally in auxin signal transduction (Gray and Estelle, 2000), jasmonic acid signal transduction (Xie et al., 1998), flower development (Samach et al., 1999), and the circadian clock (Nelson et al., 2000; Somers et al., 2000). Given the number of F-box sequences in the Arabidopsis genome, functional roles for this family of proteins undoubtedly will continue to grow.
Figure 10.
Model for ZTL Family Member Mode of Action.
Members of the ZTL gene family containing PAS, F-box, and kelch domains are represented in orange, and ubiquitin (Ub) is represented in red. See text for further details. Phy's, phytochromes; Cry's, cryptochromes.
The model presented in Figure 10 predicts that the “targets” of the ZTL protein family will be dictated by protein–protein interactions with the kelch domain, a known function of this protein motif (Adams et al., 2000). Although the combination of a kelch domain and an F-box is unique to plants, kelch domains are known to form a β-propeller structure that is very similar to the structure formed by WD40 repeats, a motif found at the C terminus of both yeast and mammalian F-box proteins (Patton et al., 1998; Winston et al., 1999), indicating that this structural organization may be conserved. Given the effects ZTL and LKP2 have on the circadian clock, it is likely that their targets are directly responsible for the changes observed in the circadian oscillator. The correlation between hypocotyl length, flowering time, and circadian clock phenotypes found in other mutants also may indicate that these same targets are responsible for all of these phenotypes. Although our analysis of LKP2 phenotypes is limited at present to overexpression studies, the striking phenotypes associated with loss-of-function ZTL and FKF1 strongly suggest that members of this gene family have distinct targets. Identification of the proteins that interact with these kelch domains and determination of their role in the circadian clock, hypocotyl length, and flowering time are under way.
The presence of a PAS domain in the ZTL gene family and the light dependence of the period length phenotype of ztl mutants suggest that these proteins may be responsive to light. PAS domains can be divided functionally into two broad categories: those that mediate small molecule (cofactor) binding and those that mediate heterotypic protein–protein interactions (Gu et al., 2000). The PAS domain in the sequence databases most closely related to the ZTL family members is the LOV domain (a known PAS sequence) in the phototropin protein of Arabidopsis, which functions as a blue light photoreceptor mediating phototropism (Christie et al., 1998). This PAS domain has been shown biochemically to bind flavin mononucleotide (FMN), a known cofactor for blue light photoreceptors (Christie et al., 1999; Salomon et al., 2000). The conservation of amino acids responsible for FMN binding in phototropin (Salomon et al., 2000) and the PAS domains of the ZTL family members strongly suggests that the ZTL family of proteins binds FMN and may act as blue light photoreceptors. A second possible function for the PAS domain in light signaling is to mediate protein–protein interactions. The red light photoreceptors in plants known as phytochromes also possess PAS domains and may mediate dimerization with other photoreceptors; this dimerization could serve to regulate the activity of the ZTL family of proteins in a light-dependent manner. The observation that both ztl mutants and LKP2-OX plants exhibit phenotypes in red and blue light predicts that both direct binding of FMN and heterodimerization with photoreceptors may occur. Regardless of the mechanism, our model predicts that the PAS domain senses light, either directly or indirectly, which then is used to modulate the activity of the ZTL protein family.
From this model, three possible mechanisms emerge for how light could modulate the activity of the ZTL family of proteins: it could alter the interaction of the F-box with the SCF ubiquitin ligase complex, it could alter the interaction of the kelch domain with its targets, or it could alter the stability of the F-box protein itself. A survey of the literature indicates precedence for at least two of these models. In all cases examined to date, the binding of F-box proteins to their targets has been shown to be dependent on the phosphorylation state of the target (Feldman et al., 1997; Skowyra et al., 1997). In Arabidopsis, Drosophila, and mammals, phosphorylation has been implicated in clock function (Price et al., 1998; Sugano et al., 1999; Lowrey et al., 2000), and it is easy to imagine how light-dependent phosphorylation of targets could confer light sensitivity to the system. Although this mechanism may operate for ZTL family members, it fails to explain the presence of a PAS domain, because the systems that require phosphorylated substrates do not use PAS domains. A model that could explain the presence of a PAS domain is light control over the stability of the F-box proteins themselves. In this model, the F-box protein would be unstable in the light and therefore unable to mediate ubiquitination of its targets, but in the dark the protein would become stable and increase the turnover of its targets. The observations that F-box proteins can be ubiquitinated themselves, possibly via an autocatalytic mechanism, and that their instability provides a control point for their activity (Galan and Peter, 1999; Smothers et al., 2000) provide support for this idea. Ectopic expression of LKP2 would be able to overcome light-dependent degradation, resulting in circadian phenotypes in both light and dark. The observation that period length is light dependent in the semidominant ztl mutants (Somers et al., 2000) also is consistent with this idea; at lower fluences, there would be more mutant protein present, which causes period lengthening, and at higher fluences, ZTL would become unstable, causing period length shortening. This model fits the phenotypes and is being tested by comparing protein stability in the light and in the dark.
In nature, organisms experience a LD cycle corresponding to day and night, and the main function of a circadian clock is to allow the organism to maintain a stable phase relationship with the LD cycle it encounters. Incorporating this concept into the model, we propose that members of the ZTL family probably act as “light switches” that would help to maintain the phase relationship between the circadian clock and the LD cycle. Targets of the ZTL family would be degraded in the dark and stable in the light, and at the transition between night and day there would be a switch from being a stable protein to being degraded rapidly. Using this type of light switch mechanism, the plant could transmit light information from the environment directly to the circadian system, maintaining a stable phase relationship between the environment and the clock. The major goal arising from this model is the identification of target proteins. A number of potential targets exist, and these are outlined in Figure 10. Photoreceptors functioning in the input pathway of the clock have been shown to affect clock function but are unlikely targets because a quadruple photoreceptor mutant was still rhythmic (Yanovsky et al., 2000). The transcription factors CCA1, LHY, and TOC1 are more likely candidates because they have been shown to affect clock function, although novel targets with clock function also may exist. These targets presumably are directly responsible for the phenotypes associated with this gene family and very likely play a role either within or very close to the central oscillator.
METHODS
Plant Growth Conditions and Rhythm Assays
Growth conditions and transformations for Arabidopsis thaliana were as described previously (Kiyosue and Wada, 2000; Somers et al., 2000). The LKP2-OX construct was introduced into the CAB2::LUC and CCR2::LUC reporter lines by crossing T2 plants containing the LKP2 overexpression construct to the reporter lines; luminescent F1 seedlings exhibiting long hypocotyls were selected for the luciferase imaging assays. Luciferase imaging for the white light and light/dark experiments was performed as described previously (Somers et al., 1998a). Imaging for the continuous dark experiments was performed using a Night Owl CCD Camera System (EGG Berthold, Berthold Technologies, Bad Wildbad, Germany), and images were processed with WinLight32 software (Berthold Technologies). The leaf movement rhythm assay was performed as described previously (Hicks et al., 1996; Somers et al., 2000).
Hypocotyl Length Assays and Flowering Time
Seed were sown on Petri plates containing 0.8% agar without sucrose. All incubations were performed at 23°C. Seed were stratified in the dark at 4°C for 3 days, given a 2-hr white light pulse to stimulate germination, transferred back to dark for 24 hr, and then transferred to specific light conditions for 3 days. Hypocotyl lengths were measured and recorded by hand using a ruler. Flowering time assays were performed as described previously (Kiyosue and Wada, 2000).
DNA Constructs and Reverse Transcriptase–Mediated Polymerase Chain Reaction
A 4.26-kb polymerase chain reaction (PCR) fragment encompassing LKP2 was generated from genomic DNA using the primers 5′-CATAAGCAAATCAATGACTAAAGAGAGTAG-3′ and 5′-GGAGACTTCGATTACCTACAGATATCAGAT-3′. This genomic PCR fragment corresponds to 44,850 to 49,110 kb of bacterial artificial chromosome F19F24 from Arabidopsis. This PCR fragment was used to screen a cDNA library as described previously (Kiyosue and Wada, 2000), and four cDNAs encoding LKP2 were isolated and sequenced. A PCR fragment corresponding to the full-length coding sequence of the cDNA was generated using the primers 5′-GAG-AGGATCCGTATGCAAAATCAAATGGAG-3′ and 5′-TCTCGGATCCGATCAAGTACTTGCAGTGGT-3′ and was cloned into the BamHI sites of the 35S expression vector pBE2113 (Mitsuhara et al., 1996). This LKP2 overexpression construct was transformed into Arabidopsis using an Agrobacterium tumefaciens–mediated transformation protocol described previously (Clough and Bent, 1998). A 1.0-kb PCR fragment encompassing the LKP2 promoter was generated using the primers 5′-GAGAGAGGATCCGCAGAGCAAAGACAAAACCGC-3′ and 5′-CTCTCTCAAGCTTCCCACAAAATAAAGCTTAAGCCC-3′ and was subcloned into the BamHI–HindIII sites of the luciferase expression cassette pZPXomegaLuc+. A 1.0-kb PCR fragment of the FKF1 promoter was subcloned similarly using the primers 5′-GAGAGGATCCCCAAAAGGGTTAGTTTTCTTGTGT-3′ and 5′-AGAGAGAAGCTTTTTCTCGCTTACAGAAATATC-3′.
Total RNA was isolated using a phenol/SDS extraction protocol (Kiyosue et al., 1992), and 2 μg was used as a template for Ready-to-Go Reverse Transcriptase PCR Beads (Amersham Pharmacia Biotech). The primers used to amplify the LKP2 cDNA were 5′-GTATGCAAAATCAAATGGAG-3′ and 5′-ATGGCCCTCTACATTGCAA-3′, which correspond to the first and second exons, respectively. LHY was amplified using the primers 5′-AATGCAACTACTGATTCGTG-3′ and 5′-GAG-ACAAGACATGGGGTAAT-3′, CCA1 was amplified using the primers 5′-TGGAAGTCTGTGTCTGACGA-3′ and 5′-AAGATTAAACAGTTTATGAT-3′, and CAB2 was amplified using the primers 5′-GGTTCACGCTCAGAGCATTTTGGCCATTTG-3′ and 5′-GAATTCAAAAGATTGAAAGAAAAGTAAATT-3′. Quantum RNA 18S Internal Standard Primers (Ambion, Austin, TX) were used to amplify the 18S rRNA controls. Reverse transcriptase–mediated PCR products were electrophoresed on agarose gels and visualized by staining with ethidium bromide.
Accession Numbers
The GenBank accession numbers for the sequences described in this article are AB038797 (LKP2) and AF216525 (FKL2).
Acknowledgments
We are grateful to Stacey Harmer and Paloma Mas for critical reading of the manuscript. This work was supported by National Institutes of Health Grant No. GM56006 to S.A.K., a grant from the Program for the Promotion of Basic Research Activities for Innovative Bioscience to M.W., a National Science Foundation Postdoctoral Fellowship in Biosciences Related to the Environment (DBI-9804249) to T.F.S., a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Sumitomo Foundation to T.K., and grants from the National Council for Scientific and Technology Research (Conicet), Antorchas Foundation, and the University of Buenos Aires to M.Y.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010332.
References
- Adams, J., Kelso, R., and Cooley, L. (2000). The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 10, 17–24. [DOI] [PubMed] [Google Scholar]
- Anderson, S.L., Somers, D.E., Millar, A.J., Hanson, K., Chory, J., and Kay, S.A. (1997). Multiple genetically defined phototransduction pathways interact with circadian clock signals to regulate CAB2 transcription. Plant Cell 9, 1727–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall, D.J., King, R.W., Whitelam, G.C., Boylan, M.T., Wagner, D., and Quail, P.H. (1995). Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 108, 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Salomon, M., Nozue, K., Wada, M., and Briggs, W.R. (1999). LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96, 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Covington, M.F., Panda, S., Strayer, C.A., Kay, S.A., and Wagner, D.R. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13, 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17, 63–71. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Feldman, R.M., Correll, C.C., Kaplan, K.B., and Deshaies, R.J. (1997). A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock–controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.M., and Peter, M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96, 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, I. (2000). Function of the ubiquitin–proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Z., Hogenesch, J.B., and Bradfield, C.A. (2000). The PAS superfamily: Sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40, 519–561. [DOI] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carré, I.A., Somers, D.E., Straume, M., Meeks-Wagner, R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274, 790–792. [DOI] [PubMed] [Google Scholar]
- Huq, E., Tepperman, J.M., and Quail, P.H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97, 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo, J.A., Capel, J., Tang, R.H., Yang, H.Q., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487–490. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., and Wada, M. (2000). LKP1 (LOV kelch protein 1): A factor involved in the regulation of flowering time in Arabidopsis. Plant J. 23, 807–815. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., Shinozaki, K., Higashi, K., Satoh, S., Kamada, H., and Harada, H. (1992). Isolation and characterization of a cDNA that encodes ECP31, an embryogenic-cell protein from carrot. Plant Mol. Biol. 19, 239–249. [DOI] [PubMed] [Google Scholar]
- Kume, K., Zylka, M.J., Sriram, S., Shearman, L.P., Weaver, D.R., Jin, X., Maywood, E.S., Hastings, M.H., and Reppert, S.M. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Lowrey, P.L., Shimomura, K., Antoch, M.P., Yamazaki, S., Zemenides, P.D., Ralph, M.R., Menaker, M., and Takahashi, J.S. (2000). Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408, 716–720. [DOI] [PubMed] [Google Scholar]
- Merrow, M., Brunner, M., and Roenneberg, T. (1999). Assignment of circadian function for the Neurospora clock gene frequency. Nature 399, 584–586. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carré, I.A., Strayer, C.A., Chua, N.-H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mitsuhara, I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37, 49–59. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y., Andersson, C.R., Kondo, T., Golden, S.S., and Johnson, C.H. (1998). Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 95, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Price, J.L., Blau, J., Rothenfluh, A., Abodeely, M., Kloss, B., and Young, M.W. (1998). double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Salomon, M., Christie, J.M., Knieb, E., Lempert, U., and Briggs, W.R. (2000). Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39, 9401–9410. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. (2000). SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Smothers, D.B., Kozubowski, L., Dixon, C., Goebl, M.G., and Mathias, N. (2000). The abundance of Met30p limits SCF(Met30p) complex activity and is regulated by methionine availability. Mol. Cell. Biol. 20, 7845–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998. a). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A., Pearson, M., and Kay, S.A. (1998. b). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Strayer, C.A., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Sugano, S., Andronis, C., Ong, M.S., Green, R.M., and Tobin, E.M. (1999). The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 12362–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Winston, J.T., Koepp, D.M., Zhu, C., Elledge, S.J., and Harper, J.W. (1999). A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Mazzella, M.A., and Casal, J.J. (2000). A quadruple photoreceptor mutant still keeps track of time. Curr. Biol. 10, 1013–1015. [DOI] [PubMed] [Google Scholar]
- Zagotta, M.T., Shannon, S., Jacobs, C., and Meeks-Wagner, D.R. (1992). Early-flowering mutants of Arabidopsis thaliana. Aust. J. Plant Physiol. 19, 411–418. [Google Scholar]
- Zagotta, M.T., Hicks, K.A., Jacobs, C.I., Young, J.C., Hangarter, R.P., and Meeks-Wagner, D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10, 691–702. [DOI] [PubMed] [Google Scholar]