Abstract
In this study, DNA microarray analysis was used to expand our understanding of the dst1 mutant of Arabidopsis. The dst (downstream) mutants were isolated originally as specifically increasing the steady state level and the half-life of DST-containing transcripts. As such, txhey offer a unique opportunity to study rapid sequence-specific mRNA decay pathways in eukaryotes. These mutants show a threefold to fourfold increase in mRNA abundance for two transgenes and an endogenous gene, all containing DST elements, when examined by RNA gel blot analysis; however, they show no visible aberrant phenotype. Here, we use DNA microarrays to identify genes with altered expression levels in dst1 compared with the parental plants. In addition to verifying the increase in the transgene mRNA levels, which were used to isolate these mutants, we were able to identify new genes with altered mRNA abundance in dst1. RNA gel blot analysis confirmed the microarray data for all genes tested and also was used to catalog the first molecular differences in gene expression between the dst1 and dst2 mutants. These differences revealed previously unknown molecular phenotypes for the dst mutants that will be helpful in future analyses. Cluster analysis of genes altered in dst1 revealed new coexpression patterns that prompt new hypotheses regarding the nature of the dst1 mutation and a possible role of the DST-mediated mRNA decay pathway in plants.
INTRODUCTION
Cells must be able to adjust their gene expression patterns quickly to respond to intracellular and extracellular stimuli; therefore, it is necessary for certain transcripts to reach new steady state levels rapidly. The steady state levels of eukaryotic mRNAs are determined by both their rate of synthesis and their rate of degradation. The control of mRNA stability is a major determinant of steady state mRNA levels in the cell and often has a great impact on the level at which a particular gene is expressed. Furthermore, mRNA stability affects the rate at which new steady state RNA levels are achieved after changes in transcription; the more unstable the mRNA, the more rapidly it reaches a new steady state (Abler and Green, 1996).
Most recent studies on mRNA stability in eukaryotes have focused on transcripts that are relatively unstable, generally with half-lives of <60 min (Ross, 1995; Johnson et al., 1998). Unstable transcripts are particularly interesting because they often correspond to genes that must be controlled rapidly or stringently, such as those involved in regulating cell growth and differentiation. Transcripts that fall into this category include phytochrome mRNA (Higgs et al., 1995) and several auxin-induced transcripts (McClure and Guilfoyle, 1989) in plants, mating-type transcripts in yeast (Peltz and Jacobson, 1992), and several proto oncogene transcripts in mammalian cells (Greenberg and Belasco, 1993).
The majority of mRNAs fall in the stable range of mRNA half-lives for a given organism (Taylor and Green, 1995; Ross, 1996). Thus, within the body of unstable mRNAs, specific sequence motifs must be present that can either act constitutively to establish the inherent instability of a particular transcript or modulate the stability of an mRNA in response to certain physiological, developmental, or environmental cues.
The most widely studied instability determinants are the mammalian AU-rich elements (AREs). AREs are found in the 3′ untranslated region (UTR) of several of the most unstable mammalian transcripts, such as lymphokine, cytokine, and protooncogene mRNAs (Chen and Shyu, 1995). Repeats of the pentamer AUUUA often are found in these AU-rich elements and are important for their instability function (Shyu et al., 1991; Vakalopoulou et al., 1991). AUUUA repeats are likely to be of broad significance in higher eukaryotes because they can target transcripts for rapid decay in plants (Ohme-Takagi et al., 1993), and recent evidence suggests that the ARE-mediated decay pathway is functional in yeast as well (Vasudevan and Peltz, 2001). It seems that all functional AREs mediate deadenylation as the first step in mRNA decay, although different classes of AREs exhibit different reaction kinetics (Chen and Shyu, 1995).
In plants, in addition to AREs, an instability determinant called DST (downstream) (Newman et al., 1993) has been studied in detail. The DST element was identified first as a highly conserved sequence in the 3′ UTRs of the soybean SAUR (for small-auxin-up-RNA) genes (McClure et al., 1989). These genes encode unstable transcripts whose half-lives have been estimated to be on the order of 10 to 50 min (McClure and Guilfoyle, 1987; Franco et al., 1990). Although the function of the SAUR proteins is unknown, the temporal and spatial expression of SAUR genes correlates with auxin-induced cell elongation (McClure and Guilfoyle, 1989). The prototype DST, from SAUR15A of soybean, is ∼45 bp in length and consists of three highly conserved subdomains separated by two variable regions (Newman et al., 1993). Mutagenesis studies have demonstrated that residues within two of the conserved subdomains, the ATAGAT and GTA regions, are necessary for the instability function (Sullivan and Green, 1996).
To gain insights into the cellular components involved in the DST-mediated mRNA degradation pathway, we isolated Arabidopsis mutants defective in their ability to recognize DST elements. dst1 and dst2 were isolated based on their ability to stabilize specific DST-containing transgene mRNAs (Johnson et al., 2000). For this strategy, two transgenes, hygromycin phosphotransferase (HPH) and β-glucuronidase (GUS), both containing a tetramer of the consensus DST element in the 3′ UTR, were introduced into Arabidopsis as selectable and screenable markers, respectively. The presence of the DST elements decreased the mRNA levels for both transgenes, HPH-DST and GUS-DST, resulting in decreased resistance to hygromycin and low GUS activity. The dst mutants were isolated as hygromycin-resistant plants with increased GUS activity because of stabilization of the corresponding mRNAs. In addition, dst mutants have increased levels of SAUR-AC1 mRNA, the only endogenous DST-containing unstable transcript characterized to date (Gil and Green, 1996; Johnson et al., 2000). The only reported phenotype of the dst mutants is the increase of these three mRNAs as a result of increased mRNA stability. Genetic analysis of two dst mutants isolated via this selection showed that they are incompletely dominant and represent two independent loci. The dst mutants exhibit no obvious morphological or developmental phenotype (Johnson et al., 2000). The main objective of the present work was to search for additional genes that were regulated, directly or indirectly, by this pathway.
Genes regulated differentially in mutant backgrounds have been identified in the past using a variety of techniques, such as differential display and subtractive hybridization (Kehoe et al., 1999). Now, with the application of plant genomics, monitoring of gene expression can be achieved on a scale much greater than was possible previously. DNA microarray technology (Schena et al., 1995) takes advantage of the vast amount of information generated by the genome-sequencing projects as well as the large number of expressed sequence tag (EST) clones isolated and sequenced from different organisms. In addition to dramatic changes in mRNA levels that can be identified by differential display or other techniques, DNA microarrays allow the detection of more subtle changes in gene expression (Kehoe et al., 1999). By comparing global patterns of gene expression in a mutant with those in the wild type, we now have the unique opportunity to identify changes in biochemical processes caused by the mutation. This approach may be especially fruitful when studying a mutation that affects the regulation of mRNA abundance and may be of particular interest when the mutation does not generate an obvious morphological phenotype.
Recently published studies have demonstrated how microarrays containing a large number of Arabidopsis genes can provide a powerful tool for plant gene discovery, functional analysis, and elucidation of genetic regulatory networks (Schenk et al., 2000; Schaffer et al., 2001; Seki et al., 2001). To date, nearly all reported plant microarray experiments have focused on gene expression in wild-type plants; there has been little work published on mutant analyses. One exception, which highlighted the promise of this approach, is a microarray study performed recently by Reymond et al. (2000) in which the coronatine-insensitive coi1-1 Arabidopsis mutant was analyzed on DNA microarrays, resulting in the classification of a large number of COI1-dependent and COI1-independent wound-inducible genes.
In this report, we describe the use of DNA microarrays containing >11,000 Arabidopsis ESTs, representing ∼7800 unique genes, to examine gene expression in the DST-mediated mRNA degradation mutant dst1. Our results indicate that DNA microarrays are a powerful tool to identify new molecular markers affected by DST-mediated mRNA decay, some of which likely are direct targets of the pathway. Furthermore, our results suggest new experimental directions in which to pursue the biological significance of the pathway.
RESULTS
Generation of a 600-Element DNA Microarray
As a first step toward analyzing the dst1 mutant for additional changes in gene expression, a DNA microarray representing ∼600 Arabidopsis genes was assembled. The clones included were mainly ESTs from the Michigan State University (MSU) collection (Newman et al., 1994). Table 1 includes a description of these genes, grouped according to function or sequence similarity. Because the dst1 mutant affects RNA metabolism, half of the clones included in the microarray were those predicted to be associated with some aspect of RNA metabolism. The criterion for the selection of many of these EST clones was their sequence similarity to potential RNA metabolism genes described previously in Arabidopsis or other organisms.
Table 1.
Genes Included in the 600-Element DNA Microarray
| Category | Number of Genes |
|---|---|
| Pathogen related | 70 |
| Lipid metabolism | 40 |
| Wounding/jasmonic response | 25 |
| Mitogen-activated protein kinase | 25 |
| Protein transport vacuole/chloroplast | 30 |
| Senescence associated | 10 |
| Auxin induced | 10 |
| Polyamine metabolism | 5 |
| RNA metabolism | 300 |
| RNA binding proteins | 70 |
| Helicases | 20 |
| Transcription/translation | 20 |
| RNases | 15 |
| Splicing factors/snoRNP | 15 |
| Nonsense decay | 8 |
| DST-containing genes | 140 |
| AU-rich genes | 10 |
| Other | 20 |
| Controls | 76 |
| Highly expressed/well studied | 17 |
| Ribosomal proteins | 15 |
| rRNA | 2 |
| Transgenes | 5 |
| Human clones | 12 |
| Other | 25 |
To date, the SAUR-AC1 transcript is the only known target for the DST-dependent mRNA degradation pathway in Arabidopsis (Johnson et al., 2000). To identify additional direct targets of this pathway, genes with possible DST elements were included. The strategy to identify genes containing potential DST elements was to combine the results of several pattern searches using different degenerate criteria, because our knowledge of what constitutes a functional DST element is still limited (Newman et al., 1993; Sullivan and Green, 1996). Because AREs also function in plants as mRNA instability elements (Ohme-Takagi et al., 1993), genes with AU-rich elements were selected in a similar manner by searching for variations of the element AUUUA in an AU-rich context in the 3′ UTR. As a positive control, the HPH and GUS coding sequences also were included so that the mutant phenotype of dst1 could be confirmed.
Approximately 300 additional clones were included in the microarray to expand coverage beyond genes associated with RNA metabolism. A complete list of the clones in this array and the raw microarray expression data can be found at http://www.bch.msu.edu/pamgreen/Perez-Amador_etal/600_list.htm.
Expression Levels for Most Genes Are Similar in dst1 and the Parental Plants
The dst1 mutant has a subtle phenotype: it exhibits a threefold to fourfold increase, relative to the parental plants, of HPH-DST and GUS-DST transgene mRNAs, which was the basis for its isolation. Each of these transgenes contains a tetramer of the prototype DST element (Newman et al., 1993; Sullivan and Green, 1996). The dst1 mutant also shows a threefold increase of the unstable SAUR-AC1 mRNA, an endogenous Arabidopsis transcript known to contain a DST element. Before this work, these were the only known dst1 phenotypes; dst1 mutant plants appear normal with respect to morphology and development compared with parental plants (Johnson et al., 2000).
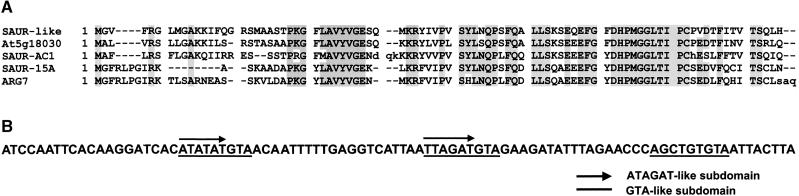
To determine whether additional molecular differences existed, poly(A)+ RNA from leaves of 40-day-old dst1 and parental plants was extracted and used in a DNA microarray experiment. After hybridization and normalization of data, several genes were identified with changes in mRNA levels >1.5-fold. As expected, HPH-DST and GUS-DST were among the genes with increased mRNA levels in dst1. SAUR-AC1 also was increased in the mutant. In addition, a DEAD box RNA helicase RH15 (At5g11200) and EST 125O9T7 were identified as being increased at the mRNA level in the dst1 mutant. When the sequence from EST 125O9T7 was compared with the TAIR database (http://www.arabidopsis.org/), the translation showed high similarity (63.7%) to the SAUR-AC1 protein and several other SAUR-like proteins, as depicted in Figure 1A. As a result, this gene was named SAUR-like1. Most interestingly, although the 3′ UTR of this gene lacks a classic DST element, it contains multiple DST-like subdomains (i.e., two ATAGAT-like and three GTA-like subdomains), as shown in Figure 1B.
Figure 1.
Analysis of EST 12509T7 Identifies It as Savr-like1.
(A) Sequence alignment of SAUR-like1 with several SAUR-like proteins. Boxes indicate amino acid identities in five of five or four of five sequences.
(B) Multiple DST subdomains in the SAUR-like1 3′ UTR. Repeats of the ATAGAT- and GTA-like subdomains are indicated by arrows and bars, respectively. The determination of the 3′ UTR was based on the 5′ and 3′ sequences of ESTs 125O9T7 and 125O9XP, respectively.
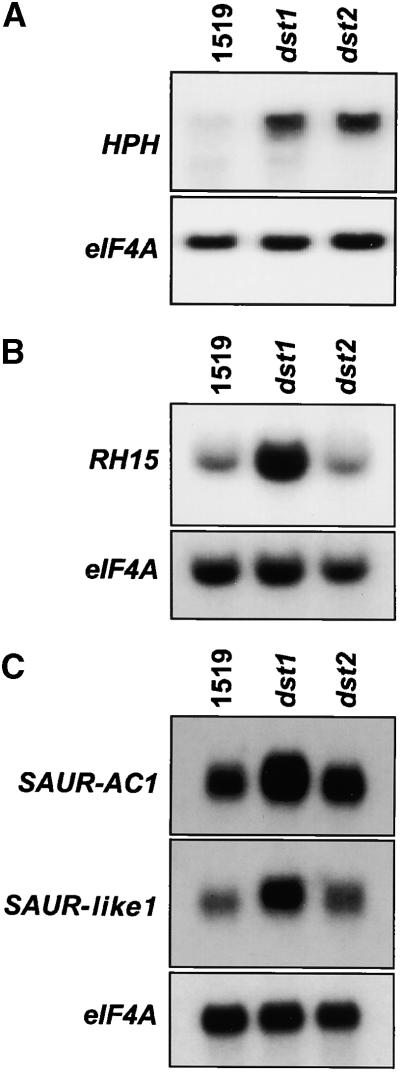
RNA gel blot analysis was used to confirm the differential regulation of these genes identified by DNA microarray analysis. In previous experiments, RNA gel blot analysis had proven an effective method to measure subtle differences in mRNA abundance between dst1 and parental plants (Johnson et al., 2000). In addition, this approach has other advantages. RNA gel blot analysis traditionally has been relied on for accurate analysis of gene expression at the level of mRNA abundance. Moreover, once differentially abundant mRNAs are identified using one combination of mutant and wild type, these mRNAs can be investigated easily in additional mutants. Finally, RNA gel blot analysis provides the means to test the biological reproducibility of data obtained in microarray experiments, because additional samples can be tested readily. Accordingly, the dst2 mutant was included in this analysis to compare the molecular phenotypes between dst1 and dst2. Like dst1, dst2 has no known phenotype other than increased HPH-DST, GUS-DST, and SAUR-AC1 mRNA levels caused by increased stability.
EST clones to be investigated were radiolabeled and used as probes on the RNA gel blots. The translation elongation factor eIF4A, which was used to normalize RNA gel blot data as described previously (Johnson et al., 2000), did not show altered mRNA levels in the dst1 mutant in DNA microarray experiments, as expected. All blots were hybridized first to labeled eIF4A probe, and then they were stripped and hybridized with the individual gene probes. Representative RNA gel blots are shown in Figure 2. HPH and SAUR-AC1 mRNAs increased in abundance in both dst1 and dst2 mutants, as expected (3.5-fold and three-fold, respectively). The mRNA levels for SAUR-like1 also were increased in both mutants, although to different extents (2.2-fold in dst1 and 1.6-fold in dst2). RH15 showed a threefold increase in mRNA level in dst1 but did not show an altered mRNA level in dst2.
Figure 2.
RNA Gel Blot Analysis to Confirm DNA Microarray Data.
Lanes contained 10 μg of total RNA extracted from wild-type (1519), dst1, and dst2 plants. Each blot was hybridized sequentially with 32P-labeled eIF4A and then with HPH (A), RH15 (B), and SAUR-AC1 and SAUR-like1 (C).
11,521-Element Microarray Reveals Additional Genes with Altered Gene Expression in the dst1 Mutant
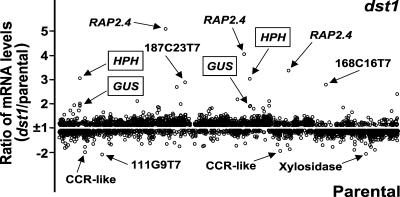
To expand on the 600-element microarray analysis, high-density arrays containing 11,521 clones (prepared at the Arabidopsis Functional Genomics Consortium DNA microarray facility at MSU) were used. Poly(A)+-selected RNA from leaves of 5-week-old dst1 and parental plants was extracted, labeled with Cy3- and Cy5-CTP, and used for the hybridization. Analysis was performed using four independently grown sample sets to control for biological variability. Each sample set consisted of the dst1 mutant and parental plants. For each set, two microarrays were used, each with reverse labeling, for a total of eight microarray slides (raw data are available at http://genome-www4.stanford.edu/cgi-bin/SMD/cluster/QuerySetup.pl, under the experimenter name Green). DNA microarray expression data were normalized as indicated in Methods to calculate the ratios of the fluorescence intensities of the two probes. The number of clones with a ratio >1.5-fold was determined. Although a relatively low cutoff ratio of 1.5 was used, the data were reproducible and were confirmed by RNA gel blot analysis (see below). When the expression data, represented as the median of eight microarray slides, were plotted, 36 clones (31 ESTs and five clones for HPH and GUS) with ≥1.5-fold increased or decreased mRNA levels could be identified (Figure 3). Most of these clones showed a difference of 1.5-fold or greater on at least seven of the eight slides analyzed. Each EST was mapped to the Arabidopsis genome by conducting a BLAST search against the Arabidopsis genome sequence database. Multiple ESTs representing the same gene also were identified in this manner, revealing that the 31 ESTs corresponded to 25 genes. The redundancy among the ESTs present in the array allowed us to verify the microarray data further, because ESTs for the same genes showed similar changes in expression. In some cases, ESTs corresponding to the same gene showed slightly different expression ratios (e.g., the four ESTs corresponding to RAP2.4 ranged from two- to five-fold higher in dst1, with a mean of 3.9-fold); this variability could be attributable to variable probe or target length, genetic redundancy, or a combination thereof. The transgenes HPH and GUS, as well as the DEAD box RNA helicase gene RH15, again were detected with increased mRNA levels in dst1.
Figure 3.
Comparison of mRNA Levels in the dst1 Mutant and Parental Plants Using the 11,521-Element DNA Microarray.
The graph was generated from the data obtained with ScanAlyze software. Ratio values less than 1 were transformed to −1/ratio to plot them as fold difference (y axis) against arbitrary clone numbers (x axis). The five-point scale indicates genes with increased/decreased mRNA levels in the dst1 mutant.
Twenty genes, including the transgenes HPH and GUS, showed increased mRNA levels, whereas seven displayed decreased levels of mRNA in the dst1 mutant compared with those of the parental plants, as listed in Tables 2 and 3, respectively. After correction for redundancy in the ESTs and excluding the transgenes HPH and GUS, 25 endogenous genes whose expression levels were altered in dst1 were identified. The probable biological functions of these genes, listed in Tables 2 and 3, were based on annotation by the Munich Information center for Protein Sequences (MIPS) Arabidopsis thaliana database. The 3′ UTRs of all of the genes identified as being regulated differentially in dst1 were analyzed for the presence of either a classic DST element or DST-like subdomains. Seven genes were identified that contain possible DST-like sequences in their 3′ UTRs (Table 4). Therefore, these genes could be hypothesized to be primary targets of the DST-mediated decay pathway that is deficient in the mutant.
Table 2.
Genes with Increased mRNA Levels in dst1 versus Parental Plants
| EST
|
|||||
|---|---|---|---|---|---|
| Gene | MIPSa Number | Number of Clones | MSU Number | Accession Number | Average ±sd |
| RAP2.4 (AP2 domain) | At1g78080 | 4 | 89I20T7 | T20915 | 3.9 ± 1.1 |
| 207H22T7 | N37644 | ||||
| 172J24T7 | AA712560 | ||||
| 272E2T7 | AA651548 | ||||
| Similar to cadmium induced (AP2 domain) | At1g22190b | 1 | 168C16T7 | R64886 | 3.4 ± 1.9 |
| HPH | — | 2 | — | 3.2 ± 1.1 | |
| Chalcone synthase | At5g13930 | 1 | 187C23T7 | R89978 | 3.0 ± 1.6 |
| Similar to chalcone flavonone isomerase | At5g05270 | 1 | 113I3T7 | T42455 | 2.8 ± 1.3 |
| Putative membrane channel | At2g28900 | 1 | 141P19T7 | T46371 | 2.4 ± 0.7 |
| Unknown | At3g56360 | 1 | 180E15T7 | H36869 | 2.2 ± 0.6 |
| Putative transcription factor | At1g56650 | 1 | 99P9T7 | T22223 | 2.1 ± 0.5 |
| Calmodulin-like | At2g41410 | 1 | F2H12T7 | AA713007 | 2.2 ± 0.8 |
| GUS | — | 2 | — | 2.0 ± 0.4 | |
| Heat shock protein 90 | At5g56010 | 1 | 204P1T7 | H76891 | 2.2 ± 1.3 |
| CCR4-associated factor1–like | At3g44260 | 1 | 222C9T7 | N38276 | 2.1 ± 0.9 |
| Unknown | At5g09850a | 1 | 120E6T7 | T43948 | 1.9 ± 0.5 |
| Unknown | At2g27830 | 1 | E8D5T7 | AA042743 | 1.9 ± 0.4 |
| Unknown | Atg68490 | 1 | 209G16T7 | N37313 | 1.9 ± 0.3 |
| Unknown | 1 | 222M14T7 | N38314 | 1.8 ± 0.5 | |
| Unknown | At3g28270a | 1 | 139I3T7 | T46268 | 1.8 ± 0.4 |
| Similar to APR2 | At1g62180 | 1 | 73F9T7 | T45380 | 1.8 ± 0.6 |
| Unknown | At3g26000 | 1 | 281D7T7 | AA650744 | 1.6 ± 0.2 |
| DEAD box RNA helicase RH15 | At5g11200 | 1 | 91P21T7 | T21481 | 1.6 ± 0.2 |
Munich Information Center for Protein Sequences.
EST sequence aligns upstream of the ATG codon of the deduced open reading frame of the indicated gene.
Table 3.
Genes with Decreased mRNA Levels in dst1 versus Parental Plants
| EST
|
|||||
|---|---|---|---|---|---|
| Gene | MIPSa Number | Number of Clones | MSU Number | Accession Number | Average ±sd |
| Putative patatin protein | At2g26560 | 1 | 111G9T7 | T42260 | 2.5 ± 1.0 |
| Senescence-associated protein | At4g35770 | 1 | 170N19T7 | R65416 | 2.0 ± 0.4 |
| Similar to stem-specific protein | At4g27450 | 1 | 201O1T7 | H76730 | 1.9 ± 0.4 |
| Xylosidase | At5g49360 | 2 | H5A10T7 | W43767 | 1.9 ± 0.5 |
| G1G9T7 | N96119 | ||||
| STP1 glucose transporter | At1g11260 | 1 | 171P10T7 | R65119 | 1.9 ± 0.6 |
| Similar to CCR protein | At3g26740 | 3 | E6E1T7 | AA042331 | 1.8 ± 0.3 |
| 245H16T7 | N97142 | ||||
| 210A12T7 | N37331 | ||||
| Unknown | At5g45510b | 1 | G4A11T7 | N96240 | 1.8 ± 0.5 |
Munich Information Center for Protein Sequences.
EST sequence aligns upstream of the ATG codon of the deduced open reading frame of the indicated gene.
Table 4.
Genes with Possible DST-like Sequences in Their 3′ UTRs
| Gene | 3′ UTRa |
|---|---|
| RAP2.4 (AP2 domain) | |
| CAF1-like | |
| Similar to APR2 | |
| Putative patatin protein | |
| Senescence-associated protein | |
| Xylosidase | |
| Similar to CCR protein |
Boxes indicate classic DST elements; arrows indicate ATAGAT-like subdomains; bars indicate GTA-like subdomains.
RNA Gel Blot Analysis Confirms DNA Microarray Data
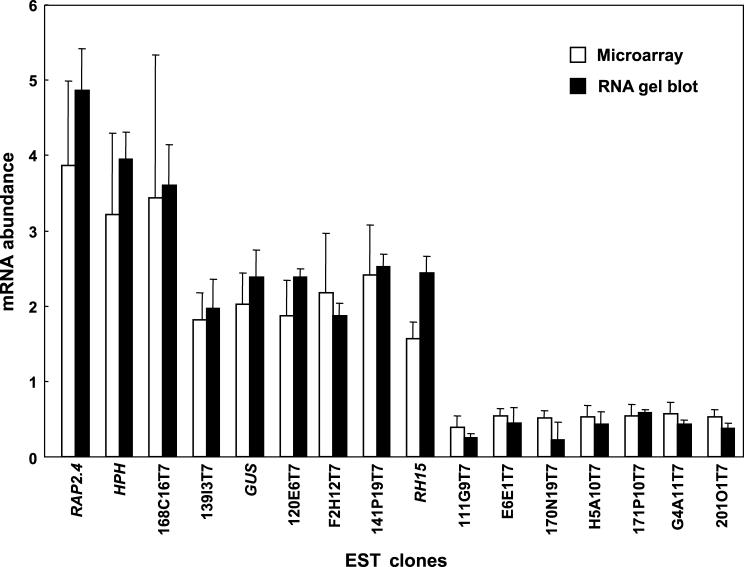
To determine whether the transcript changes identified in the dst1 mutant by microarray analysis were reliable, total RNA was obtained from the same plants that were used for each of the four microarray experiments and examined by RNA gel blot analysis. Several genes exhibiting altered mRNA levels in the dst1 mutant as determined from analysis of the 11,521 microarrays were tested. All of the genes exhibited increased or decreased mRNA abundance, as expected. Although mRNA changes were relatively small (1.5- to 3.9-fold), in all cases, differences in gene expression detected by the microarray translated into similar fold differences as determined by RNA gel blot analysis (Figure 4).
Figure 4.
Histogram Plot Comparing the Gene Expression Values Obtained Using Microarray Analysis and RNA Gel Blot Analysis.
The x axis denotes the various EST clones tested, and the y axis represents the fold difference for mRNA levels for each clone.
Error bars indicate ±sd.
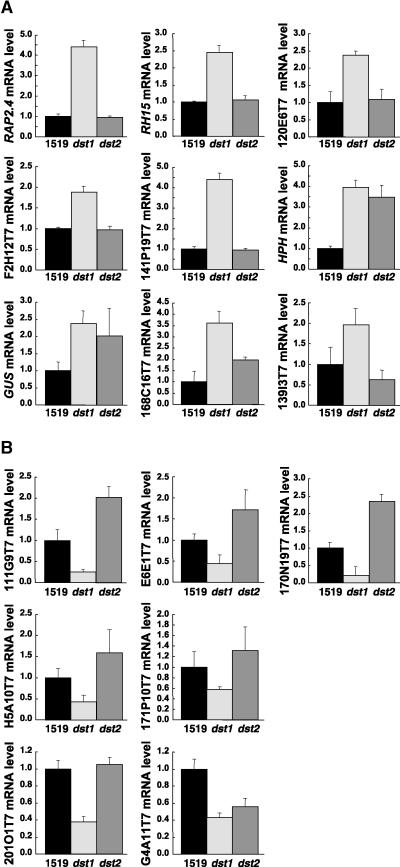
Again, RNA levels in dst2 also were monitored in these experiments. Using this approach, molecular markers specific for each dst mutant were identified. Most of the genes that had increased mRNA abundance in the dst1 mutant were unaffected in the dst2 mutant. As demonstrated in Figure 5A, RAP2.4, RH15, and ESTs 120E6T7, F2H12T7, and 141P19T7 showed mRNA levels similar to those of the parental plants in the dst2 mutant. HPH and GUS mRNA levels were increased in both mutants. The EST for one gene, 168C16T7, which shows high similarity to a cadmium-induced AP2 protein, had mRNA levels that were increased in both mutants, although the increase was greater in dst1 than in dst2 (∼3.5-fold versus two-fold compared with that of the parental type). EST 139I3T7, which encodes an unknown protein, may have marginally reduced mRNA levels in the dst2 mutant.
Figure 5.
Histograms of the Mutant (dst1 or dst2)/Parental Ratios of mRNA Expression for the Indicated Clones.
(A) Increased levels of mRNA in dst1.
(B) Decreased levels of mRNA in dst1.
The number 1519 indicates wild type. Error bars indicate ±sd.
Of particular interest was the analysis of the seven genes with lower mRNA abundance in dst1 compared with that in the parental plants. Most of these genes either were increased (ESTs 111G9T7, E6E1T7, 170N19T7, and H5A10T7) or unaffected (ESTs 171P10T7 and 201O1T7) in the dst2 mutant, as is evident from the histograms shown in Figure 5B. The only exception was EST G4A11T7; this EST codes for an unknown protein, and the mRNA levels for this gene also were diminished in dst2.
SAUR-AC1 and SAUR-like1 also were analyzed by RNA gel blot analysis and found to be increased in both the dst1 and dst2 mutants (Figure 2), although they were beneath our detection limit on the majority of the slides used for the microarray experiments. These signals were below detection because, in most experiments, the signals were too close to background to calculate a valid ratio. However, in the experiments in which we could detect a valid ratio, increase in dst1 was observed, as expected (data not shown).
Multiple Replicates Remove Nonreproducible Changes
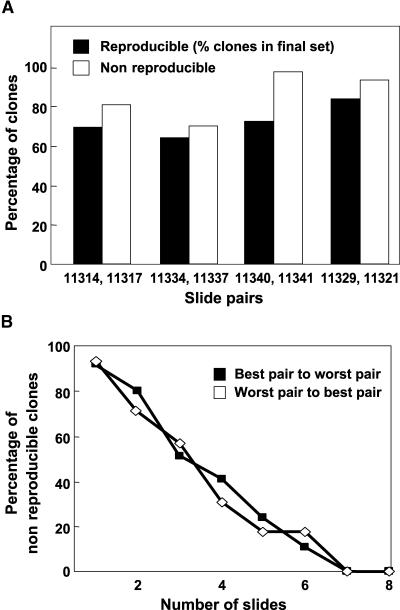
An important aspect of our analysis of the dst1 mutant was the ability to detect subtle changes in gene expression. As shown in Figure 4, using eight microarray slides, all of the changes that met our criteria were highly reproducible on independent RNA gel blots. However, it was of interest to examine the degree of reproducibility using a single pair of technical replicate slides and whether fewer than eight slides are sufficient, because the number of slides to use for microarray experiments is not yet routine. This could be particularly significant for future experiments, because the 1.5-fold cutoff used was lower than the commonly used threshold value of two-fold (DeRisi et al., 1997; Wildsmith and Elcock, 2001), although a recent study reported that the minimum detectable fold change for differential expression is 1.4-fold (Yue et al., 2001).
To monitor the reproducibility of the microarray hybridizations, for individual pairs of technical replica slides, the number of clones with a ratio >1.5 was calculated. For each, a clone was considered reproducible if it was represented in the final set of 36 clones showing ≥1.5-fold difference and nonreproducible if it showed ≥1.5-fold difference in the slide pair but was not present in the final set (see http://www.bch.msu.edu/pamgreen/Perez-Amador_etal/600_list.htm). Using a cutoff value of 1.5-fold, a histogram was plotted to compare the percentage of reproducible and nonreproducible clones. From this histogram, it is evident that such a low cutoff yields nonreproducible changes (Figure 6A). This result was expected because the cutoff value is close to the background. However, these nonreproducible changes can be identified and removed by conducting multiple experiments. When the reproducibility across repetitions was plotted as a function of the number of slides (worst to best pair and vice versa based on the percentage of visible gradient on the slide), the percentage of nonreproducible clones decreased to <20% after four slides. In our experiment, it was possible to remove virtually all nonreproducible clones using seven slides (Figure 6B).
Figure 6.
Assessment of the Reproducibility of the Microarray Data Using Fewer Replicates.
(A) Histogram plot comparing the percentage of reproducible and nonreproducible clones. The percentage of clones (y axis) is plotted against slide pairs (x axis) used for the microarray experiments. Each slide pair denotes microarray slides that were technical replicas (i.e., reciprocal labeling was performed with each pair of targets). The slide numbers denote the ExptID in the SMD.
(B) Graph showing the percentage of nonreproducible clones (y axis) plotted against the number of slides used for the microarray analysis (x axis).
Cluster Analysis of Genes with Altered Expression Levels in dst1
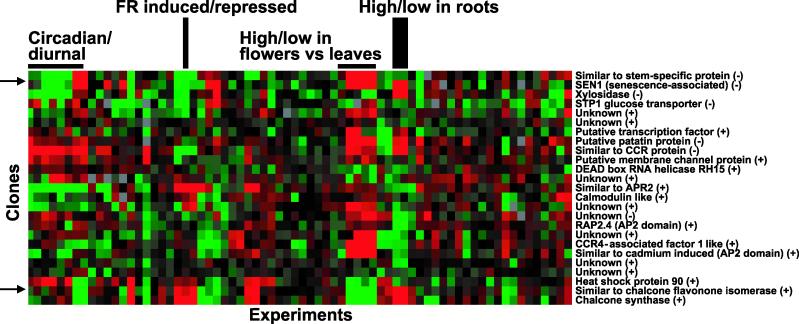
Clustering allows the grouping of genes with similar expression profiles and provides a holistic view because coexpressed genes involved in similar processes provide clues regarding the biological significance of the pathway being studied. We used cluster analysis to compare gene expression data for the 25 genes identified in this study using data from the 47 Arabidopsis microarray experiments (113 slides) available in the Stanford Microarray Database (SMD) (http://genome-www.stanford.edu/Microarray) at the time of the analysis. A portion of the resulting clusters is shown in Figure 7. As expected, when EST redundancy was not removed, ESTs representing the same gene clustered next to or in the immediate vicinity of each other, thus validating the observed pattern of gene expression (data not shown).
Figure 7.
Cluster Analysis of Genes with Altered mRNA Levels in dst1.
Data from the 25 genes were clustered with data from 47 experiments from the SMD. Each gene is represented by a single row, and each experiment is represented by a single column. Arrows indicate the predominant clusters identified. (−), genes with decreased mRNA levels in dst1; (+), genes with increased mRNA levels in dst1.
Although the cluster analysis was performed using the limited number of genes that were identified in this study, certain common expression patterns could be found. The most prominent cluster is composed of three genes that are regulated in a diurnal fashion and share several other expression characteristics. This cluster is characterized by genes whose transcripts are decreased in abundance in the dst1 mutant and include genes encoding a protein similar to a stem-specific protein, senescence-associated protein, and xylosidase. These genes show tissue-preferential expression with a high level of expression in leaves and a low level in flowers and roots, and are increased by light. Two of the three genes in this cluster, senescence-associated protein and xylosidase, have unstable transcripts (see Discussion), contain possible DST-like sequences in their 3′ UTRs, and are increased in the dst2 mutant.
Apart from the aforementioned cluster, a second cluster of coordinately expressed genes was identified. This cluster includes genes for chalcone synthase and a protein similar to chalcone flavanone isomerase, both of which show increased mRNA levels in the dst1 mutant. These genes are regulated in a diurnal fashion as well but are decreased by light and have higher levels of expression in flowers compared with leaves or roots. In addition, these mRNAs are increased in a mutant defective for protein import into chloroplasts.
No other common characteristic gene expression patterns could be seen. However, eight genes (senescence-associated protein, putative patatin protein, protein similar to CCR, putative membrane channel protein, protein similar to APR2, calmodulin-like protein, an unknown protein, and chalcone synthase) are regulated in a circadian manner, which suggests a possible connection between the DST-mediated decay pathway and circadian rhythms. Four of these eight genes also contain DST-like subdomains in their 3′ UTRs and could be direct targets of the dst1 mutation.
DISCUSSION
Microarray technology has great potential for the characterization of mutants, especially those with no visible aberrant phenotype. Here, we report on the dst1 mutant, which fits into this category. To evaluate the molecular phenotypes of this mutant, DNA microarray technology was used, which allowed the identification of primary as well as secondary effects and also revealed clues to the identification of possible relationships among genes.
Transcripts Altered in dst1 Have Predominantly Increased Levels Although Some Decrease in Abundance
The dst1 mutant is known to increase mRNA levels of HPH-DST and GUS-DST transgenes by threefold to fourfold (Johnson et al., 2000). The DNA microarray experiments described in this article were sensitive enough to detect reproducibly the increases of these transcripts in multiple replicates of the experiment. Moreover, additional changes of similar magnitudes were detected for transcripts corresponding to 25 genes from among those represented by the 11,521-clone array. Such a limited number of changes is not surprising, because the mutant does not exhibit any morphological abnormalities. It has been suggested that dst1 and dst2 correspond to weak alleles or affect genes that are part of gene families and therefore have only partial defects in mRNA stability (Johnson et al., 2000). The results of our microarray experiments are certainly consistent with this idea, because we detected only moderate changes in mRNA levels for a few genes in dst1. Increased transcript levels for some genes were expected, because dst1 is known to increase levels of DST-containing transgene mRNAs and the endogenous DST-containing SAUR-AC1, presumably as a result of a defect in a component of the cellular machinery involved in the recognition and degradation of DST-containing transcripts.
Our analysis showed that the expression of several genes decreases in dst1. Decreases in gene expression could be explained by the same defect if the DST recognition component acts as both a repressor and an activator in a context-dependent manner, similar to some transcriptional regulators. For example, Drosophila Drap1 and dCtBP are bifunctional transcription factors with distinct activation and repression functions (Phippen et al., 2000; Willy et al., 2000). Further support for this hypothesis comes from studies conducted with AUF1. It has been demonstrated that AUF1 acts as an RNA-destabilizing protein in the ARE-mediated mRNA decay pathway (Buzby et al., 1999; Loflin et al., 1999), but it also has been implicated as part of the complex that mediates the stabilization of α-globin mRNA (Kiledjian et al., 1997). Similarly, the DST recognition component could be part of different multiprotein complexes that perform separate functions.
The genes identified in this study encode proteins involved in a variety of cellular processes. The DEAD box RNA helicase, RH15, is part of a large gene family in Arabidopsis. RNA helicases are primarily RNA-unwinding enzymes and have been implicated in a variety of molecular processes, including mRNA splicing, ribosome assembly, and translation initiation (Aubourg et al., 1999). Of the 18 endogenous genes that are increased in the dst1 mutant, two encode proteins that have an amino acid motif known as the AP2 domain (Table 2). The AP2 domain is essential for APETALA2 function and contains an 18–amino acid core region that is predicted to form an amphipathic α-helix (Okamuro et al., 1997). The four ESTs that were most highly increased in dst1 correspond to the gene RAP2.4, which belongs to the RAP2 (related to AP2) gene family. The AP2 polypeptide is distinct from known fungal and animal regulatory proteins, and it has been proposed that the RAP2 proteins may function as plant sequence-specific DNA binding proteins. Another interesting EST, 222C9T7, that was increased in dst1 corresponds to a protein that is similar to the CCR4-associated factor 1 protein (CAF1). CAF1 is a transcription-associated protein and is a member of the RNase D family of 3′ to 5′ exonucleases (Moser et al., 1997). Recently, it was shown that Caf1p is a critical component of the major cytoplasmic deadenylase in yeast (Tucker et al., 2001), suggesting a potential link between mRNA deadenylation and the DST-mediated mRNA degradation pathway.
Most transcripts that are affected in the dst1 mutant relative to the parental type are expected to fall into one of two categories. One category consists of mRNAs that are the direct targets of the DST-mediated decay pathway, the primary transcripts, whereas the second category consists of transcripts, referred to as secondary transcripts, that are increased or diminished as a result of the secondary effect of changes in the primary transcripts. A number of genes showing altered levels of gene expression in dst1 may be caused by secondary effects, because these genes do not appear to contain a consensus DST element or DST-like subdomains in their 3′ UTRs. Secondary effects would be expected if altered levels of DST-containing mRNAs influence the abundance of other RNAs. This situation is not unique to dst1; secondary effects would be expected in all microarray experiments comparing mutants with the wild type. However, this study allowed us to predict the primary and secondary effects of the dst1 mutation based on the presence or absence of DST-like sequences, respectively. Although the precise sequences required for a functional DST element have not been elucidated fully, our data provide us with the advantage of identifying at least some of the most likely primary effects. For example, none of the 140 possible DST-containing sequences found by motif searching tools in the 600-element array were affected in dst1. This finding suggests that the DST element is more complex in Arabidopsis than in soybean and that it is difficult to identify this element with simple sequence search tools. It appears that multiple subdomains of the DST element are sufficient for its function as an instability determinant, because seven of the 25 genes identified contain DST-like subdomains. Earlier mutagenesis studies of individual subdomains also are consistent with this hypothesis (Feldbrügge et al., 2001). Further experiments are required to define the DST element, which will help refine our understanding of the requirements for DST recognition.
Additionally, initial microarray experiments conducted to monitor mRNA stability on a global basis have indicated that a number of transcripts identified in this study may be unstable (R.A. Gutiérrez, R.M. Ewing, J.M. Cherry, and P.J. Green, unpublished data). These results are encouraging for future studies, because primary targets of the DST-mediated decay pathway are expected to be stabilized in the dst1 mutant.
Molecular Markers to Expedite the Characterization of and Differentiation between dst1 and dst2
The most immediate utility of these findings is the identification of new molecular markers for dst1 that could be used to enhance mapping and prompt additional biological experiments. For example, the analysis of dst1 showed that RAP2.4 mRNA levels are more highly increased than are HPH or GUS mRNA levels. Thus, RAP2.4 could be a more useful marker for the detection of homozygous dst1 mutants in future experiments.
Before this study, it was difficult to distinguish between dst1 and dst2 because the known phenotypes of the two mutants were identical and F2 progeny of a cross between the mutants do not show additive increases in the abundance of DST-containing mRNAs (Johnson et al., 2000). By examining mRNAs in dst2 that show altered levels in dst1, differences between the two mutants were identified. dst1 and dst2 represent mutations in independent genes (Johnson et al., 2000). The results from these experiments indicate that some targets of the decay pathway mediated by the two genes could be different. Furthermore, there could be some degree of overlap leading to the coordinated regulation of common targets, as exemplified by SAUR-AC1 and the transgenes. The cataloging of molecular markers specific for each mutant also is significant because specific markers should allow us to identify the double mutant without relying on map positions and subsequent crosses. This emphasizes another potential use of microarrays in identifying genes that can be used to examine and compare related mutants without necessitating individual microarray experiments for each. For example, a new dst mutant was obtained recently in our laboratory by activation tagging. The molecular markers identified in this study will facilitate the characterization of this mutant as well as subsequent mutants that affect this sequence-specific mRNA degradation pathway.
Circadian Association of the dst1 Mutation
Using the technique of cluster analysis, parallel expression profiling was conducted over many experiments. The clusters generated revealed coexpression characteristics of the genes affected in the dst1 mutant that would not have been evident otherwise. Eight genes, which correspond to 32% of the 25 genes described in this analysis, were regulated in a circadian manner, whereas only 2% of 7800 genes were found to cycle with a circadian rhythm in a study conducted by Schaffer et al. (2001). In a study by Harmer et al. (2000), 6% of 8000 genes showed circadian changes in mRNA levels. On the basis of comparison with their results, four of 25 genes (16%) in our experiments exhibited a circadian pattern of expression. These data suggest a higher representation of circadian-regulated genes than would be expected by chance. Despite differences in results and experimental conditions between the two studies (e.g., only ∼1500 genes were in common), four genes with altered expression in dst1 were found to be regulated in a circadian manner in both studies, adding more credence to our data. The significance of several genes in the cluster being regulated in a circadian manner is not yet clear, but the occurrence suggests an association between the dst1 mutation and circadian rhythms. Furthermore, Harmer et al. (2000) showed that SAUR-AC1, which is a target of the DST-mediated mRNA degradation pathway, also is regulated in a circadian manner. It is known that a number of circadian-regulated transcripts are unstable and are expressed during a brief period of time. It is possible that in the dst1 mutant, the level of a potential regulatory factor or signal molecule that plays some part in the circadian clock function is altered, which leads to a cascade effect and changes the expression of several circadian-regulated genes. If this is true, the circadian effects described here would provide insight into the biological or physiological significance of the DST-mediated mRNA decay pathway. Elucidating the exact nature of this association and further testing this hypothesis requires the cloning of DST1.
Conclusions and Future Prospects
In addition to reproducing the known increase of two transgene mRNAs in dst1, 25 additional transcripts were found to increase or decrease in abundance relative to the parental plant. Many of the corresponding genes were examined subsequently by RNA gel blot analysis to confirm the dst1 microarray results and to evaluate their expression in the dst2 mutant. In this way, several interesting and useful differences were revealed between the two dst mutants. These new molecular markers should enhance subsequent analysis of the dst mutants, provide insight into their biological significance, and help identify other targets of the DST-mediated mRNA decay pathway.
As a result of the stringency of our parameters to avoid false positives, the 25 genes identified in this study are most likely an underrepresentation of the molecular phenotypes of dst1. For example, ratios derived from the microarray results from lowly expressed genes, such as SAUR-AC1 and SAUR-like1, which have low levels of expression in the absence of auxin (McClure and Guilfoyle, 1987), were significantly different from those determined by RNA gel blot analysis. The channel intensity values for these transcripts are close to the background fluorescence intensity levels and therefore are more susceptible to variation. Also, there may be additional targets of the DST-mediated decay pathway not identified because the present generation of slides do not contain all of the genes of Arabidopsis. In the future, comprehensive microarrays with improved sensitivity should result in the discovery of a greater number of molecular phenotypes for dst1.
Beyond extending our knowledge of the DST-mediated decay pathway in plants, this study provides additional general insights. Our study shows that subtle changes in gene expression can be measured reliably using multiple microarrays, which should enhance the global investigation of gene expression patterns under several conditions. This analysis demonstrates that new molecular phenotypes for mutants without a visible phenotype can be identified using DNA microarray technology. Also, with full genome microarrays, it should be possible to catalog all of the molecular phenotypes for any mutant. Furthermore, new hypotheses and associations, such as the potential link between the dst1 mutation and circadian rhythms, can be developed using the publicly available databases. Future mutant analyses via DNA microarray analysis should have even greater utility, particularly for the analysis of mutants identified by reverse genetic approaches (e.g., T-DNA insertions) that have no apparent mutant phenotype.
METHODS
Plant Material
All Arabidopsis thaliana plants described in this report are from the accession Columbia; they were grown in growth chambers under 16 hr of light and 60% relative humidity at 20°C. Tissue from the parental line (p1519-31) and dst1 and dst2 homozygous mutants (Johnson et al., 2000) were harvested from 35- to 40-day-old plants. All tissue was harvested from plants grown in parallel under the same conditions in different growth chambers. Seed from the dst mutants and the parental line are freely available upon request for basic research from the corresponding author. They cannot be distributed using Arabidopsis Biological Resource Center (Ohio State University, Columbus) accession numbers because they contain proprietary materials.
Generation of a 600-Element DNA Microarray
Expressed sequence tag (EST) clones were selected based on sequence similarity by BLAST analysis (Altschul et al., 1997) to known genes involved in RNA metabolism in Arabidopsis, in other plants, or in other systems such as bacteria, yeast, and mammals.
Selected Arabidopsis EST clones were obtained from the PRL2 EST collection (Newman et al., 1994). These ESTs were cloned in λZip-Lox (pZL1 clones; Gibco BRL, Rockville, MD) or pBluescript SK− vector (pBSK clones; Stratagene, La Jolla, CA). To amplify the ESTs by polymerase chain reaction (PCR), we designed universal primers corresponding to the 3′ and 5′ ends flanking the cloning site of each vector backbone. pZL1 clones were amplified using the 5′ primer 5′-CGACTCACTATAGGGAAAGCTGG-3′ and the 3′ primer 5′-ATTGAATTTAGGTGACACTATAGAAGAGC-3′. pBSK clones were amplified using the 5′ primer 5′-CGACTCACTATAGGGCGAATTGG-3′ and the 3′ primer 5′-GGAAACAGCTATGACCATGATTACG-3′. cDNA clones isolated in our laboratory were cloned in pBluescript SK− (primers as above) or pGEM-T (Promega, Madison, WI) vectors. To amplify clones in pGEM-T, we designed the 5′ primer 5′-CGACTC-ACTATAGGGCGAATTGG-3′ and the 3′ primer 5′-ATTTAGGTGACA-CTATAGAATACTCAAGC-3′.
Plasmid DNA from the Michigan State University (MSU) collection was diluted to a final concentration of 1 to 3 ng·μL−1 in TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). PCR reactions, in a final volume of 100 μL, contained 40 pmol of the corresponding primers, 0.2 mM of each deoxynucleotide triphosphate (dNTP), 2 to 5 ng of plasmid DNA, and 2 units of Taq DNA polymerase in 1 × reaction buffer (10 mM Tris-HCl, pH 8.0, 50 mM KCl, and 2 mM MgCl2). After PCR, DNA was precipitated in ethanol and resuspended in 20 μL of 3 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate, pH 7.0). To determine the quality and quantity of the amplified DNA, 1 μL of the final resuspension was analyzed by electrophoresis in 1.0% agarose gels. PCR products with low concentration (<200 ng·μL−1) or showing multiple bands were discarded and replaced with a new PCR amplification product derived from a different EST clone corresponding to the same gene. If no alternative EST was available, a new PCR amplification was performed using a gene-specific primer designed for the 5′ end of the EST clone along with the corresponding 3′ vector primer. In this way, we ensured that only high-quality PCR products were included on the 600-element DNA microarray. To confirm the identity of the amplified DNAs, we sequenced 12 randomly selected clones. In each case, the sequence obtained matched the EST sequence deposited in the database.
For clones obtained from genomic DNA, DNA was extracted from total aboveground tissue of mature Arabidopsis plants using the method of Dellaporta et al. (1983), and 100 ng was amplified as described for EST clones. In all cases, a single PCR product was obtained. Low-abundance PCR products were reamplified under the same conditions using 1 μL of the first PCR product as a template.
DNA from the PCR reactions was transferred to master DNA plates and stored at 4°C until printing. The final concentration of DNA for printing was estimated to be 200 to 400 ng·μL−1. DNAs were arranged as four subgrids of 12 × 13 and printed twice on poly-l-lysine–coated slides. Printing, handling, and use of the 600-element DNA microarray were performed with the same methods used for the 11,521-element MSU DNA microarray, as indicated below.
11,521-Element Arabidopsis Functional Genomics Consortium DNA Microarray
Microarrays were generated at the Arabidopsis Functional Genomics Consortium microarray facility at MSU. A total of 11,521 ESTs were spotted onto superaldehyde glass slides (Telechem International, Sunnyvale, CA). Slides were washed and blocked according to the Telechem International protocol.
Total RNA Extraction, Poly(A)+ RNA Purification, and RNA Gel Blot Hybridization
Total RNA from leaf samples was extracted as described previously (Newman et al., 1993). Poly(A)+ RNA was purified from 200 to 400 μg of total RNA using the Oligotex mRNA kit (Qiagen, Valencia, CA). RNA (10 μg of total RNA or 2 μg of poly(A)+ RNA) was analyzed by electrophoresis on 2% formaldehyde/1.2% agarose gels and blotted onto nylon membranes (Nytran Plus; Schleicher and Schuell, Keene, NH). DNA probes were labeled with α-32P-dCTP by the random primer method (Feinberg and Vogelstein, 1983) and purified from unincorporated nucleotides using probe purification columns (NucTrap; Stratagene). The RNA gel blots were hybridized as described by Taylor and Green (1991) using the indicated 32P-labeled probes. For a loading control, RNA gel blots were hybridized with a 32P-labeled cDNA probe for the Arabidopsis translation initiation factor eIF4A (Taylor et al., 1993). Blots were stripped between hybridizations in 0.1% SDS at 90 to 95°C for 1 hr. Quantification of hybridization signals was achieved using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Labeling of Poly(A)+ RNA
For first-strand cDNA synthesis, 1 μg of poly(A)+ RNA and 1 μg of oligo(dT) (Gibco BRL) in a total volume of 25 μL of diethyl pyrocarbonate–treated water was denatured at 70°C for 10 min and cooled on ice. On ice, 8 μL of 5 × reverse transcriptase buffer, 4 μL of 0.1 M DTT, 2 μL of dNTPs (10 mM each), and 1 μL of Superscript II (Gibco BRL) were added, and the mixture was incubated at 42°C for 1 hr. To remove RNA, 0.5 μL of RNase H (4 units·μL−1) was added, and the reaction was incubated for 30 min at 37°C. Single-stranded cDNA was purified in a Microcon YM-30 column (Millipore, Bedford, MA), concentrating to a volume of 10 μL of TE. Single-stranded cDNA was divided into two aliquots for Cy3- and Cy5-dCTP labeling, allowing for two hybridizations for each sample. The second-strand synthesis reaction contained 5 μL of single-stranded cDNA, 22 μL of water, 4 μL of Klenow buffer (500 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 100 mM MgCl2), and 2 μL of random hexamers (3 μg·μL−1; Gibco BRL). The mixture was denatured at 95°C for 3 min and annealed at room temperature for 5 min. Next, 4 μL of dNTP mix (250 μM dATP, dGTP, and dTTP and 90 μM dCTP), 1 μL of dCTP-Cy3 or dCTP-Cy5 (Amersham, Arlington Heights, IL), and 2 μL of Klenow (5 units· μL−1) (Gibco BRL) were added, and the reaction was incubated for 2 hr at 37°C. All incubations were performed in thin-wall 0.5-mL tubes in a RoboCycler 40 Temperature Cycler (Stratagene). Labeled double-stranded DNA was purified using a QIAquick PCR purification kit (Qiagen) and eluted in 50 μL of 2 mM Tris-HCl, pH 8.0. The probe was dried to 10 μL. To test the quality and quantity of the product, 1 μL was separated on a 1% agarose gel using a miniprotean gel electrophoresis system (Bio-Rad Laboratories, Hercules, CA). The portion of the gel containing the DNA sample was placed on a glass microscope slide, dried on a heat block at 70°C, and scanned using a ScanArray 3000 or 5000 (GSI Lumonics, Billerica, MA). The rest of the probe was used to prepare the hybridization mixture.
DNA Microarray Hybridization and Analysis
For a single DNA microarray, 30 μL of hybridization solution was prepared by mixing 5.2 μL of 20 × SSC, 4.5 μL of 2% SDS, 2.4 μL of tRNA (20 μg·μL−1), 9 μL of Cy3-labeled probe, and 9 μL of Cy5-labeled probe. The mixture was denatured at 100°C for 1 min and spun for 1 min to recover any condensate. The mixture then was hybridized to the array under a glass cover slip (24 × 40 mm; Corning) that had been washed in 95% ethanol and 0.2% SDS, and then it was rinsed in distilled water. The slide was placed in a microarray hybridization chamber (ArrayIt Hybridization Cassette; Telechem International) with 200 μL of 3 × SSC to ensure high-humidity conditions.
Hybridization was performed in a water bath at 65°C for 12 to 20 hr. After hybridization, the microarray was washed for 5 min in 1 × SSC and 0.2% SDS, 5 min in 0.1 × SSC and 0.2% SDS, and 30 sec in 0.1 × SSC without SDS. Then it was dried by centrifugation at 62g for 5 min.
The slide was scanned once in a ScanArray 3000 or 5000 (GSI Lumonics) for both channels 1 and 2 (corresponding to Cy3- and Cy5-labeled probes, respectively) at 10-μm resolution. The image files obtained were analyzed using ScanAlyze software (version 2.32; M. Eisen, Stanford University, http://genome-ww4.stanford.edu/MicroArray/SMD/restech.html). To ensure that only spots of high quality were used in the analysis, quality control measurements produced by the ScanAlyze software were used. For example, the GTB2 value represents the fraction of pixels within each spot that are >1.5-fold of the background measurement. Spots with GTB2 values <0.50 for either channel were removed and not considered for further analysis.
Data from each channel were transformed to the natural logarithm, and a Z score was calculated to normalize the channel values to account for variation in RNA labeling. For the Z score calculation [Z = (χ − μ)/σ, where χ = channel value, μ = mean of channel data, and σ = standard deviation of channel data], the trimmed mean and standard deviation using the middle 93% of the channel values were used to not bias the calculation because of extremely high or low values. The new data set has, by definition, a normal distribution with zero mean and unit variance. Values were retransformed from the natural logarithm by raising to the power e, and the channel ratio was calculated. From each of the four replica samples used with the 11,521-element MSU DNA microarray, we generated two slides, one with direct and one with reverse labeling. Ratios from reverse labeling were reversed to compare with the ratios from direct labeling. Therefore, for all slides, ratios >1 and <1 indicated increased and decreased mRNA levels, respectively, in dst1 versus parental plants. An average, standard deviation, and median of these eight ratios were determined and used as final ratio of mRNA levels.
Hierarchical clustering was performed using Cluster and Treeview software (Eisen et al., 1998; available at http://genome-www4.stanford.edu/MicroArray/SMD/restech.html).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use in noncommercial research purposes.
Accession Numbers
Acknowledgments
We thank Dr. Christoph Benning for critical reading of the manuscript and members of the Green laboratory for helpful discussions. We also thank Tom Newman for donating EST clones and the members of the microarray facility of the Arabidopsis Functional Genomics Consortium (AFGC) at MSU for technical advice. Zihua Hi is acknowledged for helping to collect some of the clones for the 600 element array. This work was funded by grants from the United States Department of Agriculture (2000-014191), the Department of Energy (DE-FG02-91ER20021), and the National Science Foundation (DBN987638) to P.J.G. M.A.P.-A. received postdoctoral fellowships from North Atlantic Treaty Organization–Spain and the Ministerio de Educatión y Ciencia, Spain.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010295.
References
- Abler, M.L., and Green, P.J. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32, 63–78. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A, Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg, S., Kreis, M., and Lecharny, A. (1999). The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 27, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby, J.S., Brewer, G., and Nugent, D.J. (1999). Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J. Biol. Chem. 274, 33973–33978. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y.A., and Shyu, A.B. (1995). AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.R. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Feldbrügge, M., Arizti, M.L., Zamore, P.D., Belasco, J.G., and Green, P.J. (2001). Comparative analysis of the plant mRNA-destabilizing element DST, in mammalian and tobacco cells. Plant Mol. Biol., in press. [DOI] [PubMed]
- Franco, A.R., Gee, M.A., and Guilfoyle, T.J. (1990). Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J. Biol. Chem. 265, 15845–15849. [PubMed] [Google Scholar]
- Gil, P., and Green, P.J. (1996). Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: The 3′ untranslated region functions as an mRNA instability determinant. EMBO J. 15, 1678–1686. [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.E., and Belasco, J.G. (1993). Control of the decay of labile protooncogene and cytokine mRNAs. In Control of Messenger RNA Stability, J. Belasco and G. Brawerman, eds (San Diego, CA: Academic Press), pp. 199–218.
- Harmer, S.L., Hogenesch, J.B., Straum, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Higgs, D.C., Barnes, L.J., and Colbert, J.T. (1995). Abundance and half-life of the distinct oat phytochrome A3 and A4 mRNAs. Plant Mol. Biol. 29, 367–377. [DOI] [PubMed] [Google Scholar]
- Johnson, M.A., Baker, E.J., Colbert, J.T., and Green, P.J. (1998). Determinants of mRNA stability in plants. A look beyond transcription. In Mechanisms Determining mRNA Stability and Translation in Plants, J. Bailey-Serres and D.R. Gallie, eds (Rockville, MD: American Society of Plant Physiologists), pp. 40–53.
- Johnson, M.A., Perez-Amador, M.A., Lidder, P., and Green, P.J. (2000). Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc. Natl. Acad. Sci. USA 97, 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe, D.M., Villand, P., and Somerville, S. (1999). DNA microarrays for studies of higher plants and other photosynthetic organisms. Trends Plant Sci. 4, 38–41. [DOI] [PubMed] [Google Scholar]
- Kiledjian, M., DeMaria, C.T., Brewer, G., and Novick, K. (1997). Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol. 17, 4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loflin, P., Chen, C.Y.A., and Shyu, A.B. (1999). Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B.A., and Guilfoyle, T. (1987). Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 9, 611–623. [DOI] [PubMed] [Google Scholar]
- McClure, B.A., and Guilfoyle, T.J. (1989). Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243, 91–93. [DOI] [PubMed] [Google Scholar]
- McClure, B.A., Hagen, G., Brown, C.S., Gee, M.A., and Guilfoyle, T.J. (1989). Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, M.J., Holley, W.R., Chatterjee, A., and Mian, I.S. (1997). The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, T.C., Ohme-Takagi, M., Taylor, C.B., and Green, P.J. (1993). DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, T.C., de Bruijn, F., Green, P.J., Keegstra, K., Kende, H., McIntosh, L., Ohlrogge, J., Raikhel, N.V., Somerville, S.C., Tomashow, M., Retzel, E., and Somerville, C.R. (1994). Genes galore: A summary of methods for accessing results from partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., Taylor, C.B., Newman, T.C., and Green, P.J. (1993). The effect of sequences with high AU content on mRNA stability in tobacco. Proc. Natl. Acad. Sci. USA 90, 11811–11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz, S.W., and Jacobson, A. (1992). mRNA stability: In trans-it. Curr. Opin. Cell Biol. 4, 979–983. [DOI] [PubMed] [Google Scholar]
- Phippen, T.M., Sweigart, A.L., Moniwa, M., Krumm, A., Davie, J.R., and Parkhurst, S.M. (2000). Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both Mad and Groucho transcriptional repression. J. Biol. Chem. 275, 37628–37637. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Dammond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. (1995). mRNA stability in mammalian cells. Microbiol. Rev. 59, 423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. (1996). Control of mRNA stability in higher eukaryotes. Trends Genet. 12, 171–175. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Accerbi, M., Simon, V.V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression patterns of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu, A.-B., Belasco, J.G., and Greenberg, M.E. (1991). Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Sullivan, M.L., and Green, P.J. (1996). Mutational analysis of the DST element in tobacco cells and transgenic plants: Identification of residues critical for mRNA instability. RNA 2, 308–315. [PMC free article] [PubMed] [Google Scholar]
- Taylor, C.B., and Green, P.J. (1991). Genes with homology to fungal and S-gene RNases are expressed in Arabidopsis thaliana. Plant Physiol. 96, 980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C.B., and Green, P.J. (1995). Identification and characterization of genes with unstable transcripts (GUTs) in tobacco. Plant Mol. Biol. 28, 27–38. [DOI] [PubMed] [Google Scholar]
- Taylor, C.B., Bariola, P.A., DelCardayré, S.B., Raines, R.T., and Green, P.J. (1993). RNS2: A senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc. Natl. Acad. Sci. USA 90, 5118–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., and Parker, R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386. [DOI] [PubMed] [Google Scholar]
- Vakalopoulou, E., Schaack, J., and Shenk, T. (1991). A 32-kilodalton protein binds to AU-rich domains in the 3′ untranslated regions of rapidly degraded mRNAs. Mol. Cell Biol. 11, 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, S., and Peltz, S.W. (2001). Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7, 1191–2000. [DOI] [PubMed] [Google Scholar]
- Wildsmith, S.E., and Elcock, F.J. (2001). Microarrays under the microscope. Mol. Pathol. 54, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy, P.J., Kobayashi, R., and Kadonaga, J.T. (2000). A basal transcription factor that activates or represses transcription. Science 290, 982–985. [DOI] [PubMed] [Google Scholar]
- Yue, H., Eastman, P.S., Wang, B.B., Minor, J., Doctolero, M.H., Nuttall, R.L., Stack, R., Becker, J.W., Montgomery, J.R., Vainer, M., and Johnston, R. (2001). An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29, E41. [DOI] [PMC free article] [PubMed] [Google Scholar]