Abstract
The HUELLENLOS (HLL) gene participates in patterning and growth of the Arabidopsis ovule. We have isolated the HLL gene and shown that it encodes a protein homologous to the L14 proteins of eubacterial ribosomes. The Arabidopsis genome also includes a highly similar gene, HUELLENLOS PARALOG (HLP), and genes for both cytosolic (L23) and chloroplast ribosome L14 proteins. Phylogenetic analysis shows that HLL and HLP differ significantly from these other two classes of such proteins. HLL and HLP fusions to green fluorescent protein were localized to mitochondria. Ectopic expression of HLP complemented the hll mutant, indicating that HLP and HLL share redundant functions. We conclude that HLL and HLP encode L14 subunits of mitochondrial ribosomes. HLL mRNA was at significantly higher levels than HLP mRNA in pistils, with the opposite pattern in leaves. This differential expression can explain the confinement of effects of hll mutations to gynoecia and ovules. Our elucidation of the nature of HLL shows that metabolic defects can have specific effects on developmental patterning.
INTRODUCTION
Developmental patterning must be coordinated precisely with growth to achieve correct organ morphology. Plant morphogenesis occurs via cell division, cell expansion, or a combination of these processes. Both processes put demands on energetic and metabolic resources for the synthesis of new proteins and other biomolecules. The Arabidopsis ovule is being used as one model to study the interaction of growth and patterning processes in plants (Gasser et al., 1998; Schneitz, 1999).
In Arabidopsis, ovule primordia arise from the placenta as cylindrical projections that can be divided conceptually into three regions by their ontogenetic fate. The distal part, the nucellus, is the site of megasporogenesis and development of the embryo sac. The central portion, the chalaza, gives rise to the two integuments. The proximal region elongates and differentiates into the funiculus, a supporting stalk (Robinson-Beers et al., 1992; Schneitz et al., 1995).
Arabidopsis genes involved in ovule growth regulation have been identified through mutant analysis. AINTEGUMENTA (ANT) and SHORT INTEGUMENTS2 (SIN2) are essential for growth of the integuments (Elliott et al., 1996; Klucher et al., 1996; Baker et al., 1997; Schneitz et al., 1998; Broadhvest et al., 2000). The SPOROCYTELESS or NOZZLE gene is essential for normal development of the funiculus, integuments, and nucellus (Schiefthaler et al., 1999; Yang et al., 1999). TSO1 (Liu et al., 1997; Hauser et al., 1998) and SIN1 (Robinson-Beers et al., 1992; Ray et al., 1996) participate in the regulation of directional cell expansion and division. INNER NO OUTER (Villanueva et al., 1999) and SUPERMAN (Gaiser et al., 1995) regulate the asymmetric growth of the outer integument. All of these genes encode putative regulatory factors (Schultz et al., 1991; Bowman et al., 1992; Jacobsen et al., 1999; Yang et al., 1999; Balasubramanian and Schneitz, 2000; Hauser et al., 2000; Song et al., 2000; T.A. Hill, personal communication). However, it has not been determined how these regulatory factors affect the fundamental processes of cellular expansion and division that are more directly responsible for morphogenesis.
The Arabidopsis huellenlos (hll) mutants exhibit defects in ovule growth and development. Integuments are highly reduced or absent, and the cells in the distal regions of the ovule primordia often collapse (Schneitz et al., 1998). Double mutant analysis of hll in combination with either sin2 or ant has shown an even more severe, synergistic reduction in ovule primordium outgrowth (Schneitz et al., 1998; Broadhvest et al., 2000). The novel phenotype of the hll mutant implicates the HLL protein in cellular growth processes, whereas the genetic interactions with other mutants indicate that hll may be useful to study the coordination between growth and development.
To accommodate the energetic and metabolic burden in areas undergoing growth, the activities of chloroplasts and mitochondria must be coordinated and upregulated. In an apparent response to these demands, mitochondria numbers and steady state levels of several nucleus-encoded mitochondrial mRNAs have been shown to increase severalfold in developing flowers (Huang et al., 1994; MacKenzie and McIntosh, 1999). Reproductive development has been shown to be particularly sensitive to changes in mitochondria. For example, altered proteins in mitochondria have been associated with disruptions in tapetal and pollen development in both petunia and maize (Levings, 1993; Conley and Hanson, 1995). Aspects of vegetative development also have been found to be sensitive to mutations that affect mitochondrial function. A maize mutant, Non-Chromosomal Stripe3, has sectors of undeveloped leaf tissue and aborted kernels resulting from somatic segregation of wild-type and mutant mitochondria that lack two mitochondrially encoded ribosomal proteins (Newton and Cole, 1986; Hunt and Newton, 1991).
We report the cloning of the HLL gene and the nature of the HLL protein. On the basis of amino acid sequence similarities and subcellular localization, we propose a role for HLL and for a paralogous Arabidopsis protein, HUELLENLOS PARALOG (HLP), in mitochondrial ribosomes.
RESULTS
Phenotype of hll Mutants
Our results confirm previous work in which the most pronounced phenotypic effects of hll mutations are observed in ovule development (Schneitz et al., 1998). As in the wild type, hll ovules initiate as cylindrical primordia, but subsequent growth is halted at early stages of ovule development. In the strong hll-1 allele shown in Figure 1B, growth arrests before or soon after initiation of the integuments (Schneitz et al., 1998). The weaker hll-2 allele arrests growth after the integuments have begun to extend around the nucellus (Schneitz et al., 1998). In both alleles, the funiculus ceases growth and has a reduced number of cells relative to the wild type (Schneitz et al., 1998; data not shown). Both alleles exhibit cell collapse and degeneration of the distal regions of the ovule primordia (Schneitz et al., 1998) (Figure 1B). Cellular degeneration has variable onset among the different ovules in a single pistil, but eventually it affects most primordia (Figure 1B; Schneitz et al., 1998). Distal regions are affected most severely by the degeneration, and degeneration of more proximal regions rarely occurs without collapse of adjacent distal regions.
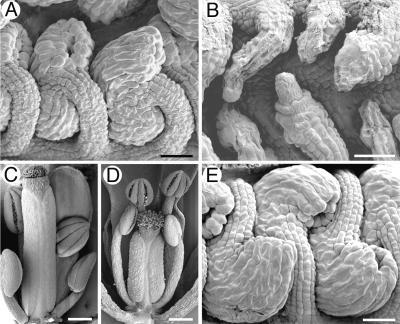
Figure 1.
Scanning Electron Micrographs of Arabidopsis Ovules and Flowers.
(A) Wild-type ovules at anthesis.
(B) hll-1 ovules at anthesis exhibit reduced or absent integuments and show variable cellular collapse.
(C) Wild-type flower just before anthesis with carpel exposed.
(D) hll-1 flower at anthesis. The pistil is significantly smaller than shown in (C), despite the more advanced age of the flower.
(E) Ovules from an hll-1 plant complemented with a genomic fragment containing the HLL gene (pDS08) show wild-type morphology.
Bars in (A), (B), and (E) = 25 μm; bars in (C) and (D) = 250 μm.
In addition to ovule defects, hll mutants also show pleiotropic effects on the gynoecium, as illustrated in Figures 1C and 1D. hll-1 and hll-2 mutants have smaller gynoecia, which may bear fewer, and sometimes misplaced, ovules relative to those of the wild type (Schneitz et al., 1998). These two effects may be linked, because a smaller gynoecium may be an effect, or a cause, of fewer ovules.
Isolation of HLL
A segregating F2 population was used to define the position of HLL to three overlapping bacterial artificial chromosome (BAC) clones (T19K13, T25P10, and T24L7) from chromosome 1 (see Methods). Cosmid subclones of these BACs were used to transform heterozygous F2 progeny of the hll-1 mutant crossed to Columbia wild-type plants. Transgenic plants were screened for homozygous hll-1 individuals (as evaluated by flanking marker analysis) to determine if the introduced sequences complemented the mutation.
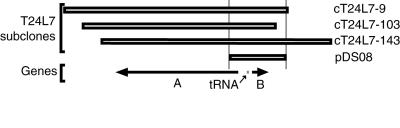
Three overlapping cosmid subclones from BAC T24L7 that complemented the hll-1 mutation are depicted in Figure 2. Segregation analysis in the progeny of these plants confirmed the complementation, because the transgenes always cosegregated with a wild-type phenotype in hll-1 homozygotes (data not shown).
Figure 2.
Genomic Clones Complementing hll.
Three cosmid subclones (cT24L7-9, cT24L7-103, and cT24L7-143) of BAC T24L7 complemented the hll-1 mutant. A total of one (cT24L7-103) plus one (cT24L7-9) plus three (cT24L7-143) transgenic plants were homozygous for hll-1 but had a wild-type phenotype. The cosmids had three genes in common: A, B, and a tRNA gene. A subclone of cT24L7-143 containing gene B and the tRNA (pDS08) also complemented the mutant (one plant).
The 16-kb region of overlap between the complementing cosmids was sequenced from cosmid T24L7-143, and two candidate protein coding regions (designated A and B) and a putative tRNA gene were identified (Figure 2). Polymerase chain reaction (PCR) products for each putative gene were amplified from hll-1 and hll-2 genomic DNA and sequenced to compare with wild-type genomic sequences of Landsberg erecta and Columbia. Mutations were found only in products for candidate gene B from both hll-1 and hll-2. Transformation with a 3.5-kb region including only gene B and the tRNA gene was shown to complement the mutant (Figure 1E). The introduction of a Cauliflower mosaic virus (CaMV) 35S::gene B cDNA transgene ‘into hll-1 plants also resulted in complementation of the mutant phenotype and no other phenotypic effects (data not shown). We conclude that candidate gene B (corresponding to At1g17560 in the Arabidopsis genome sequence) is HLL.
HLL Is Similar to L14 Ribosomal Proteins of Eubacteria
The HLL cDNA derives from three exons spanning 1 kb of genomic sequence. Figure 3A shows the sequence of the 196–amino acid protein encoded by the cDNA sequence. Sequence database searches showed that HLL was similar to L14 ribosomal proteins of eubacteria, showing up to 43% identity and 59% similarity. HLL showed even greater sequence similarity to predicted proteins from a second gene in Arabidopsis (At5g46160 on chromosome 5, which we refer to as HLP) and from expressed sequence tag sequences from other plants. In both hll alleles, C-to-T changes result in premature stop codons (Figure 3A). hll-1 has a change at Q97 to stop, resulting in a protein of 96 amino acids. hll-2 has a change from Q44 to stop, creating a shorter predicted protein of 43 amino acids.
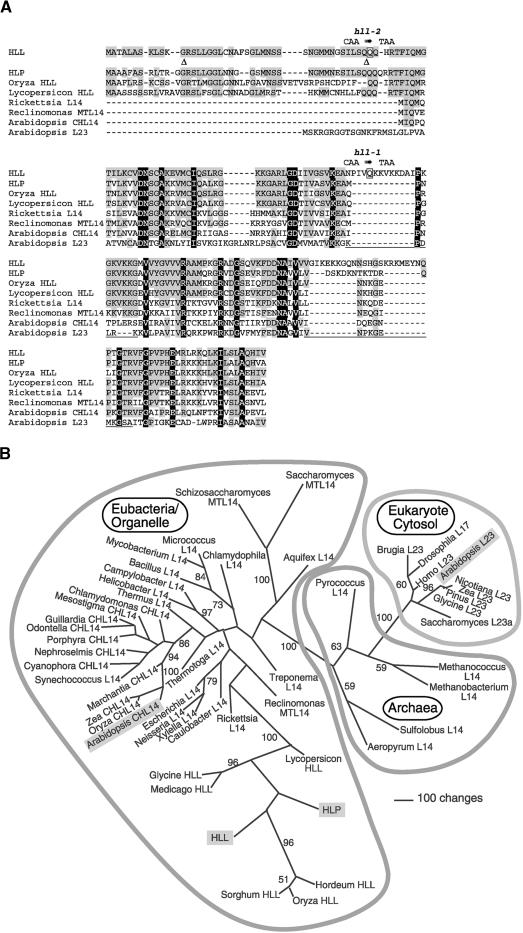
Figure 3.
Sequence Alignment of Proteins Similar to HLL and Phylogenetic Analysis of L14 Homologs from Diverse Lineages.
(A) Deduced protein sequences of HLL and HLP, aligned with those encoded by similar genes from other plants (rice [Oryza sativa] and tomato [Lycopersicon esculentum]), a eubacterial ancestor of mitochondria (Rickettsia prowazekii), the Reclinomonas americana mitochondrion (Reclinomonas MTL14), the Arabidopsis chloroplast (Arabidopsis CHL14), and the Arabidopsis cytosolic L14 homolog (Arabidopsis L23). Amino acids with black backgrounds are identical in all illustrated proteins, and gray shading denotes identity to HLL. The positions of introns in the HLL gene are indicated by triangles. Amino acids altered in the hll-1 and hll-2 alleles are shown in boxes with the DNA sequence change above. Underlines correspond to the locations of loops in the protein structure of the corresponding regions of Bacillus stearothermophilus L14 (Ramakrishnan et al., 1995).
(B) One of eight most parsimonious trees resulting from the analysis of HLL and related proteins. A selection of L14 and L23 proteins from a variety of organisms (and subcellular compartments: CH, chloroplast; MT, mitochondria; all others, cytosolic/cytoplasmic) were analyzed. Insertions and deletions that were not phylogenetically informative were omitted from analysis. The illustration shows an unrooted phylogram of one of the eight shortest trees (13,291 steps) that resulted from 500 random taxon addition replications of a heuristic search on random starting trees using the tree bisection reconnection algorithm for branch swapping. The other trees of equivalent length differed from this tree only in relationships among plant L23 proteins and among the taxa of the three-member clade that includes the Synechococcus sequence. Values from 500 bootstrap replicates (same search settings as described above, except that five random addition trials were performed for each replicate) are shown for those branches that obtained >50% support. Outlines indicate phylogenetic domains of the sequence sources. Sequences from Arabidopsis are highlighted with shaded boxes. For accession numbers see Methods.
The L14 ribosomal protein is part of a core group of rRNA binding proteins in the large ribosomal subunit (Herold and Nierhaus, 1987; Urlaub et al., 1995; Ban et al., 1999). Putative L14 orthologs are found in eubacterial, archaean, organellar, and eukaryotic cytosolic ribosomes. A phylogenetic analysis of representative sequences from each of these groups is shown in Figure 3B. HLL, HLP, and closely related plant proteins form a well-supported clade within a group of eubacterial and organellar L14 proteins. This eubacterial group was clearly separated from archaeal L14 proteins and eukaryotic L23 proteins (L14 homologs in cytosolic ribosomes). Three nearly identical L23 proteins are encoded in the Arabidopsis genome (At3g04400, At2g33370, and At1g04480), and the one included in this analysis falls within the well-supported eukaryotic clade in association with other plant L23 proteins. The Arabidopsis chloroplast L14 protein (encoded on the chloroplast genome) falls within a well-supported chloroplast clade within the eubacterial group. Thus, HLL and HLP are unlikely to be additional cytosolic or chloroplast L14-type proteins, but they could be the mitochondrial homologs of L14 ribosomal proteins. Indeed, a known mitochondrial L14 protein and the L14 protein of Rickettsia prowazekii, a relative of the putative ancestor of mitochondria (Andersson et al., 1998), appear at the base of the clade of HLL-related plant genes. Mitochondrial L14 proteins of two ascomycetes (Saccharomyces cerevisiae and Schizosaccharomyces pombe) were grouped on long branches elsewhere in the eubacterial part of the tree.
To date, Arabidopsis is the only plant species from which we have found more than one HLL-like gene by use of available sequence databases. Relationships among the different plant HLL-like proteins were not well resolved, so specific orthology of the proteins from other plants to either HLL or HLP could not be determined. It is also noteworthy that some of the longest branches in the tree are within the group of plant HLL-like proteins.
Significant differences between HLL and L14 proteins can be observed in protein alignments (Figure 3A). HLL has two insertions relative to the bacterial and chloroplast proteins: a 12–amino acid insertion at position 94 and a 17–amino acid insertion at position 156. HLP also has an insertion of 7 amino acids at the latter site. The S. cerevisiae and S. pombe mitochondrial proteins have 10– and six–amino acid insertions at the former site (data not shown). These two sites correlate with known exterior loop regions of the eubacterial proteins (Figure 3A); thus, the insertions may not alter the core structure of the proteins significantly.
A variable length (47 to 51 amino acids) N-terminal extension exists on all of the HLL-like plant proteins compared with eubacterial proteins. HLL and HLP share 77% similar amino acids in this region, but there is generally lower conservation of amino acids (∼30% similar) between diverse plant species in this region compared with the rest of the protein (∼75% similar). This N-terminal extension may serve as a transit sequence for translocation of the protein to a subcellular compartment. Mitochondrial localization signals are thought to take the form of structural motifs in N-terminal sequences and have not yet been well defined (Glaser et al., 1998; Schneider et al., 1998). Amino acid motifs associated with chloroplast import (von Heijne et al., 1989) are not present in the N-terminal extensions of plant HLL-like proteins. The mutation in hll-2 terminates the protein at the end of this putative leader sequence; therefore, no functional L14-like protein would be made. In contrast, the hll-1 mutant protein would include both the putative leader sequence and part of the mature protein.
HLL and HLP Localize to Mitochondria
To determine the subcellular location of HLL and HLP, vectors for the production of translational fusions with the green fluorescent protein (GFP) of Aequorea victoria (Haseloff et al., 1997) were constructed for both proteins. GFP was fused to the C terminus of both proteins to avoid disruption of potential N-terminal targeting signals. The chimeric coding regions, under the control of the 35S promoter of CaMV, were expressed transiently in onion epidermal cells. Figure 4A shows a control experiment using GFP alone in which GFP was observed throughout the cytoplasm and nucleus. In contrast, both the HLL-GFP and HLP-GFP fusions were restricted to small bodies distributed throughout the cytoplasm (Figures 4B and 4D). When stained with MitoTracker Red, a mitochondrial stain (Poot et al., 1996), the punctate fluorescence of the dye corresponded exactly with the GFP-fluorescing bodies. Similar results were obtained in examination of roots from Arabidopsis plants stably transformed with a 35S::HLL-GFP transgene (Figures 4F and 4G). The structural complexity of the root results in variable penetration of the MitoTracker Red stain and difficulty visualizing GFP fluorescence. Thus, not all mitochondria fluoresce equally in both Figures 4F and 4G, and there is also artifactual staining of plant cell walls by MitoTracker Red. However, the significant correspondence of punctate fluorescence between the images indicates mitochondrial localization of HLL-GFP in Arabidopsis. The results of these localization experiments demonstrate that HLL and HLP include information sufficient for protein targeting to mitochondria.
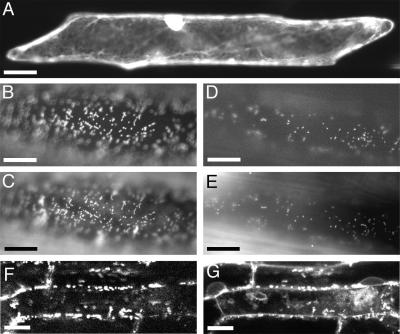
Figure 4.
Subcellular Localization of HLL-GFP and HLP-GFP Translational Fusions.
Images of onion epidermal cells transiently transformed with various constructs and Arabidopsis root cells stably transformed with a 35S::HLL-GFP transgene were recorded using filters for detection of GFP ([A], [B], [D], and [F]) or MitoTracker Red ([C], [E], and [G]).
(A) An onion cell transformed with a 35S::GFP plasmid. GFP accumulates in the cytoplasm and diffuses into the nucleus.
(B) An HLL-GFP fusion protein is restricted to small bodies in the cytoplasm in a cell transformed with a 35S::HLL-GFP plasmid.
(C) These same bodies also fluoresce with MitoTracker Red, a mitochondrial stain.
(D) An HLP-GFP fusion protein is restricted to similar small bodies in a cell carrying a 35S::HLP-GFP plasmid.
(E) These same bodies show MitoTracker Red staining.
The positions of fluorescence may not correspond exactly when comparing (B) with (C) and (D) with (E) because of cytoplasmic streaming of mitochondria during the time required for filter changes.
(F) and (G) Confocal images of Arabidopsis root cells containing a 35S::HLL-GFP transgene show punctate fluorescence in subcellular bodies that also stain with MitoTracker Red (G).
Bar in (A) = 40 μm; bars in (B) to (E) = 20 μm; bars in (F) and (G) = 5 μm.
hll Phenotype Can Be Complemented by Ectopic HLP Expression
To determine if HLP could substitute functionally for HLL in plants, a 35S::HLP transgene was introduced into heterozygous hll-1 plants. Homozygous hll-1 transgenic progeny were complemented by the transgene in 8 of 10 independent lines (data not shown). This result indicates that HLP can substitute functionally for HLL. We observed no other phenotypic effects of ectopic production of the HLP protein on the plants.
Expression Analysis of HLL and HLP
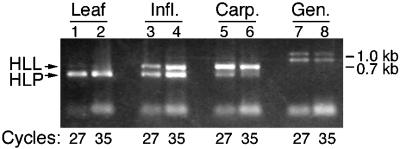
Because HLL and HLP proteins may be functionally equivalent, the expression patterns of the corresponding genes were investigated to help understand the ovule and gynoecium phenotypes of hll mutants. Using gene-specific primers, reverse transcriptase–mediated (RT)-PCR was used to compare the levels of mRNA from the two genes in wild-type leaves, inflorescences, and carpels. Amplifications were performed simultaneously for both genes in single PCR reactions to allow direct comparisons of the amount of mRNA from each gene in a sample. To ensure that the reactions had not reached saturation, product was measured at 20, 27, and 35 cycles. Figure 5 shows that products were obtained for both genes in all tissues. However, more HLL product than HLP product was produced from carpel mRNA. Conversely, more HLP product than HLL product was amplified from leaf mRNA. mRNA from whole inflorescences, including floral organs and meristems, produced similar levels for each gene.
Figure 5.
RT-PCR Analysis of HLL and HLP Shows Differential Expression in Leaves and Carpels.
PCR was performed on first-strand cDNA from rosette leaves (Leaf), inflorescences with unopened flowers (Infl.), carpels dissected from flowers before anthesis (Carp.), and on genomic DNA (Gen.). Primers for both HLL and HLP products were included in each reaction, and products were analyzed after 20, 27, and 35 cycles to enable a more quantitative comparison between the products. No product was visible after 20 cycles.
DISCUSSION
Our cloning of HLL shows that it encodes a protein similar to L14 ribosomal proteins of eubacteria. L14 ribosomal proteins are known to bind rRNA (Herold and Nierhaus, 1987; Urlaub et al., 1995; Ban et al., 1999), participate in a bridge between the large and small ribosomal subunits (Yusupov et al., 2001), and have been hypothesized to interact with regulatory factors important for controlling translation (Ban et al., 1999). L14 is an essential protein required for translation in E. coli (Neidhardt and Curtiss, 1996). Despite variation among ribosomes in both total numbers of proteins and protein sequences, L14 proteins are present and relatively well conserved in all ribosomes characterized from a variety of species (Wool et al., 1995).
We derive several lines of evidence indicating that both HLL and HLP are mitochondrial ribosomal L14 proteins. The presence of unusual insertions and the overall sequence divergence in HLL and HLP relative to other eubacterial and plastid sequences could be considered indications that HLL and HLP were derived from L14 proteins but now have distinct roles. However, alignment with L14 of B. stearothermophilus, for which the crystal structure has been solved, indicates that the insertions should be within loops and would not disrupt the overall β-barrel or α-helical structures of the proteins (Figure 3A; Davies et al., 1996). Other comparative studies have shown that mitochondrial ribosomal proteins are generally less conserved than those of cytosolic ribosomes (Goldschmidt-Reisin et al., 1998; Graack and Wittmann-Liebold, 1998; Graack et al., 1999). The unique features of HLL and HLP and the relatively long branches in this part of the phylogenetic tree may reflect simply a similar divergence. Because the other L14 and L23 genes in Arabidopsis clearly represent the chloroplast and cytosolic forms of this protein family, HLL and HLP are the only candidates to encode mitochondrial L14 proteins. The placement of the products of both genes within the eubacterial group in phylogenetic analysis is consistent with the genes deriving from mitochondria. HLL and HLP were both found to include functional mitochondrial targeting signals. The ability of HLP to substitute functionally for HLL indicates that the two proteins have similar or identical functions. Given the functional redundancy and localization of HLL and HLP, and because they are the only candidates for mitochondrial L14 proteins, we conclude that both function as this class of ribosomal protein.
Mitochondria are widely believed to have evolved from α-proteobacteria (Andersson et al., 1998; Lang et al., 1999) and retain varying amounts of the eubacterial genome, including genes encoding rRNA, tRNA, and some protein-coding genes (Gray et al., 1999). Mitochondria must have functional ribosomes for translation of the products of the retained protein-coding genes. Given the apparent role of HLL in mitochondrial ribosomes, the cell collapse phenotype of hll mutants is likely to be caused by insufficient or defective translation of the 34 mRNAs encoded by the Arabidopsis mitochondrial genome (Unseld et al., 1997). These mRNAs include those encoding other ribosomal proteins and subunits of respiratory chain enzymes (Unseld et al., 1997). The effect of this on plant cells is likely to be complex and highly damaging, because the mitochondria of higher plants are required for carbon backbone synthesis and the final steps of some amino acid biosynthesis pathways, in addition to their role in energy production. These processes are essential for the cell; thus, loss of HLL may slow growth and cause cell collapse. Cytochrome c, a component of mitochondrial membranes, has been implicated as a trigger of cell death in plants (Jones, 2000; Loeffler and Kroemer, 2000). The cellular degeneration of hll ovules, therefore, also could be a result of leakage of cytochrome c into the cytoplasm from structurally compromised mitochondria. The variability of the hll phenotype is likely to be a result of a stochastic decrease in mitochondrial function resulting from variable rates of depletion of functional L14 proteins through cell division and protein turnover during ovule development.
The reduced phenotypic severity of hll-2 relative to hll-1 is surprising because the shorter hll-2 protein comprises only the mitochondrial transit peptide and thus is likely to be a null allele. The mutational change in the hll-2 transcript, at the first base of exon 3, may disrupt splicing, but this is unlikely to yield functional product because there are no close alternative acceptor sites that would preserve the reading frame. The hll-1 protein, including both the transit peptide and part of the mature protein, may have compromised function, but still it may be able to bind rRNA or other ribosomal proteins. This could produce a weak antimorphic effect that is revealed only in the absence of competing wild-type protein and that would not be apparent in heterozygous plants.
Our demonstration that ectopic expression of HLP can compensate for the loss of HLL activity shows that the normal levels of HLP expression in ovules must not be sufficient for this purpose. Consistent with this, we found that HLL was expressed preferentially in the gynoecium relative to HLP. Thus, the relative specificity of the effects of hll mutations on ovule development could be the result of differential expression of the functionally redundant genes HLL and HLP. This hypothesis is consistent with the previous observation that small gene families of some cytosolic ribosomal proteins in Arabidopsis are under developmental regulation and, when mutated, cause tissue-specific phenotypes (Larkin et al., 1989; Van Lijsebettens et al., 1994; Williams and Sussex, 1995; Ito et al., 2000). In addition, increased numbers of mitochondria have been observed in reproductive tissue (MacKenzie and McIntosh, 1999), and transgenic plants with reduced mitochondrial citrate synthase activity show ovary-specific degeneration (Landschütze et al., 1995), indicating a high energy requirement in carpels and ovules. This enhanced sensitivity to disruptions in mitochondrial function also could contribute to the specificity of the effects of hll mutations on ovules.
The synergistic interactions exhibited by hll with ant (Schneitz et al., 1998) and sin2 (Broadhvest et al., 2000) allow speculation on the result of limiting the metabolic capacity of cells during growth and division. ANT and SIN2 are both promoters of growth of ovule primordia. ANT is expressed in all primordia (Elliott et al., 1996) and has been shown to be associated with the maintenance of meristematic activity in organs (Krizek, 1999; Mizukami and Fischer, 2000). In strong ant mutants, integument primordia do not form (Elliott et al., 1996; Klucher et al., 1996; Baker et al., 1997). The sin2 mutant has severely reduced integuments as a result of a reduction in cell divisions and subtle pleiotropic defects in other floral organs (Broadhvest et al., 2000). The ant-72F5 hll-2 double mutant lacks both integuments and a funiculus (Schneitz et al., 1998), and the sin2 hll double mutant shows even stronger synergism as ovule primordia arrest and cells collapse soon after primordia initiation (Broadhvest et al., 2000). Thus, in two cases, the effects of reduction in growth-promoting activity (as in the ant and sin2 mutants) are exacerbated by a reduction in metabolic competence, resulting in a dramatic alteration in growth and form. The observation that the effects of hll on ovule form differ in the wild type and the two mutants shows that metabolic changes can have potentially regulated effects on form.
Development is dependent on metabolism, and plants often show variation in normal development, depending on their metabolic health. An important question is whether plants exploit metabolism as an avenue for control of morphology. We observed that an apparent metabolic mutant shows specific developmental effects on its own and produces alterations in patterning when combined with mutations in developmental regulators. Although the changes we observed were associated specifically with the effects of mutations, effects of similar magnitude could result from alterations in metabolic gene expression as part of normal plant development. This raises the possibility that some developmental regulators could manifest their effects through the modulation of basic metabolic processes. Metabolic regulation thus should be considered as one possible effector pathway when constructing and evaluating models of genetic regulation of morphogenesis.
METHODS
Plant Growth and Genetic Mapping
Arabidopsis thaliana plants were grown as described previously (Kranz and Kirchheim, 1987; Robinson-Beers et al., 1992). Isolation of the hll alleles in the Landsberg erecta (Ler) ecotype has been described previously (Schneitz et al., 1997, 1998). hll was mapped previously to a region on chromosome 1 flanked by nga 63 and nga 248 using a segregating F2 population resulting from a cross between hll-1 (Ler) and wild-type Columbia (Col-3) (Schneitz et al., 1998). Further analysis showed cosegregation with a 6.5-centimorgan region between NCC1 (Konieczny and Ausubel, 1993) and g2395 (Nam et al., 1989; Hardtke and Berleth, 1996). Using these flanking cleaved-amplified polymorphic sequence markers and two additional included markers, m59 and 10F7R (Hardtke and Berleth, 1996), five informative recombinants were identified from among nearly 1,000 F2 plants. These five plants were self-fertilized, and F3 individuals homozygous for the recombinant chromosomes were used to map hll-1 to an interval contained within yeast artifical chromosome yUP10F7 (Guzmán and Ecker, 1988) between markers 10F7R and 19E6L (a left end rescue of yeast artifical chromosome yUP19E6). A contig of three overlapping bacterial artificial chromosome (BAC) clones (T25P10 [which included 10F7R], T24L7, and T19K13 [which included 19E6L]) (Choi et al., 1995) spanning the region including HLL was assembled from hybridization and fingerprint data.
Cloning and Plant Transformation
Cosmid and plasmid subclone libraries were constructed by partial digestion of BAC or cosmid DNA with Sau3AI, followed by size fractionation on a sucrose gradient (25% sucrose, 1 M NaCl, 10 mM EDTA, and 5 mM Tris-HCl, pH 8). Cosmid libraries were constructed by inserting 15- to 20-kb BAC fragments (from BACs T19K13, T24L7, and T25P10) into the BamHI site of vectors pOCA28 (Olszewski et al., 1988) or pOCA28-15 (Eshed et al., 1999). Fragments of 4 to 8 kb from complementing cosmids were cloned into the BamHI site of BJ97 (Gleave, 1992; B. Janssen, unpublished data) for sequencing and further characterization. pBJ97-13 is one such plasmid subclone of cosmid T24L7-143 that includes a 3.5-kb genomic fragment containing HLL and a tRNA gene. The insert of pBJ97-13 was removed as a NotI fragment and was cloned into pMLBART (B. Janssen, unpublished data), creating plasmid pDS08 for plant transformation.
An HLL cDNA was obtained by reverse transcriptase–mediated polymerase chain reaction (RT-PCR) (Veres et al., 1987) from flower mRNA using the primers HLLF2 (5′-CGTAAGCTTGGTGTCTTTGTGCAATCGC-3′) and HLLR1 (5′-GCCTCGAGTCACATAAAGGAGATC-3′) on first-strand oligo(dT)-primed cDNA (SuperScript II; Invitrogen, Carlsbad, CA). The resulting fragment was cloned into the EcoRV site of pLITMUS28 (New England Biolabs, Beverly, MA), creating pDS13 (confirmed by sequence determination). To make 35S::HLL constructs, the XbaI-BglII fragment from pDS13 was subcloned into the XbaI-BamHI sites of pMON999 between the 35S promoter of Cauliflower mosaic virus (CaMV) (with a duplication of the enhancer region [Kay et al., 1987]) and the nopaline synthase (nos) poly(A) addition site, creating pDS23. The NotI fragment from this plasmid, containing 35S::HLL and the nos 3′ poly(A) site, was subcloned into the NotI site of pMLBART, creating plasmid pDS24 for transfer into plants. Similarly, HLP cDNA was obtained using RT-PCR of leaf mRNA with the primers HLPF2 (5′-ATTCTAGAGCAGAAATGGCCG-CTGCTTT-3′) and HLPR1 (5′-GTACTAGTGGGTAATACTCCGAGACAG-3′). The PCR product was cloned into the EcoRV site of pBluescript KS− (Stratagene, La Jolla, CA) to make pDS29. To create a 35S::HLP construct, the XbaI-KpnI fragment from pDS29 was cloned into pMON999, creating pDS36. The NotI fragment from this plasmid was subcloned into pMLBART, creating plasmid pDS38 for transfer into plants.
For plant transformation, vectors were transferred into Agrobacterium tumefaciens strain ASE (Fraley et al., 1985) by triparental mating (Figurski and Helinski, 1979). Transformations were performed on wild-type Ler or on F2 progeny of a cross between hll-1 (Ler) and the wild type (Col-3) (in tests for complementation) by the in planta procedure (Bechtold et al., 1993). Transformed plants were selected using Basta (for pOCA28-15 and pMLBART vectors) or kanamycin (for pOCA28 vector). Plants were genotyped using the HLL-proximal cleaved-amplified polymorphic sequence markers m59 and g2395BH (g2395 modified to 5′-CTCCTTGTCTCCAGGTCCC-3′ [forward] and 5′-GGAGGTTGGTGATGGAACC-3′ [reverse]) to distinguish complemented hll-1 (Ler) homozygotes from heterozygous or wild-type plants.
Analytical RT-PCR
Poly(A)+ RNA was extracted from rosette leaves, inflorescences with unopened flowers, and isolated preanthesis pistils using the mRNA Direct kit (Dynal, Oslo, Norway) according to the manufacturer's instructions. Approximately 10 ng of RNA was used to make first- strand cDNA with an oligo(dT) primer using SuperScript II reverse transcriptase (Gibco BRL, Invitrogen). One-tenth of the product of first-strand cDNA synthesis was used in PCR reactions. Gene-specific primers for the expression analysis were as follows: HLLF2 and HLLR1 for HLL and HLPF2 and HLPR2 (5′-TTGGATCCCTCCCGC-AACGTGTTGAGCCAAAGC-3′) for HLP. Reactions of 50 μL containing all four primers at 0.2 μM each and either cDNA or genomic DNA were split into three 16-μL reactions, and PCR was performed simultaneously on all samples. One tube of each template type was removed at 20, 27, and 35 cycles, and half of each resulting product was analyzed by agarose gel electrophoresis.
Protein Localization
To enable construction of translational fusions with green fluorescent protein (GFP) (Haseloff et al., 1997), the stop codons of HLL and HLP were replaced with glycine codons by PCR. pDS13 was used as a template for modification of HLL using primers HLLF3 (5′-ACT-CGAGTCTAGAATGGCGACAGCTTTAGCTTC-3′) and HLLR2 (5′-GGC-TCAGCACATTGTTGGAGGGATCCA-3′). The product was cloned into the EcoRV site of pLITMUS28, creating pDS15. The XbaI-BamHI fragment from pDS15 was cloned into pGFP1.1.5 (Schumacher et al., 1999) to construct pDS17, resulting in a C-terminal fusion of GFP to HLL. The XbaI-SacI fragment containing the fused coding regions was cloned into the same sites of pMON999, creating pDS20 (35S::HLL:GFP). A NotI fragment containing this expression cassette was cloned into the NotI site of pMLBART, creating pDS22. Total first- strand cDNA was used as a template for direct amplification of a modified HLP cDNA using primers HLPF2 and HLPR2. Using cloning steps identical to those for the modified HLL cDNA, the 35S::HLP:GFP expression vector pDS27 was constructed.
The PDS1000/He biolistic transformation system (Bio-Rad, Hercules, CA) was used to transform onion cells for transient expression of HLL-GFP and HLP-GFP fusions. Yellow onions were obtained locally, and epidermal peels were made from the adaxial surface of the bulb leaves. Epidermis was placed abaxial (inner) side up on solid media plates containing 0.5 × Murashige and Skoog (1962) salts (Gibco BRL and Invitrogen), B5 vitamins (per liter: 10 μg of thiamine HCl, 1 μg of pyridoxine HCl, 1 μg of nicotinic acid, and 100 μg of myo-inositol), 2.5% sucrose, and 12.5 μg/mL chloramphenicol. Plasmid DNA was purified on Qiaprep Spin Miniprep columns (Qiagen, Inc., Valencia, CA) and used to coat 1.5- to 3.0-μm gold particles (Sigma) according to the Bio-Rad transformation system protocol: 50 μg of DNA was precipitated onto 3 mg of gold particles using 50 μL of 2.5 M CaCl2 and 20 μL of 100 mM spermidine. The coated beads were resuspended in 100% ethanol, and one-fifth of the beads were used in each biolistic delivery with a rupture disk pressure of 1100 p.s.i. Each construct was analyzed in at least three bombardment events from each of two separate precipitations. Samples were observed 1 to 4 days after bombardment using a fluorescein isothiocyanate filter set on a Zeiss (Oberkochen, Germany) Axioscope microscope. More than 20 transformed cells were observed in each sample. Tissue samples were stained with 400 nM MitoTracker Red CMXRos (Molecular Probes, Eugene, OR) for 5 to 10 min and viewed with tetramethylrhodamine isothiocyanate filters. Images were recorded using the Openlab system (Improvision, Inc., Lexington, MA) and were adjusted for contrast in Photoshop 6.0 (Adobe Systems, Inc., San Jose, CA).
Roots of Arabidopsis pDS22 primary transformants that expressed the HLL:GFP fusion were stained with MitoTracker Red as described in Results and observed using a Leica TCS-SP laser scanning confocal microscope (Mannheim, Germany). GFP emission was detected between 510 and 550 nm, and MitoTracker Red was detected between 580 and 600 nm, with excitation supplied by Ar (488 nm) and Kr (568 nm) lasers, respectively.
Sequence and Phylogenetic Analyses
DNA was sequenced with the ABI Prism 377 system (Applied Biosystems, Foster City, CA) and assembled using Sequencher 3.0 software (GeneCodes Corp., Ann Arbor, MI). Database searches were performed at the National Center for Biotechnology Information (http://www.ncbi.mln.nih.gov/), the Institute for Genomic Research (http://www.tigr.org/tdb/), The Arabidopsis Information Resource (http://www.arabidopsis.org), and the Monsanto Rice Genome Database (http://www.rice-research.org). Alignments were performed using Clustal X version 1.8 for Macintosh (Thompson et al., 1997) and refined by hand. Phylogenetic trees were generated using PAUP* 4.0b6 (Sinauer Associates, Sunderland, MA). The BLOSUM62 substitution matrix (Henikoff and Henikoff, 1992) was used both in assembling alignments and in parsimony analysis (as implemented for PAUP*; R.K. Kuzoff, personal communication).
Scanning Electron Microscopy
Samples were prepared and examined as described (Broadhvest et al., 2000). Digital images were adjusted using Photoshop 5.5 and 6.0 (Adobe Systems, Inc.).
Accession Numbers
The GenBank/DDBJ/EMBL accession numbers for HLL, HLP, and rice HLL are AF402993, AF402992, and AF402991, respectively.
GenBank accession numbers and/or other sources for the sequences used are as follows: eukaryote cytosol: Arabidopsis thaliana (AC002332), Brugia malayi (AAB07464), Drosophila melanogaster (AAF46914), Glycine max (AW458459), Homo sapiens (AAH10114), Nicotiana tabacum (Q07760), Oryza sativa (AAK27802), Pinus taeda (AW064722), Saccharomyces cerevisiae (NP_009476), Zea mays (NP_043060); Archaea: Aeropyrum pernix (BAA79960), Methanobacterium thermoautotrophicum (G69039), Methanococcus jannaschii (AAB98455), Pyrococcus abyssi (CAB50062), Sulfolobus solfataricus (AAK40722); eubacteria: Aquifex aeolicus (O67570), Bacillus subtilis (R5BS4B), Campylobacter jejuni (CAB73683), Caulobacter crescentus (AAK23239), Escherichia coli (P02411), Helicobacter pylori (AAD06795), Micrococcus luteus (S29882), Mycobacterium tuberculosis (E70643), Neisseria meningitidis (B81232), R. prowazekii (G71670), Synechococcus sp (BAA22459), Thermotoga maritima (B72249), Thermus aquaticus (S15437), Treponema pallidium (AAC65184), Xylella fastidiosa (AAF83972); chloroplast: A. thaliana CHL14 (P56792), Chlamydomonas reinhardtii CHL14 (R5KM14), Cyanophora paradox CHL14 (P23405), Guillardia theta CHL14 (AAC35713), Marchantia polymorpha CHL14 (NP_039336), Mesostigma viride CHL14 (AAF43807), Nephroselmis olivacea CHL14 (NP_050823), Odontella sinensis CHL14 (CAA91638), O. sativa CHL14 (R5RZ14), Porphyra purpurea CHL14 (S73225), Z. mays CHL14 (CAA60322); mitochondria: R. americana MTL14 (AAD11878), S. cerevisiae MTL14 (P35996), Schizosaccharomyces pombe MTL14 (CAA21892); HLL homologs: A. thaliana HLL (this work), A. thaliana HLP (this work), Glycine max (AW832411), Hordeum vulgare (BE558440), Lycopersicon esculentum (BG134949), Medicago truncatula (AW586632), O. sativa (this work), and Sorghum bicolor (BE363626, BG323131).
Acknowledgments
We thank Jacinto Villanueva, Bernard Hauser, and Jessica McAbee for advice and support; Theresa Hill and Robert Kuzoff for helpful comments on the manuscript; Daphne Preuss for the hll-1 mutant; Christian Hardtke, Thomas Berleth, Theresa Hill, and Robert Kuzoff for generously providing information before publication; Bart Janssen, Neil Olszewski, Yuval Eshed, and the Arabidopsis Biological Resource Center at Ohio State University for clones and cloning vectors; Lawrence Eng and Roderick Kumimoto for help with mapping; Jodi Nunnari and Sean Burgess for assistance in labeling and visualizing mitochondria; and the Monsanto Company for access to the Monsanto Rice Genome Database. This work was supported by National Science Foundation Grant No. IBN-0079434 to C.S.G. and by fellowships from the National Science Foundation Plant Cell Biology Training Program to R.J.M. and the University of California Davis Genetics Graduate Group to D.J.S.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010323.
References
- Andersson, S.G., Zomorodipour, A., Andersson, J.O., Sicheritz-Pontén, T., Alsmark, U.C., Podowski, R.M., Näslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.G. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140. [DOI] [PubMed] [Google Scholar]
- Baker, S.C., Robinson-Beers, K., Villanueva, J.M., Gaiser, J.C., and Gasser, C.S. (1997). Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145, 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, S., and Schneitz, K. (2000). NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development 127, 4227–4238. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Capel, M., Moore, P.B., and Steitz, T.A. (1999). Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature 400, 841–847. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Life Sci. 316, 1194–1199. [Google Scholar]
- Bowman, J.L., Sakai, H., Jack, T., Weigel, D., Mayer, U., and Meyerowitz, E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–615. [DOI] [PubMed] [Google Scholar]
- Broadhvest, J., Baker, S.C., and Gasser, C.S. (2000). SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 155, 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R.A. (1995). Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Plant Mol. Biol. Rep. 13, 124–128. [Google Scholar]
- Conley, C.A., and Hanson, M.R. (1995). How do alterations in plant mitochondrial genomes disrupt pollen development? J. Bioenerg. Biomembr. 27, 447–457. [DOI] [PubMed] [Google Scholar]
- Davies, C., White, S.W., and Ramakrishnan, V. (1996). The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus. Structure 4, 55–66. [DOI] [PubMed] [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q.J., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Figurski, D.H., and Helinski, D.R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley, R.T., Rogers, S.G., Horsch, R.B., Eichholtz, D.A., Flick, J.S., Fink, C.L., Hoffmann, N.L., and Sanders, P.R. (1985). The SEV system: A new disarmed Ti plasmid vector for plant transformation. Bio/Technology 3, 629–635. [Google Scholar]
- Gaiser, J.C., Robinson-Beers, K., and Gasser, C.S. (1995). The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7, 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, C.S., Broadhvest, J., and Hauser, B.A. (1998). Genetic analysis of ovule development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 1–24. [DOI] [PubMed] [Google Scholar]
- Glaser, E., Sjoling, S., Tanudji, M., and Whelan, J. (1998). Mitochondrial protein import in plants. Plant Mol. Biol. 38, 311–338. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Reisin, S., Kitakawa, M., Herfurth, E., Wittmann-Liebold, B., Grohmann, L., and Graack, H.R. (1998). Mammalian mitochondrial ribosomal proteins: N-terminal amino acid se-quencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 273, 34828–34836. [DOI] [PubMed] [Google Scholar]
- Graack, H.R., and Wittmann-Liebold, B. (1998). Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem. J. 329, 433–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graack, H.R., Bryant, M.L., and O'Brien, T.W. (1999). Identification of mammalian mitochondrial ribosomal proteins (MRPs) by N-terminal sequencing of purified bovine MRPs and comparison to data bank sequences: The large subribosomal particle. Biochemistry 38, 16569–16577. [DOI] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. (1999). Mitochondrial evolution. Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Guzmán, P., and Ecker, J.R. (1988). Development of large DNA methods for plants: Molecular cloning of large segments of Arabidopsis and carrot DNA into yeast. Nucleic Acids Res. 16, 11091–11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1996). Genetic and contig map of a 2200-kb region encompassing 5.5 cM on chromosome 1 of Arabidopsis thaliana. Genome 39, 1086–1092. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, B.A., Villanueva, J.M., and Gasser, C.S. (1998). Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics 150, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, B.A., He, J.Q., Park, S.O., and Gasser, C.S. (2000). TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 127, 2219–2226. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Henikoff, J.G. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, M., and Nierhaus, K.H. (1987). Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262, 8826–8833. [PubMed] [Google Scholar]
- Huang, J., Struck, F., Matzinger, D.F., and Levings III, C.S. (1994). Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 6, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M.D., and Newton, K.J. (1991). The NCS3 mutation: Genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Kim, G.T., and Shinozaki, K. (2000). Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 22, 257–264. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Jones, A. (2000). Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci. 5, 225–230. [DOI] [PubMed] [Google Scholar]
- Kay, R., Chan, A., Daly, M., and McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236, 1299–1302. [DOI] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kranz, A.R., and Kirchheim, B. (1987). Handling of Arabidopsis. In Genetic Resources in Arabidopsis, A.R. Kranz, ed (Frankfurt, Germany: Arabidopsis Information Service), pp. 4.1.1–4.2.7.
- Krizek, B.A. (1999). Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236. [DOI] [PubMed] [Google Scholar]
- Landschütze, V., Willmitzer, L., and Müller-Röber, B. (1995). Inhibition of flower formation by antisense repression of mitochondrial citrate synthase in transgenic potato plants leads to a specific disintegration of the ovary tissues of flowers. EMBO J. 14, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B.F., Gray, M.W., and Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Hunsperger, J.P., Culley, D., Rubenstein, I., and Silflow, C.D. (1989). The organization and expression of a maize ribosomal protein gene family. Genes Dev. 3, 500–509. [DOI] [PubMed] [Google Scholar]
- Levings, C.S.I. (1993). Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell 5, 1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Running, M.P., and Meyerowitz, E.M. (1997). TSO1 functions in cell division during Arabidopsis flower development. Development 124, 665–672. [DOI] [PubMed] [Google Scholar]
- Loeffler, M., and Kroemer, G. (2000). The mitochondrion in cell death control: Certainties and incognita. Exp. Cell Res. 256, 19–26. [DOI] [PubMed] [Google Scholar]
- MacKenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nam, H.G., Giraudat, J., Den Boer, B., Moonan, F., Loos, W.D.B., Hauge, B.M., and Goodman, H.M. (1989). Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell 1, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt, F.C., and Curtiss, R. (1996). Escherichia coli and Salmonella: Cellular and Molecular Biology. (Washington, DC: American Society of Microbiology Press).
- Newton, K.J., and Cole, E.H.J. (1986). Mitochondrial DNA changes in abnormal growth (nonchromosomal stripe) mutants of maize. Proc. Natl. Acad. Sci. USA 83, 7363–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N.E., Martin, F.B., and Ausubel, F.M. (1988). Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 16, 10765–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot, M., Zhang, Y.Z., Krämer, J.A., Wells, K.S., Jones, L.J., Hanzel, D.K., Lugade, A.G., Singer, V.L., and Haugland, R.P. (1996). Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 44, 1363–1372. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V., Davies, C., Gerchman, S.E., Golden, B.L., Hoffmann, D.W., Jaishree, T.N., Kyila, J.H., Porter, S., and White, S.W. (1995). Structures of prokaryotic ribosomal proteins: Implications for RNA binding and evolution. Biochem. Cell Biol. 73, 979–986. [DOI] [PubMed] [Google Scholar]
- Ray, A., Lang, J.D., Golden, T., and Ray, S. (1996). SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development 122, 2631–2638. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruitt, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, G., Sjöling, S., Wallin, E., Wrede, P., Glaser, E., and von Heijne, G. (1998). Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins 30, 49–60. [PubMed] [Google Scholar]
- Schneitz, K. (1999). The molecular and genetic control of ovule development. Curr. Opin. Plant Biol. 2, 13–17. [DOI] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7, 731–749. [Google Scholar]
- Schneitz, K., Hulskamp, M., Kopczak, S., and Pruitt, R. (1997). Dissection of sexual organ ontogenesis: A genetic analysis of ovule development in Arabidopsis thaliana. Development 124, 1367–1376. [DOI] [PubMed] [Google Scholar]
- Schneitz, K., Baker, S.C., Gasser, C.S., and Redweik, A. (1998). Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 125, 2555–2563. [DOI] [PubMed] [Google Scholar]
- Schultz, E.A., Pickett, F.B., and Haughn, G.W. (1991). The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell 3, 1221–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, K., Vafeados, D., McCarthy, M., Sze, H., Wilkins, T., and Chory, J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 13, 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.Y., Leung, T., Ehler, L.K., Wang, C., and Liu, Z. (2000). Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development 127, 2207–2217. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61. [DOI] [PubMed] [Google Scholar]
- Urlaub, H., Kruft, V., Bischof, O., Müller, E.C., and Wittmann-Liebold, B. (1995). Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 14, 4578–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens, M., Vanderhaeghen, R., De Block, M., Bauw, G., Villarroel, R., and Van Montagu, M. (1994). An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 13, 3378–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres, G., Gibbs, R.A., Scherer, S.E., and Caskey, C.T. (1987). The molecular basis of the sparse fur mouse mutation. Science 237, 415–417. [DOI] [PubMed] [Google Scholar]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G., Steppuhn, J., and Herrmann, R.G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180, 535–545. [DOI] [PubMed] [Google Scholar]
- Williams, M.E., and Sussex, I.M. (1995). Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 8, 65–76. [DOI] [PubMed] [Google Scholar]
- Wool, I.G., Chan, Y.L., and Glück, A. (1995). Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 73, 933–947. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., and Noller, H.F. (2001). Crystal structure of the ribosome at 5.5 angstrom resolution. Science 292, 883–896. [DOI] [PubMed] [Google Scholar]