Abstract
The Ycf3 protein is essential for the accumulation of the photosystem I (PSI) complex and acts at a post-translational level. The sequence of Ycf3 is conserved in cyanobacteria, algae, and plants and contains three tetratrico-peptide repeats (TPR). TPRs have been shown to function as sites for protein–protein interactions. The mutations Y95A/Y96A and Y142A/W143A in the second and third TPR repeats lead to a modest decrease of PSI, but they prevent photoautotrophic growth and cause enhanced light sensitivity even though the accumulated PSI complex is fully functional. This phenotype can be reversed under anaerobic conditions and appears to be the result of photooxidative damage. A temperature-sensitive ycf3 mutant, generated by random mutagenesis of a conserved region near the N-terminal end of Ycf3, was used in temperature-shift experiments to show that Ycf3 is required for PSI assembly but not for its stability. Immunoblot analysis of thylakoid membranes separated by two-dimensional gel electrophoresis and immunoprecipitations shows that Ycf3 interacts directly with the PSI subunits PsaA and PsaD, but not with subunits from other photosynthetic complexes. Thus, Ycf3 appears to act as a chaperone that interacts directly and specifically with at least two of the PSI subunits during assembly of the PSI complex.
INTRODUCTION
The thylakoid protein Ycf3 from the green alga Chlamydomonas reinhardtii is essential for the stable accumulation of photosystem I (PSI) (Boudreau et al., 1997; Ruf et al., 1997). The PSI reaction center complex mediates the electron transfer from plastocyanin to ferredoxin in oxygenic photosynthetic organisms (Golbeck, 1994; Scheller et al., 1997; Schubert et al., 1997). PSI contains the primary electron donor P700 (a chlorophyll a dimer) and the electron acceptors A0 (chlorophyll a), A1 (a phylloquinone), and three [4Fe-4S] centers, FX, FA, and FB (Brettel, 1997). The terminal acceptors FA and FB are bound to the PsaC subunit (Høj et al., 1987; Oh-oka et al., 1988), whereas the remaining electron transfer acceptors are bound to the PsaA-PsaB heterodimer (Golbeck and Cornelius, 1986; Høj and Møller, 1986). The PSI complex of plants and some green algae contains eight nucleus-encoded subunits PsaD, PsaE, PsaF, PsaG, PsaH, PsaK, PsaL, and PsaN and five chloroplast-encoded subunits PsaA, PsaB, PsaC, PsaI, and PsaJ. PSI is associated with light-harvesting complex I (LHCI), which consists of at least nine nucleus-encoded proteins in C. reinhardtii (Bassi et al., 1992). The biosynthesis of the PSI complex depends on the coordinated expression of nuclear and chloroplast genes, the targeting of subunits to their proper location within the chloroplast, the association of the various redox cofactors, and the assembly of the subunits. The proper docking of LHCI to PSI is a crucial step because a faulty connection between these two complexes would prevent the transfer of the excitation energy from LHCI to the PSI reaction center. Excess excitation energy can cause the formation of singlet oxygen (1O2) through energy transfer from excited triplet chlorophylls to ground state triplet O2 (Asada, 1994, 1996). These reactive oxygen species cause photooxidative damage especially to photosystem II (PSII), which is considered to be the primary target for photoinhibition (Barber and Andersson, 1992; Hippler et al., 2000).
To date, three thylakoid proteins involved in the stable accumulation of PSI have been identified: BtpA (Bartsevich and Pakrasi, 1997), Ycf3 (Boudreau et al., 1997; Ruf et al., 1997), and Ycf4 (Boudreau et al., 1997). Because translation of the psaA and psaB mRNAs encoding the two reaction center polypeptides is not affected in mutant strains lacking functional ycf3 and ycf4, the products of these two genes appear to act at a post-translational step of PSI biosynthesis (Boudreau et al., 1997; Ruf et al., 1997; E. Boudreau, J. Girard-Bascou, and J.-D. Rochaix, unpublished results). These genes are therefore involved either in the stabilization or in the assembly of the PSI complex. However, their exact roles remain unknown. The BtpA protein appears to act at the level of PSI stabilization (Zak and Pakrasi, 2000). It is an extrinsic membrane protein located on the cytoplasmic side of the thylakoid membrane (Zak et al., 1999). The Ycf4 protein is firmly associated with the thylakoid membrane, presumably through a transmembrane domain (Boudreau et al., 1997). Ycf4 cofractionates with a protein complex larger than PSI upon sucrose density gradient centrifugation of solubilized thylakoids (Boudreau et al., 1997). The Ycf3 protein is loosely associated with the thylakoid membrane and can be released from the membrane with sodium carbonate. This suggests that Ycf3 is not part of a stable complex and that it probably interacts transiently with its partners.
The amino acid sequence of Ycf3, in particular at its N-ter-minal end, is conserved from cyanobacteria to higher plants, and it contains three tetratrico-peptide repeat (TPR) domains. The TPR motif consists of a degenerate 34–amino acid consensus sequence that is often repeated in tandem arrays (Goebl and Yanagida, 1991). TPR domains have been observed to mediate interactions between two independent proteins and thereby assist the assembly of multi-subunit complexes (Das et al., 1998; Young et al., 1998; Prodromou et al., 1999).
Here, we have used random and site-directed mutagenesis of ycf3 to study the role of its product in PSI accumulation. The analysis of several mutants has revealed that Ycf3 is required for the assembly but not for the stabilization of PSI. Although several of these mutants accumulate at least half the amount of PSI complex compared with that of the wild type, and although these complexes are fully functional, the mutants are unable to grow photoautotrophically and are sensitive to light. Furthermore, immunoprecipitations reveal that the Ycf3 protein interacts specifically with at least two PSI subunits, PsaA and PsaD.
RESULTS
Mutagenesis of ycf3
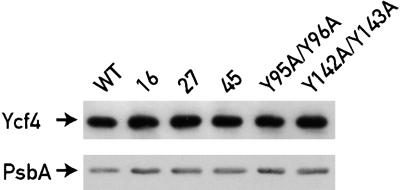
Degenerate oligonucleotides were used to mutate by polymerase chain reaction (PCR) the conserved N-terminal region upstream of the TPR domains (Figure 1). Because more than three mutations could lead to a loss of function of Ycf3 and thereby to the loss of PSI, the oligonucleotide was designed such that Ycf3 with double and triple changes should constitute 18 and 82%, respectively, of the total Ycf3 population (see Methods). The partially degenerate oligonucleotide was flanked by 18 bases on the 3′ side and nine bases on the 5′ side that match the template to allow the PCR to proceed. The amplified DNA of the mutated ycf3 genes was digested with ClaI-ApaI and inserted into a plasmid containing the C. reinhardtii chloroplast 3.6-kb XbaI-EcoRV fragment with the rps9, ycf4, ycf3, and rps18 genes (Boudreau et al., 1997) and with aadA inserted at the KpnI site 200 bp downstream of ycf3. To facilitate the chloroplast transformations with the mutant ycf3 plasmid library, we first constructed a C. reinhardtii strain lacking ycf3 (for details, see Methods). The library was introduced in the chloroplast of this strain, called ycf3Δ, by biolistic transformation (see Methods). Transformants were selected on Tris-acetate-phosphate medium (TAP) plates supplemented with spectinomycin and maintained in dim light (5 μE·m−2·sec−1). Although the Ycf4 protein was not produced in the ycf3Δ strain because the 3′ end of ycf4 is deleted, accumulation of the Ycf4 protein was restored in all transformants (Figure 2). Thus, the phenotype of these mutants is solely the result of the mutation within the ycf3 gene.
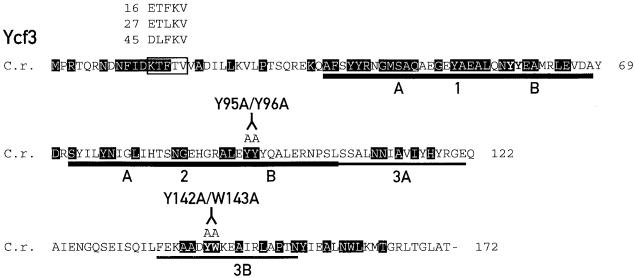
Figure 1.
Mutations within ycf3.
The sequence of Ycf3 from C. reinhardtii (C.r.) is shown. Residues conserved in Ycf3 from liverwort, tobacco, black pine, Odontella sinensis, Cyanidium caldarium, Porphyra purpurea, Cyanophora paradoxa, and Synechocstis PCC 6803 are shaded. The region subjected to degenerate oligonucelotide mutagenesis is boxed, and the changes in the mutants 16, 27, and 45 are indicated. The C. reinhardtii TPR motifs are underlined, and the changes in the TPR domains 2 (Y95A/Y96A) and 3 (Y142A/W143A) are indicated. The regions corresponding to the TPR subdomains A and B are marked, and the borders between the subdomains are indicated with arrowheads.
Figure 2.
Ycf4 Accumulates to Wild-Type Levels in Selected ycf3 Mutants.
Thylakoid protein (10 μg) from the wild type (WT) and ycf3 mutants were separated on a 12% polyacrylamide gel and probed with antibodies against Ycf4 and PsbA. Equal loading of proteins was tested by probing the blot with PsbA antibody.
Transformants were re-streaked once on selective media and were directly analyzed. Because no copy of the ycf3 gene is present in the recipient strain, only the mutated version of ycf3 is expressed, and its phenotype can be readily characterized. A total of 120 transformants were tested for PSI activity by recording fluorescence transients (Bennoun and Delepelaire, 1982). Of these, 90 transformants lacked PSI activity and were discarded. The 30 remaining transformants were tested for their ability to grow on TAP (permissive for mutants of C. reinhardtii deficient in photosynthesis) or high-salt-minimal medium (HSM) plates under different light and temperature regimes. The amounts of Ycf3 and PSI in the mutants were determined by immunoblotting. Most mutants analyzed were sensitive to light at 60 μE·m−2· sec−1. Temperature-sensitive transformants and light-sensitive transformants that accumulate at least 50% of wild-type PSI, as measured by immunoblot analysis, were retained for further characterization. C. reinhardtii cells that accumulate 50% of wild-type PSI complex are normally not light sensitive (Redding et al., 1998). It is therefore likely either that the PSI complexes in these ycf3 mutants have structural defects or that PSI assembly is impaired.
To test the importance of the TPR domains of Ycf3, the conserved tyrosines in domain B of the second TPR and the conserved tyrosine-tryptophan of the third TPR domain were mutated (Figure 1). The ycf3 genes with the mutations Y95A/Y96A and Y142A/W143A were introduced into the chloroplast of the ycf3Δ strain by biolistic transformation, and transformants were selected on TAP plates containing spectinomycin under dim light (5 μE·m−2·sec−1).
Growth Phenotypes and PSI Accumulation in ycf3 Mutants
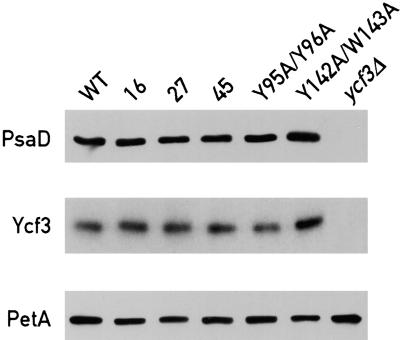
The growth patterns of several ycf3 mutants on TAP and HSM and under different light intensities are shown in Figure 3A. Growth of the TPR mutants Y95A/Y96A (strain 95) and Y142A/W143A (strain 142) is slightly impaired on HSM at a light intensity of 60 μE·m−2·sec−1. Inhibition of growth is more pronounced for the mutants 16, 27, and 45. However, normal growth of the mutants is restored under anaerobic conditions (Figure 3A). Growth of the mutants 27 and 45 is also impaired on TAP at 600 μE·m−2·sec−1 but not in the presence of DCMU or under anaerobic conditions (Figure 3A). The restoration of growth in the presence of DCMU indicates that the photooxidative damage occurs mainly in PSII under these high light conditions. When grown on minimal medium under a light intensity of 45 μE·m−2·sec−1, mutants 16, 27, 45, 95 (Y95A/Y96A), and 142 (Y142A/W143A) are temperature sensitive (Figure 3B). However, the temperature-sensitive phenotype is reversed under anaerobic conditions (Figure 3B). Figure 3C shows the growth patterns upon transfer of the mutant cells from anaerobic to aerobic conditions under medium or high light. Although such a shift did not affect cell survival at 60 μE·m−2·sec−1, it led to significant photooxidative damage at 600 μE·m−2·sec−1 (Figure 3C, lanes 2 and 3). It is interesting that at 600 μE·m−2·sec−1, the psaC K35E mutant affected in electron transfer between PSI and ferredoxin (Fischer et al., 1999) is significantly more light sensitive than are the ycf3 mutants. Figure 4 shows the immunoblot analysis of the selected ycf3 mutants probed with antibodies against PsaD, Ycf3, and PetA (cytochrome f). The resulting bands were quantified relative to the reference PetA band. All five mutants accumulate at least 50% PSI and nearly normal levels of Ycf3 as measured by phosphorimager analysis.
Figure 3.
Growth Patterns of the Wild Type and ycf3 Mutant Strains.
(A) Photosensitivity of ycf3 mutants. Cells were plated on TAP, TAP with 5 μM DCMU, or HSM plates and grown at light intensities of 60 or 600 μE·m−2·sec−1 under aerobic and anaerobic conditions.
(B) Temperature sensitivity of ycf3 mutants. Cells were plated on either TAP or HSM plates and grown at 17, 25, or 34°C at a light intensity of 45 μE·m−2·sec−1. Mutants 16, 27, and 45 are described in Figure 1. The arrangement of the mutants and wild type are indicated in the block scheme. Mutant 95, Y95A/Y96A; mutant 142, Y142A/W143A; dis.ycf3, ycf3::aadA disruption mutant; WT, wild type.
(C) Anaerobic–aerobic transfers. The same mutant cells were used except that the psaC mutant K35E was added (in [B], indicated by K surrounded by a broken circle). Cells were grown for 3 days at 60 or 600 μE·m−2·sec−1 under aerobic (lane 1) or anaerobic (lane 2) conditions. Cells growing anaerobically were then grown aerobically (lane 3) or maintained under anaerobic conditions (lane 4) for 4 additional days.
Figure 4.
PSI Accumulation in the ycf3 Mutants.
Total cell proteins (40 μg) from the wild type (WT) and the ycf3 mutants were separated on a 12% polyacrylamide gel, immunoblotted, and probed with antibodies against PsaD, Ycf3, and PetA (which was used as a loading control).
Characterization of the PSI Complexes from the ycf3 Mutants
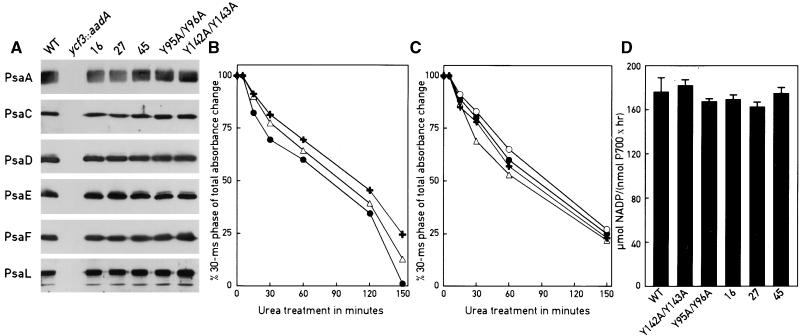
The ycf3 mutants 16, 27, 45, Y95A/Y96A, and Y142A/W143A accumulate PSI to a level that should normally support photosynthetic growth under a light intensity of 60 μE· m−2·sec−1 (Figure 4; Redding et al., 1998). However, the mutants do not grow well under these conditions, possibly because the PSI complexes are not properly assembled in these mutants and/or because their subunit composition is altered. This could impair electron transfer within PSI. Alternatively, critical steps in the PSI assembly process may be considerably slower than they are in the wild type, which could lead to the transient accumulation of assembly intermediates prone to photooxidative damage. To distinguish between these two possibilities, we tested the PSI complexes from the ycf3 mutants for alterations in their subunit composition. PSI from each of the five mutants was purified and examined by immunoblotting with antibodies against the subunits PsaA, PsaC, PsaD, PsaE, PsaF, and PsaL (Figure 5A). All the subunits are present in the mutants, at approximately the same levels as they were in the wild type. As expected, none of the PSI subunits accumulates in the ycf3::aadA mutant.
Figure 5.
Composition and Functional Stability of the PSI Complex from the ycf3 Mutants and the Wild Type Are Undistinguishable.
(A) PSI was purified from each of the mutant strains 16, 27, 45 (see Figure 1 for description), 95 (Y95A/Y96A), 142 (Y142A/W143A), and the wild type (WT). In the case of ycf3::aadA, the thylakoid membrane fraction corresponding to PSI in the wild type was used. Aliquots (5 μg of protein) of each preparation were loaded onto SDS–polyacrylamide gels and analyzed by immunoblotting with the antibodies indicated.
(B) and (C) Stability of PSI in the presence of 6.8 M urea. PSI preparations from the wild type, 16, 27, 45, Y95A/Y96A, and Y142A/W143A were analyzed by flash-induced absorption transients at 817 nm after incubation with urea. Charge recombination between electron acceptors in the PSI complex [FA/FB] −, FX−, A1−, and P700+ was recorded. Before urea treatment, charge recombination is monophasic, consisting exclusively of the [FA/FB] − to P700+ charge recombination of >30 msec. After addition of urea, charge recombination becomes biphasic, consisting of recombinations between [FA/FB] − and P700+ and between FX− and P700+. The proportion of the absorbance change corresponding to the 30-msec phase ([FA/FB] − and P700+) relative to the total absorbance change is shown. In (B), black crosses, Y95A/Y96A; open triangles, wild type; solid circles, Y142A/W143A. In (C), open triangles, 27; crosses, 16; closed circles, wild type; open circles, 45.
(D) In vitro PSI activity of the wild type (WT) and the mutant strains. NADP+ photo reduction of PSI from the indicated strains was measured on isolated PSI complexes as described in Methods. Error bars indicte ±sd.
To test whether PSI of the ycf3 mutants is affected in its functional stability, we treated PSI complexes with 6.8 M urea for increasing lengths of time and followed the intactness of the electron transfer components of PSI by flash-induced absorption changes at 817 nm. In an intact PSI complex carrying all the electron acceptors P700, A0, A1, FX, and [FA/FB], the half-time for charge recombination between [FA/FB] − and P700+ is >30 msec. When PsaC carrying [FA/FB] is dissociated by urea treatment, charge recombination between FX− and P700+ occurs in 1 msec. If FX is destroyed, the observed charge recombination from A1− to P700+ takes place in <1 msec (see Golbeck and Bryant, 1991; Brettel, 1997). The PSI complexes from the wild type and ycf3 mutants were intact before the addition of urea, as evidenced by the presence of a major kinetic component of the absorbance decay corresponding to the 30-msec phase (data not shown). After exposure to urea, the fraction of the 30-msec phase decreased with time to the same extent for the wild-type and mutant PSI (Figures 5B and 5C), thus indicating that the PSI redox cofactors are equally stable in all of these strains. However, it is still possible that there are kinetic differences in some of the intermediate electron transfer steps.
The PSI complexes were further tested by subjecting them to trypsin digestion (data not shown) and assayed for their NADP+ reducing activity (Figure 5D) and their electron transfer kinetics from plastocyanin to P700+ and from PSI to ferredoxin (data not shown). In all cases, the properties of the mutant ycf3 PSI complexes were indistinguishable from those of the wild type. Furthermore, thylakoid membranes of the Ycf3 mutants were solubilized and analyzed by native gel electrophoresis. No size difference between the mutant and wild-type PSI complexes could be detected (data not shown). These results indicate that the PSI complexes in the different ycf3 mutants are correctly assembled.
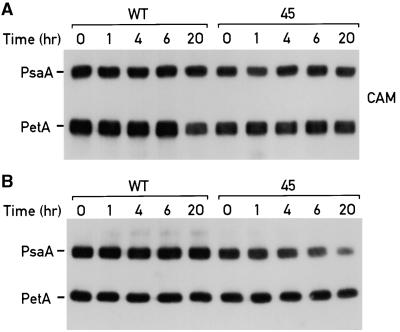
PSI Is Stable but Does Not Accumulate in the Temperature-Sensitive ycf3 Mutant 45 Grown at the Restrictive Temperature
To test further whether Ycf3 is involved in the assembly or in the stability of PSI, we exploited the temperature sensitivity of mutant 45 by performing temperature-shift experiments. Cultures of mutant 45 and the wild type were grown at 18°C with a light intensity of 45 μE·m−2·sec−1 to a cell density of 2.5 × 106 cells mL−1. The cells were pelleted, washed with HSM, and resuspended to exactly 2 × 106 cells mL−1 in HSM. The culture was divided into two parts, incubated for 2 hr at 18°C at a light intensity of 45 μE·m−2·sec−1, and shifted to the restrictive temperature, 34°C. One culture was supplemented with chloramphenicol to inhibit chloroplast protein synthesis at the time of the temperature shift. In this way the stability of PSI could be compared in mutant 45 and the wild type. Samples of cells (150 μL) were removed at 0, 1, 4, 6, and 20 hr after the shift to the restrictive temperature and analyzed by electrophoretic fractionation and immunoblotting, as shown in Figure 6A. The membranes were probed with antibodies against the PSI subunit PsaA and the PetA subunit of the cytochrome b6/f complex. PSI was found to be equally stable in the wild type and mutant 45 (Figure 6A). We therefore conclude that the growth sensitivity at 34°C of the ycf3 mutant 45 is not the result of instability of the PSI complex and that Ycf3 is not required for PSI stability.
Figure 6.
Ycf3 Is Required for PSI Assembly but Not for Its Stability.
Cultures from the wild type (WT) and the ycf3 mutant 45 were grown in 50 mL of TAP at 18°C at a light intensity of 45 μE·m−2·sec−1 until the cell density reached 2 × 106 cells mL−1. Cells were pelleted by centrifugation, washed in HSM, and resuspended to the same concentration in HSM. Each culture was split into two parts. Both cultures were incubated for 2 hr at 18°C at a light intensity of 45 μE·m−2·sec−1 and shifted to 34°C at the same light intensity.
(A) Chloramphenicol (CAM; 200 μg·mL−1) was added to one culture at the same time as the temperature shift to inhibit chloroplast protein synthesis. Samples of 150 μL were transferred to 1.5 mL of cold acetone (−20°C) at 0, 1, 4, 6, and 20 hr after the shift to 34°C. The acetone-precipitated cells were boiled in sample buffer, loaded on a 12% SDS–polyacrylamide gel, and immunoblotted.
(B) Samples (1 mL) were collected from the other culture at 0, 1, 4, 6 and 20 hr after the shift to 34°C. Samples were solubilized, the protein concentrations were measured, and sample volumes corresponding to 40 μg of protein were boiled in sample buffer, loaded onto 12% SDS–polyacryamide gels, immunoblotted, and probed with antibodies against PetA and PsaA.
The remaining cultures of the wild type and mutant 45 were grown without chloramphenicol. Aliquots of cells (1 mL) were collected at 0, 1, 4, 6, and 20 hr after the shift to 34°C. The protein concentration of the samples was determined, and samples corresponding to 40 μg of protein were fractionated by gel electrophoresis and analyzed by immunoblotting. This analysis reflects the protein accumulation in the growing cultures (Figure 6B). The amount of PSI in the mutant was lower relative to that of the wild type, especially after 20 hr, whereas the accumulation of PetA was very similar in both strains. The PSI complex synthesized before the shift to the restrictive temperature remained stable but constituted a decreasing portion of total cell proteins in a strain growing exponentially. The diminished amount of PSI in mutant 45 upon a shift to the restrictive temperature is therefore the result of impaired assembly of PSI; thus, it is concluded that Ycf3 is required for the assembly of PSI.
Ycf3 Interacts with the PSI Subunits
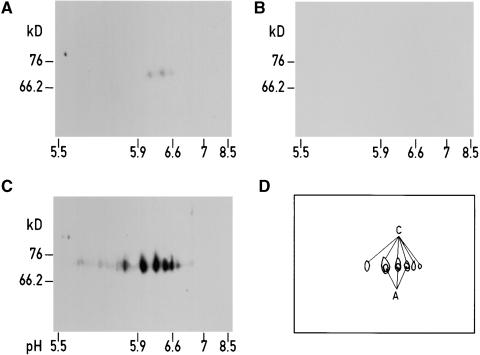
It has been shown previously that although Ycf3 is required for the accumulation of PSI, it does not cofractionate with PSI (Boudreau et al., 1997). However, if Ycf3 is involved in the assembly of PSI, one would expect that it interacts with at least some of the subunits of the PSI complex. To test this hypothesis, we solubilized thylakoid membranes, fractionated the proteins by two-dimensional gel electrophoresis, and blotted them on a membrane (for details see Methods). The membrane was first incubated with recombinant Ycf3 protein that carried a polyhistidine tag at its N-terminal end. Proteins binding to Ycf3 were identified by subsequent incubation with an antibody specific to the tag (Figure 7A). The three spots obtained comigrate with the major signals obtained by immunoblotting of a duplicate membrane with an antibody against the PsaA subunit (Figures 7A, 7C, and 7D). Incubation of the membrane with antibodies against the tag in the absence of Ycf3 did not give rise to any signal (Figure 7B). These results strongly suggest that Ycf3 interacts directly with PsaA.
Figure 7.
Ycf3 Interacts with PsaA.
Wild-type thylakoid membrane proteins (200 μg per gel) were separated by two-dimensional gel electrophoresis on four gels, which were each transferred to nitrocellulose membranes.
(A) The nitrocellulose membrane was incubated with recombinant His-tagged Ycf3 protein and subsequently reacted with His antibodies.
(B) After blotting, the membrane was reacted with His antibody.
(C) After blotting, the membrane was reacted with PsaA antibody.
(D) Superposition of (A) and (C).
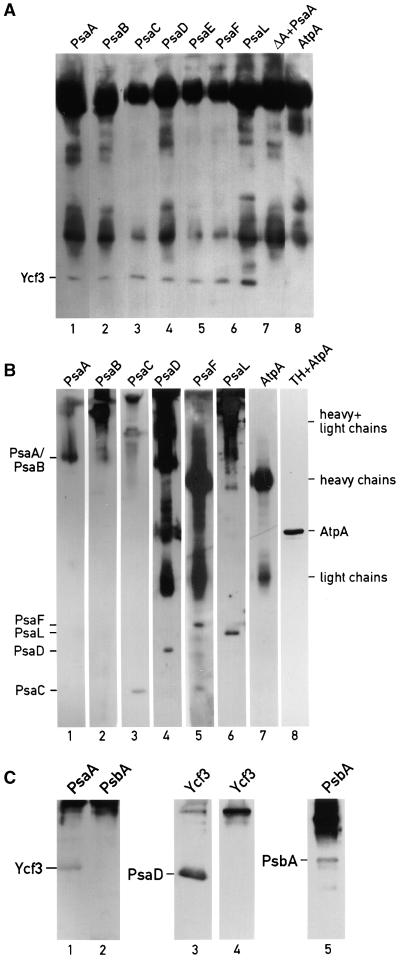
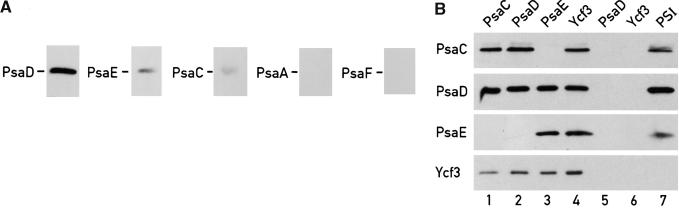
To examine this interaction further and to test whether Ycf3 interacts with other PSI subunits, we solubilized thylakoid and immunoprecipitated them with antibodies against the PSI subunits PsaA, PsaB, PsaC, PsaD, PsaE, PsaF, and PsaL cross-linked to cyanogen bromide–activated Sepharose 4B beads. Beads linked to antibodies raised against the AtpA subunit of the ATP synthase were used as control. Furthermore, thylakoids from the psaA deletion strain (ΔA) that contains wild-type levels of Ycf3 were incubated with PsaA antibody to test that no unspecific interaction occurs between Ycf3 and this PSI-deficient cell extract. The immunoprecipitates were eluted with sample buffer, fractionated by gel electrophoresis, and immunoblotted with Ycf3 antibody, as shown in Figure 8A. Ycf3 is only present in the immunoprecipitates of wild-type thylakoids obtained with the PSI antibodies, but not in the immunoprecipitate obtained with AtpA antibodies (lane 8) or in that recovered with PsaA antibodies reacted with the extract from the psaA deletion strain ΔA (lane 7). The reciprocal immunoprecipitations performed with the Ycf3 antibody and probed with the in-dividual PSI subunit antibodies are shown in Figure 8B. Thylakoid membranes were solubilized and immunoprecipitated with Ycf3 antibody cross-linked to beads. The immunoprecipitates were either eluted with 0.1 M glycine buffer, pH 2.8, leaving the antibody on the beads (lanes 1 to 3), with sample buffer which elutes the antibody from the beads (lanes 4, 5, and 7), or with Laemmli SDS sample buffer without DTT (lane 6; under these conditions, the IgG heavy and light chains do not dissociate). The immunoprecipitated proteins were fractionated by gel electrophoresis and immunoblotted with PsaA, PsaB, PsaC, PsaD, PsaF, PsaL, and AtpA antibodies (Figure 8B). Whereas each of the PSI subunits could be detected in the immunoprecipitate, the AtpA subunit was undetectable (cf. lanes 7 and 8). Additional controls included immunoprecipitations with antibodies against the PSII subunit PsbA (Figure 8C). Ycf3 was undetectable in the immunoprecipitate obtained with PsbA antibody (Figure 8C, lane 2). Furthermore, immunoprecipitates obtained with the Ycf3 antibody contain PsaD but not PsbA (Figure 8C, lanes 3 and 4). The immunoprecipitate obtained with the PsbA antibody was shown to contain the PsbA protein (Figure 8C, lane 5). Taken together, these results indicate that the interactions between Ycf3 and the PSI subunits are specific. Furthermore, thylakoids isolated from the ycf3 mutants 16, 27, 45, 95, and 142 were solubilized and immunoprecipitated with Ycf3 antibody. The immunoprecipitates were analyzed by immunoblotting and found to contain the same amount of PsaL as did wild-type thylakoids immunoprecipitated with Ycf3 antibody (data not shown). Thus, the mutated regions in the five mutants examined are not serving as important sites of interaction with PsaL.
Figure 8.
Ycf3 Co-Immunoprecipitates with PsaA, PsaB, PsaC, PsaD, PsaE, PsaF, and PsaL.
Solubilized thylakoid membranes from the wild-type strain were immunoprecipitated with antibodies cross-linked to cyanogen bromide–activated Sepharose 4B. The Sepharose beads were washed, and the immunoprecipitates eluted. The eluates were fractionated by 10 to 17% polyacrylamide gels and immunoblotted.
(A) Immunoprecipitation with PsaA, PsaB, PsaC, PsaD, PsaE, PsaF, PsaL, and AtpA antibodies. Lane 7 marked ΔA+PsaA contains solubilized thylakoids, from the psaAΔ strain that accumulates wild-type levels of Ycf3 incubated with PsaA antibody. The immunoprecipitates were eluted with Laemmli SDS sample buffer, immunoblotted, and probed with Ycf3 antibody. The antibodies used for the immunoprecipitation are indicated at the top of each lane.
(B) Immunoprecipitation with Ycf3 antibody. The immunoprecipitates were eluted with 0.1 M glycine, pH 2.8 (lanes 1, to 3), with Laemmli SDS sample buffer (lanes 4, 5, and 7), or with Laemmli SDS sample buffer without DTT (lane 6; under these conditions the IgG heavy and light chains do not dissociate). Lane 8 shows an immunoblot of thylakoid membrane proteins reacted with AtpA antibodies. Lanes 1 to 4, 5 to 7, and 8 are from separate gels. The antibodies used for the immunoblots are indicated at the top of each lane. Heavy and light chains refer to the IgG chains.
(C) Immunoprecipitation with PsaA, PsbA (D1), and Ycf3 antibodies. The antibodies used for the immunoprecipitation are indicated at the top of each lane. The immunoblots were probed with Ycf3 (lanes 1 and 2), PsaD (lane 3), and PsbA (lanes 4 and 5) antibodies as indicated.
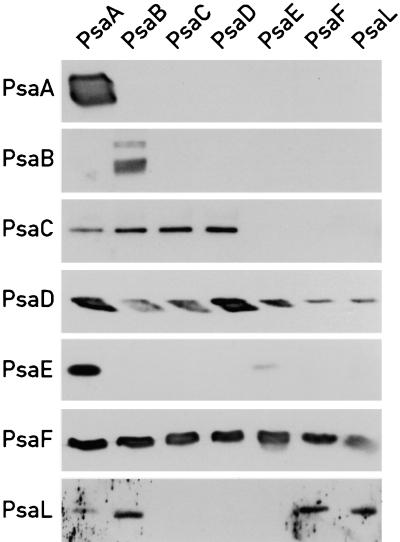
The results obtained could either be explained by an interaction of Ycf3 with the PSI complex or by specific interactions of Ycf3 with individual PSI subunits. To distinguish between these possibilities, the immunoprecipitates obtained with antibodies to individual PSI subunits were probed with the remaining PSI antibodies (Figure 9). In most cases, only a subset of the PSI subunits could be detected in the immunoprecipitate, indicating that the PSI complex is no longer intact under these conditions. In some cases the results of the immunoprecipitations were not reciprocal. As an example, PsaD is immunoprecipitated by PsaE antibody, but PsaE is not immunoprecipitated by PsaD antibody (Figure 9). One possibility is that in the latter case, the antibody binding site overlaps the site of interaction between the two subunits and displaces PsaE. The only subunits that were present in all of the immunoprecipitates were PsaD and PsaF. Thus, one possible interpretation of the results of Figures 7 to 9 is that Ycf3 interacts not only with PsaA but also with PsaD and/or PsaF. The possible interaction between Ycf3 and PsaD was investigated by extracting PSI with butanol. The water phase was shown to contain PsaC, PsaD, and PsaE but not PsaA and PsaF (Figure 10A). It was mixed with solubilized thylakoid membranes from the ΔA mutant that contains wild-type levels of Ycf3 and immunoprecipitated with PsaC, PsaD, PsaE, and Ycf3 antibodies. Ycf3 from PSI-less thylakoids was used in these experiments because the presence of these membranes enhanced the interaction between Ycf3 and the PSI subunits. The immunoprecipitates were fractionated by PAGE and immunoblotted as shown in Figure 10B. In all three immunoprecipitates obtained with PsaC, PsaD, and PsaE antibodies, both Ycf3 and PsaD were present and Ycf3 antibodies immunoprecipitated all three PSI subunits. From these results, we conclude that Ycf3 interacts directly with PsaD in addition to PsaA. Whether Ycf3 also interacts directly with PsaC and PsaE cannot be determined from these experiments because in the absence of Ycf3, immunoprecipitates obtained with PsaE and PsaC antibodies revealed that after butanol extraction, PsaC, PsaD, and PsaE form a complex (data not shown).
Figure 9.
PSI Subunit Content in Immunoprecipitations with PSI Antibodies.
Thylakoid membranes from the wild-type strain were solubilized with 0.9% dodecylmaltoside at a chlorophyll concentration of 0.8 mg·mL−1 and immunoprecipitated with antibodies cross-linked to cyanogen bromide–activated Sepharose 4B. The Sepharose beads were washed, and the immunoprecipitates were eluted with 100 mM glycine, pH 2.8. The eluates were fractionated by 10 to 17% polyacrylamide gels and immunoblotted. The antibodies used for the immunoprecipitations are indicated at the top of each lane. The antibodies used to probe the immunoblots are indicated at left.
Figure 10.
Ycf3 Interacts with PsaD.
PSI preparations were extracted with butanol. The butanol phase containing the membrane proteins of PSI was discarded, and the water phase containing the soluble stromal subunits PsaC, PsaD, and PsaE was saved.
(A) The PsaC, PsaD, and PsaE-enriched extract was fractionated on SDS–polyacrylamide gels, immunoblotted, and probed with PsaA, PsaC, PsaD, PsaE, and PsaF antibodies.
(B) Immunoprecipitation of the PsaC, PsaD, and PsaE-enriched extract. The antibodies used for immunoprecipitation are indicated at the top of the lanes. The antibodies used to probe the immunoblots are indicated at left. Samples were immunoprecipitated with antibodies cross-linked to cyanogen bromide–activated Sepharose 4B. The Sepharose beads were washed, and the immunoprecipitates were eluted with 100 mM glycine, pH 2.8. The eluates were fractionated by 10 to 17% polyacrylamide gels and immunoblotted. Lanes 1 to 4, the PsaC, PsaD, and PsaE-enriched extract was mixed with thylakoid membranes solubilized with 0.9% dodecylmaltoside at a chlorophyll concentration of 0.8 mg·mL−1 from the psaA-deficient mutant ΔA containing Ycf3 in wild-type amounts and immunoprecipitated as indicated; lane 5, solubilized thylakoid membranes from the ΔA mutant were immunoprecipitated with PsaD antibody in the absence of the psaC, PsaD, and PsaE-enriched extract; lane 6, solubilized thylakoid membranes from the ycf3::aadA strain were mixed with the the PsaC, PsaD, and PsaE-enriched extract and immunoprecipitated with Ycf3 antibody; lane 7, purified PSI complex was used for immunoprecipitation.
DISCUSSION
TPR Domains of Ycf3 Are Important
Studies with C. reinhardtii and higher plants have shown that Ycf3 is required for the stable accumulation of the PSI complex (Boudreau et al., 1997; Ruf et al., 1997). To gain new insights into the role of the Ycf3 protein in this process, we introduced changes in specific regions of Ycf3, in particular in its conserved N-terminal domain. This domain appears to play an important role because two-thirds of the random mutations in this region lead to the inactivation of PSI. Ycf3 contains three TPR domains that are thought to be involved in protein–protein interactions and in the assembly of multi-molecular complexes. These TPR domains are therefore an obvious target for mutagenesis. Accordingly, two highly conserved residues, Tyr95-Tyr96 in the second TPR domain and Tyr142-Trp143 in the third TPR domain, were altered. This led to impaired growth of the mutants on minimal medium. Thus, the TPR domains appear to be important for the function of Ycf3. The structure of the TPR domain can be overlaid with the 14-3-3 protein domain (Das et al., 1998). 14-3-3 proteins are regulatory proteins mediating protein–protein contact. This contact is provided uniquely when the 14-3-3 domains of two independent 14-3-3 proteins have dimerized (Liu et al., 1995; Xiao et al., 1995). Preliminary cross-linking studies suggest that Ycf3, like 14-3-3 proteins, forms dimers and that it interacts mostly in the dimer form with the PSI subunits PsaC and PsaD (H. Naver and J.-D. Rochaix, unpublished results). Assembly of multi-subunit complexes requires more than one assembly protein; for example, several TPR domain–containing proteins mediate the assembly of the Hsp90 complex (Prodromou et al., 1999). Thus, it is possible that unidentified chloroplast chaperone proteins assist the Ycf3-mediated PSI assembly. The TPR domains of Ycf3 could be the sites of interaction between Ycf3 and other assembly factors, especially those involved in the assembly of the redox cofactors. Ycf3 could also provide contacts with a precomplex of PSI, although such a complex has not yet been detected.
Ycf3 Mutants Accumulate PSI but Are Light sensitive
The ycf3 mutants 16, 27, 45, Y95A/Y96A, and Y142A/W143A accumulate at least 50% PSI compared with that of the wild type, which is usually sufficient to sustain growth on minimal media under a light intensity of 60 μE·m−2·sec−1 (Redding et al., 1998). Surprisingly, the mutants do not grow well under these light conditions, although they can be rescued under anaerobic conditions. It is well established that mutations affecting electron transfer through PSI can lead to photosensitivity because of the production of reactive oxygen species in PSII and/or PSI (Redding et al., 1998; Rochaix et al., 2000). One possibility, therefore, is that the PSI complex in these mutants is not properly assembled and that its subunit composition is altered. Such a change could affect electron transfer and lead to photooxidative damage. However, both the size of the PSI complexes of these ycf3 mutants as measured by non-denaturing gel electrophoresis and their subunit composition (see Figure 5) are very similar to those of the wild type. In addition, the mutant PSI complexes are as resistant, as are wild-type PSI, to trypsin digestion or 6.8 M urea treatment. Furthermore, the rates of electron transfer on the donor and acceptor side of PSI and the overall electron transfer through the entire PSI complex from plastocyanin to NADP+ are the same in the mutants and in the wild type (Figure 5). Taken together, these results strongly suggest that a fully functional PSI complex, indistinguishable from wild-type PSI, is produced in the ycf3 mutants examined. The light sensitivity of these mutants could be the result of a photosensitive step during the assembly of PSI that is exacerbated in the mutants and that leads to the formation of reactive oxygen species, which enhances the photodamage of PSII. The light sensitivity of at least two of these mutants and their inability to grow on minimal medium under aerobic but not under anaerobic conditions result from photodamage to PSII as indicated by the loss of light sensitivity in the presence of DCMU under high light (Figure 5). Under moderate light (60 μE·m−2·sec−1), the photooxidative damage appears to occur mostly during the assembly of PSI in the ycf3 mutants because a shift from anaerobic to aerobic conditions stops cell growth but does not lead to visible cell damage (Figure 3C). These results suggest that the ycf3 mutations examined in this study affect mostly the rate of assembly rather than the final conformation of PSI. However, we cannot exclude the possibility that subtle changes in the conformation of PSI that are undetectable by the methods used in this study could be the cause of the photosensitivity of the ycf3 mutants, especially under high light.
Ycf3 Is Required for PSI Assembly Rather Than for PSI Stability
One of the mutants isolated, mutant 45, displays a temperature-sensitive growth phenotype on HSM. This mutant was used to show that upon a shift of the growing mutant cells from the permissive to the restrictive temperature, the assembled PSI complex of the mutant is as stable as its wild-type counterpart. However, accumulation of newly synthesized PSI is decreased significantly relative to that of the wild type after a shift to the restrictive temperature. This suggests that Ycf3 is involved in the assembly of the PSI complex but does not play an important role for the stability of the complex.
Ycf3 Interacts with at Least Two PSI Subunits
If Ycf3 is required for the assembly of PSI, it is likely that it interacts with some of the PSI subunits. Using high-resolution two-dimensional gel electrophoresis coupled to immunoblot analysis and immunoprecipitations, it was shown that Ycf3 interacts with at least two of the PSI subunits: PsaA and PsaD. No interaction between Ycf3 and the thylakoid protein PsbA of PSII and the AtpA subunit of ATPase was observed, indicating that the observed interactions with the PSI complex are specific (Figure 8). Because PsaD is present in all the immunoprecipitations with the PSI antibodies, it is possible that PsaD mediates the interaction between Ycf3 and PSI in the Ycf3-PSI immunoprecipitate. However, this possibility does not exclude that Ycf3 is interacting with additional PSI subunits. A specific interaction of Ycf3 with all the PSI subunits appears unlikely, however, given that the assembly requirements for the different PSI subunits are diverse. The iron–sulfur clusters of PsaC are oxygen labile except when the subunit is situated on the PSI complex protected by PsaA, PsaB, and PsaD (Oh-oka et al., 1988; Mehari et al., 1991; Naver et al., 1996, 1998). This subunit is likely to require a chaperone to stabilize its conformation outside the PSI complex. It is likely that the insertion and assembly of the membrane subunits of PSI would involve a different mechanism.
Cyanobacterial and plant PSI can be assembled and remain functional in vivo without each of the following eight subunits: PsaE (Chitnis et al., 1989; Varotto et al., 2000), PsaF (Chitnis et al., 1991; Farah et al., 1995; Haldrup et al., 2000), PsaH (Naver et al., 1999), PsaI (Chitnis and Chitnis, 1993), PsaJ (Xu et al., 1994; Fischer et al., 1999), PsaK (Jensen et al., 2000), PsaL (Xu et al., 1995; Lunde et al., 2000; H. Naver and J.-D. Rochaix, unpublished results), and PsaN (Haldrup et al., 1999). Depletion or inactivation of ycf3 prevents the accumulation of the PSI complex. It is likely that Ycf3 assists the assembly of the two major essential subunits, PsaA and PsaB, that carry most of the PSI redox cofactors and that are required for the accumulation of the other PSI subunits. Earlier studies have revealed that PsaB and PsaA are strongly coupled and that PsaB could act as an anchor protein during PSI assembly (Girard-Bascou et al., 1987; Stampacchia et al., 1997). One possibility is that Ycf3 assists the proper folding and integration of this subunit into the thylakoid membrane and that it participates in the subsequent integration of PsaA and perhaps of some of the other PSI subunits associated with the core complex (Figure 10). Providing contact between different proteins by holding them together is the classical function for TPR proteins (Goebl and Yanagida, 1991; Das et al., 1998). Thus, ycf3 appears to be a PSI-specific chaperone assisting the assembly of the complex through direct interactions with some of its major subunits.
METHODS
Strains and Media
Chlamydomonas reinhardtii wild-type and mutant cells were grown as described (Harris, 1989). The ycf3::aadA mutant specifically deficient in ycf3 was used (Boudreau et al., 1997). Tris-acetate-phosphate medium (TAP) and high-salt-minimal medium (HSM) were solidified with 2% Bacto agar (Difco) and supplemented with spectinomycin (Sigma) or DCMU (ICN Biomedicals, Meckenheim, Germany). The Genebag anaerobic system (Biomérieux, Marcy l'Etoile, France) was used for anaerobic growth conditions for plates.
Nucleic Acid Techniques
Procedures for the preparation of recombinant plasmids and DNA sequencing were performed as described by Sambrook et al. (1989). The bacterial host strain used was DH5α.
Production of a C. reinhardtii ycf3Δ Strain
The pKS rps9-rps18 vector contains the 3.6-kb C. reinhardtii chloroplast XbaI-EcoRV fragment with the open reading frames rps9-ycf4-ycf3-rps18 (Boudreau et al., 1997). The vector has two HincII sites located 256 bp upstream and 10 bp downstream of the ycf3 gene. HincII digestion thus completely excises the ycf3 gene and the 3′end of the ycf4 gene. The aadA cassette which confers spectinomycin resistance to C. reinhardtii (Goldschmidt-Clermont, 1991), flanked by homologous sequences of 483 bp, which allow subsequent excision of aadA by homologous recombination (Fischer et al., 1996), was inserted into the HincII-digested vector. The resulting plasmid was introduced into the chloroplast of C. reinhardtii by biolistic transformation (Boynton et al., 1988). Transformants were selected on TAP plates supplemented with 150 μg·mL−1 spectinomycin and restreaked five times on TAP plates supplemented with spectinomycin. To promote excision of the aadA cassette, the transformants were then replated four times on TAP plates lacking spectinomycin, as described by Fischer et al. (1996). Deletion of ycf3 and the removal of aadA were verified by DNA gel blot analysis (data not shown). This strain also lacks Ycf4. Transformation of this strain with the 3.6-kb XbaI-EcoRV fragment yielded transformants with wild-type phenotype, as measured by growth tests, immunoblot, and fluorescence transient analysis (data not shown). Total DNA was isolated from the wild type and ycf3Δ strain as described by Rochaix et al. (1988).
Production of C. reinhardtii ycf3 Mutants
Degenerate oligonucleotide-directed mutagenesis was performed by polymerase chain reaction (PCR) as described (Fischer et al., 1998), using the 42-mer oligonucleotide 5′-TTTATCGAT(A/G)AA(A/C)CT(C/T)TCA(A/C)(A/G/T/C)G(A/T)(A/G/C/T)GTCGTCGACATCTTATTA-3′ which, together with an oligonucleotide corresponding to the 3′ end, results in amplification of the whole ycf3 gene. The amplified DNA fragments were gel-purified, digested with ApaI-ClaI, and cloned into the pKS rps9-rps18aadA (ApaI-ClaI) vector. This vector consists of pKS rps9-rps18, with the aadA expression cassette (Goldschmidt-Clermont, 1991) inserted at the unique KpnI site 200 bp downstream of ycf3. The vector contains unique sites for ClaI and ApaI restriction endonucleases at the 5′ and 3′ ends of the ycf3 gene.
Site-directed mutagenesis was performed using the following 27-mer oligonucleotides: 5′-TAAAGCTTGAGCAGCATATTCTAAAGC-3′ for Y95A/Y96A and 5′-GCTTCTTTCGCGGCATCTGCCGCTTTT-3′ for Y142A/W143A, which, together with an oligonucleotide corresponding to the 3′ end, result in the amplification of fragments of 297 bp and 428 bp, respectively. The fragments were used as megaprimers together with an oligonucleotide corresponding to the 5′ end of the ycf3 gene for PCR amplification of the whole ycf3 gene. These amplified DNA fragments were gel-purified, digested with ApaI-ClaI, and cloned into the pKS rps9-rps18aadA (ApaI-ClaI) vector. This vector consists of pKS rps9-rps18, with the aadA expression cassette (Goldschmidt-Clermont, 1991) inserted at the unique KpnI site 200 bp downstream of ycf3. The vector contains unique sites for ClaI and ApaI restriction endonucleases at the 5′ and 3′ ends of the ycf3 gene.
Chloroplast transformation in C. reinhardtii was performed as described by Boynton et al. (1988), with a helium-driven particle gun adapted from the one described by Finer et al. (1992). ycf3Δ cells were grown in TAP liquid at 25°C and plated on TAP plates supplemented with 150 μg·mL−1 spectinomycin. Once the plates were dry, the cells were bombarded with tungsten microprojectiles coated with the appropriate DNA. The bombarded cells were incubated for 2 weeks at 25°C at 5 μE·m−2·sec−1. Colonies were restreaked on fresh TAP plates containing 150 μg·mL−1 spectinomycin and characterized. The ycf3 mutant genes in the C. reinhardtii mutant were amplified by PCR on total DNA, purified by gel electrophoresis, and sequenced.
Chloramphenicol Treatment of Temperature-Sensitive Mutants
Cultures of the wild type and ycf3 C. reinhardtii mutant 45 were grown in 50 mL of TAP at 18°C at 45 μE·m−2·sec−1to a cell density of 2 × 106 cells mL−1. Cells were pelleted by centrifugation, resuspended in 50 mL of HSM, centrifuged again, resuspended to exactly 2 × 106 cells mL−1 in HSM, and split into two parts. The cultures were incubated 2 hr at 18°C at a light intensity of 45 μE·m−2·sec−1 and shifted to 34°C at the same light conditions. Chloramphenicol was added at the time of the temperature shift to a concentration of 200 μg·mL−1 to one of the cultures to inhibit chloroplast protein synthesis (Schmidt and Mishkind, 1983), and samples (150 μL) were transferred to 1.5 mL of acetone chilled at −20°C at 0, 1, 4, 6, and 20 hr after the shift to 34°C. The acetone-precipitated cells were centrifuged 15 min at 15,000g, dried, resuspended in SDS gel loading buffer (Laemmli, 1970), and boiled for 5 min. The samples were then electrophoresed on 12% acrylamide gels, and the resulting gels were subjected to immunoblotting. No chloramphenicol was added to the other culture, and samples (1 mL) were collected at 0, 1, 4, 6, and 20 hr after the shift to 34°C. The cells were harvested by centrifugation and frozen at −20°C. The culture samples were solubilized in 0.1 M Tris, pH 6.8, containing 4% SDS and 1 μM phenylmethylsulfonylfluoride and incubated 1 hr at 37°C. Insoluble material was pelleted, and the protein concentration of the green supernatant was determined with bicinchoninic acid. Samples corresponding to 40 μg of protein were boiled 5 min in SDS gel loading buffer and loaded onto 12% polyacryamide gels for immunoblot analysis.
Protein Preparations
The Ycf3 recombinant protein was obtained as follows. The ycf3 open reading frame was amplified by PCR using a pair of synthetic oligonucleotides to introduce an N-terminal His tag and a NcoI restriction site, 5′-AAACCATGGCTCATCACCACCACCACCACCCAAGAACGCAAAGAAATG-3′, and the 3′ restriction site BamHI, 5′-ACAGGATCCTTAAGTAGCTAAACCTGT-3′. The PCR fragment was purified and introduced into the Pet3D (NcoI-BamHI) vector (Studier et al., 1990). Isopropylthio-β-d-galactoside–induced expression was performed in the Escherichia coli strain BL21. The recombinant protein was purified with the PET His-Tag system (Novagen, Madison, WI) under denaturing conditions. The protein was refolded by dilution to a concentration of 0.1 mg·mL−1 followed by extensive dialysis against 50 mM Tricine, pH 7.6, and 10 mM KCl.
P700 concentration was determined from absorption transients at 817 nm (see below). C. reinhardtii ferredoxin and plastocyanin isolation followed the procedures of Ellefson et al. (1980) and Merchant and Bogorad (1986), respectively, with the modifications of Hippler et al. (1997). Photosystem I (PSI) was purified as described by Fischer et al. (1997). Chlorophyll concentrations were measured according to Lichtenthaler (1987).
Two-Dimensional Gel Electrophoresis
Samples containing ∼200 μg of protein were precipitated with methanol (Hippler et al., 2001) and solubilized for 1 hr in two-dimensional gel sample buffer of 20 mM Tris base containing 8 M urea, 2 M thiourea, 30 mM dithioerythritol, 4% Chaps, 0.5% bromophenolblue, and 0.17% dodecylmaltoside. The solubilized samples were loaded on nonlinear immobilized pH gradient gels (3.5 to 10.0 NL of IPG, 18 cm; Amersham Pharmacia Biotech AB, Uppsala, Sweden). The isoelectric focusing was performed on a IPGphor (Amersham Pharmacia Biotech AB). After the first dimension, the strips were loaded on 9 to 16% SDS–acrylamide-bis-acryloyl piperazine gradient gels, and gel eletrophoresis was performed as described (Hippler et al., 2001).
Urea Treatment and Flash-Induced Absorption Measurements
Flash-induced absorption transients at 817 nm were measured in a reaction mixture of 500 μL containing 50 mM Tris, pH 8.3, 2 mM sodium ascorbate, 60 μM 2,6-dichloroindophenolindophenol (DCPIP), and 100 μg·mL−1 chl. Urea was added to a final concentration of 6.8 M, and the cuvette was incubated at room temperature and subjected to flash-induced absorption measurements at different time points during the incubation, as described (Naver et al., 1999). Apart from the actinic flashes, the sample was kept in the dark during the incubation. Flash-induced absorption spectroscopy was performed as described by Hippler et al. (2000), using a single beam spectrophotometer (Occam Technologies, Cincinnati, OH). The measuring light, provided by a quartz halogen lamp, was filtered through a 817-nm (5-nm FWHM, Schott, Feldbach, Switzerland) interference filter. The signals from the photodiode were recorded by a Tektronic Oscilloscope THS 710 (Tektronix, Wilsonville, OR) and transferred to a computer. Flash excitation was provided by a Xenon flash (Occam Technologies). Sixteen individual signals were averaged for each trace at a repetition rate of 0.2 Hz.
NADP+ Photoreduction Measurements
NADP+ photoreduction activity was determined from the absorbance change at 340 nm in a 2-mL reaction mixture containing 20 mM Tricine, pH 7.5, 40 mM NaCl, 7 mM MgCl2, 2 mM sodium ascorbate, 60 μM DCPIP, 0.5 mM NADP+, 3 μM C. reinhardtii ferredoxin, 2 μM C. reinhardtii plastocyanin, 2 μM spinach ferredoxin–NADP+ reductase (Sigma), and 0.15 μM PSI. The production of NADPH was measured in a Uvikon 860 spectrophotometer (Bio-Tek Kontron Instruments, Zurich, Switzerland). The actinic light source was a Hansatech halogen lamp type LS2H (Hansatech Instruments Ltd., Norfolk, UK) connected to the spectrophotometer via fiberoptics.
Immunoprecipitation
Immunoprecipitation was performed in 0.5-mL reaction volumes containing 50 mM Tris, pH 7.4, 2 mg of thylakoids solubilized with 0.9% of dodecylmaltoside, and 100 μL of cyanogens-bromide–activated Sepharose with coupled polyclonal antibody. Samples were incubated overnight at 4°C, and the Sepharose beads were collected by centrifugation. The Sepharose pellet was washed five times with 50 mM Tris, pH 7.4, and 0.1% Triton X-100, and twice with 50 mM Tris, pH 7.4. The resulting pellet was incubated with either 0.1 M glycine, pH 2.8, to elute the immunoprecipitate without the antibody or loading buffer to elute the immunoprecipitate with the antibody. The eluates were loaded on SDS–polyacrylamide 10 to 17% gradient gels for separation and subsequent immunoblotting.
Immunoblot Analysis
Two-dimensional gels were transferred to nitrocellulose membranes, blocked with 20 mM Tris-HCl, pH 7.6, 0.137 M NaCl, 0.1% Tween 20, containing 5% skimmed milk and reacted with the Ycf3 recombinant His-tagged protein at a concentration of 0.05 mg·mL−1 in 20 mM Tris-HCl, pH 7.6, 0.137 M NaCl, and 0.1% Tween 20 with 1% skimmed milk. The membrane was washed in 20 mM Tris-HCl, pH 7.6, 0.137 M NaCl, and 0.1% Tween 20, probed with His-tag antibody and visualized by the enhanced chemiluminescence method.
The protein concentration of PSI preparations from the wild type and the Ycf3 mutants was determined by bicinchoninic acid. PSI preparations corresponding to 5 μg of protein were boiled in SDS sample buffer, and SDS-PAGE was performed as described (Laemmli, 1970). The samples were loaded onto 12% polyacrylamide gels, transferred to nitrocellulose membranes, incubated with antibodies, and visualized by the enhanced chemiluminescence method.
Acknowledgments
We thank N. Roggli for preparing the figures, M. Hippler for advice on the two-dimensional gel analysis, P. Sétif for performing flash absorption spectroscopy, F.-A. Wollman for PetA and AtpA antibodies, H.V. Scheller for PsaL antibody, and M. Goldschmidt-Clermont for helpful comments. H.N. was supported by a long-term EMBO fellowship (Grant No. LT97-509) and a fellowship from the Danish Natural Science Research Council. This work was supported by Grant No. 3100-050895.97 from the Swiss National Fund.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010253.
References
- Asada, K. (1994). Mechanisms for scavenging reactive molecules generated in chloroplasts under light stress. In Photoinhibition of Photosynthesis: From Molecular Mechanisms to the Field, N.R. Baker and J.R. Bowyer, eds (Oxford, UK: BIOS Scientific Publishers), pp. 129–142.
- Asada, K. (1996). Radical production and scavenging in the chloroplast. In Advances in Photosynthesis, N.R. Baker, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 123–150.
- Barber, J., and Andersson, B. (1992). Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 17, 61–66. [DOI] [PubMed] [Google Scholar]
- Bartsevich, V.V., and Pakrasi, H.B. (1997). Molecular identification of a novel protein that regulates biogenesis of photosystem I, a membrane protein complex. J. Biol. Chem. 272, 6382–6387. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Soen, S.Y., Frank, G., Zuber, H., and Rochaix, J.-D. (1992). Characterization of chlorophyll a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267, 25714–25721. [PubMed] [Google Scholar]
- Bennoun, P., and Delepelaire, P. (1982). Isolation of photosynthetic mutants in Chlamydomonas. In Methods in Chloroplast Molecular Biology, M. Edelman, ed (New York: Elsevier Biomedical Press), pp. 25–38.
- Boudreau, E., Takahashi, Y., Lemieux, C., Turmel, M., and Rochaix, J.-D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16, 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton, J.E., Gillham, N.W., Harris, E.H., Hosler, J.P., Johnson, A.M., Jones, A.R., Randolph-Anderson, B.L., Robertson, D., Klein, T.M., and Shark, K.B. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240, 1534–1538. [DOI] [PubMed] [Google Scholar]
- Brettel, K. (1997). Electron transfer and arrangement of the redox cofactors in photosystem I. Biochim. Biophys. Acta 1318, 322–373. [Google Scholar]
- Chitnis, V.P., and Chitnis, P.R. (1993). PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 336, 330–334. [DOI] [PubMed] [Google Scholar]
- Chitnis, P.R., Reilly, P.A., Miedel, M.C., and Nelson, N. (1989). Structure and targeted mutagenesis of the gene encoding 8-kDa subunit of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 264, 18374–18380. Errata: J. Biol. Chem. 265, 1821. [PubMed] [Google Scholar]
- Chitnis, P.R., Purvis, D., and Nelson, N. (1991). Molecular cloning and targeted mutagenesis of the gene psaF encoding subunit III of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 266, 20146–20151. [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein–protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson, L.E., Ulrich, E.A., and Krogmann, D.W. (1980). Plastocyanin. Methods Enzymol. 69, 223–228. [Google Scholar]
- Farah, J., Rappaport, F., Choquet, Y., Joliot, P., and Rochaix, J.-D. (1995). Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: Efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 14, 4976–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer, J.J., Vain, P., Jones, M.W., and Mcmullen, M.D. (1992). Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 11, 323–328. [DOI] [PubMed] [Google Scholar]
- Fischer, N., Stampacchia, O., Redding, K., and Rochaix, J.-D. (1996). Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251, 373–380. [DOI] [PubMed] [Google Scholar]
- Fischer, N., Setif, P., and Rochaix, J.-D. (1997). Targeted mutations in the psaC gene of Chlamydomonas reinhardtii: Preferential reduction of FB at low temperature is not accompanied by altered electron flow from photosystem I to ferredoxin. Biochemistry 36, 93–102. [DOI] [PubMed] [Google Scholar]
- Fischer, N., Hippler, M., Setif, P., Jacquot, J.P., and Rochaix, J.-D. (1998). The PsaC subunit of photosystem I provides an essential lysine residue for fast electron transfer to ferredoxin. EMBO J. 17, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N., Setif, P., and Rochaix, J.-D. (1999). Site-directed mutagenesis of the PsaC subunit of photosystem I. FB is the cluster interacting with soluble ferredoxin. J. Biol. Chem. 274, 23333–23340. [DOI] [PubMed] [Google Scholar]
- Girard-Bascou, J., Choquet, Y., Schneider, M., Delosme, M., and Dron, M. (1987). Characterization of a chloroplast mutation in the psaA2 gene of Chlamydomonas reinhardtii. Curr. Genet. 12, 489–495. [DOI] [PubMed] [Google Scholar]
- Goebl, M., and Yanagida, M. (1991). The TPR snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16, 173–177. [DOI] [PubMed] [Google Scholar]
- Golbeck, J.H. (1994). Photosystem I in cyanobacteria. In Molecular Biology of Cyanobacteria, D.A. Bryant, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 179–220.
- Golbeck, J.H., and Bryant, D.A. (1991). Photosystem I. In Current Topics in Bioenergetics: Light Driven Reactions in Bioenergetics, C.P. Lee, ed (New York: Academic Press), pp. 83–177.
- Golbeck, J.H., and Cornelius, J.H. (1986). Photosystem I charge separation in the absence of centers A and B. Optical characterization of center A2 and evidence for its association with a 64 kDa protein. Biochim. Biophys. Acta 681, 77–84. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldrup, A., Naver, H., and Scheller, H.V. (1999). The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 17, 689–698. [DOI] [PubMed] [Google Scholar]
- Haldrup, A., Simpson, D.J., and Scheller, H.V. (2000). Down-regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana. J. Biol. Chem. 275, 31211–31218. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego: Academic Press). [DOI] [PubMed]
- Hippler, M., Drepper, F., Farah, J., and Rochaix, J.-D. (1997). Fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii requires PsaF. Biochemistry 36, 6343–6349. [DOI] [PubMed] [Google Scholar]
- Hippler, M., Biehler, K., Krieger-Liszkay, A., van Dillewjin, J., and Rochaix, J.D. (2000). Limitation in electron transfer in photosystem I donor side mutants of Chlamydomonas reinhardtii. Lethal photo-oxidative damage in high light is overcome in a suppressor strain deficient in the assembly of the light harvesting complex. J. Biol. Chem. 275, 5852–5859. [DOI] [PubMed] [Google Scholar]
- Hippler, M., Klein, J., Fink, A., Allinger, T., and Hoerth, P. (2001). Towards functional proteomics of membrane protein complexes: Analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J., in press. [DOI] [PubMed]
- Høj, P.B., and Møller, B.L. (1986). The 110-kDa reaction center protein of photosystem I, P700- chlorophyll a-protein 1, is an iron–sulfur protein. J. Biol. Chem. 261, 14292–14300. [PubMed] [Google Scholar]
- Høj, P.B., Svendsen, I., Scheller, H.V., and Møller, B.L. (1987). Identification of a chloroplast-encoded 9-kDa polypeptide as a 2[4Fe- 4S] protein carrying centers A and B of photosystem I. J. Biol. Chem. 262, 12676–12684. [PubMed] [Google Scholar]
- Jensen, P.E., Gilpin, M., Knoetzel, J., and Scheller, H.V. (2000). The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J. Biol. Chem. 275, 24701–24708. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. [Google Scholar]
- Liu, D., Bienkowska, J., Petosa, C., Collier, R.J., Fu, H., and Liddington, R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376, 191–194. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Haldrup, A., Knoetzel, J., and Scheller, H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408, 613–615. [DOI] [PubMed] [Google Scholar]
- Mehari, T., Parrett, K.G., Warren, P.V., and Golbeck, J.H. (1991). Reconstitution of the iron–sulfur clusters in the isolated FA/FB protein: EPR spectral characterization of same-species and cross-species photosystem I complexes. Biochim. Biophys. Acta 1056, 139–148. [Google Scholar]
- Merchant, S., and Bogorad, L. (1986). Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol. Cell Biol. 6, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naver, H., Scott, M.P., Golbeck, J.H., Møller, B.L., and Scheller, H.V. (1996). Reconstitution of barley photosystem I with modified PSI-C allows identification of domains interacting with PSI-D and PSI- A/B. J. Biol. Chem. 271, 8996–9001. [DOI] [PubMed] [Google Scholar]
- Naver, H., Scott, M.P., Golbeck, J.H., Olsen, C.E., and Scheller, H.V. (1998). The eight–amino acid internal loop of PSI-C mediates association of low molecular mass iron–sulfur proteins with the P700-FX core in photosystem I. J. Biol. Chem. 273, 18778–18783. [DOI] [PubMed] [Google Scholar]
- Naver, H., Haldrup, A., and Scheller, H.V. (1999). Cosuppression of photosystem I subunit PSI-H in Arabidopsis thaliana. Efficient electron transfer and stability of photosystem I is dependent upon the PSI-H subunit. J. Biol. Chem. 274, 10784–10789. [DOI] [PubMed] [Google Scholar]
- Oh-oka, H., Takahashi, Y., Kuriyama, K., Saeki, K., and Matsubara, H. (1988). The protein responsible for center A/B in spinach photosystem I: Isolation with iron–sulfur cluster(s) and complete sequence analysis. J. Biochem. 103, 962–968. [DOI] [PubMed] [Google Scholar]
- Prodromou, C., Siligardi, G., O'Brien, R., Woolfson, D.N., Regan, L., Panaretou, B., Ladbury, J.E., Piper, P.W., and Pearl, L.H. (1999). Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR) domain co-chaperones. EMBO J. 18, 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding, K., MacMillan, F., Leibl, W., Brettel, K., Hanley, J., Rutherford, A.W., Breton, J., and Rochaix, J.-D. (1998). A systematic survey of conserved histidines in the core subunits of photosystem I by site-directed mutagenesis reveals the likely axial ligands of P700. EMBO J. 17, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.-D., Mayfield, S., Goldschmidt-Clermont, M., and Erickson, J. (1988). Molecular biology of Chlamydomonas. In Plant Molecular Biology: A Practical Approach, C.H. Shaw, ed (Oxford: IRL Press), pp. 253–275.
- Rochaix, J.-D., Fischer, N., and Hippler, M. (2000). Chloroplast site-directed mutagenesis of photosystem I in Chlamydomonas: Electron transfer reactions and light sensitivity. Biochimie 82, 635–645. [DOI] [PubMed] [Google Scholar]
- Ruf, S., Kossel, H., and Bock, R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I–related gene. J. Cell Biol. 139, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scheller, H.V., Naver, H., and Møller, B.L. (1997). Molecular aspects of photosystem I. Physiol. Plant. 100, 842–851. [Google Scholar]
- Schmidt, G.W., and Mishkind, M.L. (1983). Rapid degradation of unassembled ribulose 1,5-biphosphate carboxylase small subunits in chloroplasts. Proc. Natl. Acad. Sci. USA 80, 2632–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, W.D., Klukas, O., Krauss, N., Saenger, W., Fromme, P., and Witt, H.T. (1997). Photosystem I of Synechococcus elongatus at 4 Å resolution: Comprehensive structure analysis. J. Mol. Biol. 272, 741–769. [DOI] [PubMed] [Google Scholar]
- Stampacchia, O., Girard-Bascou, J., Zanasco, J.L., Zerges, W., Bennoun, P., and Rochaix, J.-D. (1997). A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell 9, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, W., Rosenberg, A.H., Dunn, J.J., and Dubendorf, J.W. (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89. [DOI] [PubMed] [Google Scholar]
- Varotto, C., Pesaresi, P., Meurer, J., Oelmuller, R., Steiner-Lange, S., Salamini, F., and Leister, D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22, 115–124. [DOI] [PubMed] [Google Scholar]
- Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., and Gamblin, S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376, 188–191. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Yu, L., Chitnis, V.P., and Chitnis, P.R. (1994). Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunits. J. Biol. Chem. 269, 3205–3211. [PubMed] [Google Scholar]
- Xu, Q., Hoppe, D., Chitnis, V.P., Odom, W.R., Guikema, J.A., and Chitnis, P.R. (1995). Mutational analysis of photosystem I polypeptides in the cyanobacterium Synechocystis sp. PCC 6803. Targeted inactivation of psaI reveals the function of psaI in the structural organization of psaL. J. Biol. Chem. 270, 16243–16250. [DOI] [PubMed] [Google Scholar]
- Young, J.C., Obermann, W.M., and Hartl, F.U. (1998). Specific binding of tetratricopeptide repeat proteins to the C- terminal 12-kDa domain of hsp90. J. Biol. Chem. 273, 18007–18010. [DOI] [PubMed] [Google Scholar]
- Zak, E., and Pakrasi, H.B. (2000). The BtpA protein stabilizes the reaction center proteins of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803 at low temperature. Plant Physiol. 123, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak, E., Norling, B., Andersson, B., and Pakrasi, H.B. (1999). Subcellular localization of the BtpA protein in the cyanobacterium Synechocystis sp. PCC 6803. Eur. J. Biochem. 261, 311–316. [DOI] [PubMed] [Google Scholar]