Abstract
Glucosinolates are anionic thioglucosides that have become one of the most frequently studied groups of defensive metabolites in plants. When tissue damage occurs, the thioglucoside linkage is hydrolyzed by enzymes known as myrosinases, resulting in the formation of a variety of products that are active against herbivores and pathogens. In an effort to learn more about the molecular genetic and biochemical regulation of glucosinolate hydrolysis product formation, we analyzed leaf samples of 122 Arabidopsis ecotypes. A distinct polymorphism was observed with all ecotypes producing primarily isothiocyanates or primarily nitriles. The ecotypes Columbia (Col) and Landsberg erecta (Ler) differed in their hydrolysis products; therefore, the Col × Ler recombinant inbred lines were used for mapping the genes controlling this polymorphism. The major quantitative trait locus (QTL) affecting nitrile versus isothiocyanate formation was found very close to a gene encoding a homolog of a Brassica napus epithiospecifier protein (ESP), which causes the formation of epithionitriles instead of isothiocyanates during glucosinolate hydrolysis in the seeds of certain Brassicaceae. The heterologously expressed Arabidopsis ESP was able to convert glucosinolates both to epithionitriles and to simple nitriles in the presence of myrosinase, and thus it was more versatile than previously described ESPs. The role of ESP in plant defense is uncertain, because the generalist herbivore Trichoplusia ni (the cabbage looper) was found to feed more readily on nitrile-producing than on isothiocyanate-producing Arabidopsis. However, isothiocyanates are frequently used as recognition cues by specialist herbivores, and so the formation of nitriles instead of isothiocyanates may allow Arabidopsis to be less apparent to specialists.
INTRODUCTION
Plants elaborate an enormous variety of metabolites that are generally believed to function in defense against herbivores, but it has been very difficult to demonstrate the protective roles of individual compounds under natural conditions. Modern molecular and genetic methods have enormous potential to unravel the complexity of plant defensive mechanisms and to help test their defensive function as well as to provide insight on the evolution of defense. One of the most frequently studied plant defense systems consists of the glucosinolates, a group of low-molecular-weight, sulfur-rich thioglucosides (Halkier, 1999), and myrosinases, enzymes capable of hydrolyzing the thioglucoside linkage (Bones and Rossiter, 1996; Rask et al., 2000). Found in many members of the Brassicaceae (Arabidopsis, Brassica, etc.) and other families of the Capparales, the glucosinolate/myrosinase system also includes a large number of proteins that associate with myrosinase.
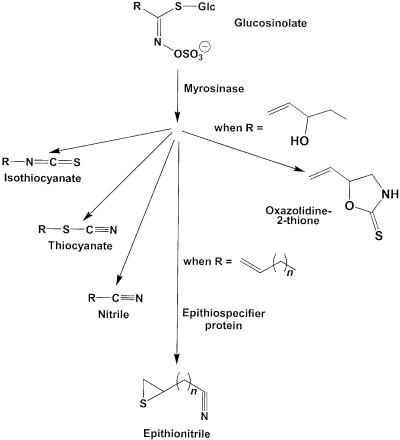
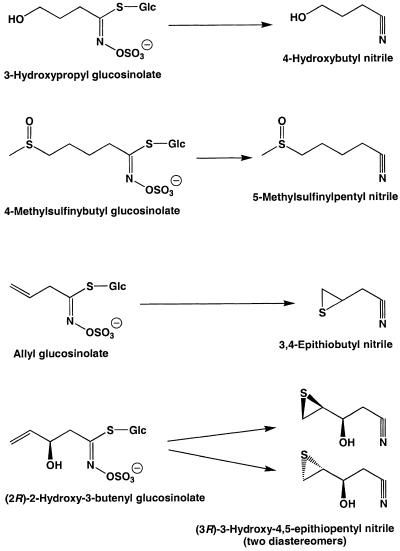
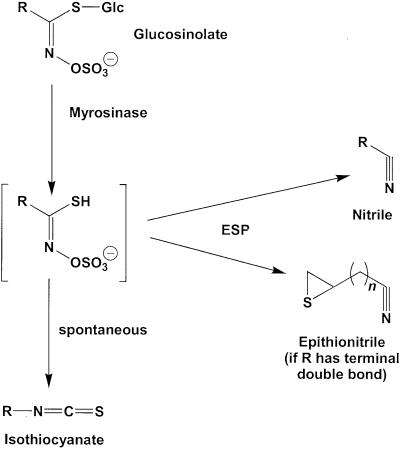
Glucosinolates and myrosinases are spatially separated within the plant (Bones and Rossiter, 1996; Kelley et al., 1998) but are brought into contact by tissue damage that yields a variety of hydrolysis products, including nitriles, isothiocyanates, thiocyanates, oxazolidine-2-thiones, and epithionitriles (Figure 1) (Bones and Rossiter, 1996; Halkier, 1999). The type of hydrolysis products formed has been shown to depend on the chemical nature of the side chain of the parent glucosinolate and the conditions of hydrolysis. For example, glucosinolates with an aliphatic side chain are generally hydrolyzed to yield isothiocyanates at a neutral pH, but an acidic pH or the presence of Fe2+ ions favors the formation of nitriles (Gil and MacLeod, 1980; MacLeod and Rossiter, 1986; Galletti et al., 2001). In some plants, specific proteins aside from myrosinase may also be involved in glucosinolate hydrolysis. For example, in the seeds of Brassica napus and other members of the Brassicaceae, a protein promoting the formation of one class of nitriles has been described (Daxenbichler et al., 1968; Tookey, 1973; MacLeod and Rossiter, 1985). Called the epithiospecifier protein (ESP), its presence during the hydrolysis of alkenyl glucosinolates leads to the formation of epithionitriles instead of isothiocyanates by transfer of the sulfur atom from the basic glucosinolate backbone to the terminal alkene residue of the side chain (Figure 1). Common in plants containing glucosinolates with a terminal alkene, this protein appears to have no catalytic activity in the absence of myrosinase. Glucosinolates without terminal alkene functions may also be converted to nitriles under various conditions (Cole, 1976; Hasapis and MacLeod, 1982; Latxague et al., 1991). However, it is unclear whether the ESP or another protein plays a role in this process.
Figure 1.
Major Hydrolysis Products of Plant Glucosinolates.
In this investigation, we describe an Arabidopsis protein that promotes the formation of both nitriles and epithionitriles upon plant damage. R, variable side chain; n = 1 or 2.
Knowledge of the factors controlling glucosinolate hydrolysis is critical for understanding the ecological roles of these compounds, because it is the hydrolysis products, and not the glucosinolates themselves, that have been found to be most active against herbivores and pathogens (Louda and Mole, 1991). For example, allyl isothiocyanate was significantly more toxic to larvae of the diamondback moth (Plutella xylostella), a crucifer specialist, than was the parent allyl glucosinolate when both compounds were added to an artificial diet (Li et al., 2000). In addition, volatile hydrolysis products are often used as cues for the feeding and oviposition by insects that specialize in glucosinolate-containing plants (Ahman, 1985; Chew and Renwick, 1994; Ekbom, 1998; Rojas, 1999). For plant pathogenic fungi, the hydrolysis products are also more important in resistance than are the intact glucosinolates (Olivier et al., 1999; Tierens et al., 2001). However, despite the importance of glucosinolate hydrolysis products in plant defense, only rarely have efforts been made to compare the effects of different types of hydrolysis products on herbivores and pathogens (Donkin et al., 1995).
In this study, we have investigated some of the biochemical and molecular controls on glucosinolate hydrolysis and evaluated the effect of different hydrolysis products on an herbivore species. Chemical analysis of 122 Arabidopsis ecotypes showed a distinct polymorphism in the nature of the predominant hydrolysis product; some ecotypes produced primarily isothiocyanates, whereas others produced primarily nitriles or epithionitriles. This polymorphism was mapped to a region containing a gene that encodes an epithiospecifier-type protein. The properties of this gene product were studied using the heterologously expressed protein, and the relative effects of isothiocyanate versus nitrile hydrolysis products were tested with the generalist lepidopteran herbivore, the cabbage looper (Trichoplusia ni).
RESULTS
Natural Variation in Glucosinolate Hydrolysis Products among Arabidopsis Ecotypes
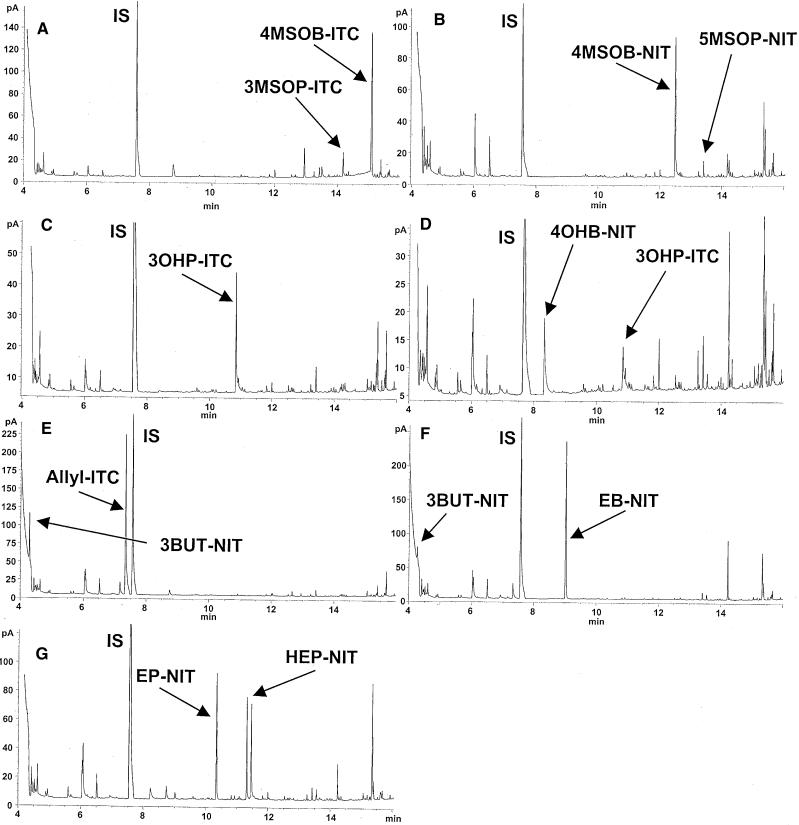
The principal glucosinolates of Arabidopsis are derived from chain-elongated derivatives of methionine (Hogge et al., 1988; Graser et al., 2000). We have previously documented extensive variation in the glucosinolate composition of ecotypes of this species, with methylsulfinylalkyl, hydroxyalkyl, and alkenyl glucosinolates being the three major classes of methionine-derived glucosinolates found (Kliebenstein et al., 2001b). To assess ecotype variation in the nature of the hydrolysis products of the methionine-derived glucosinolates, we macerated leaf samples of 122 different ecotypes in water and analyzed the resulting products by gas chromatography–mass spectrometry (GC-MS) and GC with flame ionization detector (FID). All ecotypes produced predominantly isothiocyanates or predominantly nitriles (Figure 2; see Methods for a complete list of ecotypes surveyed). For example, among ecotypes accumulating hydroxyalkyl glucosinolates, Da(1)-12 produced isothiocyanates (Figure 2C), whereas Landsberg erecta (Ler) produced predominantly nitriles (Figure 2D). Among ecotypes accumulating the alkenyl glucosinolate allyl (2-propenyl) glucosinolate, Monte Tosso (Mr-0) produced predominantly isothiocyanates (Figure 2E), whereas Eifel (Ei-2) produced predominantly nitriles (Figure 2F). In alkenyl glucosinolate–containing ecotypes that produced nitriles, the major hydrolysis products were not simple nitriles but epithionitriles (Figure 1), that is, compounds in which a sulfur atom was added to the terminal alkene function.
Figure 2.
Natural Variation in the Glucosinolate Hydrolysis Products Formed in Various Arabidopsis Ecotypes.
All ecotypes produced primarily isothiocyanates or nitriles. Depicted are GC-FID chromatograms representing each of the major glucosinolate types and each of the classes of hydrolysis products.
(A) Col-0: methylsulfinylalkyl glucosinolates, isothiocyanates.
(B) Col × Ler RIL CS 1907: methylsulfinylalkyl glucosinolates, nitriles (similar profiles were seen in Bn-0 and Da-0).
(C) Da(1)-12: hydroxyalkyl glucosinolates, isothiocyanates.
(D) Ler: hydroxyalkyl glucosinolates, nitriles.
(E) Mr-0: alkenyl glucosinolates, isothiocyanates.
(F) Ei-2: alkenyl (allyl) glucosinolate, nitriles.
(G) Tac: alkenyl (3-butenyl and 2-hydroxyl-3-butenyl) glucosinolates, nitriles.
A complete list of surveyed ecotypes is given in Methods. In comparing the nomenclature of glucosinolates and isothiocyanates with their corresponding nitriles, the numbering system must be adjusted because the nitrile carbon atom is counted as part of the alkyl chain. IS, internal standard, n-propylisothiocyanate; 3MSOP-ITC, 3-methylsulfinylpropyl isothiocyanate; 4MSOB-ITC, 4-methylsulfinylbutyl isothiocyanate; 4MSOB-NIT, 4-methylsulfinylbutyl nitrile (hydrolysis product of 3-methylsulfinylpropyl glucosinolate); 5MSOP-NIT, 5-methylsulfinylpentyl nitrile (hydrolysis product of 4-methylsulfinylbutyl glucosinolate); 3OHP-ITC, 3-hydroxypropyl isothiocyanate; 4OHB-NIT, 4-hydroxybutyl nitrile (hydrolysis product of 3-hydroxypropyl glucosinolate); Allyl-ITC, allyl isothiocyanate; 3BUT-NIT, 3-butenyl nitrile (hydrolysis product of allyl glucosinolate); EB-NIT, 3,4-epithiobutyl nitrile (hydrolysis product of allyl glucosinolate); EP-NIT, 4,5-epithiopentyl nitrile (hydrolysis product of 3-butenyl glucosinolate); and HEP-NIT, (3R)-3-hydroxy-4,5-epithiopentyl nitrile (hydrolysis product of (2R)-2-hydroxy-3-butenyl glucosinolate, both diastereomers of this compound are present).
Among the ecotypes, the nature of the hydrolysis product was significantly correlated with the major type of methionine-derived glucosinolate present (χ2 = 27.3, df = 2, P < 0.0001). Ecotypes accumulating alkenyl glucosinolates were more likely to produce nitriles than isothiocyanates, whereas ecotypes accumulating other glucosinolate types showed no correlation with the production of either isothiocyanate or nitrile products (Table 1).
Table 1.
Unequal Distribution of Hydrolysis Products among Arabidopsis Ecotypes Accumulating Different Types of Glucosinolates a
| Glucosinolate Type Accumulated | Class of Glucosinolate Hydrolysis Product
|
|||||
|---|---|---|---|---|---|---|
| Nitriles and Epithionitriles
|
Isothiocyanates
|
|||||
| Observed | Expected | χ2 | Observed | Expected | χ2 | |
| Alkenyl | 60 | 48 | 3.0 | 3 | 15 | 9.6 |
| Methylsulfinylalkyl | 16 | 24 | 2.5 | 15 | 7 | 7.9 |
| Hydroxyalkyl | 17 | 21 | 0.9 | 11 | 7 | 2.8 |
a Of the 122 ecotypes surveyed, ∼75% formed nitriles and epithionitriles and the remainder formed isothiocyanates. However, these classes of hydrolysis products were not randomly distributed among ecotypes accumulating different types of glucosinolates.
Mapping of Quantitative Trait Loci for Glucosinolate Hydrolysis in Columbia × Ler Recombinant-Inbred Lines
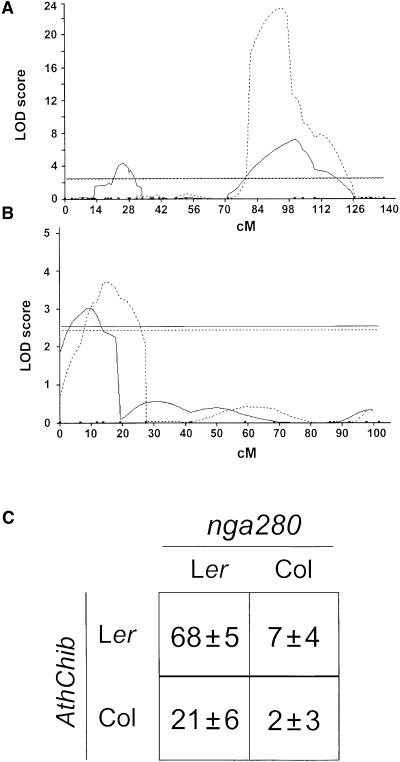
Because the Columbia ecotype (Col) produces isothiocyanates and Ler produces nitriles, we mapped the genetic loci regulating glucosinolate hydrolysis in 96 Col × Ler recombinant inbred lines (RIL) (Lister and Dean, 1993). Two loci were significantly correlated with the nature of the breakdown product: one on chromosome 1 (marker nga280) (Figure 3A) and the other on chromosome 3 (marker AthChib) (Figure 3B). When the mean percentage of nitriles versus isothiocyanates formed by each line was compared with marker data indicating whether Col- or Ler-derived chromosome segments were present at these two loci, the Ler allele was found to confer the highest percentage of nitriles for both loci (Figure 3C). The allelic status at nga280 causes nearly a 10-fold difference in the percentage of nitrile products formed, whereas the allelic status at AthChib causes an approximate threefold difference. When the Ler nga280 allele is coupled with the Col AthChib allele, the percentage of nitriles is reduced by more than half, indicating that the locus on chromosome 3 can be considered a modifier.
Figure 3.
QTLs Regulating Glucosinolate Hydrolysis and T. ni Herbivory in 96 Col × Ler.
(A) and (B) QTLs are found on chromosomes 1 (A) and 3 (B). The solid lines represent the lod scores for the amount of nitriles formed as a percentage of the total glucosinolate hydrolysis products; the dashed line represents the lod scores for the percentage of rosette leaf area damaged by T. ni. The horizontal line is the 0.05 significance level obtained after 500 permutations of the markers and data.
(C) Mean percentage of nitriles formed in Col × Ler RIL from each of four allelic classes determined by presence of alleles from either Col or Ler at the nga280 locus on chromosome 1 and the AthChib locus on chromosome 3.
Cloning of an Arabidopsis Gene That Encodes a Homolog of the B. napus ESP
In close proximity to marker nga280 is a homolog of a B. napus gene that encodes an ESP, a protein that alters glucosinolate hydrolysis by causing the formation of epithionitriles instead of isothiocyanates during hydrolysis of alkenyl glucosinolates. Bernardi et al. (2000) recently purified an ESP from B. napus seeds and obtained extensive internal amino acid sequence information. This sequence has high identity to the hypothetical protein of Arabidopsis gene F15I1.12 (annotated as the reverse complement of nucleotides 31,329 to 34,219 of bacterial artificial chromosome clone F15I1, referred to as EMBL Q39104 by Bernardi et al. [2000]). Because this gene is located within 1 Mb of nga280, it seemed a likely candidate for the gene we had mapped controlling glucosinolate hydrolysis product polymorphism in the Col × Ler cross. Several homologs of F15I1.12 are also present in the Arabidopsis genome, but none are on the same chromosome as nga280. Thus, we cloned the cDNA, measured gene expression in certain Col × Ler RIL, and investigated the catalytic properties of the gene product. The gene itself has an open reading frame (ORF) of 1023 nucleotides (341 amino acids) (Figure 4A) encoding a 37-kD protein with a pI of 5.65. It has 82% identity at the amino acid level to the purified B. napus ESP if we consider only those regions previously sequenced by Bernardi et al. (2000). No significant identity was found to any other sequence available in public databases, except to several Arabidopsis sequences that displayed 45 to 55% identity (Figure 4B), three of which are present in a tandem array.
Figure 4.
Amino Acid Sequence of Arabidopsis ESP and Some Sequence Relationships.
(A) Amino acid sequence of Arabidopsis ESP. Underlined are regions corresponding to peptides of B. napus ESP protein sequenced by Bernardi et al. (2000).
(B) Dendrogram of Arabidopsis ESP in relation to B. napus ESP and other Arabidopsis sequences.
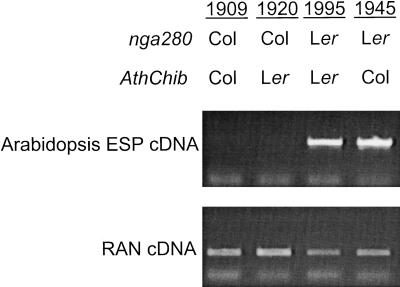
To determine if F15I1.12 expression influences the formation of nitriles versus isothiocyanates as glucosinolate hydrolysis products, we performed reverse transcription–polymerase chain reaction (RT-PCR) in Col, Ler, and 16 RIL representing all four combinations of Ler or Col status at nga280 and AthChib. The gene was found to be consistently expressed whenever nitriles were the dominant products. Expression was observed in Ler, but not Col, and in all lines with the Ler allele at nga280 (Figure 5), which is consistent with the proposal that the product of F15I1.12 controls the formation of nitriles versus isothiocyanates in Arabidopsis.
Figure 5.
Expression of Arabidopsis ESP Is Correlated with the Formation of Nitriles as Major Glucosinolate Hydrolysis Products in the Col × Ler RIL.
Shown are ethidium bromide–stained gels of semi-quantitative RT-PCR from one plant of each of the four allelic classes shown in Figure 3C. Equal amounts of cDNA were used for loading in each lane. The nga280 and AthChib allelic states are shown above the corresponding lane. Arabidopsis ESP expression correlates closely with Ler status at nga280. Relative to the Col allele, the Ler allele at nga280 increases the percentage of nitriles formed by nearly 10-fold. RAN is a loading control. The numbers at the head of each column indicate the Col × Ler RIL used.
Catalytic Properties of the Arabidopsis ESP
To demonstrate the catalytic properties of F15I1.12, the cDNA was heterologously expressed in cultures of Escherichia coli. After cell disruption, the pellet that resulted from the 3000g centrifugation was resuspended in 100 mM Mes buffer, pH 6.0, and incubated with four different purified glucosinolates. In the presence of myrosinase, 3-hydroxypropyl glucosinolate and 4-methylsulfinylbutyl glucosinolate were converted to the corresponding nitrile products, 4-hydroxybutyl nitrile (3-hydroxypropyl cyanide) and 5-methylsulfinylpentyl nitrile (4-methylsulfinylbutyl cyanide), respectively (see the note on nomenclature in the legend to Figure 2) (Figure 6). The alkenyl glucosinolates, allyl and (2R)-2-hydroxy-3-butenyl glucosinolate, were converted to the epithionitrile products, 3,4-epithiobutyl nitrile and the diastereomeric (3R)-3-hydroxy-4,5-epithiopentyl nitriles, respectively (Figure 6). Thus, the F15I1.12 gene product seemed to act as an ESP (converting alkenyl glucosinolates to epithionitriles) as well as converting nonalkenyl glucosinolates to simple nitriles. No conversion was observed in boiled control extracts, in extracts of nontransformed E. coli, or in the absence of myrosinase. Treatment with myrosinase alone (without the overexpressing E. coli extract) gave only isothiocyanate products; no nitriles were detected. When comparing reaction velocity with different substrates, we found that conversion of the alkenyl glucosinolates to their corresponding epithionitriles occurred at a rate two to three times that of the conversion of nonalkenyl glucosinolates to nitriles (Table 2).
Figure 6.
Reactions Catalyzed by Arabidopsis ESP Expressed in E. coli.
Glucosinolates with terminal alkene functions were converted to epithionitriles, and other glucosinolates were converted to simple nitriles. When comparing the nomenclature of glucosinolates with their corresponding nitriles, the numbering system must be adjusted because the nitrile carbon atom is counted as part of the alkyl chain.
Table 2.
Relative Conversion of Various Glucosinolates to Hydrolysis Products by Arabidopsis ESP Expressed in E. coli a
| Glucosinolate | Hydrolysis Product | Relative Conversion |
|---|---|---|
| 2-Hydroxy-3-butenyl | 3-Hydroxy-4,5-epithiobutyl nitrile | 100 |
| Allyl (2-propenyl) | 3,4-Epithiobutyl nitrile | 66 |
| 4-Methylsulfinylbutyl | 5-Methylsulfinylpentyl nitrile | 35 |
| 3-Hydroxypropyl | 4-Hydroxybutyl nitrile | 30 |
a Assays were performed in 100 mM Mes buffer, pH 6.0, in the presence of myrosinase. The rate of 2-OH-3-butenyl glucosinolate conversion (0.34 μmol per 30-min assay per resuspended pellet from 180 mL of bacterial culture) was set to 100.
The effect of Fe2+ concentration on the reaction was investigated, because this metal ion had previously been reported to be required for the formation of epithionitriles in crude extracts and for the stability of partially purified ESP (Tookey, 1973; MacLeod and Rossiter, 1985). However, reaction of the overexpressed Arabidopsis ESP with 3-hydroxypropyl glucosinolate proceeded at a measurable rate without any additional iron, although addition of 0.1 mM Fe2+ increased the rate threefold (Table 3). At 0.5 mM Fe2+, no increase in enzyme-catalyzed nitrile formation was measured relative to 0.1 mM Fe2+, but background nitrile formation (measured in extracts of untransformed E. coli) increased to a rate nearly equal to that arising from the extract of E. coli expressing the Arabidopsis ESP. When allyl glucosinolate was used as a substrate, the highest rate of formation of the epithionitrile, 3,4-epithiobutyl nitrile, was seen with 0.5 mM Fe2+ (Table 4). However, even without any added Fe2+, conversion was 75% of the maximum, and it was still 37% of the maximum in the presence of 1 mM EDTA. Higher concentrations of Fe2+ actually decreased the formation of the epithionitrile relative to the formation of the simple nitrile, 3-butenyl nitrile. Thus, the activity of the Arabidopsis ESP protein appears not to depend on added Fe2+, although supplemental Fe2+ can increase nitrile formation and can also support nitrile formation at neutral pH in the absence of ESP.
Table 3.
Effect of Added Fe2+ on Relative Conversion of 3-Hydroxypropyl Glucosinolate to the Corresponding Nitrile (4-Hydroxybutyl Nitrile) by Arabidopsis ESP Expressed in E. coli a
| Fe2+ Concentration (mM) | Arabidopsis ESP | Untransformed E. coli |
|---|---|---|
| 0 | 32 | 0 |
| 0.1 | 106 | 10 |
| 0.5 | 100 | 79 |
a Assays were performed as in Table 2. The rate of nitrile formation at 0.5 mM Fe2+ (72 nmol per 30-min assay per resuspended pellet from 180 mL of bacterial culture) was set to 100.
Table 4.
Effect of Added Fe2+ on Relative Conversion of Allyl Glucosinolate to Nitriles by Arabidopsis ESP Expressed in E. coli a
| Fe2+ Concentration (mM) |
Epithionitrile (3,4-Epithiobutyl Nitrile) |
Nitrile (3-Butenyl Nitrile) |
|---|---|---|
| 0 | 74 | 0 |
| 0 (1 mM EDTA) | 37 | 0 |
| 0.5 | 100 | 73 |
| 1 | 57 | 203 |
| 2 | 49 | 289 |
| 3 | 28 | 307 |
a Assays were performed as in Table 2. The rate of epithionitrile formation at 0.5 mM Fe2+ (134 nmol per 30-min assay per resuspended pellet from 180 mL of bacterial culture) was set to 100.
To confirm that Arabidopsis ESP has the capacity to promote the formation of both nitriles and epithionitriles, we analyzed the glucosinolates and glucosinolate hydrolysis products of 200 F2 progeny from a cross between the ecotypes Da(1)-12 and Ei-2 to test for linkage between nitrile and epithionitrile formation. The two parent ecotypes were chosen so that both glucosinolate type (alkenyl versus nonalkenyl) and hydrolysis-product type (nitriles versus isothiocyanates) would segregate in this cross. One hundred F2 lines were found to contain both nonalkenyl (3-hydroxypropyl) and alkenyl (allyl) glucosinolates. Upon hydrolysis, all of these lines produced both nitriles and epithionitriles: the simple nitrile, 4-hydroxybutyl nitrile, from the nonalkenyl glucosinolate and the epithionitrile, 3,4-epithiobutyl nitrile, from the alkenyl glucosinolate (data not shown). The lack of recombination between these two traits implies that the same genetic locus is responsible for both nitrile and epithionitrile production.
Regulation of ESP Gene Expression in Different Arabidopsis Ecotypes
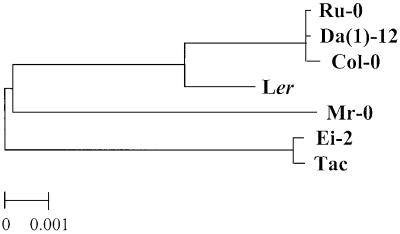
Our studies with the Col × Ler RIL demonstrated that lines that produced isothiocyanates instead of nitriles did not show detectable expression of the ESP gene by RT-PCR. To identify the critical molecular features that affect ESP gene expression, we determined the genomic sequence of F15I1.12 from seven selected ecotypes, four of which produced isothiocyanates and three of which produced nitriles. Of the isothiocyanate-producing ecotypes, three (Col, Da(1)-12, and Ru-0) did not express ESP and had sequences that clustered together on phylogenetic analysis (Figure 7), suggesting that they shared a feature responsible for lack of expression. In contrast, the isothiocyanate-producing ecotype Mr-0 did express ESP and had a sequence that clustered with the other nitrile-producing ecotypes. The lack of nitrile-producing activity in Mr-0 can probably be attributed to a deletion of 124 bases, which eliminated a splice site and 100 nucleotides of the ORF (Table 5). Thus, assuming that the ancestor of Arabidopsis possessed a functional copy of the ESP gene, there have been at least two independent losses of this nitrile-producing activity, one that knocked out gene expression and one that deleted part of the cDNA.
Figure 7.
Dendrogram of Arabidopsis ESP Genomic Sequences from Seven Ecotypes.
The tree was generated with midpoint rooting, and all branches have >65% bootstrap support. Ecotypes Ru-0, Da(1)-12, Col-0, and Mr-0 hydrolyze glucosinolates to isothiocyanates and thus do not have a functional ESP. Ru-0, Da(1)-12, and Col-0 do not express the ESP gene; Mr-0 does, but it has a deletion in the ORF.
Table 5.
Polymorphisms in the Arabidopsis ESP Genomic Sequence from Different Ecotypes and Their Correlation with Gene Expression
| Region (bp) |
Classification | In/Del (<50 bp) |
SNPs | Correlationa | Change |
|---|---|---|---|---|---|
| 0 to 290 | 3′ Noncoding | 0 | 5 | ||
| 291 to 574 | 3′ UTR | 2 | 10 | 1 | G to A |
| 575 to 1565 | ORF | 0b | 6 | ||
| 1566 to 4721 | Intron | 6c | 21 | 1 | C to T |
| 4719 to 4779 | ATG/5′ UTR | 0 | 1 | ||
| 4780 to 5780 | Promoter I | 11d | 7 | 1 | 10 bp |
| 5781 to 6780 | Promoter II | 5 | 16 | ||
| 6781 to 7780 | 5′ Noncoding | 9 | 31 | 1 | |
| 7781 to 8780 | 5′ Noncoding | 4 | 15 | 2 | |
| 8781 to 9327 | 5′ Noncoding | 2 | 8 | 2 |
a The number of polymorphisms correlated with expression versus nonexpression.
b Deletion in Mr-0 from bp 1441 to 1868.
c Deletion in Tac and Ei-2 from bp 1638 to 2388; in Da1-12, Ler, Ru-0, and Mr-0 from bp 2534 to 3232; in Da1-12, Ei-2, Ler, Ru-0, Col, and Tac, from bp 3435 to 3980.
d Deletion in Ei-2 and Tac from bp 5222 to 6070.
To investigate the loss of F15I1.12 expression in more detail, we sought single nucleotide polymorphisms (SNPs) and insertions/deletions (In/Dels) in the genomic sequence that correlated with lack of expression. A 10-bp deletion was found in the promoter sequence of all non-expressing ecotypes that removes an ABI5-like binding site (Finkelstein and Lynch, 2000) (Table 5). Other SNPs that correlated with lack of expression were found in the 3′ untranslated region (UTR), in an intron, or >2 kb upstream of the ORF in the 5′ noncoding region, and so they are unlikely to be responsible for lack of F15I1.12 expression.
Effect of Nitrile versus Isothiocyanate Hydrolysis Products on T. ni Herbivory
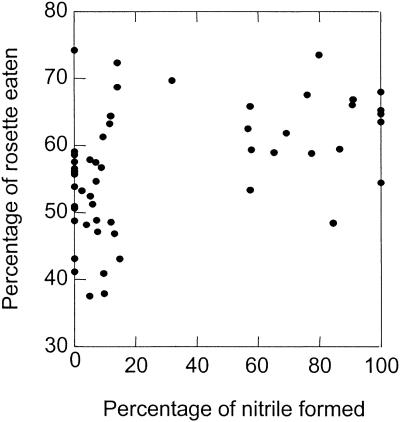
When the larvae of T. ni, a generalist lepidopteran, were allowed to feed on the Col × Ler RIL, the quantitative trait loci (QTLs) associated with feeding rate were found to closely overlap the QTLs previously shown to regulate the nature of the glucosinolate hydrolysis products (Figure 3). Lod maxima for both traits were located at nga280 and Athchib, with the Ler allele associated with increased herbivory and increased nitrile production. The genetic correlation between the percentage of nitrile formed (of the total glucosinolate hydrolysis products) and the percentage of rosette damage by T. ni on 54 RIL was statistically significant (Figure 8; rG = 0.207, P < 0.0005), indicating that nitrile-producing lines are subject to more herbivory than are the isothiocyanate-producing lines.
Figure 8.
Isothiocyanates and Nitriles as Deterrents of T. ni Herbivory.
In this graph, the percentage of nitrile formed (of total glucosinolate hydrolysis products) in 56 Col × Ler RILs is plotted against the percentage of rosette leaves damaged by T. ni in a 48-hr feeding assay. The results indicate that isothiocyanates are more efficient than are nitriles in deterring T. ni herbivory.
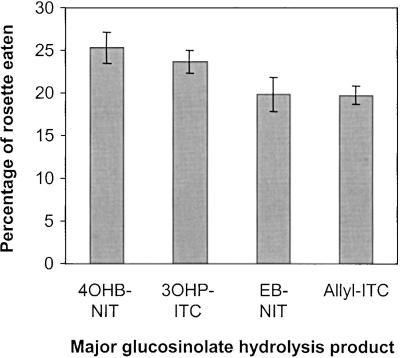
To compare the influence of individual glucosinolate hydrolysis products on herbivory, we allowed T. ni larvae to feed on 168 F2 families from Da(1)-12 × Ei-2, a cross that segregated for the type of glucosinolate (allyl versus 3-hydroxypropyl) and the type of hydrolysis product (nitriles versus isothiocyanates). ANOVA analysis showed that both factors significantly affected herbivory (Table 6). Ranking the hydrolysis products by their ability to deter T. ni herbivory gives the following: 3,4-epithiobutyl nitrile = allylisothiocy-anate > 3-hydroxypropyl isothiocyanate > 4-hydroxybutyl nitrile (Figure 9). The former two products are derived from allyl glucosinolate, whereas the latter two are derived from 3-hydroxypropyl glucosinolate.
Table 6.
Glucosinolate Hydrolysis Products Influence T. ni Herbivory in the Da(1)-12 × Ei-2 Crossa
| Source | Sum of Squares |
df | Mean Square | F-Ratio | P |
|---|---|---|---|---|---|
| Glucosinolate type | 224.978 | 2 | 112.489 | 4.457 | 0.013 |
| Hydrolysis product type | 294.492 | 2 | 147.246 | 5.834 | 0.003 |
| Error | 4113.774 | 163 | 25.237 |
a ANOVA with herbivory as the dependent variable. n = 168, r2 = 0.116.
Figure 9.
Effects of Individual Glucosinolate Hydrolysis Products on T. ni Herbivory.
T. ni larvae were allowed to feed on 168 F2 families of Da(1)-12 × Ei-2 that were classified according to the predominant glucosinolate breakdown product formed per line. 4OBP-NIT, 4-hydroxylbutyl nitrile; 3OHP-ITC, 3-hydroxypropyl isothiocyanate; EB-NIT, 3,4-epithiobutyl nitrile; Allyl-ITC, allyl isothiocyanate. The error bars indicate ±se.
DISCUSSION
Natural variation in glucosinolate hydrolysis products among Arabidopsis ecotypes has facilitated the identification of a gene whose product dramatically alters the course of hydrolysis. A survey of 122 ecotypes showed that the principal hydrolysis products formed were either nitriles or isothiocyanates (Figure 2). Because Col (isothiocyanates) and Ler (nitriles) differed in their hydrolysis products, the Col × Ler RIL (Lister and Dean, 1993) were used to map the responsible loci. The primary locus controlling nitrile versus isothiocyanate formation (Figure 3) was in close proximity to a gene (Figure 4) whose amino acid sequence has high identity to that of the B. napus ESP (Bernardi et al., 2000), a protein known to stimulate the formation of epithionitriles during myrosinase-catalyzed hydrolysis of alkenyl glucosinolates (Tookey, 1973; MacLeod and Rossiter, 1985; Foo et al., 2000). Extracts of E. coli expressing this Arabidopsis ESP analog were able to convert glucosinolates to nitriles and epithionitriles in the presence of myrosinase (Table 2, Figure 6). Furthermore, there was a perfect correlation among Col × Ler RIL between expression of the ESP gene and nitrile formation (Figure 5). Thus, Arabidopsis possesses an epithiospecifier-like protein responsible for a polymorphism in the formation of glucosinolate hydrolysis products.
The Arabidopsis ESP is more versatile than are the ESPs previously described. In the presence of myrosinase, heterologously expressed Arabidopsis ESP was able to convert nonalkenyl glucosinolates to simple nitriles as well as to convert alkenyl glucosinolates to epithionitriles (Table 2, Figure 6). In addition, genetic evidence from the Da(1)-12 × Ei-2 cross showed that the ESP locus is responsible for both simple nitrile and epithionitrile formation in vivo. Thus, the Arabidopsis ESP might be more accurately described as a nitrile-specifying protein. Proteins with similar properties could be responsible for the widespread appearance of simple nitriles upon glucosinolate hydrolysis in such genera of the Brassicaceae as Barbarea, Brassica, Lepidium, and Matthiola (Cole, 1976).
Another feature of the Arabidopsis ESP that distinguishes it from previously described members of this protein family is the lack of a strict requirement for Fe2+. Earlier work on ESP extracted from B. napus, Brassica campestris, Lepidium sativum, and Crambe abyssinica indicated that Fe2+ was necessary for ESP stability and catalysis (Tookey, 1973; MacLeod and Rossiter, 1985; Foo et al., 2000). The Arabidopsis ESP demonstrated optimal activity at an Fe2+ concentration of 0.1 to 0.5 mM, depending on the substrate, but still possessed substantial activity in the absence of added Fe2+ (with EDTA present) (Tables 3 and 4). Addition of Fe2+ above 0.1 mM dramatically increased the formation of nitriles by nonenzymatic processes. The hydrolysis of glucosinolates to nitriles in vitro rather than to isothiocyanates has long been known to be promoted by both Fe2+ and acidic (3 to 5) pH (Gil and MacLeod, 1980; MacLeod and Rossiter, 1986; Galletti et al., 2001). However, our results demonstrate that the Arabidopsis ESP is catalytically active at pH 6.0, chosen for our assays because it represents the approximate pH of fresh Arabidopsis leaf macerate, the environment in which ESP normally encounters its substrate.
The actual role of ESP in the conversion of glucosinolates to nitriles is still unclear. Because myrosinase is required for activity, ESP may simply use the aglycone produced by myrosinase as a direct substrate for nitrile formation (Figure 10) (Tookey and Wolff, 1970). In such a scenario, the active site of ESP may have to be situated very close to that of myrosinase because the aglycone is known to rearrange readily to the corresponding isothiocyanate in aqueous solution. Alternately, rather than functioning as an independent enzyme, ESP may serve instead as a proteinaceous cofactor that binds to myrosinase and alters the course of hydrolysis (Tookey, 1973). Myrosinase is reported to associate with a number of other proteins in vivo (Rask et al., 2000). In either case, the cleavage of the C–S bond of the glucosinolate aglycone leads to nitrile formation. In epithionitrile formation (where the double bond may provide assistance in reaction), the cleaved sulfur reattaches to the terminal olefin. Experiments by Brocker and Benn (1983) with 35S-labeled alkenyl glucosinolate showed that the sulfur atom from the thioglucose residue is transferred intramolecularly to the double bond of the side chain. This intramolecular addition is not stereochemically precise because products resulting from sulfur addition on either side of the double bond are found in nearly equal abundance (Figure 2G).
Figure 10.
Model for Roles of Myrosinase and Arabidopsis ESP in the Hydrolysis of Glucosinolates to Nitriles.
Myrosinase hydrolyzes the thioglucose linkage, which leads to an unstable aglycone that in the absence of other factors can spontaneously rearrange to form an isothiocyanate. However, in the presence of ESP, the aglycone is converted to a nitrile by cleavage of the C–S bond. When a terminal olefin is present in the glucosinolate side chain, the sulfur atom is recaptured as an episulfide, thus forming a three-membered thiirane ring. n = 1 or 2.
The polymorphism in glucosinolate hydrolysis products in Arabidopsis has significant impact on herbivory by T. ni (the cabbage looper), a generalist-feeding lepidopteran whose larvae attack members of the Brassicaceae as well as a host of other plant species (Shorey et al., 1962). In this study, T. ni larvae fed significantly more on Col × Ler RIL that produced nitriles than on those that produced isothiocyanates (Figure 8). In fact, the principal QTLs associated with T. ni feeding were identical to those associated with nitrile formation (Figure 3). Interestingly, one of the QTLs for T. ni feeding closely resembles a QTL reported in another investigation of T. ni herbivory on Arabidopsis that was just published. Jander et al. (2001) carefully documented differential T. ni herbivory on Col versus Ler ecotypes and then mapped these differences using the Col × Ler RIL to a locus near 85 centimorgans on chromosome 1. This locus, which they designated as TASTY, is very close to ESP (Figure 3). These authors reported no significant difference in glucosinolate content and myrosinase level between the more preferred Ler and the less preferred Col, but they did not investigate glucosinolate hydrolysis products. In experiments with Col × Ler F1 hybrids, Jander et al. (2001) demonstrated that susceptibility to feeding was dominant, which is consistent with TASTY encoding a protein, such as ESP, whose presence results in the formation of a product associated with greater feeding.
Although the effects of ESP on glucosinolate hydrolysis are clear, the role of ESP in plant defense is difficult to explain. There would seem to be little selective advantage for plants to produce a protein that increased herbivore feeding. Perhaps nitriles or epithionitriles are more effective than are isothiocyanates as deterrents for herbivores other than T. ni. Nitriles are known to be toxic to insects (Peterson et al., 2000), and epthionitriles have been shown to be cytotoxic to mammals (Wallig et al., 1988; VanSteenhouse et al., 1991).
Alternately, the role of ESP may be to block isothiocyanate formation. Isothiocyanates are often reported to be toxins or growth inhibitors for insect herbivores (Ahman, 1986; Borek et al., 1998; Li et al., 2000) and other plant enemies (Olivier et al., 1999; Tierens et al., 2001). Indeed, in this study, isothiocyanates were stronger feeding inhibitors for T. ni than were nitriles (Figure 9). However, certain isothiocyanates, such as allyl isothiocyanate, are also volatile and as such are employed as host-finding attractants by specialist Brassicaceae herbivores (Chew and Renwick, 1994; Ekbom, 1998; Rojas, 1999). Thus, in certain cases, the loss of allyl isothiocyanate may benefit the plant by making it less detectable to specialist herbivores (Chew, 1988). It may be significant that Arabidopsis ecotypes that are potentially able to release allyl or other volatile alkenyl isothiocyanates on hydrolysis (ecotypes with alkenyl glucosinolates) appear to have much higher frequencies of ESP than do ecotypes without precursors of volatile isothiocyanates (ecotypes with hydroxyalkyl and methylsulfinylalkyl glucosinolates) (Table 1). Although loss of allyl isothiocyanate could make a plant more susceptible to generalist herbivores, this may be more than offset by a reduction in pressure from specialist herbivores. Thus, temporal and spatial alterations in the relative pressure from specialist and generalist herbivores may have maintained the polymorphism for glucosinolate hydrolysis products in Arabidopsis. In this context, it is interesting to note that ESP function has been lost at least two different times in the evolutionary history of Arabidopsis, once by loss of gene expression and once by loss of a large segment of the coding region (Table 5), suggesting repeated instances in which plants may have been under selection to abolish ESP activity.
The feeding preference of T. ni larvae is influenced not only by the type of glucosinolate hydrolysis product formed but also by the nature of the parent glucosinolate itself. In the Da(1)-12 × Ei-2 cross, F2 families containing either of the hydrolysis products of 3-hydroxypropyl glucosinolate were significantly more preferred by T. ni than were those containing either of the hydrolysis products of allyl glucosinolate (Table 6, Figure 9). In contrast, when T. ni were allowed to feed on the Ler × Col RI lines, they did not differentiate between lines that contained 3-hydroxylpropyl glucosinolate and those that contained 3- and 4-methylsulfinyl glucosinolates (D.J. Kliebenstein and T. Mitchell-Olds, unpublished results). Additional studies are in progress to clarify the relative feeding deterrence of the individual glucosinolates of Arabidopsis.
Further work on the genetics and biochemistry of glucosinolate hydrolysis should increase our understanding of this process and the role of glucosinolates in plant defense. In many studies (e.g., Moyes et al., 2000), it has been difficult to show a clear correlation between glucosinolate content or myrosinase activity and the extent of herbivore damage. An improved knowledge of the actual glucosinolate hydrolysis products formed in plants may help explain patterns of herbivore choice and host resistance, because it is the hydrolysis products themselves, not the parent glucosinolates or myrosinase, that have direct physiological effects. Typical analytical methods for glucosinolates and myrosinase activity (measured by glucose release) (Poulton and Moller, 1993) provide no information on the nature of the hydrolysis products.
There is also much more to learn about the regulation of glucosinolate hydrolysis product formation in Arabidopsis. Our search for QTLs controlling glucosinolate hydrolysis identified two loci, ESP on chromosome 1 and a second locus on chromosome 3 near the marker AthChib (Figure 3). When ESP is present, the locus on chromosome 3 affects nitrile formation threefold (Figure 5). Although this locus is unlikely to encode a transcription factor, nothing else is known about it because it has no effect on ESP gene expression. Further work on this and on other genes regulating glucosinolate hydrolysis in Arabidopsis is now in progress.
METHODS
Plants and Growth Conditions
All seed stocks of Arabidopsis thaliana were obtained from the Arabidopsis Biological Resource Center (http://aims.cps.msu.edu/aims/). A list of the 122 ecotypes surveyed is given at the end of the next section. Ten plants of each ecotype were planted in 6-cm pots containing a standard vermiculite/soil mix. The plants were allowed to grow at 26°C under illumination of 60 W Cool White Deluxe GE bulbs (at a distance of 25 cm) for a 12-hr-light/12-hr-dark photoperiod. For analysis of glucosinolate hydrolysis products, leaves were harvested 4 weeks after germination.
For investigation of the 96 Col × Ler recombinant inbred lines (RIL), we planted 10 plants of each in a single cell in a 96-well flat. The plants were allowed to grow for 3 weeks postgermination, as described above, and approximately two leaves per plant (20 leaves per cell) were harvested for analysis of glucosinolate hydrolysis products.
For investigation of the Da(1)-12 × Ei-2 cross, we planted 20 F3 seeds from a single F2 line in a single cell in a 96-well flat. The plants were allowed to grow for 3 weeks postgermination, as described above, and all of the tissue from the 20 F3 plants was pooled for analysis to generate a representative sample of the hydrolysis product composition of each F2 parent.
Analysis of Glucosinolate Hydrolysis Products
Leaf samples (50 mg) of each ecotype were ground with a glass stirring rod for 15 sec in 1.2 mL of water at the bottom of a 4-mL glass tube. The tube was quickly sealed with a septum cap and left to stand for 5 min at room temperature. After the addition of 2 mL of dichloromethane through the septum, the tube was vortexed for 10 sec and centrifuged at 500g for 5 min. The dichloromethane layer was then removed, dried, and filtered by passage through a short column of anhydrous sodium sulfate held with a plug of glass wool in a Pasteur pipette. The remaining aqueous layer was re-extracted with a second 2-mL portion of dichloromethane, and the two extracts were combined and concentrated under nitrogen to 200 μL.
Samples were analyzed by gas chromatography–mass spectrometry (GC-MS) and GC–flame ionization detection (FID) using an Agilent 6890 series gas chromatograph (Agilent Technologies, Waldbronn, Germany) with an HP5MS column (30-m × 0.25-mm × 0.25-μm film), splitless injection at 200°C, and a temperature program of 35°C for 3 min, a 12°C/min ramp to 96°C, and an 18°C/min ramp to 240°C (with a 6-min final hold). For product identification by MS, we coupled the column to an Agilent 5973N quadrupole mass detector with He as the carrier gas. Parameters for electron impact sample ionization were as follows: interface temperature, 280°C; repeller, 30 V; emission, 34.6 μA; electron energy, 70 eV; and source temperature, 230°C. Peaks were identified by comparing retention times and mass spectra with those of the authentic standards, 3-butenyl nitrile and allyl isothiocyanate (both from Fluka, Taufkirchen, Germany) and with those in the literature (Spencer and Daxenbichler, 1980).
For product quantification by FID, we used hydrogen as the carrier gas and operated the detector at 300°C. Quantification was based on peak area relative to that of the internal standard, propyl isothiocyanate (Aldrich). Response factors (RF) relative to propyl isothiocyanate were experimentally determined for 3-butenyl nitrile (1.33) and allyl isothiocyanate (1.06), and they were calculated for compounds not commercially available by using the effective carbon number (ECN) concept (Scanlon and Willis, 1985): propyl isothiocyanate (ECN 4.0, RF 1.0); 4-methylsulfinylbutyl isothiocyanate (ECN 6.0, RF 0.67); 5-methylsulfinylpentyl nitrile (ECN 5.3, RF 0.75); 3-hydroxypropyl isothiocyanate (ECN 3.5, RF 1.14); 4-hydroxybutyl nitrile (ECN 2.8, RF 1.43); 2-hydroxy-3-butenyl isothiocyanate (ECN 4.4, RF 0.91); and 3-hydroxy-4-butenyl nitrile (ECN 3.7, RF 1.08).
The recovery of glucosinolate hydrolysis products during the extraction procedure was estimated by spiking samples with known amounts of the commercially available hydrolysis products mentioned above plus analogs, including butyl nitrile, pentyl nitrile, and ethyl isothiocyanate (all Fluka), and propyl isothiocyanate and phenyl isothiocyanate (both Aldrich). The overall recovery was ∼50%, with the biggest losses the result of volatilization during the concentration step. Dichloromethane was used as the extraction solvent because, of all the solvents tested (hexane, diethyl ether, and chloroform), it gave the highest recoveries and the best separation of analyte and solvent peaks in the chromatogram.
The ecotypes analyzed, grouped by their predominant hydrolysis products, are as follows:
Methylsulfinylalkyl isothiocyanates (Figure 2A): Ak-1, Achkarren, Freiburg, Germany, N938; Bsch-1, Buchschlag, Germany, N1002; Col-0, Columbia, Missouri, United States, N1092; Enkheim-D, Enkheim, Germany, N920; Ep-0, Eppenheim, Germany, N1140; Gie-0, Gieben, Germany, N1192; H-55, Relichova, Czech Republic, N923; Lan-0, Lanark, United Kingdom, N1304; Le-0, Leiden, The Netherlands, N1308; Mz-0, Merzhausen, Germany, N1382; Rld-1, N913; Si-0, Siegen, Germany, N1524; Wa-1, Warschau, Poland, N1586; Ws-0, Wassilewskija, Russia, N1602; Ws-3, Wassilewskija, Russia, N1638.
Methylsulfinylalkyl nitriles (Figure 2B): Bn-0, Ben Dhorrain, United Kingdom, N990; Bu-0, Burghaun, Germany, N1006; Da-0, Darmstadt, Germany, N1098; En-1, Enkheim, Germany, N1136; En-2, Enkheim, Germany, N1138; Enkheim-T, Enkheim, Germany, N921; Gd-1, Gudow, Germany, N1184; Ll-0, Llagostera, Spain, N1338; Me-0, Mechtshausen, Germany, N1364; Mh-0, Muhlen, Poland, N904; Mt-0, Martuba, Libya, N1380; Na-1, Nantes, France, N1384; Nle-0, Niederlauken, Germany, N1392; Nw-0, Neuweilnau, Germany, N1408; Sg-2, St. Georgen, Germany, N1520; Sp-0, Berlin, Germany, N1530.
3-Hydroxypropyl isothiocyanate (Figure 2C): Be-0, Bensheim, Germany, N964; Bl-1, Bologna, Italy, N968; Da(1)-12, Czechoslovakia, N917; Dra-0, Drahonin, Russia, N1116; Eil-0, Eilenburg, Germany, N1132; Gr-1, Graz, Austria, N1198; HL-3, Holtensen, Germany, N1232; Lm-2, Le Mans, France, N1344; No-0, Nossen, Germany, N1394; Np-0, Nieps, Germany, N1396; Sap-0, Slapy, Czech Republic, N1506.
4-Hydroxybutyl nitrile (Figure 2D): Bch-1, Buchen, Lauenburg, Germany, N956; Cen-0, Caen, France, N1066; Chi-1, Chisdra, Russia, N1074; Ct-1, Catania, Italy, N1094; Di-1, Dijon, France, N1108; Di-g, Dijon, France, N910; Dr-0, Dresden, Germany, N1114; Est-0, Estland, Russia, N1148; Ler-0, Landsberg, Germany, NW20; Lip-0, Lipowiec, Poland, N1336; Nd-0, Niederzenz, Germany, N1390; Nd-1, Niederzenz, Germany, N1636; Ra-0, Randan, France, N1480; St-0, Stockholm, Sweden, N1534; Stw-0, Stobowa, Russia, N1538; Wc-1, Westercelle, Germany, N1588; Wei-0, Weiningen, Switzerland, N3110.
Alkenyl isothiocyanates (Figure 2E): Blh-1, Bulhary, Czech Republic, N1030; Mr-0, Monte Tosso, Italy, N1372; Ru-0, Ruppachtal, Germany, N1496.
Epithionitriles (Figures 2F and 2G): Abd-0, Kingswells, Aberdeen, United Kingdom, N932; Ag-0, Argentat, France, N901; An-2, Antwerpen, Belgium, N946; Ang-0, Angleur, Belgium, N948; Ba-0, Blackmount, United Kingdom, N952; Bch-3, Buchen, Lauenburg, Germany, N958; Bla-10, Blanes, Spain, N982; Br-0, Brunn, Czech Republic, N994; Bs-1, Basel, Switzerland, N996; Bur-0, Burren, Ireland, N1028; C24, N906; Cal-0, Calver, United Kingdom, N1062; Can-0, Canary Islands, N1064; Cit-0, Citou, France, N1080; Cnt-1, Canterbury, United Kingdom, N1635; Co-1, Coimbra, Portugal, N1084; Co-2, Coimbra, Portugal, N1086; Cvi-0, Cape Verdi Islands, N902; Db-1, Tenne, Germany, N1102; Db-2, Dombachtal, Germany, N1104; Di-0, Dijon, France, N1106; Do-0, Donsbach, Germany, N1112; Edi-0, Edinburgh, United Kingdom, N1122; Ei-2, Eifel, Germany, N1124; El-0, Ellershausen, Germany, N1134; Er-0, Erlangen, Germany, N1142; Es-0, Espoo, Finland, N1144; Et-0, Etraygues, France, N1152; Ga-0, Gabelstein, Germany, N1180; Ge-0, Geneve, Switzerland, N1186; Gre-0, Greenville, Michigan, United States, N1210; Ha-0, Hannover, Germany, N1218; Hau-0, Hauniensis, Denmark, N1220; Hh-0, Hohenlieth, Germany, N1224; Hl-0, Holtensen, Germany, N1228; Hl-2, Holtensen, Germany, N1230; Hn-0, Hennetalsperre, Germany, N1234; Hodja, Khurmatov, Tadjikistan, N922; Kondara, Khurmatov, Tadjikistan, N916; Lc-0, Loch Ness, United Kingdom, N1306; Lch-0: Li-1, Limburg, Germany, N1310; Li3-3, Limburg, Germany, N1318; Loe-1, Lorrach, Germany, N1346; Loe-2, Lorrach, Germany, N1348; Lu-1, Lund, Sweden, N1352; Mir-0, Miramare, Italy, N1378; Mnz-0, Mainz, Germany, N1370; Mrk-0, Baden, Germany, N1374; Ms-0, Moscow, Russia, N905; Rd-0, Rodenbach, Germany, N1482; Rou-0, Rouen, France, N1488; Rsch-0, Rschew, Russia, N1490; Sah-0, Sierra Alhambra, Spain, N1500; Sf-1, San Feliu, Spain, N1512; Shakdara, Pamiro-Alay, Tadjikistan, N929; Sorbo, Khurmatov, Tadjikistan, N931; Su-0, Southport, United Kingdom, N1540; Tac, Tacoma, Washington, United States; Wl-0, Wildbad, Germany, N1630.
Quantitative Trait Locus Mapping in Col × Ler Recombinant Inbred Lines
Leaves of 96 Col × Ler RIL were analyzed for glucosinolate hydrolysis products, and the percentage of nitriles was computed by dividing the mass of all nitriles identified by the total mass of all hydrolysis products (isothiocyanates and nitriles) detected from methionine-derived glucosinolates. This variable was used for mapping the loci controlling nitrile formation. Quantitative trait loci (QTLs) and effects were estimated by using the family mean for each RIL in conjunction with both interval mapping and composite interval mapping in QTL cartographer (Basten et al., 1999). The genome-wide 5% significance threshold was estimated by randomly reshuffling the phenotypic data 500 times in QTL cartographer (Basten et al., 1999). Epistatic interactions were tested within SYSTAT by ANOVA, using the mean phenotypic value for each line. Only markers that were individually significant were tested for epistasis. Mapping data for the Ler × Col RIL were obtained from the Nottingham Stock Center (http://nasc.nott.ac.uk/).
Cloning of Epithiospecifier Protein and Measurement of Gene Expression with Reverse Transcription–Polymerase Chain Reaction
RNA purification, cDNA synthesis, cDNA sequencing, and reverse transcription–polymerase chain reaction (RT-PCR) were performed as previously described (Kliebenstein et al., 2001a). The cDNA was cloned from the Tac ecotype because the gene is not expressed in Col. The primers for RT-PCR of the epithiospecifier protein (ESP) gene were forward (5′-CCATGGCTCCGACTTTGC-3′) and reverse (5′-CGAGCCACATACACAACACA-3′).
Heterologous Expression of ESP in Escherichia coli
The full-length Arabidopsis ESP cDNA was amplified from the Tacoma ecotype by PCR using the forward primer (5′-ATGGCTCCGACTTTGCAAG-3′) and the reverse primer (5′-AGCTGAATTGACCGCATAGA-3′). The PCR product was then cloned into the pBAD/Thio-E Echo vector from Invitrogen (Carlsbad, CA), which places the translated polypeptide in a fusion protein with thioredoxin under the control of an arabinose-inducible promoter. Small (2 mL) pre-cultures of transformed E. coli were grown overnight in Luria-Bertani medium with antibiotic selection. On the following morning, 10 mL of pooled culture was used to inoculate 250 mL of medium in a 1-liter flask. After 2 hr of growth in a 37°C shaking incubator, 2.5 mL of 20% (w/v) arabinose dissolved in water was added to induce expression. After 4 hr of induction, 30 mL of cells was pelleted at 7000g for 2 min and the supernatant discarded. The pellet was then resuspended in 1 mL of 100 mM Mes, pH 6.0, and lysed by three freeze–thaw cycles. The lysed cells were centrifuged at 3000g at 4°C for 15 min, and the pellet was resuspended in 1 mL of 100 mM Mes. According to SDS-PAGE, virtually all of the expressed ESP fusion protein was present in the pellet. Thus, resuspended pellets corresponding to ∼180 mL of culture were pooled and preincubated with 1 mg of Sinapis alba myrosinase (Sigma) for 5 min with occasional mixing. The assay was initiated by addition of 1 mL of 100 mM Mes containing 0.4 μmol of glucosinolate substrate purified by preparative HPLC as previously described (Kliebenstein et al., 2001a); the assay mixture was incubated for 30 min at room temperature in a septum-capped 4-mL glass tube. After addition of 2 mL of dichloromethane through the septum, assay products were extracted and analyzed by GC-MS and GC-FID as described above.
Isolation of ESP Genomic Clones
The genomic sequence of the Col ESP gene was obtained from bacterial artificial chromosome F15I1 (GenBank accession number AC006577; bp 30,759 to 38,719). The six other ecotypes were then sequenced with primers designed to amplify the Col ESP in 11 segments that were ∼1 kb in size with a 200- to 400-bp overlap on both sides. The primer names and sequences were as follows: EpiWalk1-F, 5′-AGCAAGAAAACTGAAACATCCA-3′; EpiWalk1-R, 5′-AATTCT-TGGATCCCGCTTTT-3′; EpiWalk2-F, 5′-ATATGTTTTCCCGCTGTT-GC-3′; EpiWalk2-R, 5′-GTTTATGGGTTTGCGACCTC-3′; EpiWalk3-F, 5′-CTTCGGTCCACTTTTTGGAA-3′; EpiWalk3-R, 5′-GGCGAAAAA-TATGTGGGTTC-3′; EpiWalk3-S, 5′-CACACGGCATAGCCGCG-3′; EpiWalk3-S2, 5′-CCTCCAAGATATAGATCTTAG-3′; EpiWalk4-F, 5′-GTTCTGTCAAACCCGTCCTG-3′; EpiWalk4-R, 5′-AGATTTGGAAGT-TTGAATTTTATGTT-3′; EpiWalk4-S, 5′-AGTACTGTGCACAGCCATT-3′; EpiWalk5-F, 5′-TCCGTACTTGACCCCAAAAC-3′; EpiWalk5-R, 5′-TTTTAAAATAAAACGGGTGGGTA-3′; EpiWalk5-S, 5′-CGGTAGAAC-TACATACCAAC-3′; EpiWalk6-F, 5′-ATCTTGATCCACTGGCCTTG-3′; EpiWalk6-R, 5′-TTTTACGTGATTTTGTGTTTCAA-3′; EpiWalk6-S, 5′-CTTCGATCATAGTAGTGGAC-3′; EpiWalk7-F, 5′-AGTATTGTTTTT-GTTTGGAAAATGT-3′; EpiWalk7-R, 5′-TGCATTGTTTATGTTTAT-ACTTTTGAA-3′; EpiWalk7-S, 5′-GTGGGAAATGAGTAGTTGC-3′; EpiWalk8-F, 5′-CCAAGGTCCATTGTCGCTTA-3′; EpiWalk8-R, 5′-CAAACGTTTTCTCACCAAATCA-3′; EpiWalk8-S, 5′-CGGGTTCTA-TCAAGCCAC-3′; EpiWalk9-F, 5′-CTAACCAAAACCGTGCGAAG-3′; EpiWalk9-R, 5′-TGAGAAATGGTTTAGTATGAATGC-3′; EpiWalk9-S, 5′-CGTTGCGTTATGTCATAGGC-3′; EpiWalk10-F, 5′-TGACCAATAGAGAAAAGAAAAATGG-3′; EpiWalk10-R, 5′-GTGGCATGAGTTGTT-CCAGA-3′; EpiWalk10-S, 5′-AGTATCTACTCTGTACCATAC-3′; EpiWalk11-F, 5′-AACGCATTCGCAAACTTTTT-3′; and EpiWalk11-R, 5′-TACCCAAATGTTTTTAGTCAGAAA-3′. PCR products were cloned into the TA vector, and four independent clones of each segment were sequenced for each ecotype. The total sequenced portion included the entire coding region, an additional 290 bp 3′ of the polyadenylation site, and 4.9 kb 5′ of the transcription start site.
Effect of Glucosinolate Hydrolysis Products on Trichoplusia ni Herbivory
T. ni (cabbage looper) eggs were obtained from Entopath, Inc. (Easton, PA) and reared on Southland artificial diet (Entopath, Inc). When plants of Col × Ler RIL or the Da(1)-12 × Ei-2 cross were 4 weeks old, the diameter of the rosette was measured, and a single first instar T. ni larva was placed on each rosette for 48 hr. The insects were taken directly from artificial diet and placed on the plants without a starvation period. The percentage of the rosette removed by the insect was estimated visually with the aid of a transparent 1-cm2 grid. The rosette diameter was also measured as an indicator of plant size.
For feeding on the 96 Col × Ler RIL, we used a randomized complete blocks design with 10 replicates (n = 960). Each 96-cell flat contained one plant from each of the 95 lines being tested and a Col plant. Genetic differences among lines were analyzed as randomized complete blocks ANOVA, using the model HERBIVORY = CONSTANT + FLAT + LINE + SIZE*SIZE. SIZE is a covariate included to control for developmental differences that may occur between individuals of the same line as the result of size-related environmental causes. Because lines were not replicated within flats, it was necessary to assume that LINE × FLAT interactions were absent. Genetic correlations were estimated from the Pearson product-moment correlation coefficient among family means (typically as the least squares family means, controlling for flat effects in ANOVA). When traits (such as resistance to several insect species) are measured in separate experiments, rG provides an unbiased estimate of the genetic correlation (Falconer and Mackay, 1996).
For feeding on the Da(1)-12 × Ei-2 cross, the population of 192 lines was split into two sets of 96 F3 families. Each set of 96 families was replicated 16 times and then assayed for feeding damage by T. ni (n = 3072). Genetic differences among lines were analyzed as a nested ANOVA, using the model HERBIVORY = CONSTANT + SET + LINE(SET) + FLAT(SET) + SIZE. LINE × FLAT and LINE × SET interactions could not be estimated. The impact of the type of glucosinolate and type of hydrolysis product upon T. ni herbivory was estimated by ANOVA, using the model HERBIVORY = CONSTANT + GLUCOSINOLATE TYPE + HYDROLYSIS TYPE. The latter two variables are categorical variables using the genetic score at the loci GS-AlkOhp (Kliebenstein et al., 2001a) and ESP, respectively. Only the 168 lines with scores for GS-AlkOhp and ESP were used in this analysis. For clarity, Figure 9 shows only the double homozygotes.
Acknowledgments
We thank Deana Pedersen for help with the T. ni herbivory assays and Drs. Georg Jander and Fred Ausubel for sending a preprint and communicating further on their work with the TASTY locus. This work was supported by the Max Planck Society, a grant to J.G. from the German National Science Foundation (FOR 383), and a grant to T.M.-O. from the U.S. National Science Foundation (DEB-9527725).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010261.
References
- Ahman, I. (1985). Oviposition behaviour of Dasineura brassicae on a high- versus low-quality Brassica host. Entomol. Exp. Appl. 39, 247–253. [Google Scholar]
- Ahman, I. (1986). Toxicities of host secondary compounds to eggs of the Brassica specialist Dasineura brassicae. J. Chem. Ecol. 12, 1481–1488. [DOI] [PubMed] [Google Scholar]
- Basten, C.J., Weir, B.S., and Zeng, Z.-B. (1999). QTL Cartographer, version 1.13. (Raleigh, NC: Department of Statistics, North Carolina State University).
- Bernardi, R., Negri, A., Ronchi, S., and Palmieri, S. (2000). Isolation of the epithiospecifier protein from oil-rape (Brassica napus ssp. oleifera) seed and its characterization. FEBS Lett. 467, 296–298. [DOI] [PubMed] [Google Scholar]
- Bones, A.M., and Rossiter, J.T. (1996). The myrosinase–glucosinolate system, its organisation and biochemistry. Physiol. Plant. 97, 194–208. [Google Scholar]
- Borek, V., Elberson, L.R., McCaffrey, J.P., and Morra, M.J. (1998). Toxicity of isothiocyanates produced by glucosinolates in Brassicaceae species to black vine weevil eggs. J. Agric. Food Chem. 90, 109–112. [Google Scholar]
- Brocker, E.R., and Benn, M.H. (1983). The intramolecular formation of epithioalkanenitriles from alkenylglucosinolates by Crambe abyssinica seed flour. Phytochemistry 22, 770–772. [Google Scholar]
- Chew, F.S. (1988). Searching for defensive chemistry in the Cruciferae, or, Do glucosinolates always control interactions of Cruciferae with their potential herbivores and symbionts? No! In Chemical Mediation of Coevolution, K.A. Spencer, ed (New York: Academic Press), pp. 81–111.
- Chew, F.S., and Renwick, J.A.A. (1994). Host plant choice in Pieris butterflies. In Chemical Ecology of Insects II, R.T. Carde and W.J. Bell, eds (New York: Chapman and Hall), pp. 214–238.
- Cole, R. (1976). Isothiocyanates, nitriles and thiocyanates as products of autolysis of glucosinolates in Cruciferae. Phytochemistry 15, 759–762. [Google Scholar]
- Daxenbichler, M.E., VanEtten, C.H., and Wolff, I.A. (1968). Diastereomeric episulfides from epi-progoitrin upon autolysis of crambe seed meal. Phytochemistry 7, 989–996. [Google Scholar]
- Donkin, S.G., Eiteman, M.A., and Williams, P.L. (1995). Toxicity of glucosinolates and their enzymatic decomposition products to Caenorhabditis elegans. J. Nematol. 27, 258–262. [PMC free article] [PubMed] [Google Scholar]
- Ekbom, B. (1998). Clutch size and larval performance of pollen beetles on different host plants. Oikos 83, 56–64. [Google Scholar]
- Falconer, D.S., and Mackay, T.F.C. (1996). Introduction to Quantitative Genetics. (Harlow, UK: Longman).
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, H.L., Gronning, L.M., Goodenough, L., Bones, A.M., Danielsen, B.-E., Whiting, D.A., and Rossiter, J.A. (2000). Purification and characterisation of epithiospecifier protein from Brassica napus: Enzymic intramolecular sulphur addition within alkenyl thiohydroximates derived from alkenyl glucosinolate hydrolysis. FEBS Lett. 468, 243–246. [DOI] [PubMed] [Google Scholar]
- Galletti, S., Bernardi, R., Leoni, O., Rollin, P., and Palmieri, S. (2001). Preparation and biological activity of four epiprogoitrin myrosinase-derived products. J. Agric. Food Chem. 49, 471–476. [DOI] [PubMed] [Google Scholar]
- Gil, V., and MacLeod, A.J. (1980). The effects of pH on glucosinolate degradation by a thioglucoside glucohydrolase preparation. Phytochemistry 19, 2547–2551. [Google Scholar]
- Graser, G., Schneider, B., Oldham, N.J., and Gershenzon, J. (2000). The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch. Biochem. Biophys. 378, 411–419. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A. (1999). Glucosinolates. In Naturally Occurring Glycosides: Chemistry, Distribution and Biological Properties, R. Ikan, ed (New York: John Wiley), pp. 193–223.
- Hasapis, X., and MacLeod, A.J. (1982). Benzylglucosinolate degradation in heat-treated Lepidium sativum seeds and detection of a thiocyanate-forming factor. Phytochemistry 21, 1009–1013. [Google Scholar]
- Hogge, L.R., Reed, D.W., Underhill, E.W., and Haughn, G.W. (1988). HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chromatography/mass spectrometry. J. Chromatogr. Sci. 26, 551–556. [Google Scholar]
- Jander, G., Cui, J., Nhan, B., Pierce, N.E., and Ausubel, F.M. (2001). The TASTY locus on chromosome 1 of Arabidopsis affects feeding of the insect herbivore Trichoplusia ni. Plant Physiol. 126, 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, P.J., Bones, A., and Rossiter, J.T. (1998). Subcellular immunolocalization of the glucosinolate sinigrin in seedlings of Brassica juncea. Planta 206, 370–377. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D., Lambrix, V., Reichelt, M., Gershenzon, J., and Mitchell-Olds, T. (2001. a). Gene duplication and the diversification of secondary metabolism: Side chain modification of glucosinolates in Arabidopsis thaliana. Plant Cell 13, 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., and Mitchell-Olds, T. (2001. b). Genetic control of natural variation in Arabidopsis thaliana glucosinolate accumulation. Plant Physiol. 126, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latxague, L., Gardrat, C., Coustille, J.L., Viaud, M.C., and Rollin, P. (1991). Identification of enzymatic degradation products from synthesized glucobrassicin by gas chromatography–mass spectrometry. J. Chromatogr. 586, 166–170. [Google Scholar]
- Li, Q., Eigenbrode, S.D., Stringam, G.R., and Thiagarajah, M.R. (2000). Feeding and growth of Plutella xylostella and Spodoptera eridania on Brassica juncea with varying glucosinolate concentrations and myrosinase activities. J. Chem. Ecol. 26, 2401–2419. [Google Scholar]
- Lister, C., and Dean, C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4, 745–750. [DOI] [PubMed] [Google Scholar]
- Louda, S., and Mole, S. (1991). Glucosinolates: Chemistry and ecology. In Herbivores: Their Interactions with Secondary Plant Metabolites, 2nd ed, G.A. Rosenthal and M.R. Berenbaum, eds (San Diego, CA: Academic Press), pp. 123–164.
- MacLeod, A.J., and Rossiter, J.T. (1985). The occurrence and activity of epithiospecifier protein in some Cruciferae seeds. Phytochemistry 24, 1895–1898. [Google Scholar]
- MacLeod, A.J., and Rossiter, J.T. (1986). Isolation and examination of thioglucoside glucohydrolase from seeds of Brassica napus. Phytochemistry 25, 1047–1051. [Google Scholar]
- Moyes, C.L., Collin, H.A., Britton, G., and Raybould, A.F. (2000). Glucosinolates and differential herbivory in wild populations of Brassica oleracea. J. Chem. Ecol. 26, 2625–2641. [Google Scholar]
- Olivier, D., Vaughn, S.F., Mizubuti, E.S.G., and Loria, R. (1999). Variation in allyl isothiocyanate production within Brassica species and correlation with fungicidal activity. J. Chem. Ecol. 25, 2687–2701. [Google Scholar]
- Peterson, C.J., Cossé, A., and Coats, J.R. (2000). Insecticidal components in the meal of Crambe abyssinica. J. Agric. Urban Entomol. 17, 27–36. [Google Scholar]
- Poulton, J.E., and Moller, B.L. (1993). Glucosinolates. In Enzymes of Secondary Metabolism, P.J. Lea, ed (London: Academic Press), pp. 209–237.
- Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. [PubMed] [Google Scholar]
- Rojas, J.C. (1999). Electrophysiological and behavioral responses of the cabbage moth to plant volatiles. J. Chem. Ecol. 25, 1867–1883. [Google Scholar]
- Scanlon, J.T., and Willis, D.E. (1985). Calculation of flame ionization detector relative response factors using the effective carbon number concept. J. Chromatogr. Sci. 23, 333–340. [Google Scholar]
- Shorey, H.H., Andres, L.A., and Hale, R.L. (1962). The biology of Trichoplusia ni (Lepidoptera: Noctuidae). I. Life history and behavior. Ann. Entomol. Soc. Am. 55, 591–597. [Google Scholar]
- Spencer, G.F., and Daxenbichler, M.E. (1980). Gas chromatography–mass spectrometry of nitriles, isothiocyanates and oxazolidinethiones derived from cruciferous glucosinolates. J. Sci. Food Agric. 31, 359–367. [Google Scholar]
- Tierens, K.-J., Thomma, B., Brower, M., Schmidt, J., Kistner, K., Porzel, A., Mauch-Mani, B., Cammue, B., and Broekaert, W. (2001). Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125, 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tookey, H.L. (1973). Crambe thioglucoside glucohydrolase (EC 3.2.3.1): Separation of a protein required for epithiobutane formation. Can. J. Biochem. 51, 1654–1660. [DOI] [PubMed] [Google Scholar]
- Tookey, H.L., and Wolff, I.A. (1970). Effect of organic reducing agents and ferrous ion on thioglucosidase activity of Crambe abyssinica seed. Can. J. Biochem. 48, 1024–1028. [PubMed] [Google Scholar]
- VanSteenhouse, J.L., Fettman, M.J., and Gould, D.H. (1991). The effect of glutathione depletion by buthionine sulphoximine on 1-cyano-3,4-epithiobutane toxicity. Food Chem. Toxicol. 29, 153–157. [DOI] [PubMed] [Google Scholar]
- Wallig, M.A., Gould, D.H., Fettman, M.J., and Willhite, C.C. (1988). Comparative toxicities of the naturally occurring nitrile 1-cyano-3,4-epithiobutane and the synthetic nitrile n-valeronitrile in rats: Differences in target organs, metabolism and toxic mechanisms. Food Chem. Toxicol. 26, 149–157. [DOI] [PubMed] [Google Scholar]