Abstract
The plant-specific Rop family GTPases are versatile molecular switches in many processes during plant growth, development, and responses to the environment. To understand how Rop achieves its functional versatility in signaling, we performed a genome-wide identification of putative Rop targets using a combination of the yeast two-hybrid method, bioinformatic tools, and a robust functional assay in pollen. In this study, we have identified 11 Arabidopsis genes encoding novel proteins, termed RICs (for Rop-interactive CRIB motif–containing proteins), that contain a CRIB (for Cdc42/Rac-interactive binding) motif required for their specific interaction with GTP-bound Rop1. RICs are divergent and classified into five groups that share little sequence homology outside of the conserved Rop-interactive domain. Overexpression in tobacco pollen tubes of the nine Ric genes that are expressed in Arabidopsis pollen causes distinct phenotypes, implying distinct functions for various RICs. RIC3 (group III) and RIC4 (group V) both cause depolarized growth like Rop1 and display Rop1-enhanced localization to the tip of pollen tubes, suggesting that these RICs may be two distinct targets of Rop1. In contrast, RIC10 (group I) promotes pollen tube elongation but does not affect pollen tube growth polarity and shows Rop1-independent localization to the cytoplasm, suggesting that RIC10 may participate in a Rop1-independent pathway probably controlled by a different Rop. Expression of all other RICs causes various degrees of growth inhibition in pollen tubes. Furthermore, these inhibitory RICs also exhibit distinct patterns of localization in pollen tubes. Our results suggest that various RICs have evolved to interact with Rops differentially and to perform distinct functions in pollen tubes. Reverse transcriptase–mediated polymerase chain reaction analysis showed that six of the nine RICs are expressed in various parts of Arabidopsis plants. On the basis of these observations, we propose that RICs function as Rop GTPase targets that control various Rop-dependent signaling pathways in plants.
INTRODUCTION
The plant-specific family of Rho GTPases, termed Rop, has emerged as a pivotal molecular switch in plant signal transduction (Arabidopsis Genome Initiative, 2000; Zheng and Yang, 2000b). Rop is essential for pollen tube elongation, as indicated by the inhibition of pollen tube elongation by the injection of anti-Rop1 antibodies and the expression of the dominant negative (DN) mutants of Arabidopsis Rop1 and At-Rac2/Rop5 (Lin and Yang, 1997; Kost et al., 1999; Li et al., 1999). Overexpression of wild-type Rop1 or At-Rac2/Rop5 caused depolarized growth in pollen tubes (Kost et al., 1999; Li et al., 1999), as suggested by swelling of pollen tube tips or an increase in the radial expansion of pollen tubes (referred to as pollen tube expansion in this article). Expression of constitutively active (CA) mutants for these Rops induced isotropic growth (i.e., formation of a bulbous tube). These results suggest that Rop also controls the specification of a cortical site for tip growth in pollen tubes (Li et al., 1999). Evidence indicates that both polar localization of Rop and the regulation of its activity at the tip region of the plasma membrane (PM) are critical for defining the site of growth in pollen tubes (Lin et al., 1996; Kost et al., 1999; Li et al., 1999; Zheng and Yang, 2000a; G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data). Thus, Rop controls polar growth in pollen tubes probably by integrating signals for growth with polarity signals (Li et al., 1999; Zheng and Yang, 2000a). Our studies also suggest that Rop controls polar growth by regulating the dynamics of tip F-actin and the formation of tip-focused cytosolic calcium gradients (Li et al., 1999; Fu et al., 2001).
Transgenic expression of CA-rop2 and DN-rop2 mutant genes in Arabidopsis suggests that Rop signaling may control a wide range of growth and developmental processes, including embryo development, seed dormancy, seedling development, leaf morphogenesis, shoot apical dominance, lateral root initiation, and phyllotaxis (Li et al., 2001). The expression of these mutants also alters abscisic acid stimulation of seed dormancy, auxin stimulation of lateral root initiation, and brassinolide promotion of hypocotyl elongation, suggesting a role for Rop in the regulation of hormone accumulation or responses (Li et al., 2001). Several studies indicate that Rop activates H2O2 production and plant defense responses (Kawasaki et al., 1999; Potikha et al., 1999; Park et al., 2000; Ono et al., 2001). Furthermore, a Rop-like protein is localized to the tonoplast of developing vacuoles, implying a role for this Rop in vacuole development or function (Lin et al., 2001). Finally, a Rop-like protein also is associated with the active complex of the CLV1 receptor-like kinase, supporting a role for Rop in relaying receptor-like kinase signals (Trotochaud et al., 1999). The Arabidopsis genome contains 11 Rop genes, and the Rop proteins are distributed ubiquitously in various organ, tissue, and cell types in Arabidopsis (Winge et al., 2000; Zheng and Yang, 2000b; Li et al., 2001). Hence, the Rop GTPase appears to act as a universal switch in plant signal transduction. It remains a mystery how this switch regulates these diverse cellular and physiological processes.
In animals and yeast, the three major families of RHO GTPases, Cdc42, Rac, and Rho, also modulate a wide variety of cellular processes, such as actin organization, cell polarity establishment, axon guidance, gene transcription, cell cycle progression, cell-to-cell adhesion, NAPH oxidase activation, and glucan synthase activation (Mackay and Hall, 1998; Ridley, 2000). To achieve various cellular functions, GTP-bound active RHO GTPases interact with a plethora of functionally distinct target proteins or effectors (Mackay and Hall, 1998; Aspenstrom, 1999). Rho, Cdc42, and Rac each interact with a set of >10 distinct effectors in animals (Mackay and Hall, 1998; Aspenstrom, 1999). Most of the Cdc42/Rac effectors contain the conserved Cdc42/Rac-interactive binding (CRIB) motif that binds the effector domain of Cdc42/Rac GTPases in a GTP-dependent manner (Burbelo et al., 1995; Aspenstrom, 1999). The best studied examples of these types of Cdc42/Rac effectors include the P21-activated protein kinase and Wiskott-Aldrich syndrome proteins, the tyrosine kinases ACK-1 and ACK-2, and novel proteins termed Cdc42 effector proteins (CEPs) (Aspenstrom, 1999; Burbelo et al., 1999; Hirsch et al., 2001). Phylogenetic analysis suggests that Rop may have evolved from the same ancestor as Cdc42/Rac (Li et al., 1998; Zheng and Yang, 2000b). However, our preliminary search of databases, including the completed Arabidopsis genome sequence, has failed to identify any plant homologs of these animal Cdc42/Rac effectors. This suggests that the plant-specific Rop GTPase may use novel target proteins as effectors.
In this article, we report the genome-wide identification and functional characterization of a novel group of CRIB-containing proteins that interact with GTP-bound Rop GTPases from Arabidopsis. These proteins, termed RICs (for Rop-interactive CRIB-containing proteins), have been identified using database mining combined with the two-hybrid and other protein–protein interaction assays. Using overexpression and green fluorescent protein (GFP)–based protein localization in pollen tubes, we provide convincing evidence that RICs act as Rop targets that function in distinct Rop signaling pathways.
RESULTS
Identification of Active Rop-Interacting Proteins Using the Yeast Two-Hybrid Method
Because GTPase effectors typically interact with the GTP-bound active form of GTPases, we used a constitutively active Rop1 mutant (G15V/C188S) (Li et al., 1999; Wu et al., 2000) as a bait to screen an Arabidopsis seedling cDNA library. We identified 46 positive clones from ∼6 million yeast transformants. Among 20 clones sequenced, several clones encode RopGAPs (Wu et al., 2000) and one clone encodes a predicted novel polypeptide containing a CRIB motif. CRIB motifs are found in Cdc42/Rac effectors and confer their specific interaction with the active form of Cdc42/Rac GTPases in yeast and animals. Because this clone was not a full-length cDNA, we conducted a BLAST search against the Arabidopsis expressed sequence tag database and identified a second clone (RZ03e09F) encoding the full-length predicted polypeptide containing a CRIB motif. This protein was termed Rop-interactive CRIB-containing protein1 (RIC1).
To determine whether RIC1 interacts specifically with the GTP-bound Rop1, we examined the interaction of RIC1 with the mutant proteins CA-rop1 and DN-rop1 using an in vitro pulldown assay. The predicted coding sequence of RIC1 was fused to maltose binding proteins (MBP), whereas CA-rop1 and DN-rop1 were fused to glutathione S-transferase (GST). The MBP fusion protein was “pulled down” with GST–CA-rop1 or GST–DN-rop1 and was detected using an anti-MBP antibody (Wu et al., 2000). As shown in Figure 1A, the RIC1-MBP fusion protein interacted specifically with GTP-bound GST–CA-rop1 but not with GDP-bound GST–DN-rop1.
Figure 1.
CRIB Motif–Dependent Specific Interaction of RIC1 with the GTP-Bound Active Rop.
In vitro pulldown assays were performed using MBP-RIC1 and GST–CA-rop1 or GST–DN-rop1, and MBP-RICs pulled down by GST-rop1 were detected by anti-MBP antibody as described in the text.
(A) In vitro interaction of RIC1 with GTP-bound CA-rop1 and GDP-bound DN-rop1 mutant proteins.
(B) In vitro interaction of the GTP-bound CA-rop1 mutant with RIC1 and RIC mutant proteins. H37D and H37D/H40D mutations are within the CRIB motif.
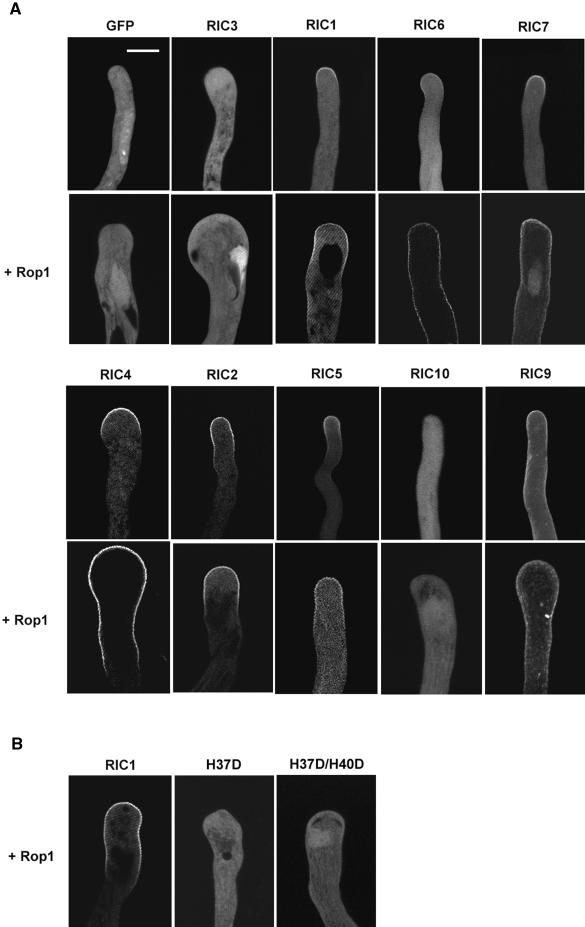
To determine whether the CRIB motif is critical for the interaction, we generated several mutations (H37D, H37D/H40D) in the conserved residues within the CRIB motif that are known to be important for interactions between Cdc42/Rac and their effectors. As shown in Figure 1B, these mutants had dramatically reduced interaction with GTP-bound CA-rop1, confirming a critical role for the CRIB motif in the interaction of RICs with Rops.
Annotation and Confirmation of the Ric Genes Encoding CRIB-Containing Proteins
The CRIB-dependent specific interaction of RIC1 with CA-rop1 is similar to the Cdc42/Rac interaction with effectors in animals. However, sequence comparison and BLAST searches reveal that RIC1 is a novel protein that shares no similarity with known Cdc42/Rac or Rho effectors or any proteins available in the animal and yeast genome databases. Thus, RIC1 may be a unique Rho target to transmit Rop signals to downstream events. Because the Arabidopsis genome contains 11 Rop genes that appear to participate in many distinct signaling pathways (Kost et al., 1999; Li et al., 1999, 2001; Zheng and Yang, 2000b), we sought to determine whether Arabidopsis contains multiple RICs that might be responsible for the versatile functions of the Rop family. To identify all Arabidopsis genes encoding CRIB motif–containing proteins, we performed a BLAST search using the RIC1 CRIB motif (Figure 2B) or nucleotide sequences encoding this CRIB motif. We identified 11 individual genomic bacterial artificial chromosome clones. Because most of these bacterial artificial chromosome clones were not annotated to contain the CRIB motif, we decided to annotate them using GeneScan software (see Methods). On the basis of the conservation of intron splicing and exon numbers combined with results from the GeneScan analysis, we predicted that all of these CRIB-related genomic sequences encode RIC proteins (Figure 2A).
Figure 2.
Comparison of Predicted Amino Acid Sequences between 11 RICs and CRIB Motifs.
(A) Comparison between 11 RICs. The predicted RIC polypeptides were identified by BLAST search using the CRIB motif from RIC1, and all except for Ric8 and Ric11 were confirmed by sequencing of cDNAs amplified by PCR as described in the text. Sequences that are identical among 11 RICs are highlighted in black (!), and conserved consensus sequences are highlighted in gray (*). Sequence alignment was performed using the GCG program (Genetics Computer Group, Madison, WI). Stretches of conserved sequences within the same group are boxed.
(B) Comparison of the CRIB motif from RICs with those from RopGAPs (Wu et al., 2000) and Cdc42/Rac effectors (Burbelo et al., 1995). The asterisk indicates the residues that distinguish between RopGAP CRIB (threonine) and RIC CRIB (glycine). A.t., Arabidopsis thaliana; D.d., Dictyo–stelium discoideum; H.s., Homo sapiens; C.e., Caenorhabditis elegans.
To confirm that the predicted RICs are in fact functional genes, we used polymerase chain reaction (PCR) to amplify Ric cDNAs from a flower cDNA library. We identified nine different Ric cDNAs. Sequencing of these cDNAs confirmed that they encode the RIC amino acid sequences predicted from the annotation. We were unable to amplify Ric8 and Ric11 from the flower cDNA library or cDNAs derived from Arabidopsis seedlings and leaves, suggesting either that they are expressed in specific stages or cell types of the plant or that they are pseudogenes.
One striking feature about RICs is their low molecular weight. As shown in Figure 2A, different RICs range from 116 to 224 amino acid residues in length. Outside of the CRIB motif, RICs share no sequence similarity with any other proteins in the database. However, different RICs share several consensus amino acid residues upstream of the CRIB motif and a consensus PSWMXDFK block downstream of the CRIB motif. RIC2 and RIC4 have distinct overall structures. They contain the CRIB motif at the C terminus and lack the PSWMXDFK block, whereas all other RICs contain the CRIB motif near the N-terminal end.
Another striking feature about RICs is the high variability in their primary structures. RICs are divided into five groups according to amino acid sequence similarities and phylogenetic analysis (Figure 3). Except for the CRIB motif (Figure 2B), the PSWMXDFK block, and several conserved residues upstream of the CRIB motif, no or little amino acid sequence similarity is found among these five groups. Members of group I, consisting of RIC9, RIC10, and RIC11, have the lowest molecular weights (116 to 156 amino acid residues in length) and are characterized by the presence of a conserved block of amino acids at the N terminus (MKMKGI/LYKG/SFK), but they are highly variable near the C-terminal regions. Group II consists of three members (RIC6, RIC7, and RIC8) and is most conserved, sharing several conserved amino acid stretches throughout the C terminus. RIC6 and RIC7 share the highest identity (66%) among all RIC pairs. Two members of group III, RIC1 and RIC3, are very similar in length and are conserved at the C terminus. In Arabidopsis, RIC5 is the lone member of group IV, sharing <20% identity with all other members outside of the CRIB motif. Unlike other group members, members of group V, consisting of RIC4 and RIC2, lack the PSWMXDFK block and contain a relatively long N terminus in front of the CRIB motif and a short C terminus. RIC4 and RIC2 share only 33% amino acid identity.
Figure 3.
Phylogenetic Analysis and Classification of Arabidopsis RICs.
Predicted amino acid sequences of RICs were aligned using the GCG PileUp program and used for the construction of an unrooted tree also using the PileUp program. On the basis of this analysis and sequence comparison, Arabidopsis RICs are divided into five groups as indicated. As shown in Figure 2, no sequence similarities are found between groups except for the conserved CRIB motif, the PSWMXDFK block, and several residues upstream of the CRIB motif.
Overexpression of RICs in Tobacco Pollen Tubes Causes Distinct Tip Growth Phenotypes
Because the function of Rops currently is best characterized in pollen tubes, we characterized the potential functions of RICs by overexpressing Ric genes in pollen tubes. Furthermore, particle bombardment–mediated transient expression has proven to be a robust method for a high-throughput characterization of gene functions in pollen (Fu et al., 2001). To facilitate monitoring of Ric expression and RIC subcellular localization, we tagged RICs with GFP. As shown in Table 1 for RIC3, for every other RIC examined, the GFP-Ric fusion gene and the Ric gene alone produced identical or nearly identical phenotypes when overexpressed under the pollen-specific LAT52 promoter, suggesting that GFP-Ric fusion genes are fully functional (data not shown).
Table 1.
Effects of RIC Expression on Pollen Tube Growtha
| GFP-RIC | Width (Mean ±sd, μm) |
Length (Mean ±sd, μm) |
Width/Length |
|---|---|---|---|
| GFP | 8.8 ± 0.24 | 297 ± 26 | 0.03 ± 0.00 |
| GFP-RIC1 | 8.5 ± 0.08 | 103 ± 2.0 | 0.08 ± 0.00 |
| GFP-RIC3 | 14.7 ± 1.26 | 107 ± 2.0 | 0.14 ± 0.01 |
| GFP-RIC2 | 9.4 ± 0.19 | 165 ± 1.9 | 0.06 ± 0.00 |
| GFP-RIC4 | 18.4 ± 0.16 | 84.8 ± 5.6 | 0.22 ± 0.01 |
| GFP-RIC9 | 8.9 ± 0.07 | 180 ± 31 | 0.05 ± 0.01 |
| GFP-RIC10 | 8.8 ± 0.07 | 440 ± 20 | 0.02 ± 0.00 |
| GFP-RIC6 | 9.6 ± 0.16 | 58.5 ± 4.2 | 0.16 ± 0.01 |
| GFP-RIC7 | 9.8 ± 0.18 | 54.5 ± 5.4 | 0.18 ± 0.01 |
| GFP-RIC5 | 6.4 ± 0.06 | 119 ± 6.5 | 0.05 ± 0.00 |
| RIC3 + GFP | 13.9 ± 1.35 | 102 ± 4.0 | 0.14 ± 0.01 |
Pollen tube length and maximum width of pollen tube tips were measured 5 hr after bombardment.
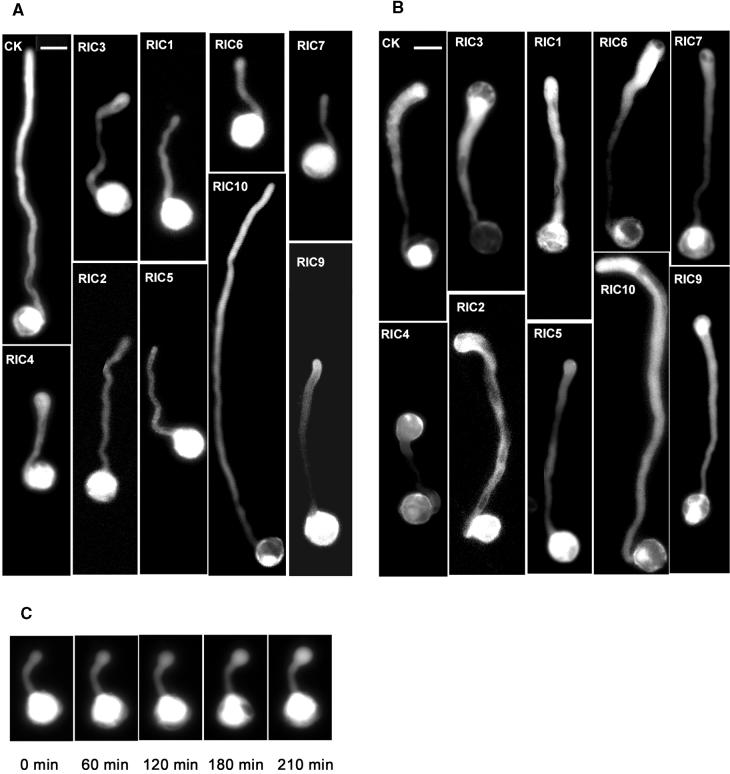
As shown in Figure 4A and Table 1, Ric overexpression phenotypes in tobacco pollen tubes can be classified into several interesting categories according to their phylogenetic groups. These results also are summarized in Table 2. First, overexpression for group III and group V induced parallel changes in pollen tube tip growth: two members within each group caused distinct phenotypes resembling the effects of Rop1 overexpression and suppression, respectively. RIC3 (group III) promoted depolarized growth, leading to the formation of a bulb at the tip of pollen tubes, similar to a moderate phenotype caused by Rop1 overexpression (Li et al., 1999). As shown in Figure 4C, time-course analysis showed that GFP-RIC3 tubes initially elongated as polarized tip growth, which was followed by tip swelling that began as soon as a detectable amount of GFP-RIC3 accumulated (3 hr after bombardment). Swelling continued for the remainder of the time series (6.5 hr after bombardment). In contrast, tubes transformed with GFP alone never produced swelling tips, even after overnight incubation (data not shown). This result also demonstrated that GFP-RIC3 tubes remained viable throughout the course of examination and that the depolarized growth was the specific effect of GFP-RIC3. In contrast to Ric3, overexpression of Ric1, the other group III member, induced a phenotype analogous to that caused by the expression of DN-rop1 and Rop1 antisense RNA. Ric1 overexpression strongly inhibited pollen tube elongation (with a 65% reduction in length) but had no effects on pollen tube expansion. In group V, Ric4 overexpression caused a slightly more severe depolarized growth than Ric3, whereas Ric2 induced an inhibition of pollen tube elongation (with a 44% reduction in length) weaker than Ric1 overexpression.
Figure 4.
Analysis of Phenotypes in Tobacco Pollen Tubes Overexpressing Ric Genes and Co-Overexpressing Ric Genes and Rop1.
Tobacco pollen grains were bombarded with different GFP-tagged constructs and germinated. The transformed tubes indicated by GFP expression were visualized using epifluorescence microscopy 5 hr after bombardment. Images were captured using a charge-coupled device camera and analyzed using MetaMorph software as described in the text. Bars = 30 μm.
(A) Pollen tubes expressing different LAT52::GFP–RICs. LAT52::GFP was used as a control (CK).
(B) Tubes coexpressing LAT52::GFP–RICs and LAT52::Rop1. The control (CK) was LAT52::Rop1 coexpressed with LAT52::GFP.
(C) Time-course series of a pollen tube expressing GFP-Ric3 showing the swelling of pollen tube tips. Time 0 indicates the beginning of imaging, which was conducted 3 hr after bombardment.
Table 2.
Summary of GFP-RIC Localization, Overexpression Phenotypes, and Functional Interaction with Rop1
| GFP-RIC | Phenotype | Localization | Effects of RICs on Rop1 Overexpression Phenotype |
Effects of Rop1 on the Localization of GFP-RICs |
|---|---|---|---|---|
| RIC1 | Reduced elongation | Apical PM | Inhibits Rop1 overexpression | Enhances apical PM |
| RIC3 | Depolarized growth | Cytoplasm | Enhances Rop1 overexpression | Enhances subcortical region |
| RIC6 | Reduced elongation | Apical PM | No effect | Shift to whole PM |
| RIC7 | Reduced elongation | Apical PM | No effect | Shift to whole PM |
| RIC4 | Depolarized growth | Apical PM | Enhances Rop1 overexpression | Enhances apical and subapical PM |
| RIC2 | Reduced elongation | Apical PM | No effect | No effect |
| RIC5 | Reduced elongation and expansion | Apical PM | Inhibits Rop1 overexpression | Slightly enhances apical PM |
| RIC10 | Increased elongation | Cytoplasm | Inhibits Rop1 overexpression | No effect |
| RIC9 | Reduced elongation | Whole PM | No effect | No effect |
Second, two members of group I, RIC9 and RIC10, affected pollen tube elongation in a completely opposite manner. Overexpression of Ric9 inhibited tube elongation by 40%. In contrast, Ric10 overexpression greatly promoted pollen tube elongation, causing an increase in pollen tube length by nearly 50%. This phenotype is similar to but distinct from that caused by weak overexpression of Rop1, which not only stimulates pollen tube elongation but also causes slight tube expansion (V. Vernoud, H. Li, and Z. Yang, unpublished data). However, Ric10 overexpression had no effects on growth polarity (i.e., pollen tube width remained unchanged).
Third, overexpression of Ric6 and Ric7 of group II induced a similar phenotype (i.e., strong inhibition of pollen tube elongation [>80% reduction in pollen tube length]). This inhibition was stronger than that caused by Ric1 overexpression. Finally, Ric5 overexpression inhibited both elongation and expansion of pollen tubes (causing a reduction in width and length by 25 and 60%, respectively). This phenotype is distinct from that caused by Ric1, Ric2, Ric6, Ric7, or Ric9, which only inhibited tube elongation and did not affect tube expansion.
Subcellular Localization of RICs in Tobacco Pollen Tubes
The resemblance of overexpression phenotypes for some RICs to those caused by either Rop1 overexpression or Rop1 inhibition indicates that these RICs may act either as a Rop1 target or its inhibitor in the Rop1 signaling pathway. To further understand the role of RICs in Rop1 signaling, we investigated the localization of RICs in pollen tubes, because Rop1 has been shown to localize to the apical region of the tube PM. To investigate the intracellular localization of RICs, tobacco pollen tubes expressing GFP-Ric fusions were examined using confocal microscopy. As shown in Figure 5A, GFP alone was distributed evenly in the pollen tube cytoplasm. GFP-RIC1 was localized to the apical region of the tube PM, a pattern similar to the localization of Rop1 (Lin et al., 1996; Kost et al., 1999; Li et al., 1999). However, GFP-RIC3 was barely detected in the tube PM but was concentrated in the cytoplasm at the tip of the pollen tube. This cytoplasmic localization was different from that of GFP alone, which was distributed uniformly throughout the cytoplasm and the nucleus. Interestingly, GFP-RIC4 was highly restricted to the apical PM region, like GFP-RIC1, although they caused essentially opposite phenotypes. GFP-RIC9 and GFP-RIC10, which induced opposite overexpression phenotypes, once again displayed distinct localization patterns; GFP-RIC10 was detected only in the cytoplasm, whereas GFP-RIC9 was localized to the whole PM of the pollen tubes, a pattern quite different from that of GFP-Rop1. RIC5, RIC6, and RIC7, which strongly inhibited tube elongation when overexpressed, were localized to the apical region of the tube PM. However, the PM localization for RIC6 was much weaker than that for RIC5 and RIC7. In contrast, GFP-RIC2, whose overexpression caused a weaker inhibition of tube elongation than RIC5, RIC6, or RIC7, was localized to both apical and subapical regions of the tube PM, a pattern distinct from Rop1 localization. These results indicate that different RICs are localized differentially in pollen tubes and that there is a correlation between localization patterns and overexpression phenotypes in tobacco pollen tubes.
Figure 5.
Subcellular Localization of GFP-RICs with and the Effect of Rop1 Expression on Their Localization in Tobacco Pollen Tubes.
Tobacco pollen grains were bombarded with various constructs and germinated as described in Figure 4. Approximately 5 hr after bombardment, GFP localization in transformed pollen tubes was analyzed using confocal microscopy.
(A) GFP-RIC localization was analyzed either in tubes expressing GFP-RIC alone (top) or coexpressing Rop1 (bottom). All images shown are 2-μm median sections. The strong fluorescence near the tips of tubes expressing GFP-RIC3 plus Rop1 most likely represents GFP-RIC3 localization to the vegetative nucleus. Bar = 20 μm.
(B) Mutations in the CRIB motif (see Figure 1B) eliminate the localization of GFP-RIC1 to the apical PM domain.
Rop1 Expression Alters the Subcellular Localization of RICs and Ric Overexpression Phenotypes
The localization patterns and overexpression phenotypes suggest that several RICs may be involved in Rop1 signaling but others are not. To test this hypothesis further, we determined which RICs are localized to the PM in a Rop1-dependent manner by coexpressing Rop1 with GFP-Ric in tobacco pollen. Results showing the effects of Rop1 co-expression on GFP-RIC localization and phenotypes are summarized in Table 2. We have shown that Rop1 overexpression induces enhanced and depolarized localization of Rop proteins to the apical region of pollen tubes, which in turn causes depolarized growth (Li et al., 1999; G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data). As shown in Figure 5A, Rop1 overexpression also caused enhanced and depolarized localization of GFP-RIC1 to the apical region of the pollen tube PM, suggesting that RIC1 localization to the PM is dependent on its association with Rop1. This was confirmed further by the observation that all mutations in RIC1 (H37D, H37D/H40D) that abolished RIC1 interaction with Rop1 (Figure 1B) also eliminated its localization to the PM (Figure 5B). Although Rop1 overexpression did not enhance the localization of RIC3 to the PM, it did change the pattern of RIC3 localization at the tip. GFP-RIC3 became concentrated in the cortex of the swelling tip or underneath the expanding PM, analogous to the localization of AtGDI1, which has been shown to interact with Rop and to dissociate Rop from the PM (Bischoff et al., 2000; G. Wu and Z. Yang, unpublished results). Thus, RIC3 appears to interact with PM-localized Rop1 in a transient and dynamic manner, as does AtGDI1.
Rop1 overexpression dramatically enhanced the PM localization of GFP-RIC4 and caused depolarized GFP-RIC4 localization extending behind the swelling tip (Figure 5A). In tubes co-overexpressing Rop1 and GFP-Ric4, no fluorescence was detected in the cytoplasm, whereas significant fluorescence was found in the cytoplasm of tubes expressing GFP-Ric4 alone. Like GFP-Rop1 localization (Li et al., 1999), however, GFP-RIC4 remained in the apical and subapical PM regions of tubes co-overexpressing Rop1 and GFP-Ric4, forming a tip-high gradient. No fluorescence was detected in the base of the tube.
Rop1 overexpression also enhanced GFP-RIC5 localization to the apical PM region, but it did not cause depolarized localization. On the contrary, Rop1 overexpression did not enhance the localization of GFP-RIC6 and GFP-RIC7 to the apical PM region, but it altered their PM localization pattern completely, causing them to be distributed evenly to the PM throughout the whole tube. This pattern is distinct from the GFP-RIC4 localization in tubes co-overexpressing Rop1 and GFP-Ric4, which does not extend beyond the subapical region. Finally, Rop1 overexpression had no effects on the localization of RIC2, RIC9, and RIC10. The Rop1-independent cytoplasmic localization of RIC10 is consistent with a role for RIC10 in a Rop1-independent pathway that promotes pollen tube elongation.
To gain further insights into the functional interaction between Rop1 and RICs, we next analyzed the phenotypes of pollen tubes co-overexpressing Ric and Rop1. We previously showed that the level of Rop1 overexpression is correlated with the severity of depolarized growth (Li et al., 1999). In the current experiments, we used a moderate level of Rop1 overexpression, which causes tubes to expand (Li et al., 1999) (Figures 4B and 5A, Table 3). The level of Rop1 overexpression was manipulated in the transient expression system using different amounts of plasmid DNA for particle bombardment (Fu et al., 2001; G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data). A moderate level of Rop1 overexpression was achieved using 0.4 to 0.5 μg of plasmid DNA in a typical bombardment experiment (G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data).
Table 3.
Effects of RICs on Rop1 Overexpression Phenotypea
| GFP-RIC | Width (Mean ±sd, μm) |
Length (Mean ±sd, μm) |
Width/Length |
|---|---|---|---|
| GFP + R1A | 18.6 ± 0.56 | 172 ± 6.8 | 0.11 ± 0.00 |
| GFP-RIC1 + R1A | 15.7 ± 0.76 | 158 ± 0.3 | 0.10 ± 0.00 |
| GFP-RIC3 + R1A | 27.9 ± 2.58 | 147 ± 5.3 | 0.19 ± 0.01 |
| GFP-RIC2 + R1A | 18.8 ± 1.09 | 221 ± 0.6 | 0.09 ± 0.01 |
| GFP-RIC4 + R1A | 26.8 ± 0.98 | 107 ± 4.9 | 0.25 ± 0.02 |
| GFP-RIC9 + R1A | 19.7 ± 0.51 | 216 ± 6.5 | 0.08 ± 0.00 |
| GFP-RIC10 + R1A | 15.9 ± 1.11 | 356 ± 17 | 0.04 ± 0.01 |
| GFP-RIC6 + R1A | 17.6 ± 0.53 | 212 ± 14 | 0.08 ± 0.00 |
| GFP-RIC7 + R1A | 15.4 ± 0.11 | 184 ± 1.1 | 0.08 ± 0.00 |
| GFP-RIC5 + R1A | 15.8 ± 0.56 | 195 ± 8.3 | 0.08 ± 0.01 |
Pollen tube length and maximum width of pollen tube tips were measured 5 hr after bombardment.
To determine the severity of depolarized growth, we measured the tube diameter (width) at the most dilated region of the tube and the tube width/length ratios. As shown in Table 3 and Figures 4B and 5A, Rop1/GFP-Ric co-overexpression phenotypes generally were consistent with the localization patterns of GFP-RICs and the effect of Rop1 on their localization. First, Rop1 expression enhanced the phenotype of Ric3 and Ric4 overexpression (i.e., co-overexpression resulted in more severe depolarized growth [26.8 or 27.9 μm in tube width] than overexpression of Rop1 [18.6 μm in tube width] or individual Ric3 and Ric4 alone [14.7 or 18.4 μm in tube width]) (Table 1). However, co-overexpression apparently did not produce an additive effect. These results are consistent with roles for RIC3 and RIC4 acting as Rop1 targets. Second, Rop1 and Ric10 co-overexpression produced a tube that was dramatically longer (356 μm in length) than control tubes (297 μm in length; Table 1) and Rop1-overexpressing tubes (172 μm in length; Table 3). Tubes co-overexpressing Rop1 and Ric10 were much wider than the wild-type control tubes and only slightly narrower than Rop1-overexpressing tubes. In accord with the GFP-RIC10 localization and overexpression phenotype, these results suggest that Rop1 and RIC10 are additive and act independently. Finally, the phenotypes induced by the co-overexpression of Rop1 with other RICs appeared to be complex. RIC1, RIC5, and RIC7, which were colocalized with Rop1 to the apical PM region, all partially suppressed tube expansion induced by Rop1 overexpression (Table 3). However, these RICs only slightly rescued tube elongation (RIC5 and RIC7) or did not rescue tube elongation (RIC1). Interestingly, RIC2, RIC6, or RIC9 did not affect tube expansion induced by Rop1 expression but slightly recovered tube elongation, although these three RICs all displayed distinct localization patterns.
Differential in Vitro Interaction between RICs and Rop1
The differential subcellular localization of RICs and the differential effect of Rop1 on their localization suggest that Rop1 interacts differentially with different RICs in pollen tubes. To assess whether such differential in vivo interactions are attributable to different capacities of RICs to bind GTP-bound active Rop1, we examined in vitro interactions between GTP-bound CA-rop1 and RICs. The GST–CA-rop1 fusion and the MBP–RIC fusion were used for pulldown assays as described in Figure 1. GST alone did not interact with any of the MBP-RICs (data not shown). As shown in Figure 6, different RICs bind GTP-bound CA-rop1 with different capacities that are largely consistent with the in vivo Rop1–RIC interactions. First, RIC6 interacted weakly with CA-rop1, in agreement with a weak localization of RIC6 to the apical PM domain that could not be enhanced by Rop1 overexpression (Figure 5A). Second, those RICs (RIC1, RIC4, and RIC5) that showed Rop1-enhanced localization to the apical PM domain interacted strongly with GTP-bound CA-rop1. Furthermore, RIC7, which was colocalized with Rop1 to the apical PM domain and was able to partially suppress the Rop1 overexpression phenotype, also bound CA-rop1 strongly. RIC2 and RIC9, which were localized to both apical and subapical PM regions, also interacted well with CA-rop1. However, the localization of RIC2, RIC7, and RIC9 to the apical PM domain was not enhanced by Rop1 overexpression (Figure 5A). These results suggest that direct binding is involved in the differential in vivo interaction between Rop1 and RICs, but other factors such as the kinetics of binding and additional proteins also may play a role in modulating the in vivo interaction between Rops and RICs.
Figure 6.
Differential in Vitro Interaction of Rop1 with Different RICs.
GTP-bound GST–CA-rop1 fusion and indicated MBP-RIC fusion proteins were expressed in E. coli, purified by affinity chromatography, and used for pulldown assays as described in Figure 1. Only seven of the nine RICs were used for this assay because we were unable to express the other two RICs (RIC3 and RIC10) as MBP fusion proteins in E. coli. At top is shown MBP-RIC fusion proteins that bind to GST–CA-rop1; at bottom is shown the loading control for MBP-RIC fusion proteins used for the pulldown assay. Numbers between the rows indicate relative signal intensities for CA-rop1–bound MBP-RICs standardized with the loading control (see text).
Accumulation of Ric Transcripts in Pollen and Vegetative Tissues
To determine whether the Ric overexpression phenotypes and RIC localization in pollen tubes reflected their function in pollen tubes or were caused by ectopic expression of Ric genes, we examined the expression of Ric genes in mature pollen. To our surprise, our reverse transcriptase–mediated (RT)-PCR results showed that all nine Ric genes tested were expressed in pollen, as shown in Figure 7A. Ric1, Ric3, Ric5, and Ric7 transcripts appeared to be more abundant that those for Ric2, Ric4, Ric9, and Ric10.
Figure 7.
RT-PCR Analysis of RIC Gene Expression in Arabidopsis Pollen and Various Tissues.
Total RNA was isolated from pollen or different parts of Arabidopsis plants and used for RT-PCR analysis as described in the text.
(A) Accumulation of various RIC transcripts in mature pollen.
(B) Organ distribution of various RIC transcripts.
An important question is whether different RICs also participate in other Rop-dependent signaling pathways during plant growth, development, and responses to the environment. If so, we would expect Ric genes to be expressed ubiquitously throughout Arabidopsis plants, as are Rop proteins (Li et al., 2001). As shown in Figure 7B, the majority of Ric genes (Ric1, Ric2, Ric4, Ric7, and Ric9) were expressed constitutively in various parts of Arabidopsis plants; only Ric3, Ric5, and Ric6 were flower specific. Interestingly, group II and group III had similar differential expression patterns between members of the same group. Ric1 and Ric7 transcripts were found in both reproductive and vegetative organs (including leaves, roots, and stems), whereas Ric3 and Ric6 transcripts were found only in flowers and inflorescences. Ric2 and Ric4 of group IV showed similar organ distribution. Ric5 showed preferential expression in flowers and inflorescences, but the overall level of Ric5 transcripts was much lower than that of Ric3 and Ric6. Ric10 transcript appeared to be more abundant in leaves and roots but was expressed weakly in flowers, inflorescences, stems, and siliques.
DISCUSSION
In this study, we have provided evidence that RICs act as direct targets of the versatile Rop GTPase switch. We believe that the genome-wide identification and characterization of Rop targets are significant for several reasons. First, RICs are the first direct G-protein targets known in plants. Second, the 11 structurally divergent RICs identified in this study may provide an important mechanism linking Rop GTPases to various distinct signaling pathways in plants, generating functional diversity for this universal switch. Third, RICs, being novel small proteins distinct from conventional Rho GTPase effectors from animals and fungi, may present a novel mechanism for Rho GTPases to regulate downstream events in plants.
RICs Act as Direct Rop Targets
Emerging evidence has implicated the Rop GTPase as a common signaling switch in many signaling pathways in plants (Lin and Yang, 1997; Kawasaki et al., 1999; Kost et al., 1999; Li et al., 1999, 2001; Potikha et al., 1999; Li and Yang, 2000; Zheng and Yang, 2000b; Lin et al., 2001; Ono et al., 2001). Like Ras and Rho GTPases in animals, the ability of Rop GTPases to control a plethora of processes may depend largely on distinct families of their target proteins. A recent study suggests that a phosphatidylinositol monophosphate kinase (PIPK) might act downstream of Rop in the control of pollen tube polar growth (Kost et al., 1999). However, no direct Rop targets had been identified unambiguously until this study.
Our conclusion that at least some RICs are direct targets of the Rop GTPase in Arabidopsis is based on several observations. First, all 11 RICs contain a conserved CRIB motif, which was discovered originally in Cdc42/Rac effectors and allows these effectors to bind to the effector domain of the Rop-related Cdc42/Rac GTPases in animals (Burbelo et al., 1995). The CRIB motif in RICs may be functionally equivalent to the animal CRIB motif in binding to the Rop effector domain. Indeed, we have shown that RIC1 exhibits CRIB-dependent specific interaction with the GTP-bound active form of Rop1 but not with GDP-bound Rop1. It is likely that the other 10 RICs also specifically bind active Rop, because of the high conservation of the CRIB motif among the 11 RICs (Figure 2B). A variant of the CRIB motif (Figure 2B) located in the N terminus of the Rop-specific Rho GTPase-activating proteins (RopGAPs) interacts with both GDP-Rop1 and GTP-Rop1 (Wu et al., 2000). An invariable glycine residue in the CRIB motif of RICs is replaced with threonine in the CRIB variant (Figure 2B). In agreement with its interaction with GDP-Rop, the N-terminal domain of RopGAPs acts as a regulator of RopGAP but not as a Rop target and does not compete with RICs in binding to Rop (Wu et al., 2000; G. Wu and Z. Yang, unpublished data).
More importantly, we have shown that overexpression of some Ric genes, including Ric3 and Ric4, induces depolarized growth, as does Rop1 overexpression, and enhances the Rop1-induced depolarized growth phenotype. Furthermore, both GFP-tagged RIC3 and RIC4 are localized to the tip of pollen tubes, and their subcellular localization can be influenced by Rop1 overexpression. Together, these in vitro and in vivo experiments provide evidence that RIC3 and RIC4 directly transmit signals from Rop1 in the control of polar growth in pollen tubes. Nonetheless, further studies should demonstrate more conclusively that RIC3 and RIC4 act as Rop1 targets. As discussed below, the existence of 11 divergent Ric genes and their expression in various parts of Arabidopsis plants support the notion that different RICs are Rop targets in distinct Rop signaling pathways.
RICs May Have Evolved to Control Various Rop-Dependent Signaling Pathways
We propose that RICs may have diverged or evolved rapidly to perform distinct functions. This hypothesis was suggested initially by the analysis of predicted RIC polypeptide sequences. On the basis of phylogenetic analysis and comparison of their predicted amino acid sequences, the 11 Ric genes are divided into four groups and an orphan member (Figures 2 and 3). Apart from the CRIB motif (Figure 2B), most RICs share a PSWMXDFK domain immediately downstream of the CRIB motif and several conserved residues immediately upstream of the CRIB motif. However, outside of this conserved region, different RIC groups have no sequence similarity, and homology is low even within members of the same group.
The potential functional diversity for variable RICs has been tested by our functional analysis of RICs using overexpression in tobacco pollen tubes. We were able to detect RNA transcripts for nine of the 11 Ric genes, and all of the nine genes are expressed in Arabidopsis pollen. Overexpression of GFP-tagged RICs in tobacco pollen tubes revealed that RICs of different groups and different RICs of the same group induce distinct phenotypes and display distinct subcellular localization patterns. As discussed above, RIC3 and RIC4, belonging to group III and group V, respectively, both cause depolarized growth and evidently are targets of Rop1. However, RIC4 is tightly associated with the apical PM domain, whereas RIC3 is primarily cytoplasmic but appears to be associated transiently with the apical PM domain, as does AtGDI1, which dissociates Rop from PM (G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data). Furthermore, RIC3 and RIC4 are very distinct structurally. Notably, RIC4 (and RIC2) contain the CRIB motif at the C terminus and lack the PSWMXDFK domain, unlike all other RICs. These observations suggest that RIC3 and RIC4 may be two distinct Rop1 targets that regulate different pathways downstream of Rop1. Circumstantial evidence supports the existence of two distinct Rop1 downstream pathways that regulate actin assembly and calcium accumulation at the tip, both of which appear to be essential for pollen tube tip growth (Li et al., 1999; Fu et al., 2001). It will be interesting to determine if RIC3 and RIC4 in fact control these two different pathways.
Our results suggest that RIC10 may participate in a pathway independent of Rop1. Overexpression of Ric10 only stimulates pollen tube elongation, and Ric10/Rop1 coexpression produces an additive effect. Furthermore, RIC10 is localized in the cytosol throughout the whole pollen tube, and this localization pattern is completely independent of Rop1. Therefore, RIC10 controls pollen tube elongation apparently independent of cell polarity development, a role distinct from that of the tip-localized Rop1 that couples growth control with polarity development (Li et al., 1999). Nonetheless, it cannot be excluded that RIC10 is a Rop1 target that is involved specifically in the regulation of pollen tube elongation. But if RIC10 acts in a Rop1-independent pathway, which Rop targets RIC10? Our previous RT-PCR analysis showed that three of six Rop genes examined are expressed in pollen (Li et al., 1998). However, these three Rops (Rop1, Rop3, and Rop5/AtRac2) are essentially identical in amino acid sequences, and Rop5/AtRac2 has been shown to play a role identical to that of Rop1 (Li et al., 1998; Kost et al., 1999; Li et al., 1999). Thus, these three Rops are expected to be functionally redundant in pollen (Zheng and Yang, 2000a). In contrast, our recent RT-PCR analysis showed that three other Rops (Rop8, Arac8, and Arac10) also are expressed in Arabidopsis pollen (Y. Gu and Z. Yang, unpublished results). Thus, it is likely that one or more of these Rops act upstream of RIC10 to specifically control pollen tube elongation. Further studies using knockouts for relevant genes are necessary to test this hypothesis.
All of the other six Ric genes expressed in pollen inhibit pollen tube growth to various degrees when overexpressed in tobacco pollen tubes. RIC1, RIC2, and RIC9 are particularly interesting, because their sister members (RIC3, RIC4, and RIC10, respectively) positively control either polar growth or tube elongation and thus show overexpression phenotypes opposite of those discussed above. Whether these six RICs play an inhibitory role in RIC3-, RIC4-, and RIC10-dependent pathways in pollen tubes requires further functional analysis using knockout mutants. Nonetheless, these observations suggest that each RIC has evolved or diverged to perform distinct functions by interacting with distinct partners. Thus, the growth inhibition phenotypes induced by the overexpression of those six Ric genes can be explained by their DN effects on RIC3, RIC4, and/or RIC10. The six inhibitory RICs likely competitively bind active Rop1 and/or other Rops expressed in pollen tubes via the CRIB motif, but they failed to properly activate specific RIC3, RIC4, and/or RIC9 downstream targets. However, other explanations remain possible. For example, these RICs might act as the targets of a Rop that might function to inhibit pollen tube growth. Alternatively, the inhibitory effect of Ric overexpression could be caused by the sequestering of a rate-limiting RIC-interactive partner, although the endogenous RIC may act as a target of Rop1 or another Rop in wild-type tubes.
Given the evidence that RIC3, RIC4, and RIC10 act as Rop targets in several distinct Rop-dependent pathways in pollen tubes, it is tempting to speculate that various RICs may function as Rop targets to control different Rop-dependent pathways during other parts of the plant life cycle. This notion is supported by the observation that Ric genes are expressed in various vegetative and reproductive parts of the Arabidopsis plant. In this regard, RICs would be analogous to the plethora of functionally distinct effectors for Cdc42, Rac, and Rho in animals and yeast (Aspenstrom, 1999). As discussed below, the functional specificity of RICs may involve both their specific interactions with distinct downstream effectors via the variable C or N termini of RICs and their differential interactions with different Rops.
Functional Specificity of RICs May Involve Differential in Vivo Interactions between Rops and RICs
Our results strongly support the notion that different RICs interact differentially with Rops in pollen tubes. Several lines of evidence suggest that RIC1, RIC4, and RIC5 interact strongly with Rop1. First, GFP-tagged RIC1, RIC4, and RIC5 are colocalized with Rop1 to the apical PM region. Second, Rop1 overexpression enhances their localization to the apical PM region. Third, RIC4 enhances the Rop1 overexpression phenotype, implying its role as a direct Rop1 target. In contrast, RIC1 and RIC5 both partially suppress Rop1 overexpression phenotypes, suggesting that they may compete with Rop1 targets in binding to active Rop1. Overexpression phenotype analysis suggests that RIC3 also is a Rop1 target. However, its distinct subcellular localization at the tip suggests that the interaction between RIC3 and Rop1 is very transient and dynamic. The distinct kinetics of Rop1 interaction with RIC3 and RIC4 may allow Rop1 to activate two distinct targets and two different downstream pathways simultaneously, as discussed above.
RIC2 is localized to both the apical and subapical regions of the tube PM, a localization pattern quite distinct from that of Rop1 and RIC4. Furthermore, RIC2 localization to the PM is not affected by Rop1 overexpression, suggesting that it may not interact well with Rop1 in pollen tubes. RIC10 is localized uniformly in the cytoplasm, suggesting that it may not interact with Rop1 in pollen tubes either. The apparent lack of interaction between the cytoplasmic RIC10 and the tip-localized Rop1 is indicated further by the inability of Rop1 overexpression to alter the subcellular distribution of RIC10 (see above). RIC9 is localized to the PM throughout the whole tube, and its localization also is unaffected by Rop1 overexpression, suggesting that it interacts with a different Rop (e.g., Arac8, Arac10, or Rop8) that is distributed in the same manner as RIC9. Arac8 and Arac10 are related most closely to maize ROP6, which is localized constitutively to the PM in maize leaf epidermal cells (Ivanchenko et al., 2000). RIC9 may interact with such a PM-localized Rop (likely Arac8 or Arac10).
Interestingly, both RIC6 and RIC7 are localized to the apical PM region and the cytoplasm, like Rop1, but Rop1 overexpression causes them to shift from the apical PM domain and the cytosol to the whole PM. In particular, RIC6 becomes localized strictly to the PM. A simple explanation for this complex localization pattern is that although RIC6 and RIC7 are able to interact with Rop1, they interact preferentially with a different Rop (e.g., Rop8, Arac8, or Arac10) that may work in a pathway independent of Rop1. Rop1 overexpression may release the cytosolic sequestration of this Rop, presumably by a GDI (which may sequester multiple distinct Rops), allowing RIC6 and RIC7 to localize to the PM of the whole tube along with this Rop. This explanation also is consistent with the fact that RIC6 and RIC7 overproduction causes much more severe inhibition of pollen tube elongation than the overproduction of RIC1 and RIC5, which show Rop1-enhanced localization to the apical PM region.
Together, these localization and co-overexpression phenotype analyses strongly imply that RICs can regulate distinct Rop-dependent pathways through their differential interactions with Rops in pollen tubes. With different Rops and RICs expressed in various parts of the plant, differential interactions between Rops and RICs most likely play a key role in determining the functional specificity of Rops and RICs in various Rop-dependent signaling pathways in plants.
Our in vitro pulldown assays suggest that differential binding between Rop1 and different RICs contributes partly to the differential interactions between RICs and Rops. Rop1-RIC binding capacity is correlated largely with the in vivo Rop1 interaction with different RICs that was assessed based on their colocalization with Rop1, the dependence of their localization on Rop1, and their ability to affect Rop1 overexpression phenotypes, as discussed above. It has been shown that the CRIB motif is required and sufficient for Cdc42/Rac effectors to interact with Cdc42/Rac; however, amino acid sequences immediately adjacent to this motif also are involved in the interaction (Thompson et al., 1998). Thus, the CRIB motif and its flanking sequences of RICs likely play a major role in determining their interaction specificity with Rops. However, RIC-Rop1 in vitro binding capacities are not correlated strictly with their in vivo interaction. For example, RIC2 and RIC9 bind Rop1 well, although their localization in pollen tubes is not affected by Rop1 overexpression. These results indicate that in vivo interactions between Rops and RICs are more complicated and may involve binding kinetics between these two types of proteins and other interactive partners.
RICs May Act as Adaptors to Link Rops to Specific Effector Proteins
Apart from the conserved CRIB motif, the RIC family of proteins is characterized by its high sequence variability, sequence novelty, and low molecular weights. For example, predicted polypeptides for groups I and V are only 116 to 167 amino acid residues in length. It seems unlikely that the variable regions of RICs can function as enzymes to regulate downstream events. Nonetheless, our results suggest that at least some RICs, if not all of them, function as Rop target proteins. Thus, it is reasonable to speculate that RICs act as adaptor proteins linking Rops to effector proteins that in turn activate specific downstream events. Such a linker function would allow the generation of a greater functional diversity for the Rop GTPase switch and would present a novel mechanism for the activation of G protein effectors. Typically, G proteins directly activate an effector that is an enzyme producing second messengers, a protein kinase, an ion channel, or a regulator of actin binding proteins. This potential unconventional mechanism of Rop GTPase signaling is consistent with the fact that Arabidopsis apparently lacks homologs of animal and yeast RHO effectors (see Introduction).
All three types of Cdc42/Rac effectors (kinases [P21-activated protein kinase or activated Cdc42 kinase (ACK)], Wiskott-Aldrich syndrome proteins, and Cdc42 effector proteins [CEPs]) have been shown to regulate Cdc42-dependent actin assembly (Aspenstrom, 1999; Burbelo et al., 1999; Hirsch et al., 2001), whereas no homologs for these Cdc42 effectors have been identified in plants. Our recent studies suggest that Rop1 also promotes actin assembly in the control of polar growth in pollen tubes (Fu et al., 2001). Because of our evidence that RIC3 and RIC4 are Rop1 targets, it is likely that either or both them are involved in the regulation of actin assembly in pollen tubes. Thus, the Rop1 regulation of actin organization appears to be dependent on a novel mechanism distinct from the Cdc42/Rac regulation of actin organization in animals and yeast. Identification of RIC functional partners should provide insights into the mechanism by which Rop regulates actin organization and other cellular processes in plants.
METHODS
Yeast Two-Hybrid Method
The constitutively active Rop1At mutant (G15V/C188S; pDP1S) was used as bait to screen an Arabidopsis thaliana seedling cDNA library, as described previously (Wu et al., 2000). The plasmids from putative positive clones were rescued in Escherichia coli and reintroduced into the yeast strain containing pDP1S to confirm the interaction. The confirmed clones then were sequenced, and the sequences were used for the BLAST search of the GenBank and Arabidopsis databases. One clone encoding a novel polypeptide containing a CRIB motif was identified. The polypeptide was designated RIC1 (for Rop-interactive CRIB-containing protein).
Database Search and Bioinformatics
To identify additional genes encoding CRIB-containing proteins, the CRIB motif or the RIC1 cDNA sequence was used as a query for a BLAST search of the National Center for Biotechnology Information database and The Arabidopsis Information Resource (TAIR) database. From this search, we identified one expressed sequence tag (EST) clone and many bacterial artificial chromosome clones from Arabidopsis and several EST sequences from other plant species. Because most of the RIC1-related genomic sequences were not annotated, we used a combination of several World Wide Web–based programs and visual examination to predict CRIB-containing proteins. Genomic sequences were extracted and analyzed using DNAStrider1.2 (http://www.mbshortcuts.com/software/) to predict the frame encoding the CRIB. We then predicted the full-length open reading frames encoding CRIB motif–containing polypeptides from genomic sequences using GeneScan (http://genes.mit.edu/GENSCAN.html). The motif search was performed using scan prosite tools (http://www.isrec.isb-sib.ch/software/PFSCAN_form.html). Sequence alignment and phylogenetic analysis were performed using the GCG PileUp programs (Genetics Computer Group, Madison, WI).
DNA Manipulation and Plasmid Construction
To confirm that the annotated RIC sequences are in fact expressed, we amplified RIC cDNA from a flower cDNA library (a gift from Hong Ma, Pennsylvania State University, University Park) using polymerase chain reaction (PCR) primers covering the predicted RIC coding sequences (Table 4). A BamHI site was created at the predicted translation initiation codon of RIC coding sequences for all annotated RICs, except that a BglII site was introduced into RIC3. Amplified RIC3 cDNA was then cut with BglII, and the eight other RIC PCR fragments were cut with BamHI and ligated into a pBluescript SK+ BamHI-SmaI site. The nine resulting RIC clones then were sequenced. All plasmids used for transient expression in pollen were constructed in a derivative of the pBI221 vector (Clontech, Palo Alto, CA), termed pLAT52, in which 35S::β-glucuronidase from Cauliflower mosaic virus was replaced with the LAT52 promoter (Twell et al., 1991). The enhanced green fluorescent protein (GFP) mutant (S65C) gene was cloned downstream of the LAT52 promoter in pLAT52 to create pLAT52::GFP, as described previously (Li et al., 1999). To generate various pLAT52::GFP–RIC constructs, RIC DNA fragments were cut using BamHI-KpnI from the corresponding pBluescript SK+ constructs and cloned as a translational fusion to the C terminus of GFP in pLAT52::GFP. To generate pLAT52::RIC constructs, the GFP cDNA sequence was removed from pLAT52::GFP–RICs by BamH1 digestion followed by self-ligation.
Table 4.
Primers for RIC cDNA Amplification and RT-PCR
| Genes | Sense Primers | Antisense Primers |
|---|---|---|
| RIC1 | 5′-GGGGGATCCATGGCGACGACAATGAAGG | 5′-GTTTCTCAGATAATATCGTTACAGG |
| RIC6 | 5′-GGGATCCATGTCAAGCTCCAAGATG | 5′-GTCGTCTCTCGAGTCAAAAATCATCTC |
| RIC2 | 5′-AAGATCCATGAAAGATCGGATGGAGCG | 5′-AATCAGACGACGGTGCCGGTGAGATGG |
| RIC3 | 5′-AAGATCTATGGCGACCGTGAAAGGCCTTC | 5′-CCATGATGTGTTTTACTCTTTGTCACTG |
| RIC7 | 5′-GGGATCCGGAATGTCAAGTACCAAGATG | 5′-AATCAAAACCCATTTCTGTCGTC |
| RIC5 | 5′-AGGATCCATGACCTCACCTATGAAGGG | 5′-CTTTCATCTTCCTGTAGAACTTCC |
| RIC4 | 5′-GGGATCCATGAGAGATAGAATGGAG | 5′-GAGGGAGTGGATTATAAAGTTGG |
| RIC10 | 5′-GGGATCCATGTCAATGAAAATGAAGGGC | 5′-GGTCGACTAGATTAATCGTGATACAG |
| RIC9 | 5′-AGGATCCATGGCTACAAGATTCAAGGGG | 5′-CCTAGTAAAACCATCCCAATATCATTGCG |
Site-Directed Mutagenesis of RIC1
To create point mutations for RIC1, we used PCR-based site-directed mutagenesis. The primer RIC1F (5′-AAGATCTATGGCGACGACAATGAAGGGTCTTCTTAAGGGCCTT-3′) and a mutant primer containing the H37D mutation (5′-CGGATCCAATGTCGGCAACATGCTTTACATCGG-3′) were used to amplify mutant RIC1 fragment A from pBS-RIC1. RIC1 fragment A was ligated into pZErO-1.1 (Invitrogen, Carlsbad, CA) EcoRV sites and was sequenced to confirm the proper mutation. RIC1 fragment B was amplified using primers 42GF (5′-TGGATCCGACGGTCCAACCAA-3′) and RIC1R (5′-GTTTCTCAG-ATAATATCGTTACAGG-3′) and then ligated into pZErO-1.1 EcoRV sites (p42GF). RIC fragment B was cut with BamHI-KpnI and then ligated with individual RIC1 fragment A BamHI-KpnI fragments. H37D/H40D was generated similarly.
In Vitro Protein–Protein Interaction Assays
To demonstrate direct interaction between RICs and Rops, we used RICs fused with maltose binding protein (MBP) and Rops fused with glutathione S-transferase (GST) for pulldown assays, as described previously (Wu et al., 2000). Approximately 10 μg of GST–CA-rop1 or GST–DN-rop1 fusion proteins were bound to glutathione-conjugated agarose beads, and similar amounts of MBP-RIC fusion proteins were used in each assay. The MBP fusion proteins were detected using a polyclonal antibody against MBP (New England Biolabs, Beverly, MA) and the BM chemiluminescence protein gel blot kit (Boehringer Mannheim, Basel, Switzerland). Loading controls were examined by protein gel blot analysis of 2 μL of a total of 500 μL of reaction mixtures from each interaction assay using the anti-MBP antibody. To compare the relative ability of different MBP-RICs to interact with GST–CA-Rop1, interaction signals were standardized with the loading control. Films were scanned, and the total intensity of signals from each band was measured using MetaMorph software, version 4.5 (Universal Imaging, West Chester, PA). The ratio of the intensity from the pulldown assay to the intensity from the corresponding loading control was calculated. The lowest ratio was designated arbitrarily as 1, and all other values shown in Figure 6 are relative to this value.
Reverse Transcription and PCR Analysis of RIC Transcripts
Total RNA was isolated from different Arabidopsis tissues using the Trizol reagent (Gibco BRL). For silique and stem RNAs, two additional chloroform extractions were performed. Reverse transcription and PCR amplification were performed as described previously (Li et al., 1998). For all nine RIC genes, 30 cycles of PCR amplification (94°C for 30 sec, 54°C for 30 sec, and 72°C for 1 min) were performed using the primers shown in Table 4. As PCR amplification and loading controls, the same template cDNA was amplified using primers for the constitutive Act2 gene (An et al., 1996). Five microliters of each PCR product was loaded onto a 0.8% agarose gel to visualize the amplified cDNAs.
Particle Bombardment–Mediated Transient Expression in Tobacco Pollen
Tobacco (Nicotiana tabacum) plants were grown in growth chambers at 22°C under a light regimen of 12 hr of darkness and 12 hr of light. Pollen grains collected from these plants were used for transient expression using a particle bombardment procedure described previously (Fu et al., 2001). All plasmid DNAs were amplified in the E. coli strain Top 10 and purified using Plasmid Midi or Mini Kits according to the manufacturer's instructions (Qiagen, Valencia, CA). Routinely, 0.5-mg gold particles were coated with 0.5 μg of pLAT52::GFP–RIC DNA or a mixture of 0.5 μg of pLAT52::GFP–RIC DNA with 0.5 μg of pLAT52::Rop1. As a control, the pLAT52::GFP vector DNA was bombarded into tobacco pollen. Bombarded pollen grains were washed into Petri dishes with 0.5 mL of germination medium right after the bombardment (Fu et al., 2001). For expression of RICs, pLAT52:GFP was cobombarded with pLAT52::RIC to facilitate the identification of transformed tubes. The pollen grains then were incubated for 4 to 6 hr before observation under a Nikon (Tokyo, Japan) TE300 inverted microscope equipped with a Hamamatsu (Shizuoka, Japan) cooled charge-coupled device camera (model C4742-95) or confocal microscope as described below.
Analyses of RIC Localization and Morphology
Tubes expressing pLAT52::GFP–RIC were identified using epifluorescence microscopy and observed as described previously (Fu et al., 2001). We measured the width and length of pollen tubes exactly 5 hr after pollen grains were bombarded. Images of fluorescent tubes were recorded rapidly through a cooled charge-coupled device camera (model C4742-95; Hamamatsu) attached to an Eclipse inverted microscope (model TE300; Nikon). The images were analyzed using the MetaMorph version 4.5 measurement function. The degree of depolarized growth was determined by measuring the diameter of the widest region of the tube, and the degree of polar growth was determined by measuring the length of pollen tubes. For each treatment, data were collected from three independent experiments (40 to 80 tubes).
To determine subcellular localization, tubes expressing GFP fusion proteins were analyzed using laser scanning confocal microscopy with a Nikon Optiphot upright microscope equipped with a MRC 600 confocal laser scanning device (Bio-Rad, Hercules, CA). One-micrometer optical sections were scanned and captured using Comos software (Bio-Rad). Confocal images were analyzed using MetaMorph version 4.5 and processed using Photoshop version 5.5 (Adobe Systems, Mountain View, CA).
Accession Numbers
GenBank accession numbers for various annotated RICs are as follows: RIC1, AC002332; RIC2, AC004557; RIC3, AC000104; RIC4, AB005242; RIC5, AB015474; RIC6, AC007109; RIC7, AL021749; RIC8, AC003027; RIC9, AC005882; RIC10, AF128393; RIC11, AL035527.
Acknowledgments
We thank members of the Yang laboratory for helpful discussions and Ying Fu for technical assistance with confocal microscopy. This work was supported by grants to Z.Y. from the National Science Foundation (MCB-0096026 and MCB-0111082) and the Department of Energy (DE-FG03-00ER15060).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010218.
References
- An, Y.-Q., McDowell, J.M., Huang, S., Mckinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aspenstrom, P. (1999). Effectors of the Rho GTPases. Curr. Opin. Cell Biol. 11, 95–102. [DOI] [PubMed] [Google Scholar]
- Bischoff, F., Vahlkamp, L., Molendijk, A., and Palme, K. (2000). Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI from Arabidopsis. Plant Mol. Biol. 42, 515–530. [DOI] [PubMed] [Google Scholar]
- Burbelo, P.D., Drechsel, D., and Hall, A. (1995). A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J. Biol. Chem. 270, 29071–29074. [DOI] [PubMed] [Google Scholar]
- Burbelo, P.D., Snow, D.M., Bahou, W., and Spiegel, S. (1999). MSE55, a Cdc42 effector protein, induces long cellular extensions in fibroblasts. Proc. Natl. Acad. Sci. USA 96, 9083–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, D.S., Pirone, D.M., and Burbelo, P.D. (2001). A new family of Cdc42 effector proteins, CEPs, function in fibroblast and epithelial cell shape changes. J. Biol. Chem. 276, 875–883. [DOI] [PubMed] [Google Scholar]
- Ivanchenko, M., Vejlupkova, Z., Quatrano, R.S., and Fowler, J.E. (2000). Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. Plant J. 24, 79–90. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96, 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Yang, Z. (2000). Rho GTPase and the actin cytoskeleton. In Actin: A Dynamic Framework for Multiple Plant Cell Functions, C. J. Staiger, F. Baluska, D. Volkmann, and P. Barlow, eds (Dordrecht, The Netherlands: Kluwer Academic Press), pp. 301–321.
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to the tip-localized calcium influx. Plant Cell 11, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Shen, J., Zheng, Z., Lin, Y., and Yang, Z. (2001). The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 126, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., and Yang, Z. (1997). Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell 9, 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., Wang, Y., Zhu, J., and Yang, Z. (1996). Localization of a rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 8, 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., Seals, D.F., Randall, S.K., and Yang, Z. (2001). Dynamic localization of Rop GTPases to the tonoplast during vacuole development. Plant Physiol. 125, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, J.G., and Hall, A. (1998). Rho GTPases. J. Biol. Chem. 273, 20685–20688. [DOI] [PubMed] [Google Scholar]
- Ono, E., Wong, H.L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., Choi, H.J., Lee, S., Lee, T., Yang, Z., and Lee, Y. (2000). Rac-related GTP-binding protein in elicitor-induced reactive oxygen generation by suspension-cultured soybean cells. Plant Physiol. 124, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potikha, T.S., Collins, C.C., Johnson, D.I., Delmer, D.P., and Levine, A. (1999). The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. (2000). Rho. In GTPases, A. Hall, ed (Oxford, UK: Oxford University Press), pp. 89–136.
- Thompson, G., Owen, D., Chalk, P.A., and Lowe, P.N. (1998). Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry 37, 7885–7891. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., Wing, R.A., Ushiba, J., and McCormick, S. (1991). Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 5, 496–507. [DOI] [PubMed] [Google Scholar]
- Winge, P., Brembu, T., Kristensen, R., and Bones, A.M. (2000). Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 156, 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Li, H., and Yang, Z. (2000). Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 124, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.-L., and Yang, Z. (2000. a). The Rop GTPase switch turns on polar growth in pollen. Trends Plant Sci. 5, 298–303. [DOI] [PubMed] [Google Scholar]
- Zheng, Z.-L., and Yang, Z. (2000. b). The Rop GTPase: An emerging signaling switch in plants. Plant Mol. Biol. 44, 1–9. [DOI] [PubMed] [Google Scholar]