Abstract
Proliferating cell nuclear antigen (PCNA) is an essential component in the eukaryotic DNA replication machinery, in which it works for tethering DNA polymerases on the DNA template to accomplish processive DNA synthesis. The PCNA also interacts with many other proteins in important cellular processes, including cell cycle control, DNA repair, and an apoptotic pathway in the domain Eucarya. We identified three genes encoding PCNA-like sequences in the genome of Aeropyrum pernix, a crenarchaeal archaeon. We cloned and expressed these genes in Escherichia coli and analyzed the gene products. All three PCNA homologs stimulated the primer extension activities of the two DNA polymerases, polymerase I (Pol I) and Pol II, identified in A. pernix to various extents, among which A. pernix PCNA 3 (ApePCNA3) provided a most remarkable effect on both Pol I and Pol II. The three proteins were confirmed to exist in the A. pernix cells. These results suggest that the three PCNAs work as the processivity factor of DNA polymerases in A. pernix cells under different conditions. In Eucarya, three checkpoint proteins, Hus1, Rad1, and Rad9, have been proposed to form a PCNA-like ring structure and may work as a sliding clamp for the translesion DNA polymerases. Therefore, it is very interesting that three active PCNAs were found in one archaeal cell. Further analyses are necessary to determine whether each PCNA has specific roles, and moreover, how they reveal different functions in the cells.
Proliferating cell nuclear antigen (PCNA) is implicated in many DNA metabolic processes, including cell cycle control, DNA replication, nucleotide excision repair, and postreplication mismatch repair in the eukaryotic cells (21, 24, 25, 47). Among PCNA’s roles, its function as the elongation (processivity) factor of DNA polymerases δ and ɛ (Pol δ and Pol ɛ), which are replicative enzymes of genomic DNA, is the most understood function (52). PCNA works as a homotrimer, which forms a doughnut-shaped structure that can encircle double-stranded DNA. PCNA functions by tethering the DNA polymerase on the DNA strand; therefore, it is called a sliding clamp. In the DNA replication process in Eucarya, replication factor C (RFC) is required as a clamp loader to open the PCNA ring and to introduce the DNA strand into the ring efficiently in an ATP-dependent manner. Then the DNA polymerase binds to PCNA to form a holoenzyme capable of processive DNA replication on the DNA strands. Bacteria constitute a different domain of life. However, a common mechanism exists for facilitating processive DNA replication of the genome by the cooperative work of DNA polymerase and its clamp. PCNA shares a similar three-dimensional structure with the β subunit of Escherichia coli DNA polymerase III (Pol III), although there is little sequence homology between the two proteins (13, 29, 30, 35). The β subunit works for tethering Pol III core proteins on the DNA strand (26).
Archaea, the third domain of life (56), resemble the Bacteria in cellular ultrastructure. However, the archaeal proteins related to DNA transactions are more similar to those in Eucarya than to those in Bacteria (38). The similarity of archaeal and eukaryotic DNA replication became evident when haloarchaeal growth was found to be inhibited by aphidicolin, a specific inhibitor of the eukaryotic Pol α-like DNA polymerases (12, 46). It was subsequently confirmed that Archaea contain a family B (α-like) DNA polymerase with an amino acid sequence similar to that of the large subunit of eukaryotic DNA replicases α, δ, and ɛ (reviewed in reference 40). In addition, after the total genome sequences of several archaeal organisms were determined, it was confirmed that several genes encoding proteins sharing sequence similarities with eukaryotic DNA replication proteins are present in all archaeal genomes (5, 10, 15, 32). Among them, we characterized DNA polymerases (14, 16, 27, 49, 50), PCNA (6, 18, 35), RFC (7, 36, 39), replication protein A (28), and primase (1, 33) from a hyperthermophilic archaeon, Pyrococcus furiosus. While the Euryarchaeota, a subdomain of Archaea, contain one family B DNA polymerase, their relatives in the crenarchaeotic subdomain possess at least two homologs of the family B DNA polymerase (4, 48). A heterodimeric DNA polymerase from the euryarchaeota was found in earlier research (3, 17, 50). This polymerase is composed of a small (DP1) subunit and a large (DP2) subunit and has higher processivity than the family B polymerase in vitro (16, 50). The DP1 subunit has some sequence similarity with the second subunit of eukaryotic Pol α, δ, and ɛ (3, 34). On the other hand, DP2, the catalytic subunit, lacks similarity to any other DNA polymerases, and it was named a family D DNA polymerase (5).
Including the archaeal family B and the family D DNA polymerases, in general, all known replicative DNA polymerases require a sliding clamp, the β subunit for E. coli, gp45 for bacteriophage T4, and PCNA for eucarya and archaea. Interestingly, two PCNA-like proteins were found in the genome of a crenarchaeon, Sulfolobus solfataricus (9). Both of them stimulated the DNA-synthesizing activity of a family B DNA polymerase from S. solfataricus. However, a direct link between these plural PCNAs and the archaeal plural DNA polymerases has yet to be demonstrated. We now describe the cloning and characterization of three PCNA homologs from Aeropyrum pernix and show that these proteins interact with two DNA polymerases (Pol I and Pol II) which were previously identified in this organism (4). The difference of the band mobilities on the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) between the recombinant and native proteins detected by the Western blot analysis suggests that A. pernix PCNA 2 (ApePCNA2) used in this study may be a truncated form. However, ApePCNA2 affected Pol I and Pol II activities. These interactions augmented DNA synthesis by both DNA polymerases. Our results, showing the distinct features of the aeropyral PCNAs, provide evidence for the existential significance of plural PCNAs in crenarchaeota and contribute to our understanding of the DNA replication system in the domain Archaea.
MATERIALS AND METHODS
Cloning and sequencing of A. pernix PCNA genes.
PCR was used to amplify the genes encoding homologs of PCNA from the A. pernix genome. Six primers were designed as follows: ApePC1F (5′-GCGAACATATGTCCTCTGAGGCCACCCTA-3′), ApePC1R (5′-CGCTTAAGCTTACTAACCTGTCGAGG-3′), ApePC2F (5′-GCGA-ACATATGGTCGCCTCTATCGAG-3′), ApePC2R (5′-CATATGGATCCTTACTACT-CGATCTTC-3′), ApePC3F (5′-GCGAACATATGTTCAGACTAGTATAG-3′), and ApePC3R (5′-CATATGGATCCTTACTAGCCAGCTAG-3′), which have NdeI and either HindIII (ApePC1R) or BamHI (ApePC2R and ApePC3R) sites (underlined), respectively. The PCR conditions involved 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. Pfu DNA polymerase (Stratagene) was used to maintain the accuracy of amplification. The PCR product was cloned into a TA cloning vector (pT7Blue; Novagen), and the nucleotide sequences of the inserted DNAs were determined from several independent clones by a capillary sequencer (ABI Prism 310 Genetic Analyzer; Applied Biosystems). The putative genes for the A. pernix PCNAs were excised by digestion with NdeI and HindIII or BamHI. These DNA fragments were ligated into the pET21a vector (Novagen), which had been digested by the corresponding restriction enzymes. The constructs were designated pAP-PCNA1, pAP-PCNA2, and pAP-PCNA3.
Production and purification of recombinant ApePCNAs.
E. coli BL21-CodonPlus(DE3)-RIL (Stratagene) cells harboring the pAP-PCNAs were grown at 37°C in 1 liter of Luria-Bertani medium containing ampicillin (100 μg/ml) and chloramphenicol (20 ng/ml) to an optical density of 0.5 at 600 nm. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to the culture to a final concentration of 1.0 mM, and growth was continued for 5 h. Cells were harvested, suspended in 25 ml of buffer A (50 mM Tris-HCl, pH 8.0; 0.1 mM EDTA; 0.5 mM dithiothreitol; and 10% glycerol), and lysed by sonication. Cell debris was removed by centrifugation at 30,000 × g for 15 min at 4°C. E. coli proteins were removed by heat treatment at 75°C for 15 min. To the supernatant after centrifugation, polyethyleneimine and NaCl were added to final concentrations of 0.2% and 0.5 M, respectively, and the mixture was stirred for 30 min at 4°C. The proteins in the supernatant (30 ml) were precipitated by adding 16.8 g of ammonium sulfate (80% saturation). The precipitates were dissolved in buffer B (50 mM Tris-HCl, pH 8.0; 0.1 mM EDTA; 0.5 mM dithiothreitol; 10% glycerol; and 0.1 M NaCl) and were dialyzed against the same buffer. The dialysates were applied to an anion-exchange column (HiTrap Q, 5 ml; Pharmacia Biotech) fitted to a high-pressure liquid chromatography apparatus (AKTA explorer 10S; Pharmacia Biotech). The chromatography was developed with a 50-ml linear gradient of 0.1 to 1.0 M NaCl in buffer A at a flow rate of 2 ml/min. ApePCNA1 and ApePCNA3 eluted at a salt concentration of 0.4 to 0.6 M. After the anion-exchange chromatography, the fractions containing PCNA proteins were pooled, diluted in buffer A, and applied to an affinity column (HiTrap heparin, 1 ml; Pharmacia Biotech). The chromatography was developed with a 0.1 to 1.0 M NaCl gradient in buffer A at a flow rate of 1 ml/min. ApePCNA1 and ApePCNA3 eluted at a salt concentration range of 0.25 and 0.6 M, respectively. The purified ApePCNA1 and ApePCNA3 proteins were stored at −20°C as 50% glycerol stock solutions after dialysis against buffer A. In the case of ApePCNA2, the supernatant obtained after sonication was not heat treated. Instead, polyethyleneimine and NaCl were added to concentrations of 0.2% and 1.0 M, respectively, and the mixture was stirred for 30 min at 4°C. The protein in the supernatant was applied to a hydrophobic column (HiTrap Phenyl-Sepharose HP, 1 ml; Pharmacia Biotech), and the chromatography was developed with a 20-ml gradient of 1.5 to 0 M NaCl in buffer A at a flow rate of 1 ml/min. All of the eluted fractions were combined, and this pool was applied to an anion-exchange column (HiTrap Q, 5 ml; Pharmacia Biotech), which was developed with a 50-ml linear gradient of 0.1 to 1.0 M NaCl in buffer A at a flow rate of 2 ml/min. The ApePCNA2 protein eluted at a salt concentration range of 0.6 to 0.7 M. After anion-exchange chromatography, the fractions containing PCNA proteins were pooled, diluted in buffer A, and applied to an affinity column (HiTrap heparin, 1 ml; Pharmacia Biotech), and the chromatography was developed with a 0.1 to 1.0 M NaCl gradient in buffer A at a flow rate of 1 ml/min. The ApePCNA2 protein eluted at a salt concentration of 0.3 M. The purified ApePCNA2 protein was stored at −20°C as a 50% glycerol stock solution after dialysis against buffer A.
Purification of recombinant A. pernix Pol I and Pol II.
The preparation of the A. pernix Pol I and Pol II proteins was basically performed as described earlier (4) with some modifications. The expression plasmids pAPP1 (for Pol I) and pAPP2 (for Pol II) were used to transform E. coli CodonPlus-RIL (Stratagene) cells. The genes were expressed by incubating the transformed cells in Luria-Bertani broth supplemented with ampicillin (100 μg/ml) and chloramphenicol (20 ng/ml) at 37°C for 5 h. One-liter cultures of cells harboring the pol genes were harvested by centrifugation at 5,000 × g for 20 min. The cell pellets were suspended in 30 ml of buffer A and were lysed by sonication. The lysates were each centrifuged at 30,000 × g for 20 min, and the supernatant from each preparation was heated at 75°C for 15 min followed by recentrifugation. To the supernatant after centrifugation, polyethyleneimine and NaCl were added to concentrations of 0.1% and 0.5 M, respectively, and the mixture was stirred for 30 min at 4°C. The proteins in the supernatant (30 ml) were precipitated by adding 16.8 g of ammonium sulfate (80% saturation). The precipitates were dissolved in buffer B and dialyzed against the same buffer. The dialysate containing Pol I was applied to an anion-exchange column (HiTrap Q, 5 ml; Pharmacia Biotech), and the chromatography was developed with a 50-ml linear gradient of 0.1 to 1.0 M NaCl in buffer A at a flow rate of 2 ml/min. Pol I eluted at a salt concentration range of 0.2 to 0.3 M. After anion-exchange chromatography, the fractions containing the Pol I proteins were pooled, diluted in buffer A, and applied to an affinity column (HiTrap heparin, 1 ml; Pharmacia Biotech), and the chromatography was developed with a 0.1 to 1.0 M NaCl gradient in buffer A at a flow rate of 1 ml/min. Pol I eluted at a salt concentration range of 0.45 to 0.55 M. In the case of Pol II, after ammonium sulfate precipitation, the dialysate containing Pol II was applied to a cation-exchange column (HiTrap SP, 5 ml; Pharmacia Biotech), and the chromatography was developed with a 50-ml linear gradient of 0.1 to 1.0 M NaCl in buffer A at a flow rate of 2 ml/min. Pol II eluted at a salt concentration range of 0.2 to 0.25 M. After cation-exchange chromatography, the fractions containing the Pol II proteins were pooled, diluted in buffer A, and applied to an affinity column (HiTrap heparin, 1 ml; Pharmacia Biotech). The column was developed with a 0.1 to 1.0 M NaCl gradient in buffer A at a flow rate of 1 ml/min. Pol II eluted at a salt concentration range of 0.25 to 0.35 M. The purified Pol I and Pol II proteins were stored at −20°C as 50% glycerol stock solutions after dialysis against buffer A.
N-terminal amino acid sequencing.
The purified PCNAs were fractionated by electrophoresis on an SDS-10% polyacrylamide gel, electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Sequi-Blot PVDF, 0.2 μm; Bio-Rad), stained with Coomassie brilliant blue R250 (0.02% in 40% methanol), and destained with 5% methanol. The protein bands were excised and subjected to automated Edman degradation in an Applied Biosystems model 492 protein sequencer.
DNA polymerase nucleotide incorporation assay.
The nucleotide incorporation abilities of Pol I and Pol II, in the absence or presence of ApePCNA1, ApePCNA2, or ApePCNA3, were investigated by using two templates: linearized M13 single-stranded DNA (ssDNA) and circular M13 ssDNA (Takara Shuzo). The oligomer, 5′-ATTCGTAATCATGGTCATAGCTGTTTCCTG-3′, which is complementary to positions 6204 to 6233 of M13mp18 DNA (57), was annealed to linearized and circular M13 ssDNA. To create the linearized M13 ssDNA, an oligonucleotide, 5′-GCCAAGCTTGCATGCCTGCAGGTCGACTCT-3′, complementary to positions 6260 to 6289 (within the multicloning site), was initially annealed to the circular M13 ssDNA to create a double-stranded region containing some restriction enzyme recognition sites. The DNA was then digested overnight with PstI. The product was purified by ethanol precipitation. The assay was carried out as described below. To anneal the primer to the templates, 0.1 μg of M13 ssDNA per test and 1.0 pmol of the primer in DNA polymerase reaction buffer were boiled for 3 min to ensure denaturation. The mixture was then gradually cooled to room temperature, and 0.5 pmol of the respective DNA polymerase was added. Twenty microliters from this mixture was aliquoted into the respective tube containing either PCNA or buffer. The reaction was initiated by adding deoxyribonucleotide triphosphates to a concentration of 0.1 mM, including 227 nM [methyl-3H]TTP (Amersham). Each 25-μl assay mixture contained 20 mM Tris-HCl (pH 8.8), 5.0 mM MgCl2, and 2 mM 2-mercaptoethanol. The reaction was carried out at 70°C for 5 min for Pol I and for 20 min for Pol II. After the reaction, 20 μl of each reaction mixture was spotted onto DE81 filters (Whatman). The filters were washed three times with a 5% Na2HPO4 solution, and the radioactivity incorporated into the DNA strands was counted by a scintillation counter.
Primer extension analysis.
The primer elongation abilities of Pol I and Pol II in the absence or presence of ApePCNA1, ApePCNA2, and ApePCNA3 were also investigated by using two templates. 5′ 32P-labeled oligomer, 5′-CTGTGTGAAATTGTTATCCGCTCACAATTC-3′, complementary to positions 6177 to 6206 of the M13mp18 genome, was annealed to linearized and circular M13 ssDNA. To anneal the primers to the template, 0.1 μg of M13 ssDNA and 1.0 pmol of 32P-labeled primer in DNA polymerase reaction buffer were boiled for 3 min to ensure denaturation. The mixture was then gradually cooled to room temperature, and 0.5 pmol of the respective DNA polymerase was added. Seven microliters from this mixture was aliquoted into the respective tubes containing either PCNA or buffer. The reaction was initiated by adding deoxyribonucleotide triphosphates to a concentration of 0.2 mM. Each 10-μl assay mixture contained 20 mM Tris-HCl (pH 8.8) and 5.0 mM MgCl2. The reaction was carried out at 70°C for 5 min for Pol I and for 20 min for Pol II. After the reaction, 3 μl of stop solution (98% deionized formamide, 1 mM EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue) was added, and 2.6 μl of each reaction was analyzed on a 1.0% alkaline agarose gel in 50 mM sodium sulfate and 1 mM EDTA.
Western blotting.
A. pernix K1 cells were cultivated as described earlier (44). Ten grams of bacterial cells was suspended in buffer A. The cells were disrupted by a French press, and the supernatant was recovered by centrifugation for 15 min at 25,000 × g. For immunoblot analysis, SDS-10% polyacrylamide gels were electroblotted onto a PVDF membrane (Bio-Rad). The blots were blocked with 5% (wt/vol) skim milk (Wako) in phosphate-buffered saline (PBS) containing 0.1% (vol/vol) Tween 20 (PBS-T) for 1 h and then were incubated for over 1 h at room temperature with a 1:1,000 dilution of rabbit anti-PCNA (−1, −2, and −3) antiserum in 5% skim milk in PBS-T. Membranes were washed using PBS-T and then were incubated for 1 h at room temperature with a 1:10,000 dilution of horseradish peroxidase-conjugated anti-rabbit immunoglobulin G in 5% skim milk in PBS-T. Membranes were washed using PBS-T, and the secondary antibody was detected with the enhanced chemiluminescence Western blotting analysis system (Amersham Pharmacia Biotech).
Immunoprecipitation of ApePCNAs and immunoblotting.
One milligram of total protein was diluted into 250 μl of TBS-T buffer (10 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.1% Triton X-100) and was incubated with the anti-PCNA antibodies at room temperature for 30 min. Immunocomplexes were collected with protein A-Sepharose (Amersham Pharmacia Biotech) for 30 min at room temperature with mixing. Pellets were washed three times with TBS-T buffer, boiled in SDS sample buffer, and loaded onto SDS-12% polyacrylamide gels. Proteins were transferred to a PVDF membrane and were analyzed by Western blotting as described above.
Chemical cross-linking.
Purified ApePCNAs were subjected to a chemical cross-linking experiment, using sulfoethylene glycol-bis (succinimidyl succinate) (sulfo-EGS; Sigma) as described earlier (58). The reaction mixtures (10 μl) contained 5 μg of ApePCNA per ml and 50 μM EGS in 20 mM sodium phosphate (pH 7.0) and 150 mM NaCl. The products were analyzed by Western blotting.
RESULTS
Identification of three PCNA-like sequence in the A. pernix genome.
From the total genome sequencing project, three genes encoding sequences homologous to the eukaryotic PCNA were found in A. pernix (23). The genes encode proteins of 263, 233, and 251 amino acids, and these proteins have estimated molecular masses of 29.4, 25.9, and 28.6 kDa, respectively. The amino acid sequences deduced from the genes were compared with those of other archaeal and eukaryotic homologs (Table 1). The identities within the archaeal PCNA homologs ranged from 16 to 38%, with the average being 24.5%. From the sequence alignment, the three proteins in A. pernix may form a homotrimeric, ring-shaped structure and work as sliding clamps for DNA synthesis.
TABLE 1.
A sequence comparison of eukaryotic and archaeal PCNAsa
| Organism | % Amino acid identity
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A. pernix 1 | A. pernix 2 | A. pernix 3 | P. furiosus | M. jannaschii | A. fulyidus | M. thermoauto-trophicum | H. sapions | S. cerevisiae | D. melano-gaster | |
| A. pernix 1 | ||||||||||
| A. pernix 2 | 27 | |||||||||
| A. pernix 3 | 25 | 20 | ||||||||
| P. furiosus | 31 | 28 | 26 | |||||||
| Methanococcus jannaschii | 27 | 22 | 23 | 38 | ||||||
| Archaeoglobus fulgidus | 20 | 18 | 16 | 23 | 26 | |||||
| Methanobacterium thermo-autotrophicum | 20 | 22 | 19 | 29 | 30 | 24 | ||||
| Homo sapiens | 15 | 20 | 16 | 22 | 21 | 20 | 26 | |||
| Saccharomyces cerevisiae | 14 | 16 | 14 | 23 | 26 | 19 | 22 | 34 | ||
| Drosophila melanogaster | 17 | 19 | 16 | 23 | 21 | 19 | 22 | 71 | 35 | |
Cloning, expression, and purification of recombinant ApePCNAs.
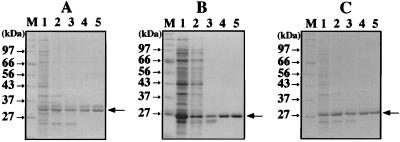
The three genes encoding PCNA-like sequence were directly amplified by PCR from the A. pernix DNA and were cloned into the expression vector pET21a. The cloned genes were expressed in E. coli, and the PCNA homologs, designated ApePCNA1, ApePCNA2, and ApePCNA3, were produced. In the cases of ApePCNA1 and ApePCNA3, the recombinant proteins of ca. 29.4 and 28.6 kDa detected by SDS-PAGE were purified to homogeneity through heat treatment to denature the majority of the host proteins, polyethyleneimine treatment to remove the DNA, ammonium sulfate precipitation, anion-exchange chromatography, and heparin affinity chromatography (Fig. 1A and C). In the case of ApePCNA2, because its thermostability was weaker than that of the other two PCNAs, the recombinant protein of ca. 25.9 kDa detected by SDS-PAGE was purified to homogeneity through polyethyleneimine treatment, ammonium sulfate precipitation, hydrophobic chromatography, anion-exchange chromatography, and heparin affinity chromatography (Fig. 1B). The amino acid sequences of the purified proteins were confirmed to match the initiation region of the open reading frames deduced from the genomic DNA sequences.
FIG. 1.
Purification of recombinant ApePCNA homologs from E. coli cells. All samples of each step were fractionated on SDS-10% polyacrylamide gels, which were stained with Coomassie brilliant blue. The sizes of the molecular mass markers (Protein Marker, Broad Range; New England BioLabs, Inc.) are indicated on the left of each figure. The arrows on the right side of the panels show the obtained proteins. (A) Purification steps of recombinant ApePCNA1. Lanes: M, molecular mass markers; 1, crude cell extract after sonication; 2, supernatant from heat treatment (75°C, 15 min.); 3, supernatant after polyethyleneimine treatment; 4, HiTrap Q fraction; and 5, HiTrap heparin fraction. (B) Purification steps of recombinant ApePCNA2. Lanes: M, molecular mass markers; 1, crude cell extract after sonication; 2, supernatant after polyethyleneimine treatment; 3, HiTrap Phenyl Sepharose HP fraction; 4, HiTrap Q fraction; 5, and HiTrap heparin fraction. (C) Purification steps of recombinant ApePCNA3. All lanes are the same as in the case of ApePCNA1.
Detection of PCNAs in A. pernix cell extract.
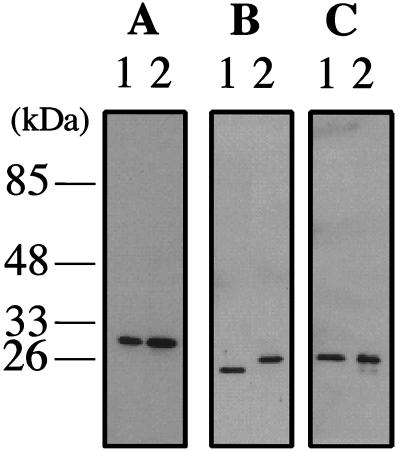
To investigate whether the PCNA-like proteins were actually produced in A. pernix cells and also whether their sizes corresponded to those of the recombinant proteins produced in E. coli, polyclonal antibodies were prepared by using the purified proteins and Western blot analysis was performed. As shown in Fig. 2A and C, two proteins with sizes corresponding to those of the recombinant ApePCNA1 and ApePCNA3 proteins were detected in the crude cell extract of A. pernix by each specific antibody. On the other hand, although a specific band that reacted with anti-ApePCNA2 was detected in A. pernix cells, the protein was slightly bigger than the recombinant ApePCNA2 protein that reacted with the anti-ApePCNA2 shown in Fig. 2B). These results indicate that all three genes are expressed and work in some processes in A. pernix cells. It is conceivable that the differences in the electrophoretic mobility of ApePCNA2 between the recombinant and native proteins reflect different modifications of that protein in E. coli and A. pernix cells. Another possibility is discussed below.
FIG. 2.
Identification of ApePCNAs in A. pernix cells. (A) Recombinant ApePCNA1 (lane 1, 40 ng) and A. pernix cell extracts (lane 2, 30 μg) were separated by SDS-10% PAGE and then were analyzed by Western blotting with anti-ApePCNA1 antiserum. (B) Recombinant ApePCNA2 (lane 1, 80 ng) and A. pernix cell extracts (lane 2, 30 μg) were analyzed with anti-ApePCNA2 antiserum. (C) Recombinant ApePCNA3 (lane 1, 10 ng) and A. pernix cell extracts (lane 2, 15 μg) were analyzed with anti-ApePCNA3 antiserum.
Effect of ApePCNAs on processivity of DNA polymerases.
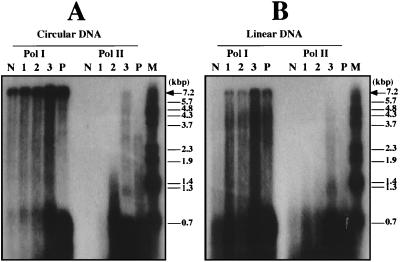
One of the biological functions of these purified PCNA homologs was investigated through their effect on the primer elongation activities of the DNA polymerases found in A. pernix. Using A. pernix Pol I and Pol II, a nucleotide incorporation assay was performed. All three PCNAs enhanced the DNA synthesis activities of Pol I and Pol II from the incorporation assay of radioactive thymidines into the DNA strands (data not shown). To confirm that the increased incorporations of radioactive nucleotides were derived from the stimulating effect of the PCNA homologs on the primer elongation abilities of these DNA polymerases, the synthesized products were visualized by denaturing gel electrophoresis by using circular and linear M13 ssDNA as substrates. As shown in Fig. 3, longer products were increased for both Pol I and Pol II in the presence of each PCNA homolog, compared to the cases without them. As with our previous observation of P. furiosus PCNA (PfuPCNA), the stimulation of the PCNA homologs in A. pernix was found not only with the linear DNA substrate but also with the circular DNA. The stimulation effect of ApePCNA3 on both Pol I and Pol II under these conditions was remarkably higher than those of the other two PCNAs in the cases of M13 circular and linear ssDNA as templates. The PfuPCNA surprisingly enhanced the nucleotide incorporation by A. pernix Pol I and Pol II, with almost the same efficiency as ApePCNA3 (data not shown). However, PfuPCNA strongly paused at a specific site of the template (at around 0.7 kb), as observed in the reactions of PfuPCNA with P. furiosus Pol I and Pol II (6). The pauses may be due to the PfuPCNA-DNA polymerase complex encountering secondary template structures. Similar transient pauses were observed with eukaryotic Pol δ in the presence of PCNA (11, 42). Under physiological conditions, replicative helicases and ssDNA binding protein are likely to prevent or alleviate this condition. This pausing occurred, but to a lesser extent, in the presence of all three ApePCNAs (Fig. 3). The reason why the pauses were weaker in the ApePCNA-DNA polymerase complexes remains unclear at this stage. Further analyses, including measurement of the strength of each PCNA-polymerase interaction and direct loading and unloading assays of these archaeal PCNAs, are necessary to understand the different features of the in vitro reaction products.
FIG. 3.
Detection of the reaction products by Pol I and Pol II. The primer elongation abilities of Pol I and Pol II in the absence and presence (0.3 μg) of ApePCNA1 (lane 1), ApePCNA2 (lane 2), and ApePCNA3 (lane 3) were investigated by using two templates: circular M13 ssDNA (panel A) and linearized M13 ssDNA (panel B). Lanes N and P indicate reactions without PCNA and with PfuPCNA, respectively, in both panels. The products were analyzed by 1.2% alkaline agarose gel electrophoresis, and the reaction products were visualized by autoradiography. The sizes indicated on the right side were from BstPI-digested lambda DNA labeled by polynucleotide kinase with [γ-32P]ATP. The arrow shows the position of the product with full-length M13 DNA.
Native molecular weight of ApePCNAs.
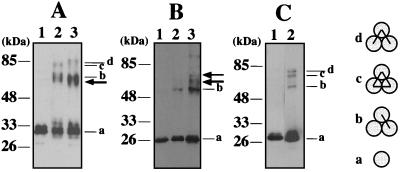
To investigate whether these PCNA homologs form a ring-shaped, trimeric structure, the purified recombinant proteins were subjected to a chemical cross-linking experiment. As shown in Fig. 4, sulfo-EGS rapidly cross-linked ApePCNA1 and ApePCNA3, which caused the complex to migrate as larger molecules in SDS-PAGE. This result supports the existence of multimeric forms of the two PCNAs. Three major cross-linked species appeared, which can be postulated to be the products from a single cross-linked dimer, a double cross-linked trimer (linear), and a triple cross-linked trimer (circular) estimated from the molecular weights of each molecule by the band mobilities of cross-linked proteins as shown earlier with the gp45 protein from T4 phage (20). Distinctly smaller amount of cross-linked products were detected from ApePCNA2 under the same conditions, indicating that the efficiency of multimer formation of ApePCNA2 is much lower than that of the other two PCNAs.
FIG. 4.
Association states of ApePCNAs assessed by chemical cross-linking. ApePCNA1 (A), ApePCNA2 (B), and ApePCNA3 (C) were treated with buffer (lane1) and sulfo-EGS (lane 2) for 2 min and were analyzed by SDS-10% PAGE, followed by Western blotting using anti-ApePCNA1, anti-ApePCNA2, and anti-ApePCNA3, respectively. Lanes 3 in panel A and B include equal amounts of ApePCNA1 and ApePCNA2. The cross-linked products from the mixture of ApePCNA1 and ApePCNA2 are indicated by a thick arrow. Each band is derived from a PCNA monomer (a), a single cross-linked dimer (b), three cross-linked circled trimers (c), and two cross-linked linear trimers (d), as indicated for the T4 gp45 protein (19). The indicated molecular sizes were derived from the prestained protein markers (APRO Science, Inc.).
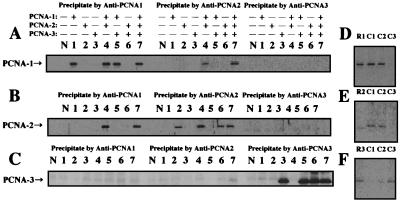
Interactions between ApePCNAs.
To examine whether the PCNA homologs physically interact with each other, an immunoprecipitation experiment was done using antibodies raised against all three ApePCNA homologs. As shown in both experiments using the purified proteins (Fig. 5A to C) and a crude cell extract of A. pernix (Fig. 5D to F), ApePCNA1 and ApePCNA2 were coprecipitated (lanes 4 and 7 in Fig. 5A and B). Conversely, ApePCNA3 was coprecipitated with neither ApePCNA1 nor ApePCNA2. These results suggest that ApePCNA3 works by itself, while ApePCNA1 and ApePCNA2 may work together in some cases. Chemical cross-linking experiments using a mixture of ApePCNA1 and ApePCNA2 also showed some complexes of the two molecules, although it is very difficult to interpret the composition of each band (Fig. 4A and B). According to these results, the primer extension assay including both ApePCNA1 and ApePCNA2 simultaneously was carried out. However, there was no difference between the reactions with both of the PCNAs and those with either ApePCNA1 or ApePCNA2 (data not shown). Therefore, the formation of the ApePCNA1 and ApePCNA2 complex does not seem likely to contribute specifically to the increase in the processivities with Pol I and Pol II. Further investigations with better quality are necessary to identify the most suitable structure of the heterologous PCNA complex in addition to know its physiological function.
FIG. 5.
Analysis of heterologous interactions of ApePCNAs. (A to C) Immunoprecipitation analysis was done using purified ApePCNAs and anti-ApePCNA1 (left part of each panel), anti-ApePCNA2 (middle part), and anti-ApePCNA3 (right part). The immunocomplexes were captured with protein A-Sepharose and were subjected to SDS-12% PAGE, followed by Western blot analysis using anti-PCNA1 (A), anti-PCNA2 (B), and anti-PCNA3 (C). Lanes: N, no protein; 1, ApePCNA1; 2, ApePCNA2; 3, ApePCNA3; 4, ApePCNA1 + ApePCNA2; 5, ApePCNA1 + ApePCNA3; 6, ApePCNA2 + ApePCNA3; and 7, ApePCNA1 + ApePCNA2 + ApePCNA3. (D to F) Total cell extract of A. pernix was reacted with anti-ApePCNA1 (lane C1), anti-ApePCNA2 (lane C2), and anti-ApePCNA3 (lane C3). These complexes were captured with protein A-Sepharose, and the immunoprecipitates were analyzed as described above, using anti-ApePCNA1 (D), anti-ApePCNA2 (E), and anti-ApePCNA3 (F). As a positive control, purified ApePCNAs precipitated by anti-PCNAs were loaded onto each gel (lanes: R1, ApePCNA1; R2, ApePCNA2; and R3, ApePCNA3).
DISCUSSION
We cloned the genes encoding homologs of PCNAs, the sliding clamps of DNA polymerases, from A. pernix, and characterized the gene products. All of the PCNA homologs actually stimulated the primer extension abilities of both Pol I and Pol II from A. pernix. These effects suggested the possibility that all three of the PCNA homologs work as the sliding clamp for DNA polymerases in this organism. It was important to study how the three PCNAs are separately used in different situations. The most plausible idea is that there is a specific DNA polymerase-and-PCNA combination. Therefore, the specificity of the stimulating effects of the ApePCNAs for Pol I and Pol II was carefully investigated. In the cases of both Pol I and Pol II, the stimulation specificity of the ApePCNA3 in the primer extension activities was evident: remarkably, ApePCNA3 enhanced Pol II activity by more than 200-fold. These results suggest that ApePCNA3 may be the authentic processivity factor for Pol I and Pol II in the cells. This idea is consistent with the results that ApePCNA3 is the most easily cross-linked to the trimeric structure (Fig. 4) and that it does not seems likely to interact directly with the other two PCNAs (Fig. 5). These results were not predicted, because the amino acid sequence of ApePCNA3 is the least similar among the three to those of other archaeal PCNA homologs (Table 1). All of the archaeal PCNAs compared here are from euryarchaeal organisms, and only one PCNA-like sequence has been found in each of them. Very recently, the total genome sequencing of two Sulfolobus organisms, S. solfataricus (45) and Sulfolobus tokodaii (22), has been completed. These crenarchaeal organisms also have three PCNA-like genes. Therefore, the deduced amino acid sequences were compared with the three PCNAs from A. pernix. As shown in Table 2, S. solfataricus 0405 and S. tokodaii 0387, S. solfataricus 1047 and S. tokodaii 0944, and S. solfataricus 0397 and S. tokodaii 0397 are the respective orthologs. However, the similarity of these sequences to those of ApePCNA1, ApePCNA2, and ApePCNA3 is very low (less than 25% identity). More detailed phylogenetic analysis is now under way to understand the relationships of the three PCNA genes found in Crenarchaeota. Recently, two PCNA-like sequences in a crenarchaeote, Sulfurisphaera ohwakuensis, have been reported (19). The third PCNA-like sequence may also be discovered soon from this crenarchaeote.
TABLE 2.
A sequence comparison of crenarchaeal PCNAs
| Organism | % Amino acid identity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A. pernix 1 | A. pernix 2 | A. pernix 3 | S. solfataricus 0397 | S. solfataricus 0405 | S. solfataricus 1047 | S. tokodaii 0387 | S. tokodaii 0397 | S. tokodaii 0944 | |
| A. pernix 1 | |||||||||
| A. pernix 2 | 27 | ||||||||
| A. pernix 3 | 25 | 20 | |||||||
| S. solfataricus 0397 | 15 | 19 | 23 | ||||||
| S. solfataricus 0405 | 22 | 22 | 24 | 19 | |||||
| S. solfataricus 1047 | 23 | 22 | 19 | 18 | 20 | ||||
| S. tokodaii 0387 | 23 | 23 | 27 | 18 | 61 | 23 | |||
| S. tokodaii 0397 | 20 | 21 | 25 | 52 | 21 | 22 | 20 | ||
| S. tokodaii 0944 | 19 | 24 | 22 | 18 | 24 | 48 | 24 | 21 | |
To explain the difference between the size of ApePCNA2 found in A. pernix cells and the size of that found in recombinant protein from E. coli cells, in addition to the idea that the different modifications occurs in the two cells, perhaps the translation initiation codon actually used in A. pernix is different from that which we selected. We checked the DNA sequence around the gene for ApePCNA2 and found a TTG codon 48 bases upstream of the ATG that we used in this study. If this TTG codon is actually used as the start codon in A. pernix, then the gene product would be 16 amino acids longer, and it should be appear as a band with the same size as that of the native protein on the SDS-PAGE. We are presently purifying the longer ApePCNA2 protein to compare the two ApePCNA2s.
From a study of the DNA polymerase activities of the Pol δ catalytic subunit from yeast and human, a consensus motif, GX4GX8GX3YFY, in which the subunit was proposed to be important for binding to PCNA (59), was put forth. The A. pernix Pol II has a sequence resembling this consensus motif in its N-terminal region, while Pol I does not. In several eukaryotic proteins that are known to interact with PCNA, including the cell cycle checkpoint protein p21, and the endonucleases FEN1 and XPG, an octapeptide sequence, referred to as the PCNA-interacting protein box, is conserved (53–55). Like the consensus sequence described above, Pol II of A. pernix has a sequence resembling the PCNA-interacting protein motif (QSTLLDFM) in its C-terminal region, while this box is not included in Pol I. Consistent with these results, the A. pernix Pol II has a higher requirement for stimulation by PCNA than does Pol I (Fig. 3). Our previous study showed that Pol I has a much more efficient primer extension ability by itself than Pol II in vitro (4); therefore, it is likely that the actual effect of the PCNAs on Pol I may be much less than that on Pol II, resulting from the weaker interaction. Quantitative analyses are necessary to discuss the strength of each DNA polymerase-PCNA interaction.
As previously found in a P. furiosus study (6), the DNA synthesis reactions by the two A. pernix DNA polymerases were stimulated by the ApePCNAs, even when a circular DNA template was used. It is well known that a molecule called the “clamp loader” (γ complexes in Bacteria and RFC in Eucarya) is required for the opening and loading of the ring-shaped sliding clamp onto circular DNA. One possible explanation is that the subunit-subunit interaction of archaeal PCNA from its predicted ring structure may be less stable than those of other sliding clamp molecules, which cause the ring of PCNA to open spontaneously in vitro at a high reaction temperature. In the crystal structures of yeast and human PCNA, a stable interface connection is produced through seven and eight clustered main chain hydrogen bonds, respectively (13, 30). However, there are only four hydrogen bonds between the subunit main chains of PfuPCNA (35). A multiple sequence alignment of the PCNAs showed that there are some deletions in the loop constituting the interfaces of the archaeal PCNAs (Fig. 6). ApePCNA1 and ApePCNA2 have deletions in the first interface region, and ApePCNA3 also has some deletions in the second interface. Due to the weak connections of the subunits, these archaeal PCNAs may be able to load onto the DNA strand without the clamp-loading activity of RFC, which is usually required to load PCNA onto DNA in the eukaryotic system. However, there is a sequence of an RFC homolog in the A. pernix genome. As in the cases of P. furiosus (7) and S. solfataricus (41), the clamp-loading process is possibly much more efficient in the presence of RFC in A. pernix cells. There is only one set of sequences corresponding to the small and large subunits of RFC in A. pernix. The relationship between the RFC and the three PCNAs should be analyzed.
FIG. 6.
Amino acid sequence alignment of the three ApePCNAs, P. furiosus, yeast, and human PCNAs. Identical and similar amino acid residues are boxed with black and gray, respectively. Important regions required for the function and oligomerization of PCNA, predicted from previous studies, are indicated with the alignment. Ape, A. pernix; Pfu, P. furiosus; Sce, Saccharomyces cerevisiae; Hsa, Homo sapiens.
Recently, an interesting model was proposed, in which three checkpoints, Rad1, Hus1, and Rad9, form a PCNA-like toroidal structure and Rad17 forms a clamp loader complex with four small subunits of RFC, which may be the specific clamp loader for the heterotrimeric circle of Rad1-Hus1-Rad9 (2, 8, 43, 51). The structural models imply that these proteins are specific partners for the damage-specific DNA polymerases, such as Pol η. From this analogy, we carefully analyzed whether the three PCNA homologs interact with each other and work as the sliding clamps for Pol I and Pol II. Our results showed that ApePCNA1 and ApePCNA2 may work together, but the heterocomplex is probably not a clamp for either Pol I or Pol II. ApePCNA1 and ApePCNA2 may be a specific clamp for a DNA polymerase with translesion activity, which has not been identified in A. pernix. We searched for a sequence in the A. pernix genome that is similar to the DNA polymerases belonging to the DinB/UmuC/Rev1/Rad30 superfamily (the name of family Y has been recently acknowledged [37] for DNA polymerases in this family), but no distinct similarity was found. However, a gene encoding a sequence homologous to that of DinB protein exists in Sulfolobus (31), and the gene product can bypass an abasic site with its nonprocessive DNA polymerase activity (60). A. pernix may have translesion DNA polymerases with very divergent sequences from those of family Y. Crenarchaeal organisms may have commonly three PCNA-like proteins; therefore, including the subject of the possibility as the sliding clamp for a translesion DNA polymerase, further studies are necessary to explain the biological meaning of the three PCNAs in A. pernix. The information obtained from this study would contribute to a better understanding of the DNA replication mechanism in Crenarchaeota.
Acknowledgments
We thank S. Ishino, K. Komori, S. Matsumiya, and K. Morikawa for discussions. We are grateful to Y. Shimura, the director of the Biomolecular Engineering Research Institute, for continuous encouragement.
REFERENCES
- 1.Bocquier, A. A., L. Liu, I. K. O. Cann, K. Komori, D. Kohda, and Y. Ishino. 2001. Archaeal primase: bridging the gap between RNA and DNA polymerase. Curr. Biol. 11:452–456. [DOI] [PubMed] [Google Scholar]
- 2.Cai, R. L., Y. Yan-Neale, M. A. Cueto, H. Xu, and D. Cohen. 2000. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275:27909–27916. [DOI] [PubMed] [Google Scholar]
- 3.Cann, I. K. O., K. Komori, H. Toh, S. Kanai, and Y. Ishino. 1998. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl. Acad. Sci. USA 95:14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann, I. K. O., S. Ishino, N. Nomura, Y. Sako, and Y. Ishino. 1999. Two family B DNA polymerases from Aeropyrum pernix, an aerobic hyperthermophilic crenarchaeote. J. Bacteriol. 181:5984–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cann, I. K. O., and Y. Ishino. 1999. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics 152:1249–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann, I. K. O., S. Ishino, I. Hayashi, K. Komori, H. Toh, K. Morikawa, and Y. Ishino. 1999. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 181:6591–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cann, I. K. O., S. Ishino, M. Yuasa, H. Daiyasu, H. Toh, and Y. Ishino. 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:2614–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspari, T., M. Dahlen, G. Kanter-Smoler, H. D. Lindsay, K. Hofmann, K. Papadimitriou, P. Sunnerhagen, and A. M. Carr. 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 74:1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFelice, M., C. W. Sensen, R. L. Charlebois, M. Rossi, and F. M. Pisani. 1999. Two DNA polymerase sliding clamps from the thermophilic archaeon Sulfolobus solfataricus. J. Mol. Biol. 291:47–57. [DOI] [PubMed] [Google Scholar]
- 10.Edgell, D. R., and W. F. Doolittle. 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89:995–998. [DOI] [PubMed] [Google Scholar]
- 11.Eissenberg, J. C., R. Ayyagai, X. V. Gomes, and P. M. J. Burgers. 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol. 17:6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forterre, P., C. Elie, and M. Kohiyama. 1984. Aphidicolin inhibits growth and DNA synthesis in halophilic archaebacteria. J. Bacteriol. 159:800–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulbis, J. M., Z. Kelman, J., M. O’Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87:297–306. [DOI] [PubMed] [Google Scholar]
- 14.Imamura, M., T. Uemori, I. Kato, and Y. Ishino. 1995. A non-α-like DNA polymerase from the hyperthermophilic archaeon Pyrococcus furiosus. Biol. Pharm. Bull. 18:1647–1652. [DOI] [PubMed] [Google Scholar]
- 15.Ishino, Y., and I. K. O. Cann. 1998. The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: fact or farce? Genes Genet. Syst. 73:323–336. [DOI] [PubMed] [Google Scholar]
- 16.Ishino, Y., and S. Ishino. 2001. DNA polymerases from Euryarchaeota. Methods Enzymol. 334:249–260. [DOI] [PubMed] [Google Scholar]
- 17.Ishino, Y., K. Komori, I. K. O. Cann, and Y. Koga. 1998. A novel DNA polymerase family found in Archaea. J. Bacteriol. 180:2232–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishino, Y., T. Tsurimoto, S. Ishino, and I. K. O. Cann. 2001. Functional interactions of an archaeal sliding clamp with mammalian clamp loader and DNA polymerase δ. Genes Cells 6:699–706. [DOI] [PubMed] [Google Scholar]
- 19.Iwai, T., N. Kurosawa, Y. H. Ito, and T. Horiuchi. 2000. Phylogenetic analysis of archaeal PCNA homologues. Extremophiles 4:357–364. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis, T. C., L. S. Paul, and P. H. von Hippel. 1989. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J. Biol. Chem. 264:12709–12716. [PubMed] [Google Scholar]
- 21.Jonsson, Z. O., and U. Hubscher. 1997. Proliferating cell nuclear antigen: more than a clamp for DNA polymerase. Bioessays 19:967–975. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123–140. [DOI] [PubMed] [Google Scholar]
- 23.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A., Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, K. Kubota, Y. Nakamura, N. Nomura, Y. Sako, and H. Kikuchi. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83–101. [DOI] [PubMed] [Google Scholar]
- 24.Kelman, Z. 1997. PCNA: structure, functions, and interactions. Oncogene 14:629–640. [DOI] [PubMed] [Google Scholar]
- 25.Kelman, Z., and J. Hurwitz. 1998. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biochem. Sci. 23:236–238. [DOI] [PubMed] [Google Scholar]
- 26.Kelman, Z., and M. O’Donnell. 1995. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64:171–200. [DOI] [PubMed] [Google Scholar]
- 27.Komori, K., and Y. Ishino. 2000. Functional interdependence of DNA polymerizing and 3′→5′ exonucleolytic activities in Pyrococcus furiosus DNA polymerase I. Protein Eng. 13:41–47. [DOI] [PubMed] [Google Scholar]
- 28.Komori, K., and Y. Ishino. 2001. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 276:25654–25660. [DOI] [PubMed] [Google Scholar]
- 29.Kong, X.-P., R. Onrust, M. O’Donnell, and J. Kuriyan. 1992. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69:425–437. [DOI] [PubMed] [Google Scholar]
- 30.Krishna, T. S. R., X.-P. Kong, S. Gary, P. M. J. Burgers, and J. Kuriyan. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79:1233–1243. [DOI] [PubMed] [Google Scholar]
- 31.Kulaeva, O. I., E. V. Koonin, J. P. McDonald, S. K. Randall, N. Rabinovich, J. F. Connaughton, A. S. Levine, and R. Woodgate. 2001. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res. 357:245–253. [DOI] [PubMed] [Google Scholar]
- 32.Leipe, D. D., L. Aravind, and E. V. Koonin. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, L., K. Komori, S. Ishino, A. Bocquier, I. Cann, D. Kohda, and Y. Ishino. 2 October 2001. The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. M106391200. [DOI] [PubMed]
- 34.Makiniemi, M., H. Pospiech, M. Jokela, M. Vihinen, and J. E. Syvaoja. 1999. A novel family of DNA-polymerase-associated B subunits. Trends Biochem. Sci. 24:14–16. [DOI] [PubMed] [Google Scholar]
- 35.Matsumiya, S., Y. Ishino, and K. Morikawa. 2001. Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 10:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayanagi, K., T. Miyata, T. Oyama, Y. Ishino, and K. Morikawa. 2001. Three-dimensional electron microscopy of the clamp loader small subunit from Pyrococcus furiosus. J. Struct. Biol. 134:35–45. [DOI] [PubMed] [Google Scholar]
- 37.Ohmori, H., E. C. Friedberg, R. P. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7–8. [DOI] [PubMed] [Google Scholar]
- 38.Olsen, G. J., and C. R. Woese. 1997. Archaeal genomics: an overview. Cell 89:991–994. [DOI] [PubMed] [Google Scholar]
- 39.Oyama, T., Y. Ishino, I. K. O. Cann, S. Ishino, and K. Morikawa. 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell 8:455–463. [DOI] [PubMed] [Google Scholar]
- 40.Perler, F. B., S. Kumar, and H. Kong. 1996. Thermostable DNA polymerases. Adv. Protein Chem. 48:377–435. [DOI] [PubMed] [Google Scholar]
- 41.Pisani, F. M., M. De Felice, F. Carpentieri, and M. Rossi. 2000. Biochemical characterization of a clamp-loader complex homologous to eukaryotic replication factor C from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Mol. Biol. 301:61–73. [DOI] [PubMed] [Google Scholar]
- 42.Podust, V. N., L. M. Podust, F. Muller, and U. Hubscher. 1995. DNA polymerase δ holoenzyme: action on single-stranded DNA and on double-stranded DNA in the presence of replicative DNA helicases. Biochemistry 34:5003–5010. [DOI] [PubMed] [Google Scholar]
- 43.Rauen, M., M. A. Burtelow, V. M. Dufault, and L. M. Karnitz. 2000. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J. Biol. Chem. 275:29767–29771. [DOI] [PubMed] [Google Scholar]
- 44.Sako, Y., N. Nomura, A. Uchida, Y. Ishida, H. Morii, Y. Koga, T. Hoaki, and T. Maruyama. 1996. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100°C. Int. J. Syst. Bacteriol. 46:1070–1077. [DOI] [PubMed] [Google Scholar]
- 45.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, de J. I. Heikamp, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schinzel, R., and K. J. Burger. 1984. Sensitivity of halobacteria to aphidicolin, an inhibitor of eukaryotic a-type DNA polymerases. FEMS Microbiol. Lett. 25:187–190. [Google Scholar]
- 47.Tsurimoto, T. 1998. PCNA, a multifunctional ring on DNA. Biochim. Biophys. Acta 1443:23–39. [DOI] [PubMed] [Google Scholar]
- 48.Uemori, T., Y. Ishino, H. Doi, and I. Kato. 1995. The hyperthermophilic archaeon Pyrodictium occultum has two α-like DNA polymerases. J. Bacteriol. 177:2164–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uemori, T., Y. Ishino, H. Toh, K. Asada, and I. Kato. 1993. Organization and nucleotide sequence of the DNA polymerase gene from the archaeon Pyrococcus furiosus. Nucleic Acids Res. 21:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uemori, T., Y. Sato, I. Kato, H. Doi, and Y. Ishino. 1997. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells 2:499–512. [DOI] [PubMed] [Google Scholar]
- 51.Venclovas, C., and M. P. Thelen. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 13:2481–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721–751. [DOI] [PubMed] [Google Scholar]
- 53.Warbrick, E. 1998. PCNA binding through a conserved motif. Bioessays 20:195–199. [DOI] [PubMed] [Google Scholar]
- 54.Warbrick, E. 2000. The puzzle of PCNA’s many partners. Bioessays 22:997–1006. [DOI] [PubMed] [Google Scholar]
- 55.Warbrick, E., D. P. Lane, D. M. Glover, and L. S. Cox. 1997. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to coordinate DNA replication and repair. Oncogene 14:2313–2321. [DOI] [PubMed] [Google Scholar]
- 56.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanisch-Perron, C., J. Vierra, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, P., S.-J. Zhang, Z. Zhang, J. F. Woessner, Jr., and M. Y. W. T. Lee. 1995. Expression and physicochemical characterization of human proliferating cell nuclear antigen. Biochemistry 34:10703–10712. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, S.-J., X.-R. Zeng, P. Zhang, N. L. Toomey, R.-Y. Chuang, L.-S. Chang, and M. Y. W. T. Lee. 1995. A conserved region in the amino terminus of DNA polymerase δ is involved in proliferating cell nuclear antigen binding. J. Biol. Chem. 270:7988–7992. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, B., J. D. Pata, and T. A. Steitz. 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8:427–437. [DOI] [PubMed] [Google Scholar]