Abstract
In the plant-pathogenic bacterium Erwinia chrysanthemi production of pectate lyases, the main virulence determinant, is modulated by a complex network involving several regulatory proteins. One of these regulators, PecS, also controls the synthesis of a blue pigment identified as indigoidine. Since production of this pigment is cryptic in the wild-type strain, E. chrysanthemi ind mutants deficient in indigoidine synthesis were isolated by screening a library of Tn5-B21 insertions in a pecS mutant. These ind mutations were localized close to the regulatory pecS-pecM locus, immediately downstream of pecM. Sequence analysis of this DNA region revealed three open reading frames, indA, indB, and indC, involved in indigoidine biosynthesis. No specific function could be assigned to IndA. In contrast, IndB displays similarity to various phosphatases involved in antibiotic synthesis and IndC reveals significant homology with many nonribosomal peptide synthetases (NRPS). The IndC product contains an adenylation domain showing the signature sequence DAWCFGLI for glutamine recognition and an oxidation domain similar to that found in various thiazole-forming NRPS. These data suggest that glutamine is the precursor of indigoidine. We assume that indigoidine results from the condensation of two glutamine molecules that have been previously cyclized by intramolecular amide bond formation and then dehydrogenated. Expression of ind genes is strongly derepressed in the pecS background, indicating that PecS is the main regulator of this secondary metabolite synthesis. DNA band shift assays support a model whereby the PecS protein represses indA and indC expression by binding to indA and indC promoter regions. The regulatory link, via pecS, between indigoidine and virulence factor production led us to explore a potential role of indigoidine in E. chrysanthemi pathogenicity. Mutants impaired in indigoidine production were unable to cause systemic invasion of potted Saintpaulia ionantha. Moreover, indigoidine production conferred an increased resistance to oxidative stress, indicating that indigoidine may protect the bacteria against the reactive oxygen species generated during the plant defense response.
During their life span, pathogenic microorganisms encounter different types of environments. They successively alternate saprophytic and pathogenic phases that could be assimilated to “diet” and “copiotrophic” periods. The success of pathogenic microbes depends on their ability to colonize host tissues and counter host defense mechanisms. Erwinia chrysanthemi is a plant-pathogenic enterobacterium responsible for soft rot diseases in a wide range of plant species. These symptoms result from the disorganization of the plant cell wall caused by a set of extracellular enzymes such as pectinases, cellulase, and proteases. Among these degradative enzymes, pectate lyases play a predominant role in plant tissue maceration (10). During their interactions with plants, bacteria are exposed to plant-generated H2O2, organic peroxides, and superoxides, which are important components of the plant defense response (23, 45). Microbial defense against oxidative stress involves both primary detoxification of the stress and secondary repair processes. It has been recently established that peptide methionine sulfoxide reductase (MsrA), an enzyme repairing oxidative damage caused to protein, and a manganese-superoxide dismutase (SodA) are important in the virulence of E. chrysanthemi (13, 39).
In E. chrysanthemi, production of pectate lyases is tightly regulated and responds to various environmental conditions, such as the presence of pectin, iron starvation, temperature, etc. (18). The characterization of regulatory mutants allowed the identification of several regulators (KdgR, PecS, CRP, PecT, Fur, ExpR) which modulate pectate lyase synthesis (15, 30, 34–36, 44). One of these regulators, PecS, that belongs to the MarR family, was shown to also control the production of a blue 3,3′-bipyridyl pigment identified as indigoidine (36). Indigoidine production has been reported both in phytopathogenic bacteria, such as Clavibacter michiganensis subsp. insidiosum and E. chrysanthemi (43), and in saprophytic bacteria, such as Arthrobacter atrocyaneus and Vogesella indigofera (22). However, nothing is known about the enzymes and precursors involved in the production of this pigment. Studies on the control of indigoidine production have shown that the media composition affects indigoidine production. For example, E. chrysanthemi produces more indigoidine when grown on potato medium (43). In an Arthrobacter species, addition of glutamic acid and pyridoxine was shown to enhance production of a reduced form of indigoidine: the leucoindigoidine (17). Our previous study of the PecS-mediated regulation demonstrated that there are common regulatory links between the production of indigoidine and the synthesis of extracellular macerating enzymes (36). The coordinate production of virulence factors and indigoidine in E. chrysanthemi led us to question whether indigoidine could play a role in E. chrysanthemi pathogenicity. With regard to its chemical structure, one of our hypotheses is that indigoidine has the potential to scavenge oxygen radicals and that it may be active in protecting bacteria during plant infection. Indeed, due to the presence in its structure of carbon-carbon double bonds conjugated with a carbonyl group, indigoidine can act as a radical scavenger (16). After capture of a radical by one of these double bonds, the newly formed radical will be substituted by both an electron-attracting (carbonyl) and an electron-releasing (amine) group and will be particularly stabilized from mutual reinforcement of the two substituent effects (47). Consequently, such a process would be thermodynamically favored.
In order to address the role of indigoidine in pathogenicity, we have initiated a characterization of indigoidine biosynthetic genes. In this report, we describe three structural genes, indA, indB, and indC, involved in indigoidine synthesis that have been located in the vicinity of the pecS-pecM regulatory locus. Results obtained showed that mutants deficient in indigoidine production were altered in their ability to induce systemic disease on Saintpaulia ionantha.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains, phage, and plasmids are described in Table 1. E. chrysanthemi and Escherichia coli cells were grown at 30 and 37°C, respectively, in Luria-Bertani (LB) medium or M63 minimal medium (28) supplemented with a carbon source (0.2% except for polygalacturonate [0.4%]) and, when required, with amino acids (40 μg/ml) and antibiotics at the following concentrations: ampicillin, 100 μg/ml; kanamycin and chloramphenicol, 50 μg/ml; tetracycline, 20 μg/ml. A crude S. ionantha extract was prepared by homogenization of Saintpaulia leaves and filtration through 0.45-μm-pore-size filters. This extract was used at the final concentration of 1% (vol/vol).
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| NM522 | Δ(lac-proAB) thi hsd-5 supE [F′ proAB+lacIqlacZΔM15] | Stratagene |
| S17-1 | Tpr SmrrecA thi pro hsdR−M+/RP4::2-Tc::Mu::KmTn7 | 40 |
| E. chrysanthemi | ||
| 3937 | Wild-type strain isolated from S. ionanthia | 21 |
| A350 | lmrT(Con) lacZ2 | 19 |
| A1524 | lmrT(Con) lacZ2 pecS::MudIIPR13 | 36 |
| A3471 | lmrT(Con) lacZ2 pecS::MudIIPR13 ind::Tn5-B21 | This work |
| A3478 | lmrT(Con) lacZ2 pecS::MudIIPR13 indB::Tn5-B21 | This work |
| A3843 | lmrT(Con) lacZ2 pecS::MudIIPR13 indA::uidA Km | This work |
| A3517 | lmrT(Con) lacZ2 pecS::MudIIPR13 indB::uidA Km | This work |
| A3812 | lmrT(Con) lacZ2 pecS::MudIIPR13 indC::uidA Km | This work |
| A3661 | lmrT(Con) lacZ2 indA::uidA Km | This work |
| A3516 | lmrT(Con) lacZ2 indB::uidA Km | This work |
| A3677 | lmrT(Con) lacZ2 indC::uidA Km | This work |
| A3953 | pecS::MudIIPR13 | This work |
| A3954 | indA::uidA Km | This work |
| A3956 | pecS::MudIIPR13 indA::uidA Km | This work |
| Plasmids | ||
| pTn5-B21 | pSUP102-Gm Tn5-B21 lacZ promoter probe transposon Tcr | 41 |
| pBR322 | Apr Tcr | 7 |
| pBR325 | Apr Tcr Cmr | 6 |
| pUC18 | AprlacZ′ | 48 |
| pULB108 | RP4::Mu3A Apr Kmr | 46 |
| pBLM2 | RP4 derivative with IS21 duplication, Kmr | 26 |
| pR’S1 | pBLM2 derivative harboring pecS::MuCmr insertion; argG, pecM, and indABC genes of E. chrysanthemi 3937 | 36 |
| pUIDK1 | pBR322 derivative harboring uidA Kmr cassette | 3 |
| pE71 | pUC18 with a 6.7-kb EcoRI fragment corresponding to the Tn5-B21-flanking DNA region from strain A3471 | This work |
| pE78 | pUC18 with a 7.4-kb EcoRI fragment corresponding to the Tn5-B21-flanking DNA region from strain A3478 | This work |
| pSR996 | pBR325 derivative harboring pecS, pecM, indA, and indB genes of strain 3937 | 36 |
| pSR1158 | pBR322 derivative harboring indC gene of strain 3937 | This work |
| pSR1180 | pBR322 derivative harboring the region downstream of indC | This work |
| pSR1972 | pUC18 derivative with a 1.6-kb Eco47III fragment harboring indA gene of E. chrysanthemi 3937 | This work |
| Phage PhiEC2 | E. chrysanthemi generalized transducing phage | 33 |
Genotype symbols are according to Berlyn (4). lmrT(Con) indicates that the transport system encoded by the gene lmrT, which mediates entry of lactose, melibiose, and raffinose into the cells, is constitutively expressed. lacZ′ indicates that the 3′ end of this gene is truncated.
Liquid cultures were grown in a shaking incubator (220 rpm). Semianaerobic conditions were achieved without shaking in liquid cultures layered with paraffin oil, using M63 minimal medium supplemented with fumarate (2.5% wt/vol) as the electron acceptor. Nitrogen starvation was performed in the following medium: M63 deprived of (NH4)2SO4 but supplemented with arginine (200 μg/ml) as the nitrogen source.
To test oxidative stress resistance, bacteria were inoculated into 5 ml of LB medium and grown for 16 h at 30°C. Under such conditions, bacterial cultures reached stationary growth phase. H2O2 (10 mM), which corresponds to a lethal concentration, was then added to the culture, and the cells were incubated for an additional 2 h at 30°C. The number of CFU was determined, by serial dilution and plating on LB agar, just before H2O2 treatment, just after the beginning of the challenge, and then every 20 min during 2 h. Survival of the H2O2-treated cells was normed to the number of CFU at the beginning of the challenge. This experiment was repeated three times.
Genetic techniques.
Mutagenesis of the E. chrysanthemi pecS strain A1524 was performed using the transposon Tn5-B21, which carries a promoterless lacZ gene and a tetracycline resistance gene (41). This transposon is present on the mobilizable E. coli vector pSUP102-Gm, which does not replicate in E. chrysanthemi. Equal volumes (0.2 ml) of a late-logarithmic-phase culture of E. coli S17-1 carrying pTn5-B21 and a late-logarithmic-phase culture of E. chrysanthemi A1524 were mixed together onto a M63 agar plate without a carbon source. After 5 h at 30°C, the cells were suspended in 500 μl of M63 medium. This suspension was diluted 100-fold, and 100-μl aliquots of the dilution were spread onto M63 agar plates containing glycerol as the carbon source and both chloramphenicol and tetracycline. After 48 h at 30°C, white colonies that no longer produce indigoidine were selected among the blue colonies.
Chromosomal localization was realized using the RP4 derivative plasmid pULB108, which can mediate the transfer of the host chromosome to a recipient bacterium (46). Transductions with phage PhiEC2 were performed as previously described (33).
Recombinant DNA techniques.
Preparations of plasmid DNA, restriction digestions, ligations, DNA electrophoresis, transformations, and electroporations were all performed using standard molecular biology techniques (38). Nucleotide sequencing was performed by Genome Express SA (Grenoble, France). Nucleotide and derived amino acid sequences were assembled and initially analyzed using the MAC Molly TETRA program (SoftGene, Berlin, Germany). Database comparisons were performed with the Basic Local Alignment Search Tool programs (1) using the server at the National Center for Biotechnology Information.
Primer extension analysis.
Total RNA was extracted from the E. chrysanthemi strain A1524 by the method using frozen phenol described by Maes and Messens (24). RNA concentration was estimated spectrophotometrically and by electrophoresis on formaldehyde denaturing 1% agarose gel. For primer extension, aliquots of about 10 and 20 μg of total RNA were annealed in S1 hybridization solution with about 6 × 104 cpm of 32P-end-labeled oligonucleotide purchased from MWG Biotech. The sequences of the primers used in this work are as follows: indA specific transcript, 5′-GGAAGCTTGACATAATATTCTCTCATCC-3′; indC specific transcript, 5′-GGAAGCTTGTTGTTATCCATTACAATCCTCG-3′. The 3′ extremity of these primers hybridize −15 and −11 nucleotides upstream from the translation start codon of indA and indC, respectively.
Band shift assays.
The regulatory regions of indA and indC were amplified by PCR using the primers indA1 (5′-GCTCTAGAGCTGGAACAATCTGACTGTG-3′) and indA2 (5′-GGAAGCTTGACATAATATTCTCTCATCC-3′) and primers indC1 (5′-GCTCTAGACTTAGTCGTTTTATTGCCCC-3′) and indC2 (5′-GGAAGCTTGTTGTTATCCATTACAATCCTCG-3′), respectively. In each pair of primers a HindIII and an XbaI site were included at the 5′ extremity (underlined nucleotides). After digestion with HindIII and XbaI, the PCR fragments were labeled by incorporating [α-32P]dCTP (3,000 Ci · mmol−1) with the Klenow fragment of DNA polymerase. These labeled fragments were further purified using the Qiaquick extraction kit from Qiagen. Band shift assays were conducted as described (31). A typical assay contains, in 20 μl, the following: 12 mM HEPES-NaOH, pH 7.0; 4 mM Tris-HCl, pH 7.0; 75 mM KCl; 5 mM CaCl2; 10 mM MgCl2; 1 mM dithiothreitol; 10% (vol/vol) glycerol; 0.1% Nonidet P-40; 5 μg of bovine serum albumin; and 1 μg of poly(dI-dC)-(dI-dC) as bulk carrier DNA. After addition of the DNA probe (80,000 cpm) and of various amounts of purified PecS protein, the reaction mixtures were incubated 30 min at 30°C and then loaded onto a 4% nondenaturing polyacrylamide gel and electrophoresed in low-ionic-strength buffer at pH 7.4 (2). Gels were then dried and exposed to Amersham MP film.
In vitro construction of ind::uidA fusions.
The uidA-Km cassettes (3) include a promoterless uidA gene that conserves its Shine-Dalgarno sequence. Insertion of this cassette in a gene in the correct orientation generates a transcriptional fusion. In addition, insertion of this cassette can cause polar effects on downstream genes in an operon through transcription termination. The EcoRI and SalI fragments from plasmids pSR996, pSR1158, and pSR1180, containing the region downstream of pecM, were mutagenized by introducing uidA-Km cassettes into various restriction sites (Fig. 1B). These different insertions were introduced into the E. chrysanthemi pecS chromosome by marker exchange recombination between the chromosomal allele and the plasmid-borne mutated allele. The recombinants were selected after successive cultures in low-phosphate medium in the presence of kanamycin, conditions under which pBR322 derivatives are very unstable (37). The indigoidine phenotype of the resulting mutants was then analyzed. We noticed that the indA::uidA mutant is also deficient in indB due to the polar effect of uidA-Km cassette insertion. Indeed, the indA mutation cannot be complemented by plasmid pSR1972, which harbors only the indA gene whereas it can be complemented in trans by the plasmid pR’S1 carrying indA-B-C.
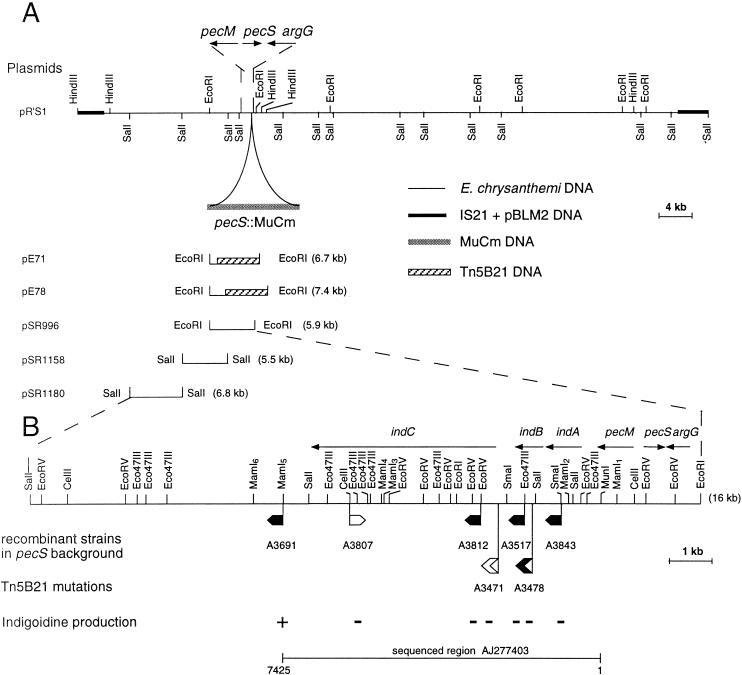
FIG. 1.
(A) Restriction map of plasmid pR’S1 harboring the argG, pecS::MudIIPR13, pecM, and indABC region of E. chrysanthemi 3937. (B) Insertion mutagenesis of the indABC region of E. chrysanthemi 3937. The sites of insertion of uidA-Km cassettes are indicated by small flags. Localization of the Tn5-B21 insertions is indicated by larger flags. In the sequenced region AJ277403, the exact positions of Tn5-B21 insertions are at nucleotide 1619 for A3478 and nucleotide 2286 for A3471. These positions were deduced from sequences obtained from plasmids pE71 and pE78 using primer Tn5 (5′GGGAAAGGTTCCGTTCAGGACG3′), which hybridizes at the end of transposon Tn5-B21. The transcription direction of the reporter gene is shown by the orientation of the flags. Where the reporter (uidA or lacZ) gene is expressed, the flag is black. The phenotypes of the mutants obtained after recombination of these insertions into the E. chrysanthemi pecS chromosome is described by the + or − signs. The deduced transcriptional organization of this DNA region is indicated by arrows.
Enzyme assays.
Assays of β-glucuronidase and β-galactosidase were performed on cell extracts treated with toluene. β-Glucuronidase activity was measured by monitoring the degradation of p-nitrophenyl-β-d-glucuronide into p-nitrophenol that absorbs at 405 nm (3). β-Galactosidase activity was measured by monitoring the degradation of o-nitrophenyl-β-d-galactoside into o-nitrophenol, at 420 nm (28). Specific activity for both enzymes is expressed as nanomoles of product liberated per minute per milligram (dry weight) of bacteria. For growth in synthetic medium, bacterial concentration was estimated by measuring turbidity at 600 nm given that an optical density at 600 nm of 1 corresponds to 109 bacteria · ml−1 and to 0.47 mg of bacteria (dry weight) · ml−1. For in planta expression of ind::uidA fusions, β-glucuronidase activities were determined in bacterial cells collected from inoculated plant leaves when the maceration appears in the inoculated area, as described previously (27). Concurrent numeration of the bacterial population present in infected leaves was performed by plating on LB agar medium supplemented with the appropriate antibiotic. To compare in planta expression and expression in synthetic medium, specific activity was also expressed as nanomoles of product liberated per minute per milligram (dry weight) of bacteria, given that 109 bacteria correspond to 0.47 mg of bacteria (dry weight).
Pathogenicity tests.
Pathogenicity tests on potted S. ionantha cv. Blue Rhapsody were performed as reported by Expert and Toussaint (14), with modifications. Bacterial cells grown on LB agar medium for 24 h at 30°C were suspended in an NaCl solution at 9 g/liter to give an optical density at 600 nm of 0.6. The number of viable bacteria in each suspension was determined by serial dilution and plating. About 100 μl of the resulting suspension (approximately 6 × 107 bacteria) was inoculated to one leaf per plant. Twelve plants were tested for each bacterial strain. Progression of the symptoms was scored daily for 14 days. The assay was carried out in triplicate.
Nucleotide sequence accession number.
The EMBL accession number for the indA-indB-indC sequence is AJ277403.
RESULTS AND DISCUSSION
Identification and localization of E. chrysanthemi indigoidine biosynthetic genes.
The pecS derivatives of the E. chrysanthemi strain 3937 produce indigoidine and appear blue on LB plates. The pecS strain A1524 was mutagenized with transposon Tn5-B21, which carries a promoterless lacZ gene and a tetracycline resistance gene (41). Out of 5,000 Tn5-B21 insertion mutants we selected two strains, A3471 and A3478, that appeared as white colonies and no longer synthesized indigoidine. Southern blotting with a Tn5 probe indicated that each mutant contains a single Tn5-B21 insertion (data not shown). These mutants were assayed for β-galactosidase activity. β-Galactosidase synthesis by strain A3478 indicated that the corresponding Tn5-B21 insertion resulted in a lacZ fusion. In contrast, the other mutant, A3471, lacked β-galactosidase activity. These two insertions were located on the chromosomal map of strain 3937 using plasmid pULB108, an RP4-derivative that can mediate the transfer of the host chromosome to polyauxotrophic recipient strains. The two insertions were found to be linked to the xyl-1 marker on the chromosomal map of strain 3937. This localization is close to the pecS-pecM locus. Using the generalized transducing phage PhiEC2, we detected 92% of cotransduction between these Tn5-B21 insertions and a pecS mutation. This result demonstrated that the indigoidine biosynthetic genes, ind, are in the vicinity of the pecS regulatory gene.
To identify DNA flanking Tn5-B21 insertions, chromosomal DNA of the two strains A3471 and A3478 was digested with EcoRI and then cloned into pUC18. After transformation into E. coli, plasmids from tetracycline-resistant colonies that contain a portion of Tn5-B21 were selected. Plasmids pE71 and pE78, originating from strains A3471 and A3478, respectively, were used for sequencing Tn5-flanking DNA. Tn5-B21 insertion from strain A3478 is situated in a gene whose product shows similarity to phosphatases involved in antibiotic synthesis. The sequence derived from Tn5-B21 insertion of strain A3471 showed no significant similarity to any protein in the databases. Since these two ind mutations were located near the pecS-pecM locus, we probed digests of the R-prime pR’S1 plasmid with the EcoRI fragments from pE71 and pE78. These hybridizations allowed us to localize the Tn5-B21 from strains A3478 and A3471, 1.5 and 2.2 kb downstream of pecM, respectively (Fig. 1A). To determine the extent of the indigoidine biosynthetic cluster, uidA-Kmr cassettes were introduced into various restriction sites of plasmids pSR996, pSR1158, and pSR1180 (Fig. 1B). These insertions were then recombined into the chromosome of the pecS strain A1524 by marker exchange, and the resulting mutants were analyzed for indigoidine production. All the insertions situated over a distance of 6.5 kb downstream of the pecM gene resulted in an indigoidine negative phenotype. The first insertion that did not abolish indigoidine production was localized at 7.3 kb, in the MamI5 site (Fig. 1B). Moreover uidA-Kmr insertions expressing β-glucuronidase, thus generating transcriptional ind::uidA fusions, were oriented in the same direction as pecM.
Sequence analysis of the ind genes.
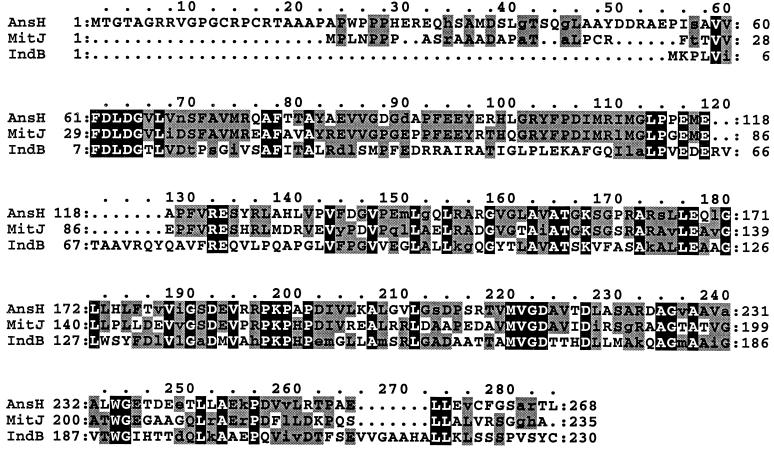
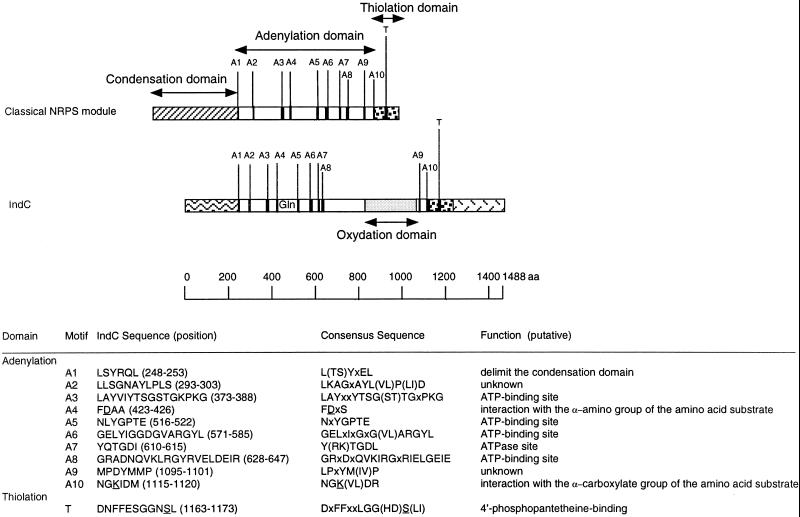
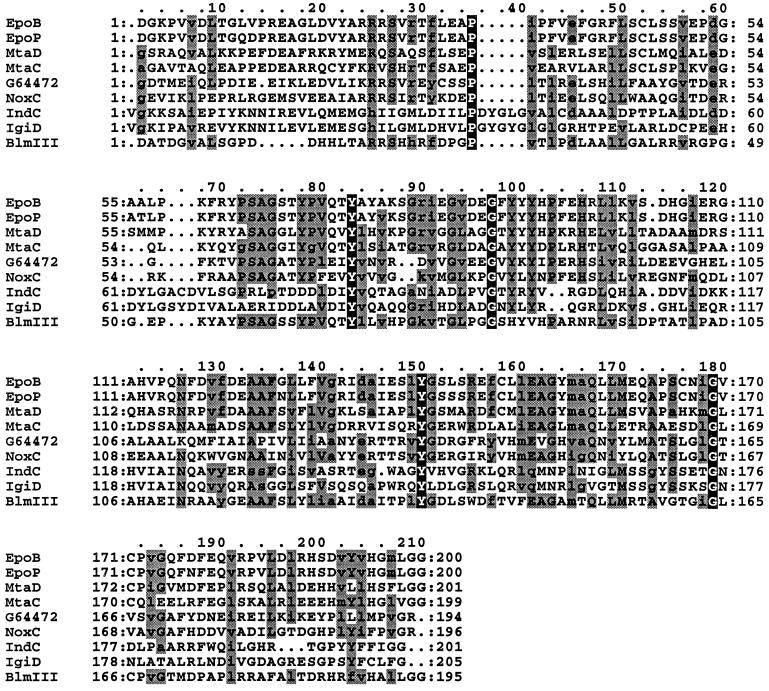
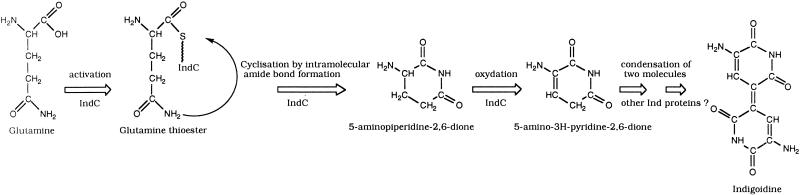
Analysis of the DNA sequence of the 7.4-kb MunI-MamI5 fragment revealed three open reading frames (ORFs) designated indA, indB, and indC situated downstream of pecM (EMBL entry AJ277403). The Tn5-B21 insertion from strain A3478 was located in the indB gene, whereas the Tn5-B21 insertion from strain A3471 was located in the indC regulatory region (Fig. 1B). During the transposition mechanism, this last insertion provoked a small deletion since Southern analysis revealed that the SmaI restriction site immediately upstream of the Tn5-B21 insertion was lost in strain A3471. The indA ORF is 948 nucleotides long and encodes a protein of 316 amino acids. The ATG initiation codon (position 463) is localized 345 nucleotides after the pecM stop codon (position 117) and is preceded by a putative ribosome-binding site (GAGAGAA) at nucleotide 452. A 46% amino acid sequence identity and 67% similarity were observed between IndA and the protein of unknown function, YeiN of E. coli (P33025). No other significant similarity could be detected with any protein in the databases searched. Consequently we could not attribute a function to IndA. The second ORF corresponding to indB is 690 nucleotides long and encodes a protein of 230 amino acids. indB is separated by 11 nucleotides from indA and probably constitutes an operon with indA. The ATG initiation codon (position 1425) is preceded by a putative ribosome-binding site (AAGGA) at nucleotide 1414 that overlaps the IndA stop codon. For IndB, the highest similarities, 51 and 48%, were found with two phosphatases, AnsH (AF131879) and MitJ (AF127374), respectively (Fig. 2). These phosphatases are involved in the synthesis of antibiotics: naphthomycin and ansatrienin in Streptomyces collinus and mitomycin C in Streptomyces lavendulae. The third ORF, corresponding to indC, is 4,464 nucleotides long and encodes a protein of 1,488 amino acids. The indB-indC intergenic region is 341 nucleotides long. The ATG initiation codon (position 2459) is preceded by a putative ribosome-binding site (GAGGA) at nucleotide 2449. IndC shows similarity to a wide variety of nonribosomal peptide synthetases (NRPSs). The highest score is obtained with the V. indigofera IgiD protein (AF088856), which is also involved in indigoidine biosynthesis (45% identity and 61% similarity). The involvement of a peptide synthetase in indigoidine biosynthesis suggests that an amino acid is the precursor of this pigment. The NRPSs are large proteins with one or more modules that employ the thiotemplate mechanism to activate and condense amino acids in an ordered manner (20). Each module represents a functional unit and acts as an independent enzyme to catalyze the sequential activation and condensation of one amino acid into the peptide product. The specific linear order of the modules within the NRPS determines the final sequence of the peptide product. These enzymes have been identified in bacilli, actinomycetes, and filamentous fungi. They catalyze the synthesis of peptides with interesting physiochemical or pharmacological characteristics, including biosurfactant, siderophore, antibiotic, antiviral, cytostatic, anticancer, and immunosuppressive properties (25). The most common module of NRPS characterized in bacteria is composed of an adenylation domain, a thiolation domain, and a condensation domain (Fig. 3). Amino acid recognition and activation occur by reaction with ATP within the adenylation domain. The resulting amino-acyl-adenylate is then covalently linked as its thioester to the enzyme-bound 4′-phosphopantetheine located within the thiolation domain. Then, it is transferred to another amino-acyl intermediate linked to the adjacent downstream module. This transfer is mediated by the condensation domain, which forms a peptide bond. IndC contains only one module with an adenylation and a thiolation domain. Within these domains a set of highly conserved signature sequences, known to be of functional significance for all investigated peptide synthetases, can be found (20) (Fig. 3). The sites responsible for discriminating the amino acid substrate are located between the motifs A4 and A5 of the adenylation domain (9, 42). The crystal structure of the adenylation domain from the gramicidin synthetase (GrsA), with l-phenylalanine and AMP bound, allowed the identification of 10 residues that interact with phenylalanine (11). Among these 10 residues, the D residue of the A4 motif interacts with the α amino group of the substrate amino acid and the K residue of the A10 motif interacts with the α carboxylate group of the amino acid. These two residues are highly conserved in all NRPSs. The other residues lining the substrate binding pocket are highly variant and are responsible for the specific amino acid recognition. Prediction of the eight amino acids lining the substrate binding pocket of IndC was performed at the web site at http://raynam.chm.jhu.edu/∼nrps/ (9). These amino acids are D424, A425, W428, C467, F491, G493, L516,and I524 and recognize glutamine. In the IgiD protein of Vogesella, the predicted amino acids are D, A, W, Q, F, G, L, and I, which also recognize glutamine. These signatures are very similar to that found within the glutamine adenylation domain DAWQFGLI from the second module of the tyrocidine synthetase TycC (9). Thus, the role of IndC or IgiD in indigoidine synthesis seems to be an activation of glutamine as its thioester. IndC does not contain a typical condensation domain, but alignment of IndC with various NRPSs reveals the presence of an additional region between the adenylation motifs A8 and A9. When this region of IndC (amino acids 860 to 1060) is used in a BLAST search, five NRPSs responsible for thiazoline to thiazole oxidation (EpoB and EpoP from Sorangium cellulosum, AF217189 and AF210843; MtaD and MtaC from Stigmatella aurantiaca, AF188287; BlmIII from Streptomyces verticillus, AF210249) and two proteins thought to be NADH oxidases (NoxC from Pyrococcus abyssi, E75214, and a hypothetical protein from Methanococcus jannaschii, G64472) are identified as having significant amino acid identity (Fig. 4). The corresponding domain of BlmIII was shown to contain one molar equivalent of noncovalently bound FMN as a prosthetic group (12), suggesting that this region corresponds to an oxidation domain. Based on these observations, we propose that IndC is responsible for glutamine cyclization and oxidation generating 5-amino-3H-pyridine-2,6-dione (Fig. 5). The condensation of two such molecules, probably by other Ind proteins (IndA-IndB or other unidentified Ind proteins), could then generate indigoidine.
FIG. 2.
Alignment of E. chrysanthemi IndB protein with phosphatases involved in antibiotic synthesis. The proteins represented are AnsH from S. collinus (AF131879) and MitJ from S. lavendulae (AF127374). The residues conserved in all the three proteins are in black boxes. The residues conserved in two of the three proteins are in grey boxes. Conservative substitutions are in lowercase letters. Dots denote gaps.
FIG. 3.
Structural organization of the IndC peptide synthetase. The relative locations of the highly conserved signature sequences found in adenylation (A1 to A10) and thiolation (T) domains are marked as stripes. Their amino acid sequences, in one-letter code, and their putative functions are indicated. Alternative amino acids for a particular position are shown in parentheses (x, any amino acid). The residues D and K underlined in motifs A4 and A10, respectively, mediate electrostatic interactions with the α-amino and α-carboxylate group of the amino acid substrate. The residue S underlined in motif T serves as the covalent attachment point for the 4′-phosphopantetheine cofactor of NRPS. This attachment is catalyzed by enzymes belonging to the superfamily of 4′-phosphopantetheine transferases. These enzymes promote the nucleophilic attack of the invariant serine hydroxy group to the pyrophosphate bridge of coenzyme A, resulting in a transfer of the 4′-phosphopantetheine cofactor to the T domain and a liberation of 3′,5′-ADP.
FIG. 4.
Amino acid comparison of putative oxidation domains. The amino acid positions of each putative oxidation domain are 1023 to 1222 for EpoB and EpoP from S. cellulosum, 1042 to 1242 for MtaD and 1083 to 1281 for MtaC from S. aurantiaca, 861 to 1061 for IndC from E. chrysanthemi, 665 to 869 for IgiD from V. indigofera, 734 to 928 for BlmIII from S. verticillus, 37 to 232 for NoxC from P. abyssi, and 2 to 195 for the hypothetical protein G64472 from M. jannaschii. The residues conserved in all proteins are in black boxes. The residues conserved in 60% of the proteins are in grey boxes. Conservative substitutions are in lowercase letters. Dots denote gaps.
FIG. 5.
Proposed model for indigoidine biosynthesis. IndC is responsible for glutamine activation as its thioester. Glutamine is then cyclized by the formation of an intramolecular amide bond. The resulting molecule, 5-aminopiperidine-2,6-dione, is then dehydrogenated, probably by the oxidation domain of IndC, to generate 5-amino-3H-pyridine-2,6-dione. Condensation of two such molecules by other Ind proteins could then generate indigoidine.
Comparison of indigoidine biosynthetic clusters of E. chrysanthemi and V. indigofera.
To our knowledge, no indigoidine biosynthesis pathway has been described in the literature. However, the nucleotide sequence of genes involved in indigoidine biosynthesis in V. indigofera are deposed in the EMBL database (AF088856). In V. indigofera, the indigoidine locus is composed of five genes, igiA, igiB, igiC, igiD, and igiE. Based on sequence similarities, it was proposed that the igiA gene encodes a putative 4′-phosphopantetheine transferase, igiB encodes a putative glutamine dehydrogenase, igiC encodes a putative N-carboxymaleamide decarboxylase, igiD encodes a product that corresponds to the peptide synthetase, and igiE encodes an ATPase component of an ABC transporter probably involved in indigoidine export.
The indigoidine loci of E. chrysanthemi and V. indigofera (AF088856) are rather different. Except for the peptide synthetase gene (indC and igiD), no other common genes have been found. Neither the phosphatase gene nor the indA homologue has been described in the V. indigofera indigoidine locus. This suggests that either the indigoidine biosynthesis pathways are different in these two bacteria or that the same set of genes exists in both bacteria but is organized differently, in separate loci. The latter hypothesis is highly possible since we have not performed a saturating mutagenesis of the E. chrysanthemi chromosome. Furthermore, the R-prime plasmid pR’S1 that contains the indA-indB-indC genes does not confer to E. coli the ability to produce indigoidine, suggesting that other genes required for pigment production are not present in this region or alternatively that ind genes are not efficiently transcribed in E. coli. V. indigofera also contains a regulatory couple (AF088857), homologous to the pecS-pecM system of E. chrysanthemi, suggesting that the control of indigoidine production is conserved between these two bacteria.
Modulation of indigoidine gene transcription.
Expression of the indA::, indB::, and indC::uidA transcriptional fusions was analyzed both in the E. chrysanthemi wild-type and pecS backgrounds and under various environmental conditions (e.g., carbon source, presence of Casamino Acids or plant extract, under oxidative stress, etc.) (Table 2). In the wild-type background, indC expression is very low. This low expression is probably responsible for the absence of indigoidine production in the wild-type strain. The expression of indC is increased by a factor of about 2,000 in the pecS mutant. The indA and indB genes are moderately expressed in the wild-type strain and derepressed 30-fold in the pecS mutant. The fact that both indA and indB show the same level of deregulation in the pecS background strengthened the hypothesis that these two genes are transcribed in an operon. On the contrary, the very low basal level of expression of indC and its high level of derepression in the pecS background suggest that indC constitutes a distinct transcriptional unit. As expected from the blue phenotype of the pecS mutant, the ind genes are thus strongly repressed by the PecS regulator. Expression of the ind genes was also determined during bacterial growth in glycerol M63 medium. We found that the expression of ind genes was roughly constant throughout the bacterial growth, both in the wild-type strain and in the pecS mutant (data not shown).
TABLE 2.
Expression of indA::, indB::, and indC::uidA fusions in wild-type and pecS backgroundsa
| Carbon source | Additional compound | β-Glucuronidase sp. act. (U) in fusion
|
|||||
|---|---|---|---|---|---|---|---|
|
indA::uidA
|
indB::uidA
|
indC::uidA
|
|||||
| Wild type | pecS background | Wild type | pecS background | Wild type | pecS background | ||
| Glycerol | 230 | 7,500 | 75 | 2,000 | 1 | 2,100 | |
| Galactose | 120 | 3,100 | 45 | 1,300 | 1 | 1,200 | |
| Glucose | 80 | 3,100 | 35 | 1,300 | 1 | 800 | |
| PGA | 135 | 2,400 | 25 | 800 | 1 | 1,200 | |
| Glycerol | Plant extract | 250 | 7,050 | 50 | 1,600 | 1 | 1,800 |
| Glycerol | Casamino Acids | 185 | 7,900 | 95 | 2,200 | 1 | 2,200 |
| Glycerol | Gln + B6 | 205 | 7,800 | 80 | 2,000 | 1 | 2,000 |
| Glycerol | Paraquat | 970 | 12,000 | 250 | 3,600 | 4 | 4,500 |
Cultures were grown in M63 minimal medium in the presence of various carbon sources and under different physiological conditions. Cultures were normally grown at 30°C. Carbon sources were added at 2 g · liter−1 except polygalacturonate (PGA), which was added at 4 g · liter−1. The following other compounds were added at the concentrations indicated: plant extract, 1% (vol/vol); Casamino Acids, 40 μg · ml−1; glutamine (Gln), 40 μg · ml−1, plus pyridoxine (vitamin B6), 200 μg ml−1; paraquat, 4 μM. The results reported are the average of three independent experiments; the standard deviation was less than 15% in each case except for low activities (<10 U), for which the standard deviation was greater (able to reach 40%).
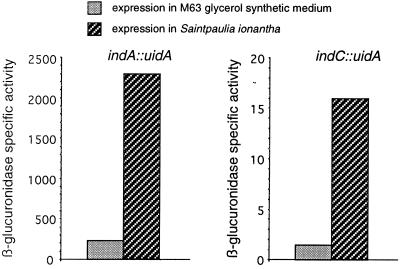
Analysis of indigoidine gene expression under various environmental conditions revealed that pigment production is influenced by the nature of the carbon source. Among the various carbon sources tested, glycerol allowed for the highest expression. The presence of an easily metabolizable carbon source, such as glucose or galactose, led to a two- to threefold repression of ind transcription. No induction of ind expression was observed in the presence of pectinase-inducing compounds, such as polygalacturonate, or plant extract. Similarly, no variation of expression was observed in the presence of Casamino Acids, and particularly in the presence of glutamine and pyridoxine. Moreover, addition of these two compounds to the growth medium of the pecS mutant did not increase indigoidine production (data not shown) in contrast to observations made from some Arthrobacter species (17). Finally, expression of ind genes is stimulated fourfold under oxidative stress, achieved by the addition of paraquat. Addition of paraquat also resulted in slow growth, and it seems possible that a low level of pigment production could be a general response to growth difficulty. The weak induction of indABC transcription observed under oxidative stress suggests that indigoidine could be produced during infection when bacteria must overcome the plant-generated reactive oxygen species to colonize the host (39). It has actually been reported that incompatible infection of tobacco cells by E. chrysanthemi strain 3937 leads to the secretion of a blue pigment (5). Expression of indA and indC genes was also analyzed during infection of Saintpaulia plants (Fig. 6). Transcription of these two genes was induced approximately 10-fold in planta. However, even during infection, the expression obtained in the wild-type background did not reach the high levels observed in the pecS mutant. Since PecS negatively autoregulates its own gene (32), the relief of PecS repression in the presence of the inducing signal(s) will also result in an increased production of PecS which, in turn, represses the target genes. This autoregulation of PecS could explain why the high ind expression levels observed in the pecS mutant can never been obtained in the wild-type background.
FIG. 6.
Expression of indA::uidA and indC::uidA fusions in Saintpaulia plants and in M63 glycerol synthetic medium. For in planta expression, β-glucuronidase activities were determined in bacterial cells collected from inoculated plant leaves when the first symptoms occurred. The data represent one of two separate experiments which gave similar results.
Direct interaction of the PecS regulator with the indA and indC regulatory regions.
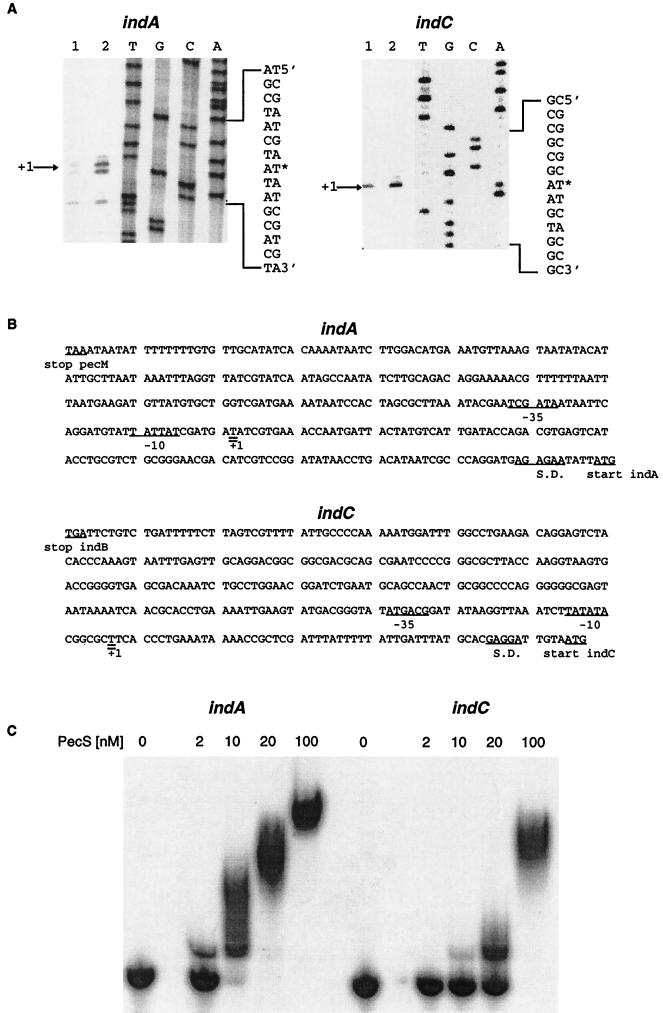
The differential regulation of indA and indC genes by the PecS repressor suggested that they constituted distinct transcriptional units. To confirm this hypothesis, we performed primer extension experiments to identify the promoter regions of these two genes (Fig. 7A). Because the level of ind transcription is low in the 3937 wild-type strain, mRNA was extracted from the pecS mutant, A1524, which exhibits increased ind transcription (Table 2). A single transcription start site was observed both for indA and indC (Fig. 7A). The 5′ ends of the indA and indC mRNAs were situated at positions −117 and −58 relative to the ATG start codon, respectively (Fig. 7B). Upstream of the indA transcriptional start site, we detected a −10 hexamer (TATTAT) presenting five out of six conserved positions with the −10 consensus of a ς70 promoter and a −35 region (TCGATA) matching the −35 consensus at four of the six positions (Fig. 7B). Similarly, sequences matching the ς70 consensus promoter were found immediately upstream of the indC transcriptional start site. The −10 region (TATATA) matches the −10 consensus at four of the six positions, and the −35 region (ATGACG) matches the −35 consensus at four of the six positions also (Fig. 7B).
FIG. 7.
(A) Identification of the E. chrysanthemi indA and indC transcription start site. RNAs (10 μg [lane 1] or 20 μg [lane 2]) from E. chrysanthemi A1524 cells grown on LB medium were submitted to primer extension analysis using indA- and indC-specific primers. DNA sequencing ladders were generated with the same primers (lanes A, C, G, and T). The nucleotide sequences of both the coding and noncoding strands are shown on the right. Arrows indicate the position of the specific transcription initiation sites (on the left). (B) Nucleotide sequences of the indA and indC regulatory regions. Sequences are numbered from the transcription start sites. The stop codon of the preceding gene, initiation codon, the ribosome-binding sites (S.D.), and regions corresponding to the −10 and −35 promoter sites are underlined. The transcriptional start sites are double underlined. (C) Interaction of the purified PecS protein with the E. chrysanthemi indA and indC regulatory regions. End-labeled and purified DNA fragments containing the indA and indC regulatory regions were incubated with various concentrations of the PecS protein.
Using gel shift assays, we demonstrated that PecS interacts with the indA and indC regulatory regions with high affinity (estimated Kd of 5 and 20 nM for indA and indC, respectively) (Fig. 7C). On the basis of previous quantitative titrations performed on genes whose expression is in vivo controlled or not by PecS (31), we feel confident in stating that the PecS interaction with ind genes is specific. Taken together, these results indicated that indA and indC constitute distinct transcriptional units and that the PecS repression of ind genes occurs via a direct mechanism.
Characterization of the oxidative stress tolerance of E. chrysanthemi pecS and ind mutants.
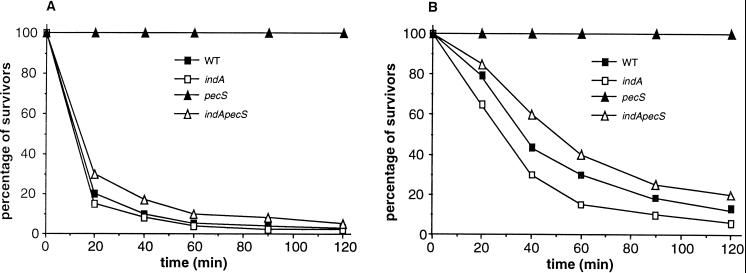
Based on the chemical structure of indigoidine, we speculated that it can act as a radical scavenger. This hypothesis is reinforced by the fact that indigoidine was discolored when submitted to the action of hydrogen peroxide in the presence of ferric sulfate, which generates hydroxyl radical (OH·) by the Fenton reaction. This result can be interpreted as the capture by indigoidine of the hydroxyl radical generated in the reaction medium resulting in the destruction of the conjugated II system which confers its blue color to indigoidine. To determine whether indigoidine could confer increased resistance to oxidative stress, we analyzed the survival of pecS and ind mutants under oxidative stress generated by the addition of 10 mM H2O2 to the LB medium (Fig. 8A). We found that the survival of the pecS mutant, in which indigoidine synthesis is derepressed, was significantly higher than that of the wild-type strain. Moreover the survival of the double pecS-indA mutant, which no longer produces indigoidine, was similar to that of the wild-type strain. These data demonstrate that indigoidine production increases tolerance to oxidative stress. We also analyzed the survival of ind mutants to oxidative stress after preinduction with sublethal concentrations of either paraquat or H2O2 (Fig. 8B). Globally, preinduction resulted in a weaker sensitivity to hydrogen peroxide of the wild-type, indA, and indA-pecS strains, indicating that the synthesis of different factors involved in bacterial defense against such a stress is induced in these three backgrounds. However, the indA mutant has lower tolerance than the wild-type strain, demonstrating that the weak induction of indigoidine synthesis under oxidative stress confers a significant advantage to the wild-type strain. Finally, the indA-pecS mutant displayed a higher resistance than the indA mutant, indicating that PecS exerts a regulatory influence over other genes involved in the resistance to oxidative stress.
FIG. 8.
Hydrogen peroxide sensitivities of the wild-type E. chrysanthemi 3937, pecS mutant A3953, indA mutant A3954, and the double indA-pecS mutant A3956. (A) Experiments were performed by incubating 3 × 109 bacteria with 10 mM hydrogen peroxide for the times indicated and determining viable cells by serial dilution and plating on LB agar. Survival of the H2O2-treated cells was normalized to the number of CFU at the beginning of the challenge. (B) The same experiments were performed with bacteria preincubated with either H2O2 (500 μM) or paraquat (40 μM). The data presented were obtained with H2O2 preinduction, but similar results were obtained with paraquat preinduction (data not shown). The data represent one of three separate experiments which gave similar results.
Pathogenicity of pecS and ind mutants.
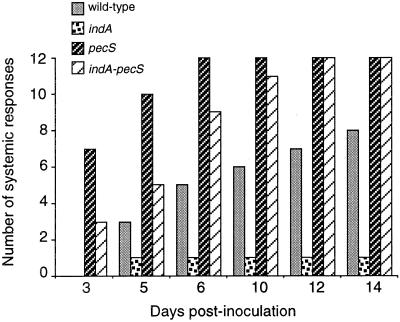
To determine whether production of indigoidine has a role in pathogenicity, we compared the ability of the wild-type strain (3937), the indA mutant (A3954), the pecS mutant (A3953) and the double pecS-indA mutant (A3956) to cause systemic disease on potted S. ionantha. Lesions were considered as systemic when at least one leaf with its petiole was macerated. We found that the timing of symptom evolution may fluctuate from one experiment to another, but similar results were obtained in all assays. In the assay presented (Fig. 9), the wild type produced systemic lesions in 8 of 12 plants inoculated after 14 days, whereas only 1 systemic response was observed with the indA mutant. These results indicate that indigoidine contributes to aggressiveness. Under the same conditions, the pecS mutant is much more aggressive than the wild-type strain since it produced systemic lesions in all the plants after 6 days only. The high aggressiveness of the pecS mutant probably results from the overproduction of extracellular enzymes and perhaps other virulence factors. With the double pecS-indA mutant, evolution of the symptoms was delayed compared to the pecS mutant, indicating that indigoidine production contributes to the aggressiveness of the pecS mutant. Finally, based on these results and those of oxidative stress tolerance, it seems reasonable to assess that indigoidine confers a significant benefit for E. chrysanthemi growth in planta by scavenging oxygen radicals. However, it is also possible that indigoidine displays other functions in pathogenicity.
FIG. 9.
Development of symptoms caused by E. chrysanthemi 3937 (wild type) and its indA, pecS, and indA-pecS derivatives on S. ionantha. The y axis indicates the number of plants (of 12 tested) with at least complete maceration of the inoculated leaf with its petiole. One representative experiment as described in Materials and Methods is shown. Other details are indicated in the text.
The increased pathogenicity of the pecS mutant more likely reflects several independent phenotypes, including extracellular macerating enzyme overproduction, higher resistance to oxidative stress and perhaps other unidentified factors also controlled by the PecS regulator. This protein belongs to a family of transcriptional regulators that includes MarR and EmrR, which regulate the expression of membrane efflux systems that confer resistance to a wide range of toxic compounds and also exert some regulatory influence over genes involved in resistance to oxidative stress (29). Interestingly, SlyA, another regulator of the MarR family whose corresponding gene was originally identified as being required for Salmonella enterica serovar Typhimurium virulence, was also shown to be required for resistance to oxidative stress encountered within the macrophage environment (8). Thus, it is tempting to speculate that PecS regulates critical genetic determinants which are required for adaptation to environmental stimuli encountered in the infected plants. The further characterization of the components of the PecS regulon promises to provide important insights into the different functions involved in host-pathogen interactions.
Acknowledgments
We thank N. Cotte-Pattat, V. Shevchik, and G. Condemine for their critical opinions and V. James for reading the manuscript. We are grateful to J. Ravel for help in determining the amino acids lining the substrate binding pocket of IndC. We thank the chemists A. Doutheau, C. Deshayes, and B. Chantegrel for helpful discussion about the chemical mechanism responsible for the radical scavenger function of indigoidine.
This work was supported by grants from the Centre National de la Recherche Scientifique and from the Ministère de l’Education Nationale et de la Recherche.
REFERENCES
- 1.Altschul, A. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 3.Bardonnet, N., and C. Blanco. 1992. uidA antibiotic resistance cassettes for insertion mutagenesis, gene fusion and genetic constructions. FEMS Microbiol. Lett. 93:243–248. [DOI] [PubMed] [Google Scholar]
- 4.Berlyn, M. K. B. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boccara, M., S. Tandon, and A. d’Harlingue. 1994. Studies of Erwinia chrysanthemi interactions with plant tissue culture cells, abstr. 176, p.58. In Proceedings of the Seventh International Symposium on Molecular Plant-Microbe Interactions, Edinburgh, Scotland, 26 June to 1 July 1994.
- 6.Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI-generated recombinant DNA molecules. Gene 4:121–136. [DOI] [PubMed] [Google Scholar]
- 7.Bolivar, F., R. L. Rodriguez, P. J. Green, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113. [PubMed] [Google Scholar]
- 8.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211–224. [DOI] [PubMed] [Google Scholar]
- 10.Collmer, A., and N. T. Keen. 1986. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24:383–409. [Google Scholar]
- 11.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16:4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, L., M. Chen, C. Sanchez, and B. Shen. 2000. An oxidation domain in the BlmIII non-ribosomal peptide synthetase probably catalyzing thiazole formation in the biosynthesis of the anti-tumor drug bleomycin in Streptomyces verticillus ATCC15003. FEMS Microbiol. Lett. 189:171–175. [DOI] [PubMed] [Google Scholar]
- 13.el Hassouni, M. E., J. P. Chambost, D. Expert, F. Van Gijsegem, and F. Barras. 1999. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. USA 96:887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Expert, D., and A. Toussaint. 1985. Bacteriocin-resistant mutants of Erwinia chrysanthemi: possible involvement of iron acquisition in phytopathogenicity. J. Bacteriol. 163:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franza, T., C. Sauvage, and D. Expert. 1999. Iron regulation and pathogenicity in Erwinia chrysanthemi 3937: role of the Fur repressor protein. Mol. Plant-Microbe Interact. 12:119–128. [DOI] [PubMed] [Google Scholar]
- 16.Giese, B. 1986. Radicals in organic synthesis. Pergamon Press, Oxford, United Kingdom.
- 17.Heumann, W., D. Young, and C. Gottlich. 1968. Leucoindigoidine formation by an Arthrobacter species and its oxidation to indigoidine by other micro-organisms. Biochim. Biophys. Acta 156:429–431. [DOI] [PubMed] [Google Scholar]
- 18.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213–257. [DOI] [PubMed] [Google Scholar]
- 19.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1985. Lactose catabolism in Erwinia chrysanthemi. J. Bacteriol. 162:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39–R48. [DOI] [PubMed] [Google Scholar]
- 21.Kotoujansky, A., M. Lemattre, and F. Boistard. 1982. Utilization of thermosensitive episome bearing transposon Tn10 to isolate Hfr donor strain of Erwinia carotovora subsp. chrysanthemi. J. Bacteriol. 150:122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn, R., M. P. Starr, D. A. Kuhn, H. Bauer, and H. J. Knackmuss. 1965. Indigoidine and other bacterial pigments related to 3, 3′-bipyridyl. Arch. Microbiol. 51:71–84. [DOI] [PubMed] [Google Scholar]
- 23.Levine, A., R. Tenhaken, R. Dixon, and C. Lamb. 1994. H2O2 from oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79:1–20. [DOI] [PubMed] [Google Scholar]
- 24.Maes, M., and E. Messens. 1992. Phenol as grinding material in RNA preparations. Nucleic Acids Res. 20:4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in non-ribosomal peptide synthesis. Chem. Rev. 97:2651–2673. [DOI] [PubMed] [Google Scholar]
- 26.Marrs, B. 1981. Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J. Bacteriol. 146:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masclaux, C., and D. Expert. 1995. Signalling potential of iron in plant-microbe interactions: the pathogenic switch of iron transport in Erwinia chrysanthemi. Plant J. 7:121–128. [Google Scholar]
- 28.Miller, J. H. 1972. Experiment in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441–448. [DOI] [PubMed] [Google Scholar]
- 30.Nasser, W., M. Bouillant, G. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 29:1391–1405. [DOI] [PubMed] [Google Scholar]
- 31.Praillet, T., W. Nasser, J. Robert-Baudouy, and S. Reverchon. 1996. Purification and functional characterization of PecS: a regulator of virulence factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 20:391–402. [DOI] [PubMed] [Google Scholar]
- 32.Praillet, T., S. Reverchon, and W. Nasser. 1997. Mutual control of the PecS/PecM couple, two proteins regulating virulence-factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 24:803–814. [DOI] [PubMed] [Google Scholar]
- 33.Resibois, A., M. Colet, M. Faelen, E. Schoonejans, and A. Toussaint. 1984. PhiEC2, a new generalised transducing phage of Erwinia chrysanthemi. Virology 137:102–112. [DOI] [PubMed] [Google Scholar]
- 34.Reverchon, S., D. Expert, J. Robert-Baudouy, and W. Nasser. 1997. The cyclic AMP receptor protein is the main activator of the pectinolysis genes in Erwinia chrysanthemi. J. Bacteriol. 179:3500–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1991. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol. Microbiol. 5:2203–2216. [DOI] [PubMed] [Google Scholar]
- 36.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1994. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 11:1127–1139. [DOI] [PubMed] [Google Scholar]
- 37.Roeder, D. L., and A. Collmer. 1985. Marker-exchange mutagenesis of pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Santos, R., T. Franza, M. L. Laporte, C. Sauvage, D. Touati, and D. Expert. 2001. Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol. Plant-Microbe Interact. 14:758–767. [DOI] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784–791. [Google Scholar]
- 41.Simon, R., J. Quandt, and W. Klipp. 1989. New derivatives of transposon Tn5 suitable for mobilisation of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene 80:161–169. [DOI] [PubMed] [Google Scholar]
- 42.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493–505. [DOI] [PubMed] [Google Scholar]
- 43.Starr, M. P., G. Cosens, and H. J. Knackmuss. 1966. Formation of the blue pigment indigoidine by phytopathogenic Erwinia. Appl. Microbiol. 14:870–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surgey, N., J. Robert-Baudouy, and G. Condemine. 1996. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J. Bacteriol. 178:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutherland, M. W. 1991. The generation of oxygen radicals during host plant response to infection. Physiol. Mol. Plant Pathol. 39:79–93. [Google Scholar]
- 46.Van Gijsegem, F., and A. Toussaint. 1982. Chromosome transfer and R-prime formation by an RP4::mini-Mu derivative in Escherichia coli, Salmonella typhimurium. Klebsiella pneumoniae and Proteus mirabilis. Plasmid 7:30–44. [DOI] [PubMed] [Google Scholar]
- 47.Viehe, H. G., R. Merenyi, L. Stella, and Z. Janousek. 1979. Capto-dative substituent effects in syntheses with radicals and radicophiles. Angew. Chem. Int. Ed. Engl. 18:917–932. [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]