Abstract
Proteome analysis of Bacillus subtilis cells grown at low and high salinity revealed the induction of 16 protein spots and the repression of 2 protein spots, respectively. Most of these protein spots were identified by mass spectrometry. Four of the 16 high-salinity-induced proteins corresponded to DhbA, DhbB, DhbC, and DhbE, enzymes that are involved in the synthesis of 2,3-dihydroxybenzoate (DHB) and its modification and esterification to the iron siderophore bacillibactin. These proteins are encoded by the dhbACEBF operon, which is negatively controlled by the central iron regulatory protein Fur and is derepressed upon iron limitation. We found that iron limitation and high salinity derepressed dhb expression to a similar extent and that both led to the accumulation of comparable amounts of DHB in the culture supernatant. DHB production increased linearly with the degree of salinity of the growth medium but could still be reduced by an excess of iron. Such an excess of iron also partially reversed the growth defect exhibited by salt-stressed B. subtilis cultures. Taken together, these findings strongly suggest that B. subtilis cells grown at high salinity experience iron limitation. In support of this notion, we found that the expression of several genes and operons encoding putative iron uptake systems was increased upon salt stress. The unexpected finding that high-salinity stress has an iron limitation component might be of special ecophysiological importance for the growth of B. subtilis in natural settings, in which bioavailable iron is usually scarce.
The soil bacterium Bacillus subtilis often experiences fluctuations in the osmolality of its habitat due to the drying and flooding of the upper layers of the soil (7, 40). A rise in the external salinity and osmolality triggers the outflow of water from the cell, resulting in a reduction in turgor and dehydration of the cytoplasm. To cope with these unfavorable osmotic conditions, B. subtilis initiates a two-step adaptation response (7, 32). Initially, K+ is rapidly taken up (56) and subsequently replaced in part by proline (55), a member of the so-called compatible solutes (19). These osmolytes can be accumulated to high levels through either de novo synthesis or uptake of preformed osmoprotectants from the environment without interfering with central cellular functions. For B. subtilis, proline serves as the primary endogenously synthesized compatible solute (55), and large quantities are produced via a dedicated osmostress-responsive synthesis pathway (J. Brill and E. Bremer, unpublished results). In addition, B. subtilis can efficiently scavenge a wide variety of compatible solutes from environmental sources by means of five osmoregulated transport systems (OpuA to OpuE) (29–31, 41, 54) and can acquire choline for the production of the osmoprotectant glycine betaine (4, 30). The accumulation of compatible solutes offsets the detrimental effects of high osmolality on cell physiology and permits growth over a wide range of osmotic conditions (3, 36).
Under situations where the osmotic stress is so strong that growth is no longer permitted, a general, unspecific, and preemptive stress response system is engaged to ensure the survival of B. subtilis. Induction of this large general stress regulon is governed by the alternative transcription factor SigB (21, 26, 43). Salt (5, 52) and osmotic stress (A. Völker, unpublished results) are among the environmental cues that cause SigB activation and transient induction of the entire SigB-regulon. The survival of a sigB mutant that lacks the general stress response is severely impaired during growth-preventing salt stress (53), but it is unknown which member(s) of the SigB regulon are responsible for the protection from this environmental challenge. Interestingly, there is an overlap between the SigB-controlled general stress response system and the stress adaptation reactions that deal specifically with osmotic stress. Two (OpuD and OpuE) of the five osmoprotectant uptake systems are members of the SigB regulon, and their osmotically controlled transcription is partially dependent on promoters recognized by SigB (49, 54; F. Spiegelhalter and E. Bremer, unpublished results).
The combination of specific and general stress responses enables B. subtilis to cope with both growth-restricting and growth-preventing osmotic and high-salinity stress. Both responses are important for the survival of B. subtilis, since its prime defensive strategy, sporulation, is severely impaired by high salinity (35, 47). The cellular response to high osmolality and salinity is not limited to the adaptation systems described so far because high salinity exerts pleiotropic effects on the physiology of B. subtilis. Mutants lacking the alternative transcription factor SigM are sensitive to high salt concentrations (28), but this may be an indirect phenotype related to the major cell wall defects exhibited by such mutants. Increases in salinity affect the phospholipid composition of the cytoplasmic membrane (37), the properties of the cell wall (38), and the synthesis of the cell-wall-associated protein WapA (13). In addition, the production of several extracellular degradative enzymes is regulated in a DegS/DegU-dependent manner at high salinity (35). Finally, under such growth conditions one observes changes in the supercoiling of reporter plasmids (1, 33) and the transient induction of the ftsH gene, which encodes an ATP-dependent, membrane-associated protease (14).
The adaptation of the B. subtilis cell to salt stress and osmotic challenges apparently has many facets. Insights into the specific adaptation to high-osmolality environments were mainly achieved by a genetic and function-based approach that led to the identification of genes encoding systems involved in K+ uptake and the synthesis and transport of compatible solutes (7, 32). In contrast, most of the members of the general stress regulon have been identified by global approaches, namely proteome analysis and transcriptional profiling (26, 43). The latter strategy yielded a large number of proteins and genes with undefined functions but provided a global view of the general stress regulon and thereby opened the possibility of a detailed functional analysis of each of its members.
Employing such a global strategy for the characterization of the adaptational network of B. subtilis for high salinity and osmolality is likely to reveal new insights into the constraints that are imposed on the cell by such growth conditions. Utilizing a two-dimensional electrophoresis approach, we have compared the protein profile of cells continuously growing under various saline conditions and found that high salinity causes iron limitation in B. subtilis, thereby triggering the derepression of a variety of iron-controlled genes.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Strain JH642 (trpC2 pheA1 sfp0; BGSC 1A96; a kind gift from J. Hoch) is a derivative of the wild-type B. subtilis strain 168. Strain TMB1 was constructed by transforming the fur::kan allele from strain HB2501 (fur::kan trpC2 attSPβ; J. Helmann, unpublished results) into JH642. Strain KE10 [Φ(yckH-comS-erm-ycxA) sfp+] is a derivative of the B. subtilis strain ATCC 21332 and has been described (15). Bacteria were grown in Spizizen’s minimal medium (SMM) with 0.5% (wt/vol) glucose as the carbon source and supplemented with l-tryptophan (20 mg/liter), l-phenylalanine (18 mg/liter), and a solution of trace elements (25). For experiments that required a defined iron concentration, the modified minimal medium (MM) described by Chen et al. (11) was used, and iron was supplied to the cells from a freshly prepared 20 mM stock solution of FeCl3 (Merck; Darmstadt, Germany) to a final concentration indicated in the individual experiments. Cells were routinely grown aerobically at 37°C in 100-ml Erlenmeyer flasks with a culture volume of 20 ml in a shaking water bath set at 220 rpm. The growth of the bacterial cultures was monitored spectrophotometrically at a wavelength of 578 nm (OD578). The salinity of bacterial cultures propagated in MM was raised by adding appropriate volumes of NaCl from a 5 M stock solution. The osmolality values of these media were determined with a vapor pressure osmometer (model 5500; Wescor); the osmolality value of SMM was 340 mosmol/kg of water, and that of the modified MM was 260 mosmol/kg of water. Cultures used for the preparation of cell extracts for two-dimensional (2D) gel analysis were grown in 250 ml of SMM of various osmolalities in 1-liter Erlenmeyer flasks on a rotary shaker set at 220 rpm at 37°C; the cells were harvested by centrifugation when they had reached an OD578 of approximately 0.5. These cultures were inoculated from overnight cultures preadapted to a particular salinity. Some of the cultures contained a 1 mM concentration of the osmoprotectant glycine betaine as a supplement.
Siderophore assay.
For the quantitation of 2,3-dihydroxybenzoate (DHB), bacteria were grown at 37°C overnight in 3 ml of iron-free modified MM containing different amounts of FeCl3 in 15-ml glass tubes. Siderophore levels were determined in the supernatant of the overnight cultures by using a previously described assay (11). The concentration of DHB was normalized to cell mass by dividing the measured DHB level (OD510) by the density of the culture (OD600). All results presented are the averages for two to four independent experiments.
2D protein gel electrophoresis.
After harvesting the bacteria by centrifugation, cells were washed in TE (10 mM Tris, 1 mM EDTA [pH 7.5]) and the cell pellet was resuspended in the same buffer. Cells were disrupted by several passages through a French pressure cell, and cell debris was removed by centrifugation at 4°C and 20,000 × g for 30 min. The protein concentration of the supernatant fraction was assayed according to the method of Bradford (6). For analytical and preparative 2D protein gel electrophoresis, 100 or 400 μg of crude protein extract was solubilized in a rehydration solution containing 8 M urea, 2 M thiourea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 28 mM dithiothreitol (DTT), 1.3% (vol/vol) Pharmalytes, pH 3 to 10, and bromphenol blue. After rehydration in the protein-containing solution for 24 h under low-viscosity paraffin oil, Immobilon dry strip gels (IPG) (Amersham Pharmacia Biotech) covering a pH range from 4 to 7 were subjected to isoelectric focusing. The following voltage/time profile was used: a linear increase from 0 to 500 V for 1,000 Vh, 500 V for 2,000 Vh, a linear increase from 500 to 3,500 V for 10,000 Vh, and a final phase of 3,500 V for 35,000 Vh. IPG strips were consecutively incubated for 15 min each in equilibration solutions A and B (50 mM Tris-HCl [pH 6.8[, 6 M urea, 30% [vol/vol] glycerol, 4% [wt/vol] sodium dodecyl sulfate, and 3.5-mg/ml DTT [solution A] or 45-mg/ml iodoacetamide instead of DTT [solution B]). In the second dimension, proteins were separated on 12.5% sodium dodecyl sulfate-polyacrylamide gels with the Investigator System (Perkin-Elmer Life Sciences, Cambridge, United Kingdom) at 2 W/gel. Analytical and preparative gels were stained with silver nitrate according to the method of Bloom et al. (2) or with PhastGel BlueR according to the manufacturer’s instructions (Amersham Pharmacia Biotech), respectively. After scanning was done, analysis of the 2D polyacrylamide gel electrophoresis images was performed with the Melanie3 software package (Bio-Rad Laboratories GmbH). Three separate gels of each condition were analyzed, and only spots displaying the same pattern in all parallels were selected for further characterization.
Protein identification by peptide mass fingerprinting.
Protein spots were excised from PhastGel BlueR stained 2-D gels, destained, and digested with trypsin (Promega GmbH); peptides were then extracted according to the method of Otto et al. (42). Peptide mixtures were purified with C18 tips according to the manufacturer’s instructions (Millipore GmbH) and eluted with 50% (vol/vol) acetonitril−0.1% (vol/vol) trifluoroacetic acid. For mass spectrometric analysis, peptide solutions were mixed with an equal volume of saturated α-cyano-3-hydroxycinnamic acid solution in 50% (vol/vol) acetonitril−0.1% (vol/vol) trifluoroacetic acid and applied to a sample template of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometer. Peptide masses were determined in the positive ion reflector mode in a Voyager DE RP mass spectrometer (Applied Biosystems) with internal calibration. Mass accuracy was usually in the range between 30 to 100 ppm. Peptide mass fingerprints were compared to databases (http://prospector.ucsf.edu) using the program MS-Fit. The searches considered oxidation of methionine, pyroglutamic acid formation at the N-terminal glutamine, and modification of cysteine by carbamidomethylation as well as partial cleavage, leaving a maximum of one internal site uncleaved.
Transcriptional analysis.
Total RNA from cells of strain JH642 grown to mid-exponential phase (OD578 of approximately 0.8) was isolated according to the acidic phenol method (52). One microgram of total RNA was dotted onto a nylon membrane (Nytran N; Schleicher & Schuell) using a dot blot apparatus (Bio-Rad Laboratories GmbH). For the detection of specific B. subtilis genes, the dot blotted total RNA was hybridized to digoxigenin-labeled RNA probes and the membranes were processed and developed according to the procedures provided by the manufacturer (Roche Diagnostics GmbH). Hybridization was performed at 68°C using a hybridization solution containing 50% (vol/vol) formamide. Digoxigenin-labeled RNA antisense probes were prepared by runoff transcription of PCR templates containing the T7-RNA polymerase promoter sequence using a kit purchased from Ambion Inc. The following oligonucleotide primers were used for PCR template preparation: dhbA (5′-GGCTGCCAAGGAATAGG-3′, 5′-TAATACGACTCACTATAGGGAGGCAAGCTCAAGGCCAAGGC-3′); dhbF (5′-CCGGGGCGCAAACTGG-3′, 5′-TAATACGACTCACTATAGGGAGGGAGACCCAAAGGAACGGC-3′); feuA (5′-CGCGCTGACGGCGGC-3′, 5′-TAATACGACTCACTATAGGGAGGGTCAGCTGGGCAAGAAGC-3′); fhuD (5′-GCAGCGCTGGCAGCC-3′, 5′-TAATACGACTCACTATAGGGAGGTTTATCCCACTTGGCCAGCC-3′); fhuB (5′-GGCGCGGTCATCGTCC-3′, 5′-TAATACGACTCACTATAGGGAGGGTAACTGCCGTACCCGCC-3′); yfmC (5′-CTGCCTCATTGTATCCGGC-3′, 5′-TAATACGACTCACTATAGGGAGGGCCGAGAACGATGTTGCGG-3′). The intensity of the hybridization signal was quantified using a Storm860 fluorescence imager and the ImageQuant software package (Amersham Pharmacia Biotech).
Computer-aided searches for Fur boxes.
Fur binding sequences have been identified in several microorganisms by footprinting experiments. These experiments and further in vitro studies with synthetic binding sites have revealed that Fur recognizes the short hexamer GATAAT. Several copies of this sequence are required for effective binding of Fur to DNA, but their orientation seems to be unimportant (17, 18, 24). The ATAAT motive appears at high frequencies in naturally occurring Fur sites, but the intervening G or C residues are less conserved (16). Therefore, the B. subtilis genome sequence (34) was searched for the occurrence of adjacent pentamers described above with the program MotivFinder from Decodon GmbH (Greifswald, Germany). This search was restricted to a region from bp −200 to +100 with respect to the initiation codon of open reading frames indicated in the annotated version of the B. subtilis genome sequence (http://genolist.pasteur.fr/SubtiList/). Potential −10 (TATAAT) and −35 (TTGACA) regions of SigA-type promoters (27) in the vicinity of potential Fur boxes were identified after visual inspection of the DNA sequence. The promoter sequence and Fur-binding regions of the dhb operon have previously been experimentally characterized (8, 11).
RESULTS
Salt-induced changes in the protein profile of B. subtilis.
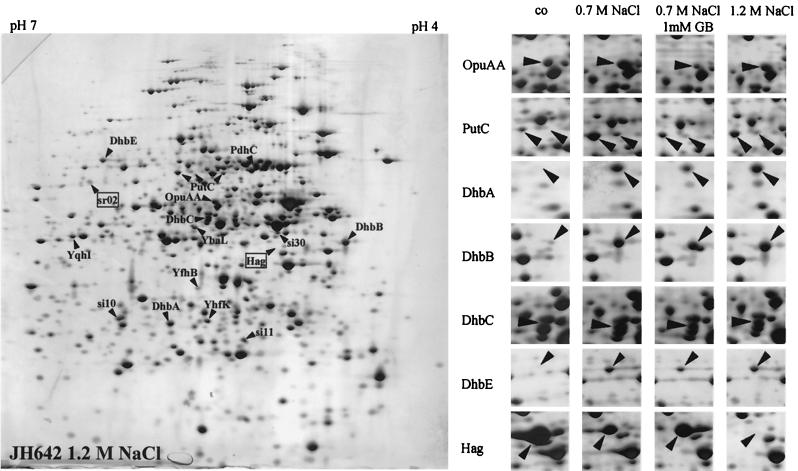
We prepared crude protein extracts of soluble proteins from B. subtilis strain JH642 grown either in SMM or in minimal medium with increased salinity and osmolality (0.7 or 1.2 M NaCl) and separated them by 2D gel electrophoresis. A comparison of the protein profiles of the silver-stained gels identified 16 protein spots that were present in significantly increased amounts when cells where grown in high-salt medium. Two protein spots were found that were reduced under high-salt growth conditions. An example of such 2D gels is shown in Fig. 1. All of these protein spots were selected for further analysis, excised from the gel, and subjected to peptide mass fingerprinting as detailed in the Materials and Methods section. All but four protein spots could be identified, and a summary of their identification is presented in Table 1. Among the high-salinity-induced proteins was the ATPase (OpuAA) of the osmotically regulated OpuA ABC transport system for the uptake of the compatible solute glycine betaine (31). We also found three different isoforms of an enzyme (YcgN/PutC) involved in proline degradation. The ycgN (putC) structural gene is part of the ycgMNO (putBCP) operon, whose products are responsible for the uptake (PutP) of proline and its enzymatic degradation (PutBC) to glutamate. Although PutC levels are increased in cultures grown under high-salinity conditions, increased PutC production is not the result of high salinity but rather is a consequence of proline leaking from B. subtilis cells grown under high-osmolality conditions (S. Moses and E. Bremer, unpublished results). The group of high-osmolality-induced proteins with known function included PdhC, a subunit of the glycolytic enzyme pyruvate dehydrogenase. Furthermore, four proteins (YbaL, YfhB, YhfK, and YqhI) with thus far undefined physiological function were identified as high-salinity-induced polypeptides (Table 1). In agreement with the transient induction of the SigB-dependent general stress response (5, 52) the level of general stress proteins did not significantly differ between the B. subtilis cells grown at high and low salinity.
FIG. 1.
Influence of salinity on the protein profile of the B. subtilis strain JH642. Bacteria were grown in SMM or SMM supplemented with either 0.7 M NaCl, 1.2 M NaCl, or 0.7 M NaCl and 1 mM glycine betaine (GB). Crude protein extracts were prepared and separated by 2D gel electrophoresis. After being stained with silver nitrate, the gels were scanned with an imaging system and analyzed with the Melanie 3.0 software package. Protein spots induced or repressed by high salinity are marked with arrowheads or boxes, respectively. Proteins that were identified by peptide mass fingerprinting are labeled with their gene names. The image in the left section of the figure displays a gel obtained with an extract from cells grown in the presence of 1.2 M NaCl. The right part of the figure displays selected regions of gels prepared with extracts from cells grown under the conditions indicated above the columns. sr, salt-repressed protein; si, salt-induced protein; co, control.
TABLE 1.
Summary of salt-responsive proteins identified by peptide mass fingerprint matching
| Protein | pIa | mwa (kDa) | Function | SwissProt acc. no.b | MOWSE scorec | Sequence coverage (%)c |
|---|---|---|---|---|---|---|
| DhbA | 5.4 | 27.5 | 2,3-Dihydro-2,3-dihydroxybenzoate dehydrogenase | P39071 | 2.0e+04 | 27 |
| DhbB | 4.6 | 35.1 | Isochorismate lyase/peptidyl carrier protein | P45743 | 4.6e+12 | 48 |
| DhbE | 5.7 | 59.9 | 2,3-Dihydroxybenzoate adenylase | P40871 | 9.0e+07 | 38 |
| Hag | 4.9 | 32.6 | Flagellin | P02968 | 4.5e+08 | 50 |
| OpuAA | 5.2 | 46.4 | Glycine betaine ABC transporter (ATP-binding protein) | P46920 | 2.4e+06 | 30 |
| PdhC | 5.0 | 47.5 | Pyruvate dehydrogenase (dihydrolipoamide acetyltransferase E2 subunit) | P21883 | 2.5e+09 | 33 |
| PutC | 5.5 | 56.5 | 1-Pyrroline-5-carboxylate dehydrogenase | 6.9e+07 | 42 | |
| YbaL | 5.3 | 38.6 | Similar to ATP-binding Mrp-like protein | P50863 | 2.1e+04 | 40 |
| YfhB | 5.3 | 32.1 | Similar to proteins of unknown function | 4.0e+06 | 53 | |
| YhfK | 5.3 | 22.8 | Similar to proteins of unknown function | 5.7e+07 | 80 | |
| YqhI | 5.8 | 39.8 | Similar to aminomethyltransferase | P54378 | 9.5e+06 | 33 |
The theoretical isoelectric point and molecular mass were calculated with the Compute pI/mw tool of the proteomics tools collection at the ExPASy Molecular Biology Server (http://www.expasy.ch/tools/pi tool.html).
acc., accession.
MOWSE score and sequence coverage were indicated in order to facilitate judgement of the reliability of the data.
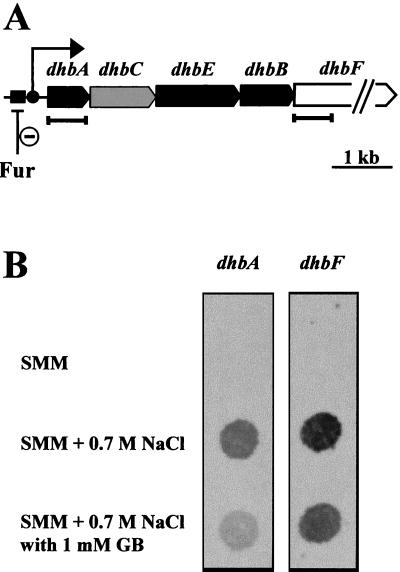
Three of the spots (DhbA, DhbB, and DhbE) whose intensity increased most significantly in cells grown in high salinity (Fig. 1) turned out to be encoded by three genes of the five-member dhbACEBF operon (Fig. 2A), which serves for the synthesis of the iron siderophore bacillibactin (39, 45, 46). A fourth product (DhbC) of this gene cluster was subsequently identified by comparison of its migration position with the current version of the 2D-protein index of B. subtilis (10). DhbC corresponded to one of the remaining high-salinity-induced proteins that were not identified by peptide mass fingerprinting in this study (Fig. 1). The fifth gene product (DhbF) of the dhb operon (Fig. 2A) was not identified, most likely due to its large molecular mass (39).
FIG. 2.
Induction of the dhb operon by salt. (A) Structure of the dhbACEBF operon. Genes whose products were identified by peptide mass fingerprinting are marked in black. The dhbC (labeled in gray) gene product was identified by comparison with a 2D reference map of B. subtilis (10); the dhbF gene product was not detected on our 2D gels. Segments of the dhbA and dhbF genes used as probes in hybridization experiments are indicated. (B) Transcription of the dhbA and dhbF genes in response to increased salinity in the absence or presence of the osmoprotectant glycine betaine (GB).
One of the two protein spots whose amount was reduced in cells propagated in high-salinity medium was characterized as the hag gene product, the structural protein of the flagellum. This protein was almost absent at very high salinity (SMM with 1.2 M NaCl) (Fig. 1), implying that the motility of B. subtilis cells is severely impaired under high-saline growth conditions.
Induction of dhb transcription by increased salinity.
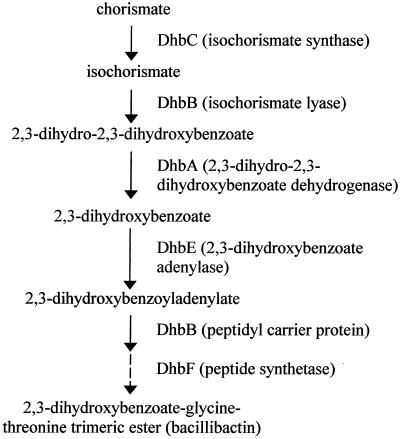
A surprising finding of the 2D gel analysis of B. subtilis cells was the considerable accumulation of the DhbA, DhbB, DhbC, and DhbE proteins upon growth at high salinity and osmolality (Fig. 1). These enzymes together with DhbF constitute the synthesis pathway for DHB from chorismate, its modification with the amino acids glycine and threonine, and the esterification of three of these intermediates into the iron chelator bacillibactin (Fig. 3) (39). These later reactions are performed by a modular nonribosomal peptide synthetase complex (DhbE, DhbB, and DhbF). For the functioning of this complex, phosphopantetheinyl residues are loaded onto the DhbB and DhbF enzymes by the Sfp protein, and the modified DhbB and DhbF enzymes then serve as carriers for DHB modification and its esterification into bacillibactin (39). The involvement of bacillibactin in iron acquisition is underscored by the regulation of the dhb operon by the central iron control protein Fur (Fig. 2A) (8). Therefore, expression of the dhb operon should be repressed in cells grown in SMM, since this minimal medium contains sufficient amounts of iron (approximately 5 μM) to satisfy the cells’ need for this trace metal (11). Indeed, the DhbA, DhbB, DhbC, and DhbE proteins were produced in small amounts under these growth conditions; however, they were present in substantial quantities when the cells were subjected to salt stress (Fig. 1). These observations therefore suggest that salt-stressed B. subtilis cells somehow experience an iron limitation.
FIG. 3.
Synthesis pathway of the iron siderophore bacillibactin. Production of bacillibactin starts with chorismate and proceeds through the enzymatic actions of the DhbC, DhbB, and DhbA proteins to DHB. This intermediate has a weak iron siderophore activity (48) and is activated by DhbE-mediated adenylation. A modular peptide synthetase then modifies the resulting 2,3-dihydroxybenzoyladenylate through the addition of glycine and threonine residues and finally esterifies three of these intermediates to form bacillibactin (39).
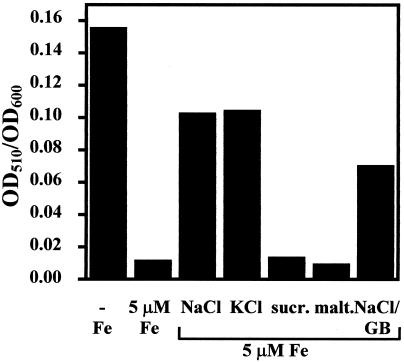
We assayed dhb expression in response to salt stress by monitoring the mRNA levels of the first (dhbA) and the last (dhbF) gene of the dhbACEBF operon in dot blot experiments. Salt stress triggered a strong increase in the level of the transcription of both genes. The presence of a 1 mM concentration of the osmoprotectant glycine betaine in the growth medium moderately decreased dhbA and dhbF expression (Fig. 2B), consistent with the protein profile we observed on the 2D gels (Fig. 1). We also quantitated DHB in culture supernatants of cells that were subjected to either iron limitation or salt stress. Iron limitation triggered a strong induction (14-fold) of DHB production (Fig. 4), a finding that is in full agreement with previously published data and the known control of dhb expression by Fur (8, 11). DHB production was also strongly enhanced (11-fold) by the addition of 0.7 M NaCl to a minimal medium containing 5 μM iron, a concentration that is sufficient to strongly repress DHB synthesis in non-salt-stressed B. subtilis cells (Fig. 4). Derepression of DHB production was not specific to NaCl, since iso-osmolar concentrations of KCl (Fig. 4), Na2SO4, and K2SO4 (data not shown) triggered the accumulation of similar levels of DHB in culture supernatants of strain JH642. While various salts led to increased DHB synthesis, the addition of iso-osmolar concentrations of the sugars sucrose and maltose were no inducers for DHB production (Fig. 4). Hence, derepression of the dhb operon is a salt-specific effect rather than an osmotic effect and appears to be independent of the cation or anion used to increase the salinity of the growth medium. As observed for the transcription of the dhbA and dhbF genes (Fig. 2B), the presence of the osmoprotectant glycine betaine in NaCl-stressed cultures moderately down-regulated the salt-dependent derepression of DHB production (Fig. 4).
FIG. 4.
Salt-induced synthesis of DHB. Supernatants of overnight cultures of strain JH642 were analyzed for DHB. The cells were grown either in iron-free medium or in iron-free medium supplemented with 5 μM FeCl3. The cells were grown in either the absence or presence of iso-osmotic concentrations of NaCl (0.7 M), KCl, sucrose (sucr.), maltose (malt.), or NaCl with 1 mM glycine betaine (NaCl/GB). The data presented are the averages for two independent experiments.
Derepression of DHB production in salt-stressed cultures is reversed by excess iron.
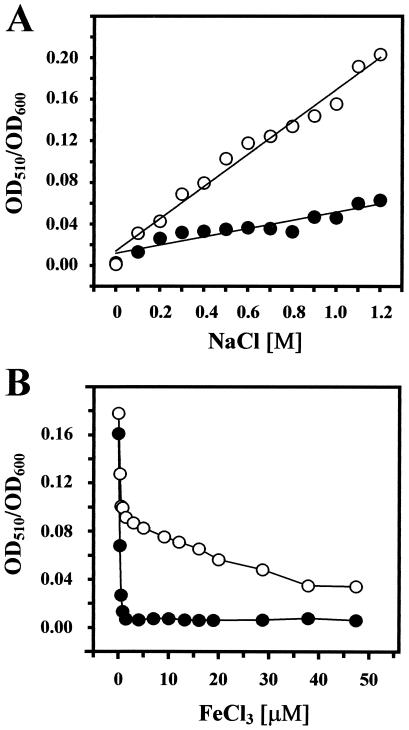
To investigate the derepression of the dhb operon by salt stress in more detail, we monitored DHB accumulation in supernatants of cultures of strain JH642 whose salinity was systematically varied. Increasing the salinity of the growth medium that contained 5 μM FeCl3 triggered an increase in DHB levels, resulting in a linear relationship between the concentration of salt in the growth medium and the amount of DHB produced (Fig. 5A). Derepression of DHB production by salt was largely reversed by the addition of excess iron (50 μM FeCl3) to the salt-containing medium (Fig. 5A). We subsequently investigated the influence of iron on DHB production by varying the iron concentration in the growth medium of cells grown in the absence or presence of salt. As observed previously (11), approximately 2 μM FeCl3 was sufficient to repress the synthesis of DHB to a basic level of 4% of the fully induced level. In contrast, in the presence of 0.7 M NaCl in the growth medium, the same iron concentration resulted in only a twofold repression (Fig. 5B). Taken together, these data attest to a finely tuned derepression of DHB production by increased salinity and demonstrate that excess iron can reverse the derepressing effect on DHB synthesis under high-salt growth conditions.
FIG. 5.
Modulation of DHB production by salt and iron. (A) Cells of strain JH642 were grown overnight in the modified MM of Chen et al. (11) containing either 5 μM FeCl3 (open circles) or 250 μM FeCl3 (closed circles), and DHB was assayed in the culture supernatant. (B) Cells of strain JH642 were grown in modified MM containing the indicated concentrations of FeCl3 in the absence (closed circles) or the presence (open circles) of 0.7 M NaCl, and culture supernatants were subsequently assayed for DHB.
The growth defect of salt-stressed cells is partially corrected by excess iron.
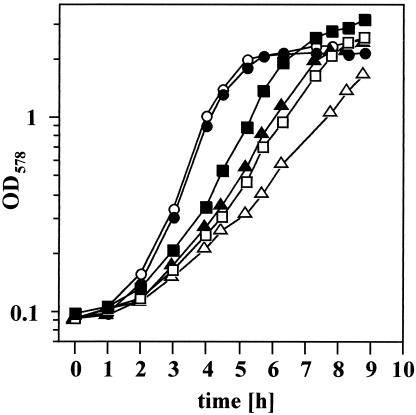
The data presented so far strongly suggest that high-salinity-stressed B. subtilis cells experience an iron deficiency. This predicts that the growth retardation exhibited by these cultures (3) should be at least partially compensated for by a substantial increase in the iron concentration of the growth medium. To test this hypothesis, we monitored the growth of the B. subtilis strain JH642 at high salinity (modified MM with 0.7 M NaCl) in the presence of 5 μM FeCl3 and 250 μM FeCl3. An ample supply of iron in high-salinity media resulted in substantially better growth of the cells in comparison with the culture that received the standard amount (5 μM FeCl3) of iron (Fig. 6). The generation time decreased from 1.6 h in the presence of 5 μM FeCl3 to 1.15 h in the presence of 250 μM FeCl3. This improvement in growth is substantial and is similar to that caused by the addition of the potent osmoprotectant glycine betaine to the salt-stressed culture that grew with a generation time of 1.17 h (Fig. 6). The growth stimulation by glycine betaine could be further enhanced by the simultaneous addition of high concentrations of iron (250 μM), resulting in a generation time of 0.87 h (Fig. 6).
FIG. 6.
Growth of salt-stressed B. subtilis cells in the presence or absence of excess iron. Cultures of strain JH642 were grown in modified MM containing the following salt, glycine betaine, and iron concentrations: 5 μM FeCl3 (○); 5 μM FeCl3, 1 mM glycine betaine (•); 5 μM FeCl3 and 0.7 M NaCl (▵); 5 μM FeCl3, 1 mM glycine betaine, and 0.7 M NaCl (▴); 250 μM FeCl3 and 0.7 M NaCl (□); and 250 μM FeCl3, 1 mM glycine betaine, and 0.7 M NaCl (▪).
Salt stress causes a derepression of genes encoding iron uptake systems.
High-salinity-induced iron limitation of B. subtilis cells not only should cause a derepression of the Fur-controlled dhb operon but also should affect other genes that are involved in iron homeostasis. This group of genes should include systems that encode transporters for the acquisition of exogenous iron. With the exception of the ABC transport system (Fhu) for the iron-hydroxamate ferrichrome (48), there is little physiological information available about iron transport systems of B. subtilis. We therefore consulted the annotation of the B. subtilis genome sequence (34) for potential iron transporters and also used a consensus-directed search for potential Fur recognition sequences (16, 24) to discern candidates for iron uptake systems. This information led us to focus our further experiments on the divergently described fhuB and fhuD genes, on the feuA gene encoding a potential binding protein for the Feu ABC transporter, which exhibits homology to the ferrienterobactin uptake system Fep of Escherichia coli, on yfiY, which encodes a protein with similarity to iron(III) dicitrate transport permeases, and on yfmC, the gene for the putative binding protein for the ferrichrome ABC transport system YfmCDEF. Each of these genes is preceded by potential Fur recognition sequences that are located in the vicinity of potential −10 and −35 sequences of SigA-dependent promoters (Fig. 7).
FIG. 7.
Consensus-directed search for Fur boxes. The B. subtilis genome sequence was searched for the occurrence of Fur boxes (ATAAT) with the program Motif-Finder from Decodon GmbH. Several copies of this pentamer are required for effective recognition by Fur (16–18, 24). The search revealed multiple Fur-box-like sequences in front of the translation initiation codons of dhbA, fhuB, fhuD, feuA, yfiY, and yfmC that are indicated in bold uppercase letters in the sequences. The −10 and −35 regions of potential SigA-type promoters are indicated with lines and shading. The genes (dhbACBEF; fhuD) marked by an asterisk have been shown experimentally to be under Fur control (8, 48).
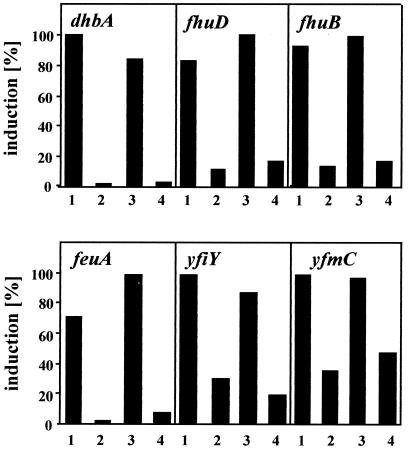
To test whether the fhuD, fhuB, feuA, yfiY, and yfmC genes were indeed controlled by iron limitation and induced by salt stress, we prepared hybridization probes for each of these genes and tested their transcription in dot blot experiments (Fig. 8). The transcription profile of the fhuD, fhuB, feuA, yfiY, and yfmC genes followed the pattern that we had found for the dhb operon (Fig. 2B and 4). First, there was a strong induction of transcription of each gene under iron-limiting conditions, implying that this entire group of genes and operons is regulated by the central iron regulatory protein Fur (Fig. 8). Second, all tested genes responded to salt stress (0.7 M NaCl) with a derepression of transcription in growth media that contained 5 μM iron. Furthermore, an increase in the iron concentration to 50 μM by and large prevented the derepression the fhuD, fhuB, feuA, yfiY, and yfmC genes (Fig. 8). Consequently, high salinity results in a derepression not only of the dhb operon encoding proteins for the synthesis of the iron chelator bacillibactin but also of a considerable variety of genes and operons whose products seemed to be involved in the acquisition of iron from external sources.
FIG. 8.
Salt-induced induction of iron-controlled B. subtilis genes. Total RNA was isolated from cells of strain JH642 grown in modified MM in the absence (lanes 1) or presence of either 5 μM FeCl3 (lanes 2 and 3) or 50 μM FeCl3 (lanes 4). In addition, 0.7 M NaCl was added to the cultures whose RNA was used for the experiments displayed in lanes 3 and 4. The RNA was dot blotted onto a nylon membrane and hybridized to gene-specific antisense RNA probes labeled with digoxigenin. The signal intensity was quantified using a Storm860 fluorimager and the software ImageQuant. For each gene investigated, the highest level of gene expression was set to 100%.
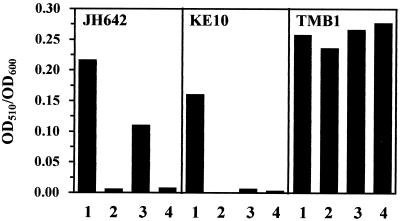
If derepression of the genes for the various iron acquisition systems by high salinity was solely mediated by the central iron-regulatory protein Fur, one would expect that DHB production would no longer be stimulated by high salinity in a fur mutant. We therefore constructed a fur::kan derivative of JH642, strain TMB1, and tested its iron limitation- and high salinity-induced DHB production. Consistent with the known regulation of the dhb operon by Fur (8), DHB production was derepressed in the fur::kan mutant TMB1 in the presence of 5 μM Fe, a concentration which was sufficient to fully repress the synthesis of this compound in the fur+ parent strain JH642 (Fig. 9). Neither iron limitation nor high salinity could trigger a further increase in the level of DHB in strain TMB1, whereas both treatments caused increased DHB production in its parent strain, JH642 (Fig. 9). These findings support the notion that the derepression of dhb expression in salt-stressed cells is indeed mediated by Fur.
FIG. 9.
DHB production in a B. subtilis fur mutant and a bacillibactin-producing strain. DHB production was assayed in culture supernatants of strain JH642 (fur+ sfp0), TMB1 (fur::kan sfp0), and KE10 (fur+ sfp+) grown in modified MM in the absence (lanes 1) or presence of either 5 μM FeCl3 (lanes 2 and 3) or 50 μM FeCl3 (lanes 4). In addition, 0.7 M NaCl was added to the culture used for the DHB assays displayed in lanes 3 and 4. The bacillibactin producer KE10 has an undefined auxotrophy and fails to grow in defined MM. Therefore, we included 0.02% Casamino Acids in the precultures of all strains.
It should be noted that the strain background (JH642) used in this study contains a mutation (sfp0) (20) that prevents or strongly reduces the phosphopantetheinylation of DhbB and DhbF. Consequently, no bacillibactin or very low amounts of bacillibactin are produced in strain JH642 and its derivatives (39). However, these strains can still produce DHB (Fig. 3), which exhibits a low level of iron-chelating activity (39, 48). We therefore tested the bacillibactin producer, KE10 (sfp+), for the regulation of DHB production by iron availability and high salinity. Limiting concentrations of iron triggered DHB synthesis in the B. subtilis strain KE10, whereas an increase in the salinity of the growth medium had only a marginal effect on the level of DHB (Fig. 9). Hence, a strain that effectively synthesizes the iron chelator bacillibactin scavenges sufficient amounts of iron even under high-salinity growth conditions so that Fur-regulated genes remain repressed. As expected from this observation, high-salinity-mediated growth retardation of the bacillibactin producer KE10 could not be rescued by increases in the iron concentration of the growth medium (data not shown), in contrast to what was observed for the commonly used laboratory strain JH642 (Fig. 6).
DISCUSSION
Through proteome analysis and subsequent more detailed transcriptional studies, we found a derepression of genes encoding the synthesis pathway of the iron chelator bacillibactin and uptake systems for iron in high-salinity-grown B. subtilis cultures. Iron limitation and high-salinity stress have thus far been regarded as separate growth-limiting factors. Our findings provide data showing that this is not the case and demonstrate that the ionic component of salt stress, rather than its osmotic component (Fig. 4), exerts indirect effects on an adequate iron supply for the cell.
Iron is an important trace element for essentially every bacterial species. In many aerobic, neutral, or alkaline environments, Fe2+ is present in only suboptimal concentrations due to its low solubility. Microorganisms have therefore developed elaborate systems for the scavenging of iron from environmental sources that frequently involve the synthesis of high-affinity iron chelators, their excretion into the environment, and the recapturing of the iron-loaded chelator via high-affinity transport systems (18, 22, 24). Although iron is required for growth, excess iron in the presence of oxygen is undesirable for the cell because it stimulates the formation of toxic hydroxyl radicals via the Fenton reaction which then damage DNA and membranes (50). Iron homeostasis is subject to tight genetic control. This is accomplished by preventing expression of iron acquisition systems through binding of the central iron regulatory protein Fur (8, 23) to its cognate DNA-binding sites in the presence of excess iron and by relieving this repression upon iron limitation (18, 22, 24). The level of transcription of genes encoding systems for the biosynthesis of iron chelators and iron transporters can therefore be regarded as molecular beacons that sensitively reflect the iron supply status of the cell.
Derepression of B. subtilis genes encoding the synthesis of the iron chelator bacillibactin and iron uptake systems in salt-stressed cells occurred despite the fact that the growth medium contained enough iron (5 μM) to fully repress these genes in non-salt-stressed cells (Fig. 1, 4, and 8). Derepression of transcription of each of these genes could be counteracted by the addition of excess iron (Fig. 8). Salt stress raised the expression of dhb, fhu, feu, yfiY, and yfmC to levels comparable to those observed in iron-limited cultures (Fig. 8). These data therefore provide strong evidence that high salinity has an iron limitation component. In support of this notion, the growth of salt-stressed B. subtilis cultures was considerably improved when the cells were provided with excess iron (250 μM). This improvement in growth is substantial, since it is similar to that caused by the potent osmoprotectant glycine betaine. Glycine betaine and excess iron both compensate for different deficiencies experienced by B. subtilis under salt stress because their growth-stimulating effects are additive (Fig. 6). Regardless of the cation or anion present at increased concentrations, various salts (NaCl, KCl, Na2SO4, and K2SO4) led to gene induction, whereas induction did not occur when iso-osmolar concentrations of nonionic osmolytes (sucrose and maltose) were used to raise the osmolality of the growth medium (Fig. 4). This probably also explains the failure of glycine betaine to fully compensate for the high-salinity-triggered growth retardation because it counteracts only the osmotic component of high-salt stress.
Each of the iron-regulated genes and operons which we investigated in more detail contains potential Fur-box-like sequences (Fig. 7), implying that the dhb, fhu, feu, yfiY, and yfmC loci are all controlled by the central iron regulator Fur of B. subtilis (9). Indeed, a regulation of the dhb operon by Fur has already been demonstrated experimentally by Bsat et al. (8) and was verified in this study (Fig. 9). We therefore expect that the high-salinity-mediated induction of iron-regulated genes we have studied will extend to other members of the B. subtilis Fur regulon as well. The absence of further stimulation of DHB production in a fur mutant (Fig. 9) implies that the observed high-salinity-induced transcription of dhb and of those genes and operons which we have additionally investigated is indeed mediated by Fur.
Our data clearly demonstrate a high-salinity induction of iron-controlled genes of B. subtilis, but the actual sequence of events that leads to iron limitation in the cell is not understood. When one considers possible explanations for high-salt-triggered iron limitation, it is important to recall that this effect can be caused by various salts but not by nonionic osmolytes (Fig. 4). In E. coli, a sudden and severe salt shock exerts strong negative effects on the activities of various carbohydrate transport systems (44). It is thus possible that transporters for the acquisition of iron in B. subtilis are partially inhibited at high salinity, thereby preventing a proper iron supply to the cell. Alternatively, the effective complexation of iron by iron chelators could be negatively affected by the high ionic strength of the growth medium, resulting in a suboptimal presentation of iron-loaded chelators to their corresponding uptake systems. Iron limitation could also be a consequence of limited iron availability following the formation of insoluble iron-phosphate complexes in high-salt media.
Whatever the underlying molecular and biochemical mechanisms for the high-salinity-mediated iron limitation, our findings have implications for understanding the growth pattern of B. subtilis in natural settings. Bioavailable iron is scarce in soil (51), necessitating the formation of high-affinity iron-scavenging systems in both microorganisms (22) and plants (12). Desiccation of the soil increases the salinity and thereby further decreases the amount of iron available for the bacterial cell. Simultaneously, B. subtilis has to adapt to the increased osmolality of its habitat (7). Consequently, the concerted action of osmo-adaptive mechanisms and the induction of iron-scavenging systems will be required to sustain growth in drying soil.
Acknowledgments
We thank K. Altendorf, V. Braun, and K. Hantke for helpful discussions and greatly appreciate the communication of data on the synthesis of bacillibactin prior to publication by M. Marahiel. We are grateful to J. Helmann and M. Marahiel for providing bacterial strains. We appreciate the help of V. Koogle in editing the manuscript and the expert technical assistance of J. Sohn
Financial support for this study was provided by the Deutsche Forschungsgemeinschaft through SFB-395 (Teilprojekte B1 and B12), the Max-Planck Society, and the Fonds der Chemischen Industrie (to U.V. and E. B.).
REFERENCES
- 1.Alice, A. F., and C. Sanchez-Rivas. 1997. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr. Microbiol. 35:309–315. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, H., H. Beier, and H. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99. [Google Scholar]
- 3.Boch, J., B. Kempf, and E. Bremer. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boch, J., B. Kempf, R. Schmid, and E. Bremer. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 7.Bremer, E., and R. Krämer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p.79–97. In G. Storz, and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 8.Bsat, N., and J. D. Helmann. 1999. Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo. J. Bacteriol. 181:4299–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189–198. [DOI] [PubMed] [Google Scholar]
- 10.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mäder, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908–2935. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L., L. P. James, and J. D. Helmann. 1993. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 175:5428–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curie, C., Z. Panaviene, C. Loulergue, S. L. Dellaporta, J. F. Briat, and E. L. Walker. 2001. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346–349. [DOI] [PubMed] [Google Scholar]
- 13.Dartois, V., M. Débarbouillé, F. Kunst, and G. Rapoport. 1998. Characterization of a novel member of the DegS/DegU regulon affected by salt stress in Bacillus subtilis. J. Bacteriol. 180:1855–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deuerling, E., B. Paeslack, and W. Schumann. 1995. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J. Bacteriol. 177:4105–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppelmann, K., S. Doekel, and M. A. Marahiel. 2001. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 276:34824–34831. [DOI] [PubMed] [Google Scholar]
- 16.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537–547. [DOI] [PubMed] [Google Scholar]
- 17.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 2000. Evidence of an unusually long operator for the Fur repressor in the aerobactin promoter of Escherichia coli. J. Biol. Chem. 275:24709–24714. [DOI] [PubMed] [Google Scholar]
- 18.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galinski, E. A., and H. G. Trüper. 1994. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol. Rev. 15:95–108. [Google Scholar]
- 20.Grossman, T. H., M. Tuckman, S. Ellestad, and M. S. Osburne. 1993. Isolation and characterization of Bacillus subtilis genes involved in siderophore biosynthesis: relationship between B. subtilis sfpo and Escherichia coli entD genes. J. Bacteriol. 175:6203–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172–177. [DOI] [PubMed] [Google Scholar]
- 23.Hantke, K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 210:135–139. [DOI] [PubMed] [Google Scholar]
- 24.Hantke, K., and V. Braun. 2000. The art of keeping low and high iron concentrations in balance, p.275–288. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 25.Harwood, C. R., and A. R. Archibald. 1990. Growth, maintenance and general techniques, p.1–26. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., Chichester, United Kingdom.
- 26.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35–91. [DOI] [PubMed] [Google Scholar]
- 27.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis ςA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsburgh, M. J., and A. Moir. 1999. ςM, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32:41–50. [DOI] [PubMed] [Google Scholar]
- 29.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC-transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203–216. [DOI] [PubMed] [Google Scholar]
- 31.Kempf, B., and E. Bremer. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701–16713. [DOI] [PubMed] [Google Scholar]
- 32.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high osmolality environments. Arch. Microbiol. 170:319–330. [DOI] [PubMed] [Google Scholar]
- 33.Krispin, O., and R. Allmansberger. 1995. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol. Lett. 134:129–135. [DOI] [PubMed] [Google Scholar]
- 34.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. [DOI] [PubMed] [Google Scholar]
- 35.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, Y., and J. N. Hansen. 1995. Characterization of a chimeric proU operon in a subtilin-producing mutant of Bacillus subtilis 168. J. Bacteriol. 177:6874–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez, C. S., H. Heras, H. Garda, S. Ruzal, C. Sanchez-Rivas, and E. Rivas. 2000. Biochemical and biophysical studies of Bacillus subtilis envelopes under hyperosmotic stress. Int. J. Food Microbiol. 55:137–142. [DOI] [PubMed] [Google Scholar]
- 38.Lopez, C. S., H. Heras, S. M. Ruzal, C. Sanchez-Rivas, and E. A. Rivas. 1998. Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr. Microbiol. 36:55–61. [DOI] [PubMed] [Google Scholar]
- 39.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209–7217. [DOI] [PubMed] [Google Scholar]
- 40.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50:101–136. [DOI] [PubMed] [Google Scholar]
- 41.Nau-Wagner, G., J. Boch, J. A. Le Good, and E. Bremer. 1999. High-affinity transport of choline-O-sulfate and its use as a compatible solute in Bacillus subtilis. Appl. Environ. Microbiol. 65:560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otto, A., B. Thiede, E. C. Muller, C. Scheler, B. Wittmann-Liebold, and P. Jungblut. 1996. Identification of human myocardial proteins separated by two-dimensional electrophoresis using an effective sample preparation for mass spectrometry. Electrophoresis 17:1643–1650. [DOI] [PubMed] [Google Scholar]
- 43.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related Gram-positive bacteria, p.179–197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 44.Roth, W. G., M. P. Leckie, and D. N. Dietzler. 1985. Osmotic stress drastically inhibits active transport of carbohydrates by Escherichia coli. Biochem. Biophys. Res. Commun. 126:434–441. [DOI] [PubMed] [Google Scholar]
- 45.Rowland, B. M., T. H. Grossman, M. S. Osburne, and H. W. Taber. 1996. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178:119–123. [DOI] [PubMed] [Google Scholar]
- 46.Rowland, B. M., and H. W. Taber. 1996. Duplicate isochorismate synthase genes of Bacillus subtilis: regulation and involvement in the biosyntheses of menaquinone and 2,3-dihydroxybenzoate. J. Bacteriol. 178:854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruzal, S. M., C. López, E. Rivas, and C. Sánchez-Rivas. 1998. Osmotic strength blocks sporulation at stage II by impeding activation of early sigma factors in Bacillus subtilis. Curr. Microbiol. 36:75–79. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, R., and K. Hantke. 1993. Iron-hydroxamate uptake systems in Bacillus subtilis: Identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8:111–121. [DOI] [PubMed] [Google Scholar]
- 49.Spiegelhalter, F., and E. Bremer. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the ςA- and ςB-dependent stress-responsive promoters. Mol. Microbiol. 29:285–296. [DOI] [PubMed] [Google Scholar]
- 50.Storz, G., and M. Zheng. 2000. Oxidative stress, p.47–59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 51.van Overbeek, L., and J. Van Elsas. 1997. Adaptation of bacteria to soil conditions: applications of molecular physiology in molecular microbiology, p.441–479. In J. van Elsas, J. Trevoors, and E. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 52.Völker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Völker, R. Schmidt, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741–752. [DOI] [PubMed] [Google Scholar]
- 53.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the ςB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175–187. [DOI] [PubMed] [Google Scholar]
- 55.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527–2535. [DOI] [PubMed] [Google Scholar]
- 56.Whatmore, A. M., and R. H. Reed. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136:2521–2526 [DOI] [PubMed] [Google Scholar]